Abstract
Adeno-associated virus serotype 9 (AAV9) vectors show promise for gene therapy of a variety of diseases due to their ability to transduce multiple tissues, including heart, skeletal muscle, and the alveolar epithelium of the lung. In addition, AAV9 is unique compared to other AAV serotypes in that it is capable of surpassing the blood-brain barrier and transducing neurons in the brain and spinal cord. It has recently been shown that AAV9 uses galactose as a receptor to transduce many different cell types in vitro, as well as cells of the mouse airway in vivo. In this study, we sought to identify the specific amino acids of the AAV9 capsid necessary for binding to galactose. By site-directed mutagenesis and cell binding assays, plus computational ligand docking studies, we discovered five amino acids, including N470, D271, N272, Y446, and W503, which are required for galactose binding that form a pocket at the base of the protrusions around the icosahedral 3-fold axes of symmetry. The importance of these amino acids for tissue tropism was also confirmed by in vivo studies in the mouse lung. Identifying the interactions necessary for AAV9 binding to galactose may lead to advances in vector engineering.
INTRODUCTION
Adeno-associated virus (AAV) vectors show great potential for use in gene therapy applications due to their ability to transduce many different types of tissues and provide stable, long-term gene expression (41). Numerous serotypes of AAV have been characterized, and more than 120 other variants have been isolated from both human and nonhuman primate tissues (13–15, 27, 35, 35a). Each of these variants has its own unique properties providing a large pool of potential vectors for gene therapy. Different AAV serotypes display distinctive tissue tropism and transduction efficiency. For example, AAV1 demonstrates efficient muscle transduction, AAV6 shows promise in targeting the conducting airway epithelium of the lung, and AAV8 provides impressive liver transduction (6, 15, 18, 24, 46).
The three-dimensional structure of the capsid of many AAV serotypes has been determined by X-ray crystallography (7, 16, 22, 28, 29, 47, 48). The core of the virion consists of a highly conserved eight stranded β-barrel motif with large loops inserted between these β-strands that contain defined variable regions (VRs) (16, 28, 29). The VRs are present at the surface of the capsid structure and are therefore readily available for cellular interactions. The high diversity of the VRs between AAV serotypes likely defines their respective phenotypes in regard to tropism, transduction efficiency, and antigenicity. Most of the VRs are located at or near the icosahedral 3-fold axes of symmetry of the capsid, forming protrusions around these axes (16, 28, 29).
Receptor binding is a proximal step necessary for AAV vector transduction. Interaction with cell surface receptors allows internalization of the virion and subsequent trafficking to the nucleus where gene expression occurs (8). Receptors for many AAV serotypes have been identified. AAV2 has been shown to bind to heparan sulfate (HS) proteoglycans and AAV1, -4, -5, and -6 all use specific linkages of sialic acid as a receptor (20, 40, 43, 44). The HS binding motif on the AAV2 capsid has been mapped by site-directed mutagenesis and determined to consist of five basic amino acid residues including arginines 484, 487, 585, and 588 and lysine 532, with arginines 585 and 588 being the most important for HS interaction (21, 31). This site has also been confirmed by structural studies using cryo-electron microscopy and image reconstruction (23, 30). This basic patch is formed by the interaction of symmetry-related monomers on the inner surface of the protrusions surrounding the icosahedral 3-fold axes of symmetry of the capsid.
The AAV serotype with the most interesting properties as a vector is that based on the human-derived virus, AAV9. After intravenous injection, AAV9 shows widespread systemic transduction, including efficient targeting of heart, liver, and skeletal muscle (4, 19, 32, 42, 49). Also, AAV9 is able to surpass the blood-brain barrier and transduce cells such as neurons in the brain and spinal cord (9, 11). Recently, through a series of in vitro binding and transduction assays, AAV9 has been shown to use galactose as a cellular receptor (1, 38). This observation was also confirmed by in vivo studies in the mouse airway, in which the abundance of free galactose residues was enhanced by enzymatic removal of the galactose-bound sialic acid residues from the surface of airway cells. The increase in terminal galactose saccharides available for binding led to robust AAV9 transduction of cells of the conducting airway epithelium, as well as enhanced transduction of nasal epithelium (1, 38).
In the present study, we sought to identify the galactose binding motif on the AAV9 capsid. We generated a series of alanine mutants to determine the necessity of specific amino acids for galactose binding both in vitro and in vivo. This led to the discovery of a galactose binding pocket at the base of the protrusions that surround the 3-fold axes of symmetry of the AAV9 capsid, which was also confirmed by molecular docking studies.
MATERIALS AND METHODS
Cells lines.
All cell lines were obtained from the American Type Culture Collection. Three different Chinese hamster ovary (CHO) cell lines were used in binding and transduction experiments, including the parental cell line Pro-5, the sialic acid-deficient cell line Lec-2, and the galactose-deficient cell line Lec-8. These cells were cultured in α-minimum essential medium (α-MEM) supplemented with ribonucleosides and deoxyribonucleosides (Invitrogen) with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin.
