Abstract
There is a growing need for novel antiviral therapies that are broad spectrum, effective, and not subject to resistance due to viral mutations. Using high-throughput screening methods, including computational docking studies and an interferon-stimulated gene 54 (ISG54)-luciferase reporter assay, we identified a class of isoflavone compounds that act as specific agonists of innate immune signaling pathways and cause activation of the interferon regulatory factor (IRF-3) transcription factor. The isoflavone compounds activated the ISG54 promoter, mediated nuclear translocation of IRF-3, and displayed highly potent activity against hepatitis C virus (HCV) and influenza virus. Additionally, these agonists efficiently activated IRF-3 in the presence of the HCV protease NS3-4A, which is known to blunt the host immune response. Furthermore, genomic studies showed that discrete innate immune pathways centered on IRF signaling were regulated following agonist treatment without causing global changes in host gene expression. Following treatment, the expression of only 64 cellular genes was significantly induced. This report provides the first evidence that innate immune pathways dependent on IRF-3 can be successfully targeted by small-molecule drugs for the development of novel broad-spectrum antiviral compounds.
INTRODUCTION
RNA viruses represent an enormous public health problem. Although drug therapy is essential to mitigate the significant morbidity and mortality associated with these viruses, the number of antiviral drugs is limited and many are poorly effective. Most current antiviral drugs, and those in development, are direct-acting antiviral molecules that specifically target viral proteins. Such drugs are narrow in spectrum and vulnerable to the rapid emergence of viral resistance. In addition, because most RNA viruses have small genomes, with many encoding fewer than a dozen proteins, viral targets are few. For these reasons, treating virus infection by stimulating or manipulating the host innate antiviral response, or by targeting cellular factors required for the viral life cycle, could prove advantageous in comparison to the traditional approach of targeting viral proteins (25).
The innate antiviral response is initiated when cellular pathogen-recognition receptor proteins, such as RIG-I, MDA5, and certain Toll-like receptors (TLRs), recognize and engage specific pathogen-associated molecular patterns present in viral proteins and nucleic acids (15, 28). This initiates intracellular signaling cascades that result in the activation of transcription factors, including interferon (IFN) regulatory factors (IRFs) and NF-κB, that regulate the expression of IFN, IFN-stimulated genes (ISGs), and genes encoding inflammatory cytokines and chemokines that direct the modulation of the adaptive immune response.
In the current study, we developed a novel high-throughput screening strategy to identify small molecules that stimulate RIG-I-mediated innate immune signaling. RIG-I is a cytosolic pathogen-recognition receptor that is essential for triggering immunity to a wide range of RNA viruses (14, 16, 19, 23, 30). By binding to motifs within the RNA virus genome, RIG-I initiates downstream signaling that results in the activation of IRF-3 and the induction of IFN-dependent gene expression (9, 20). Nucleic acid-like molecules are the only ligands of RIG-I that have been identified to date (4, 10, 20, 24). Activation of RIG-I through ligand binding induces a conformational change that relieves RIG-I signaling repression by an autologous repressor domain, thus allowing RIG-I to signal downstream through its tandem caspase activation and recruitment domains (CARDs) (19, 20). RIG-I signaling is transduced through IPS-1 (also known as Cardif, MAVs, and VISA), an essential adaptor protein that resides in the outer mitochondrial membrane (11, 18, 21, 29). IPS-1 recruits a macromolecular signaling complex that stimulates the downstream activation of IRF-3.
Compounds that trigger RIG-I signaling directly, or