Animals.
Male C57BL/6 mice (6 to 8 weeks old) were purchased from Charles River Laboratories and housed in the Animal Facility of the Translational Research Laboratories at the University of Pennsylvania. All animal procedures were approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania.
AAV9 mutagenesis and small-scale vector preparation.
Specific charged or polar amino acids of the AAV9 capsid sequence were chosen for mutation to nonpolar alanine, as indicated in Fig. 1. Mutagenesis was performed using a QuikChange Lightning site-directed mutagenesis kit from Agilent Technologies. For subsequent small-scale production of mutant vectors for use in in vitro binding and transduction assays, triple transfection of HEK293 cells in six-well plates (9.6 cm2) was performed using a plasmid expressing the AAV2 rep gene and the mutant AAV9 cap gene, as well as plasmids expressing the firefly luciferase (ffLuc) transgene expressed from a cytomegalovirus (CMV) promoter flanked by the AAV2 inverted terminal repeats, and an adenovirus helper plasmid (pAdΔF6). Cells and supernatant were harvested after 72 h in 2 ml of total medium, subjected to three freeze-thaw cycles, and then centrifuged at 3,500 × g for 30 min to remove cell debris. The preparations were then treated with DNase to remove any contaminating plasmid, and the titers of the vectors (genome copies [GC]/ml) were determined by quantitative PCR.
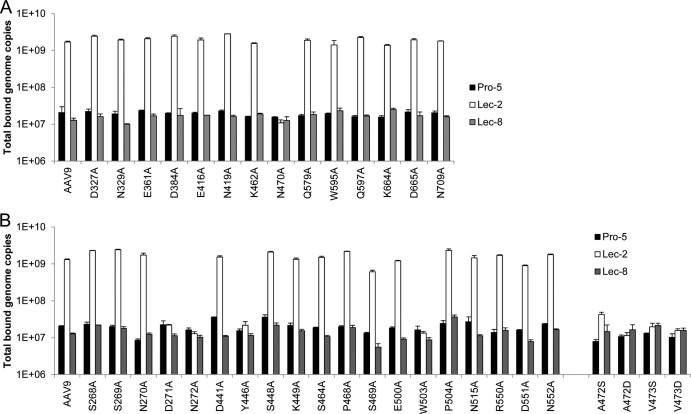
Fig 1.
AAV9 capsid amino acids required for galactose binding. (A) After comparing the capsid amino acid sequence of AAV9 to that of other serotypes, specific amino acids unique to AAV9 that contain charged or polar side chains were mutated to alanine to identify those responsible for galactose binding. Fourteen mutant vectors were constructed and were named according to the native amino acid and its specific position followed by the new amino acid: alanine. The mutants were tested for binding to Pro-5, Lec-2, and Lec-8 cells, compared to AAV9. Vector was added (5 × 109 GC) to each cell line and incubated at 4°C for 1 h. After washing, total DNA was isolated to determine bound vector GCs by quantitative PCR. N470 was determined to be necessary for AAV9 galactose binding. (B) Subsequently, 19 amino acids located in close proximity to N470 were mutated to alanine to examine the effect on AAV9 galactose binding. In addition, four other mutants were produced containing mutations of the nonpolar amino acids A472 or V473 to either serine or aspartic acid. The binding of these mutant vectors was then assessed as described above. D271, N272, Y446, and W503, as well as A472 and V473, were found to be important for AAV9 galactose binding. The data are shown as means + the standard deviation (SD).
Vector production and purification for in vivo studies.
AAV vectors for use in vivo were produced and purified by Penn Vector (http://www.med.upenn.edu/gtp/vector_core/production.shtml). A plasmid expressing nLacZ from a chicken β-actin promoter and flanked by AAV2 inverted terminal repeats was packaged by triple transfection of HEK293 cells with plasmids encoding the AAV2 rep gene and the AAV9 or mutant cap gene, and an adenovirus helper plasmid (pAdΔF6). Vectors were purified by iodixinol gradient centrifugation, and titers were determined by quantitative PCR.
Cell binding and transduction assays.
For binding assays, Pro-5, Lec-2, and Lec-8 cells were scraped from 150-cm2 flasks and seeded at 5 × 105 cells/well in 96-well plates in 100 μl of cold serum-free α-MEM. Vectors were added at 5 × 109 GC/well in 100 μl of cold α-MEM, followed by incubation at 4°C for 1 h. The cells were then washed three times with 200 μl of α-MEM and resuspended in 200 μl of phosphate-buffered saline (PBS). Total DNA was extracted by using a QIAamp DNA minikit (Qiagen), and cell-bound vector GCs were determined by quantitative PCR. For transduction assays, 105 cells/well were seeded in black-walled, clear-bottom 96-well plates overnight. After removal of the medium, AAV vector expressing ffLuc (109 GC) was added to the cells in 100 μl of complete media and incubated at 37°C for 48 h. ffLuc expression was then determined by adding 150 μg/ml of d-luciferin substrate per well in 100 μl of α-MEM and measuring the relative light units (RLU)/s using a luminometer.
Transduction of mouse lung.
As previously described (1), mice were anesthetized with ketamine-xylazine and given an intranasal instillation of 100 mU of neuraminidase (NA) in 30 μl of PBS. One hour later, 1011 GC of AAV9 or mutant vector expressing nLacZ was delivered intranasally in 50 μl of PBS. At 21 days after administration, the lungs were inflated and removed, and the β-galactosidase (β-Gal) expression was examined by previously described methods (2). Lung images were taken at ×100 magnification. For quantification of the β-Gal expression in conducting airways, nLacZ-positive cells were counted per ×200 field of view.
Evaluation of amino acids located on the AAV9 capsid surface near N470.
The program RIVEM (radial interpretation of viral electron density maps) (45) was utilized to generate a roadmap of amino acids that are on the surface of the AAV9 capsid using the high-resolution structure of AAV9, PDB accession no. 3UX1 (7). Based on this visualization, additional amino acids on the surface of the AAV9 capsid in close proximity to N470 were selected for mutation to identify additional amino acids that play a role in AAV9 receptor binding.
Prediction of the galactose binding site on the AAV9 capsid by molecular docking studies.
Molecular docking was performed to predict the galactose binding site on the AAV9 capsid using the molecular docking webserver PatchDock (http://bioinfo3d.cs.tau.ac.il/PatchDock/index.html) (36). The X-ray crystal structure for galactose bound to human galectin-3, PDB accession no. 1A3K was utilized to provide the coordinates for galactose (37). The coordinates for the AAV9 VP monomer X-ray crystal structure (PDB accession no. 3UX1 [7]) was submitted to the VIPERdb Oligomer generator (http://viperdb.scripps.edu/oligomer_multi.php) (5) to generate a trimer by icosahedral symmetry matrix manipulation. The output PDB file for the AAV9 trimer was modified to remove conserved internal residues (that would be inaccessible to cell surface glycan binding) and only include amino acids that were surface accessible (and thus more plausible binding sites) and maintained the external surface features of the viral capsid. The amino acids included in the modified AAV9 trimer PDB file as the input file for molecular docking were N262 to Y277, L435 to G475, and F501 to M559. Molecular docking was performed in an undirected fashion to evaluate the site on the AAV9 capsid that was likely to interact with galactose. Docking was also performed using the truncated AAV9 trimer and sialic acid. The X-ray crystal structure for equine rhinitis A virus in complex with its sialic acid receptor, PDB accession no. 2XBO, was utilized to provide the coordinates for sialic acid (12).
RESULTS
N470 is necessary for AAV9 galactose binding.
To identify potential amino acids of the AAV9 capsid that could be involved in galactose binding, we aligned the capsid amino acid sequence of AAV9 to that of other serotypes in closely associated clades that do not bind galactose, such as AAVs 1, 2, 6, 7, and 8, to determine which amino acids in distinct capsid positions are unique to AAV9 (see Fig. S1 in the supplemental material). Of the amino acids specific to AAV9, we identified those that have charged, polar, or aromatic side chains because these residues would likely be responsible for interaction with the sugar. Based on this alignment, 14 representative amino acids were chosen for mutation to nonpolar alanine to examine the effect on AAV9 binding (Fig. 1A). These mutant capsid constructs were used to make vector by small-scale preparation methods and yielded similar titers compared to wild-type AAV9.
The mutant vectors were then added to three different CHO cell lines to assess their binding capabilities (Fig. 1A). The parental CHO cell line Pro-5, as well as somatic cell glycosylation mutants of this cell line, Lec-2 and Lec-8, were used in these experiments (33). Lec-2 cells are deficient in CMP-sialic acid Golgi transporter and therefore lack sialic acid residues on their surface glycans, which allows exposure of the most common penultimate saccharide, galactose. Lec-8 cells are deficient in UDP-galactose Golgi transporter and are consequently devoid of galactose saccharides on their surface glycan structures. When examining vector binding to these CHO cell lines, the expected results were observed for AAV9 (1, 38). A low level of binding was observed on Pro-5 cells but was shown to increase 100-fold on Lec-2 cells, which have terminal galactose residues free for AAV9 binding. Binding to Lec-8 cells was decreased back to baseline level due to their lack of cell-surface galactose. Thirteen of fourteen of the mutant vectors demonstrated this trend of increased binding on Lec-2 cells, suggesting that they retained the ability to bind galactose. However, mutation of the asparagine residue at position 470 to alanine (N470A) reduced binding to Lec-2 cells to the level observed on Pro-5 and Lec-8 cells. This indicated that N470 is required for AAV9 binding to galactose.
Additional amino acids located in close proximity to N470 are required for AAV9 galactose binding.
Additional AAV9 mutants were generated to determine whether any of the amino acids structurally surrounding N470 are also contributing to galactose binding based on information of the capsid surface that surrounds this residue (see Fig. S2 in the supplemental material). Nineteen amino acids with polar, charged, or aromatic side chains located near N470 were selected for mutation to alanine (Fig. 1B) and vectors were produced by small-scale methods. As before, all of the mutants yielded high-titer vector similar to the AAV9 control, suggesting none of the mutations had a negative effect on capsid assembly. The mutant vectors were then tested for their ability to bind Pro-5, Lec-2, and Lec-8 cells (Fig. 1B). Again, AAV9 showed the expected result of a 100-fold increase in binding to Lec-2 cells compared to Pro-5 and Lec-8 cells. While most of the mutants tested also exhibited this phenotype, four mutants demonstrated a loss of binding to Lec-2 cells. Mutation of D271, N272, Y446, or W503 all abolished the ability of AAV9 to bind galactose.
Alongside these 19 alanine mutants, four other AAV9 site-directed mutations were performed. The nonpolar amino acids A472 and V473 are located directly next to N470 and could potentially play a role in galactose binding (see Fig. S2 in the supplemental material). To test this idea, A472 and V473 were both mutated to either the polar amino acid serine or the charged amino acid aspartic acid. Construction of these vectors by small-scale methods was successful and titers were achieved similar to wild-type AAV9. When examining the binding of these vectors to CHO cells, it was observed that all four of these mutants (A472S, A472D, V473S, and V473D) lost the ability to bind Lec-2 cells (Fig. 1B). This suggests that these nonpolar residues are essential to the formation of the galactose binding region because adding either polarity or charge to this region disrupted interaction with galactose.
Galactose binding of AAV9 mutants is indicative of in vitro transduction efficiency.
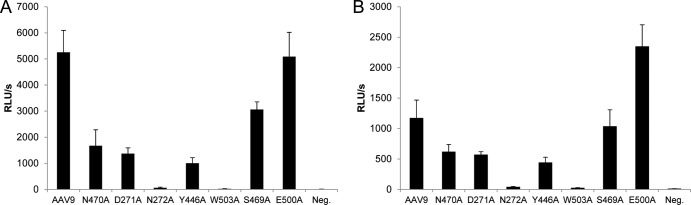
To assess the transduction of AAV9 mutants in vitro, vectors expressing ffLuc were added to Pro-5 and Lec-2 cells. The five mutants that lost galactose binding (N470A, D271A, N272A, Y446A, and W503A), as well as two mutants that retained galactose binding (S469A and E500A) and an AAV9 control vector were added to the cells (MOI = 104) and evaluated for ffLuc expression 48 h later. As expected, the mutants that lack galactose binding ability demonstrated a 3-fold or greater decrease in transduction of Lec-2 cells compared to AAV9, S469A, and E500A (Fig. 2A). W503A and N272A showed the lowest transduction levels, just above background. The overall transduction on Pro-5 cells was 2- to 5-fold lower than on Lec-2 cells because these cells lack abundant terminal galactose residues for binding (Fig. 2B). However, a similar trend in transduction was still observed as that determined for Lec-2 cells. The mutants deficient in galactose binding showed the lowest transduction and the two mutants that retained galactose binding (S469A and E500A), as well as the AAV9 control vector, demonstrated higher levels of transduction. E500A, however, showed ∼2-fold-higher expression than AAV9 on Pro-5 cells, suggesting that this vector may have favorable intracellular interactions. To eliminate the possibility that the AAV9 point mutations may be affecting capsid stability, we heated the mutant vectors at various temperatures, followed by DNase treatment and titer by quantitative PCR, and observed that the mutants demonstrate similar stability compared to the AAV9 control vector (see Fig. S3 in the supplemental material).
Fig 2.
In vitro transduction efficiency of mutant vectors. The five mutants that lost the ability to bind to galactose (N470A, D271A, N272A, Y446A, and W503A), as well as two mutants that retained galactose binding (S469A and E500A) and an AAV9 control vector, all expressing ffLuc, were added to Lec-2 (A) or Pro-5 (B) cells at 109 GC/well (MOI = 104) and ffLuc expression was measured 48 h later. The data are shown as means + the SD. RLU, relative light units.
Mutant vectors deficient in galactose binding do not transduce conducting airway epithelium.
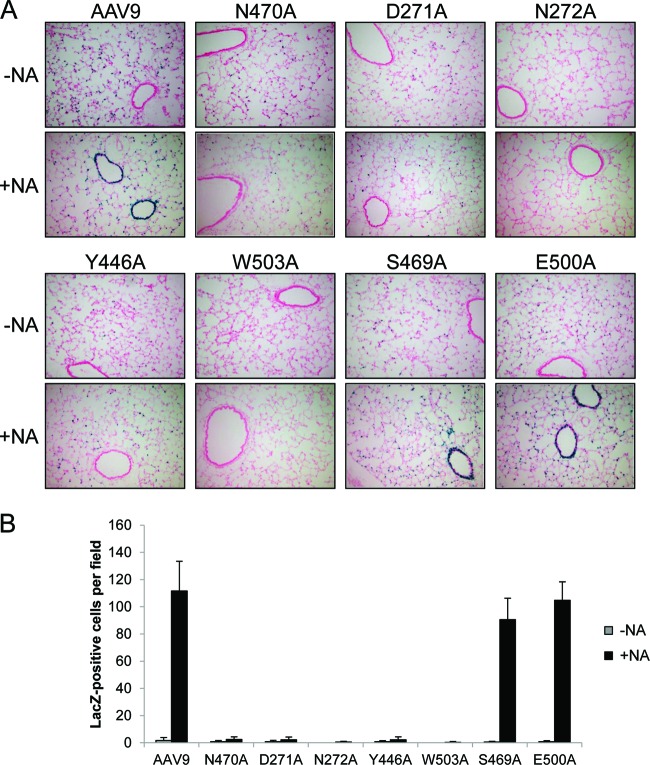
We have observed previously that delivering NA intranasally to mice prior to intranasal administration of AAV9 vector allows efficient AAV9 transduction of conducting airway epithelium of the lung (1). This occurs because NA cleaves terminal sialic acid residues from the surface glycans of the airway cells, which exposes the underlying galactose residues and allows AAV9 binding and transduction. We sought to determine whether the mutant vectors that are deficient in galactose binding can transduce conducting airway epithelium in vivo. Vectors (1011 GC) expressing nuclear-targeted LacZ (nLacZ) were delivered intranasally to mice 1 h after instillation of 100 mU of NA, and then 21 days later the lungs were removed and stained for β-Gal expression (Fig. 3A). The AAV9 control vector demonstrated efficient conducting airway transduction, as expected based on our previous results (1). The five mutants that lost the ability to bind galactose (N470A, D271A, N272A, Y446A, and W503A) were unable to transduce conducting airway epithelium, further demonstrating their inability to bind galactose in vivo. The S469A and E500A mutants, which retained galactose binding, displayed efficient conducting airway transduction similar to AAV9 (Fig. 3). Identical studies performed with these mutants in mice who were not pretreated with NA yielded the expected results in that no capsid conferred transduction in conducting airway, with variable transduction of alveolar cells (Fig. 3).
Fig 3.
Mutant vector transduction of conducting airway epithelium. C57BL/6 mice were either given an intranasal instillation of 100 mU NA (+NA) or left untreated (−NA), 1 h prior to administration of AAV9 or mutant vectors expressing nLacZ (1011 GC). After 21 days, the lungs were inflated, removed, and stained for β-Gal expression. (A) Representative images of lung expression showing conducting airway and alveolar epithelium transduction. Images were taken at ×100 magnification. (B) Quantification of nLacZ-positive conducting airway epithelial cells per ×200 field of view. The data are shown as means + SD.
The amino acids necessary for binding form a pocket on the AAV9 capsid that supports interaction with galactose.
Using knowledge of the structure of AAV9 (7, 25), we sought to determine the location of the amino acids important for galactose binding on the surface of the capsid. A molecular docking approach was utilized to identify the galactose binding sites on an AAV9 VP trimer. The top 100 docking results were evaluated, and docking solutions that placed galactose on the inner surface of the capsid were discarded. The remaining 13 solutions all dock galactose in close contact with N470, with 12 of 13 solutions docking galactose in between N470 and W503. Figure 4A and B show an AAV9 VP trimer (colored differently for each monomer) and the galactose binding site, with the amino acids found to be necessary for galactose binding by mutagenesis highlighted in specific colors on each monomer. N470, D271, N272, Y446, and W503 form a pocket at the base of the protrusions surrounding the 3-fold axes of symmetry (Fig. 4). The pocket is formed by the outside surface of the protrusions facing the 2- and 5-fold axes and a small surface protrusion between the 2- and 5-fold axes formed by VR I. Figure 4C and D show a close-up view of the binding pocket where it is further illustrated how the galactose residue fits into this region. The nonpolar A472 and V473 amino acids found to be important for galactose binding are located at the base of the binding pocket and therefore likely allow successful insertion of the galactose saccharide (Fig. 4C). An interesting feature of the galactose binding site is that the amino acids that have been shown to be important for binding are from two different monomers, where Y446, N470, A472, and V473 make up the floor of the binding pocket and are from the monomer shown in purple in Fig. 4C, and the amino acids that make up the roof of the binding pocket, D271, N272, and W503, are from another contributing monomer (shown in brown in Fig. 4C). This may provide a mechanism to ensure that only properly assembled capsids can bind to the receptor. Significantly, following identification of the galactose binding pocket, an attempt to dock a sialic acid molecule into the same capsid region showed significant clashes with W503 for one of the solutions, suggesting that due to steric hindrance this glycan may not be accommodated (see Fig. S4 in the supplemental material). This observation is consistent with the lack of sialic acid binding by AAV9.
Fig 4.
Structure of the AAV9 galactose binding pocket. Galactose was docked onto an AAV9 trimer using the molecular docking webserver PatchDock (36). (A) AAV9 trimer surface showing the location of the galactose binding pockets. Galactose (light green) is shown docked at one of the binding sites. (B) Side view of an AAV9 trimer depicting the galactose binding pocket. (C) Close-up view from the side of the AAV9 galactose binding pocket. (D) Structure of the galactose binding pocket indicating the distances between the amino acid side chains and galactose. N470, yellow; D271, red; N272, white; Y446, blue; W503, orange; A472, teal; V473, gray; and galactose, light green. Images were generated using the PyMOL Molecular Graphics System, version 1.3 (Schrödinger, LLC).
DISCUSSION
Previous experiments studying the cell surface glycan interactions of AAV9 demonstrated that this vector uses galactose as a cellular receptor (1, 38). Analysis of AAV9 binding and transduction of multiple cell lines, including Pro-5, HEK293, and Huh-7 cells, revealed that the enzymatic removal of terminal sialic acid residues from cell surface glycans using NA led to a significant increase in AAV9 binding. This increase was due to the exposure of the underlying galactose saccharides, which facilitated AAV9 binding and transduction. Further studies using the CHO glycosylation mutant cell lines Lec-2 and Lec-8, as well as lectin competition assays, confirmed this role of galactose. In addition, NA delivered intranasally to mice led to an increase in terminal galactose residues on the surface of airway cells and therefore an increase in AAV9 transduction (1, 38).
The goal of the present study was to identify the amino acids of the AAV9 capsid required for galactose binding, which we determined to include N470, D271, N272, Y446, and W503. Comparing the galactose binding site of AAV9 to that of other galactose binding proteins, similarities were discovered in regard to amino acid composition. The interaction of sugars with aromatic residues is common in the binding site of carbohydrate-binding proteins and has been observed for galactose-specific proteins (10, 39). These residues form hydrophobic interactions with the saccharide and are also involved in the discrimination of galactose from other sugars, such as glucose (10, 39). This is consistent with our mapped binding site for AAV9, which contains two aromatic residues, Y446 and W503. In addition, many galactose binding proteins contain polar and charged amino acids, including asparagine, aspartic acid, and glutamine, that have been shown to contribute to galactose binding by forming hydrogen bonds with the hydroxyl groups of the sugar (10, 26). It is likely that N470, D271, and N272 of the AAV9 capsid are forming similar interactions with galactose.
In a study by Pulicherla et al. (34), amino acids implicated in AAV9 liver transduction were identified through the generation of a random AAV9 capsid library. These researchers discovered a cluster of residues that may be involved in liver tropism, including N498, W503, P504, and Q590. They found that mutating W503 to arginine led to a vector that detargeted the liver. In our study, we found that when mutating W503 to alanine, the resultant vector lost the ability to bind to galactose, which is consistent with this amino acid being important for AAV9 tropism. We found, however, that mutating P504 to alanine had no effect on AAV9 galactose binding. N498 and Q590 are located on the peaks surrounding the 3-fold axes of symmetry of the capsid and are not located within the defined galactose binding pocket.
The location of the receptor binding domain of other AAV serotypes has also been determined. The HS binding site of AAV2 was mapped to five amino acids: R484, R487, R585, R588, and K532 (21, 23, 30, 31). These amino acids form a basic patch on the inside of each 3-fold symmetry-related protrusion of the capsid. AAV6, which has also been shown to bind to HS, contains a similar basic stretch of amino acids in analogous positions as observed for AAV2, including R485, R488, K528, and K533 (29). In addition, through mutagenesis studies, K493, K459, R576, and K531, have all been shown to be necessary for AAV6 HS binding and are located in neighboring positions (48). The galactose binding domain of AAV9, located at the base of the protrusions surrounding the 3-fold axes, is on the opposite side of this structure compared to the AAV2 HS binding region, which is located on the inside surface facing the 3-fold axis. The AAV6 HS binding residues are also located on the outside wall of the protrusions, but on the opposite wall closer to VR III and the icosahedral 2-fold axis compared to the AAV9 galactose binding residues. These observations further illustrate the importance of the amino acids and structures that assemble the protrusions surrounding the 3-fold axes and the residues adjacent to this region in receptor recognition for different AAV serotypes. It also supports a proposal that the common structural variability of the AAV capsids, particularly the surface exposed protrusions, have evolved to enable the utilization of different surface molecules for successful infection, which is an important determinant of their respective transduction phenotypes.
AAV9 shows unique characteristics that could potentially be explained by receptor interactions. For example, after intravenous injection, AAV9 can surpass the blood-brain barrier and transduce motor neurons in the spinal cord of adult and neonatal mice and cats, as well as neurons in the neonatal mouse brain and astrocytes in the adult mouse brain (9, 11). Also, when tested in nonhuman primates, AAV9 transduced motor neurons in the spinal cord and primarily glial cells in the brain (3, 17). By fluorescent lectin staining, we have previously observed abundant galactose expression on the surface of blood vessels in the mouse brain (1). AAV9 could possibly be using galactose to facilitate transcytosis across the vasculature, allowing entry into the central nervous system. Engineering the galactose binding domain onto the capsid of other AAV serotypes may allow the transfer of AAV9's attractive phenotypes and the development of novel vectors for gene therapy.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by National Institutes of Health grants T32 DK007748 (C.L.B.), R01 GM082946 (M.A.-M.), and P01 HL059407 (J.M.W.) and by a sponsored research agreement from ReGenX Bioscience (J.M.W.).
J.M.W. is a consultant to ReGenX Holdings and is a founder of, holds equity in, and receives a grant from affiliates of ReGenX Holdings; in addition, J.M.W. is an inventor on patents licensed to various biopharmaceutical companies, including affiliates of ReGenX Holdings.
Footnotes
Published ahead of print 18 April 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Bell CL, et al. 2011. The AAV9 receptor and its modification to improve in vivo lung gene transfer in mice. J. Clin. Invest. 121:2427–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bell P, et al. 2005. An optimized protocol for detection of Escherichia coli β-galactosidase in lung tissue following gene transfer. Histochem. Cell Biol. 124:77–85 [DOI] [PubMed] [Google Scholar]
- 3. Bevan AK, et al. 2011. Systemic gene delivery in large species for targeting spinal cord, brain, and peripheral tissues for pediatric disorders. Mol. Ther. doi:10.1038/mt.2011.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bish LT, et al. 2008. Adeno-associated virus (AAV) serotype 9 provides global cardiac gene transfer superior to AAV1, AAV6, AAV7, and AAV8 in the mouse and rat. Hum. Gene Ther. 19:1359–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carrillo-Tripp M, et al. 2009. VIPERdb2: an enhanced and web API enabled relational database for structural virology. Nucleic Acids Res. 37:D436–D442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Conlon TJ, et al. 2005. Efficient hepatic delivery and expression from a recombinant adeno-associated virus 8 pseudotyped α1-antitrypsin vector. Mol. Ther. 12:867–875 [DOI] [PubMed] [Google Scholar]
- 7. Dimattia M, et al. 2012. Structural insight into the unique properties of adeno-associated virus serotype 9. J. Virol. 86:6947–6958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ding W, Zhang L, Yan Z, Engelhardt JF. 2005. Intracellular trafficking of adeno-associated viral vectors. Gene Ther. 12:873–880 [DOI] [PubMed] [Google Scholar]
- 9. Duque S, et al. 2009. Intravenous administration of self-complementary AAV9 enables transgene delivery to adult motor neurons. Mol. Ther. 17:1187–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Elgavish S, Shaanan B. 1997. Lectin-carbohydrate interactions: different folds, common recognition principles. Trends Biochem. Sci. 22:462–467 [DOI] [PubMed] [Google Scholar]
- 11. Foust KD, et al. 2009. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat. Biotechnol. 27:59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fry EE, et al. 2010. Crystal structure of equine rhinitis A virus in complex with its sialic acid receptor. J. Gen. Virol. 91:1971–1977 [DOI] [PubMed] [Google Scholar]
- 13. Gao G, et al. 2003. Adeno-associated viruses undergo substantial evolution in primates during natural infections. Proc. Natl. Acad. Sci. U. S. A. 100:6081–6086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gao G, et al. 2004. Clades of adeno-associated viruses are widely disseminated in human tissues. J. Virol. 78:6381–6388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gao GP, et al. 2002. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. U. S. A. 99:11854–11859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Govindasamy L, et al. 2006. Structurally mapping the diverse phenotype of adeno-associated virus serotype 4. J. Virol. 80:11556–11570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gray SJ, et al. 2011. Preclinical differences of intravascular AAV9 delivery to neurons and glia: a comparative study of adult mice and nonhuman primates. Mol. Ther. 19:1058–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Halbert CL, Allen JM, Miller AD. 2001. Adeno-associated virus type 6 (AAV6) vectors mediate efficient transduction of airway epithelial cells in mouse lungs compared to that of AAV2 vectors. J. Virol. 75:6615–6624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Inagaki K, et al. 2006. Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Mol. Ther. 14:45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaludov N, Brown KE, Walters RW, Zabner J, Chiorini JA. 2001. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J. Virol. 75:6884–6893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kern A, et al. 2003. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 77:11072–11081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lerch TF, Xie Q, Chapman MS. 2010. The structure of adeno-associated virus serotype 3B (AAV-3B): insights into receptor binding and immune evasion. Virology 403:26–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Levy HC, et al. 2009. Heparin binding induces conformational changes in adeno-associated virus serotype 2. J. Struct. Biol. 165:146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Limberis MP, Vandenberghe LH, Zhang L, Pickles RJ, Wilson JM. 2009. Transduction efficiencies of novel AAV vectors in mouse airway epithelium in vivo and human ciliated airway epithelium in vitro. Mol. Ther. 17:294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mitchell M, et al. 2009. Production, purification, and preliminary X-ray crystallographic studies of adeno-associated virus serotype 9. Acta Crystallogr. Sect. F Struct. Biol. Crystallogr. Commun. 65:715–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Montfort W, et al. 1987. The three-dimensional structure of ricin at 2.8 Å. J. Biol. Chem. 262:5398–5403 [PubMed] [Google Scholar]
- 27. Mori S, Wang L, Takeuchi T, Kanda T. 2004. Two novel adeno-associated viruses from cynomolgus monkey: pseudotyping characterization of capsid protein. Virology 330:375–383 [DOI] [PubMed] [Google Scholar]
- 28. Nam HJ, et al. 2007. Structure of adeno-associated virus serotype 8, a gene therapy vector. J. Virol. 81:12260–12271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ng R, et al. 2010. Structural characterization of the dual glycan binding adeno-associated virus serotype 6. J. Virol. 84:12945–12957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. O'Donnell J, Taylor KA, Chapman MS. 2009. Adeno-associated virus-2 and its primary cellular receptor: cryo-EM structure of a heparin complex. Virology 385:434–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Opie SR, Warrington KH, Jr, Agbandje-McKenna M, Zolotukhin S, Muzyczka N. 2003. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J. Virol. 77:6995–7006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pacak CA, et al. 2006. Recombinant adeno-associated virus serotype 9 leads to preferential cardiac transduction in vivo. Circ. Res. 99:e3–e9 [DOI] [PubMed] [Google Scholar]
- 33. Patnaik SK, Stanley P. 2006. Lectin-resistant CHO glycosylation mutants. Methods Enzymol. 416:159–182 [DOI] [PubMed] [Google Scholar]
- 34. Pulicherla N, et al. 2011. Engineering liver-detargeted AAV9 vectors for cardiac and musculoskeletal gene transfer. Mol. Ther. 19:1070–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rutledge EA, Halbert CL, Russell DW. 1998. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J. Virol. 72:309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a. Schmidt M, et al. 2008. Adeno-associated virus type 12 (AAV12): a novel AAV serotype with sialic acid- and heparan sulfate proteoglycan-independent transduction activity. J. Virol. 82:1399–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. 2005. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 33:W363–W367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seetharaman J, et al. 1998. X-ray crystal structure of the human galectin-3 carbohydrate recognition domain at 2.1-Å resolution. J. Biol. Chem. 273:13047–13052 [DOI] [PubMed] [Google Scholar]
- 38. Shen S, Bryant KD, Brown SM, Randell SH, Asokan A. 2011. Terminal N-linked galactose is the primary receptor for adeno-associated virus 9. J. Biol. Chem. 286:13532–13540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sujatha MS, Sasidhar YU, Balaji PV. 2005. Insights into the role of the aromatic residue in galactose-binding sites: MP2/6-311G++** study on galactose- and glucose-aromatic residue analogue complexes. Biochemistry 44:8554–8562 [DOI] [PubMed] [Google Scholar]
- 40. Summerford C, Samulski RJ. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 72:1438–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Van Vliet KM, Blouin V, Brument N, Agbandje-McKenna M, Snyder RO. 2008. The role of the adeno-associated virus capsid in gene transfer. Methods Mol. Biol. 437:51–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vandendriessche T, et al. 2007. Efficacy and safety of adeno-associated viral vectors based on serotype 8 and 9 versus lentiviral vectors for hemophilia B gene therapy. J. Thromb. Haemost. 5:16–24 [DOI] [PubMed] [Google Scholar]
- 43. Walters RW, et al. 2001. Binding of adeno-associated virus type 5 to 2,3-linked sialic acid is required for gene transfer. J. Biol. Chem. 276:20610–20616 [DOI] [PubMed] [Google Scholar]
- 44. Wu Z, Miller E, Agbandje-McKenna M, Samulski RJ. 2006. α2,3 and α2,6 N-linked sialic acids facilitate efficient binding and transduction by adeno-associated virus types 1 and 6. J. Virol. 80:9093–9103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xiao C, Rossmann MG. 2007. Interpretation of electron density with stereographic roadmap projections. J. Struct. Biol. 158:182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xiao W, et al. 1999. Gene therapy vectors based on adeno-associated virus type 1. J. Virol. 73:3994–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xie Q, et al. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. U. S. A. 99:10405–10410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xie Q, Lerch TF, Meyer NL, Chapman MS. 2011. Structure-function analysis of receptor-binding in adeno-associated virus serotype 6 (AAV-6). Virology 420:10–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. 2008. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 16:1073–1080 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.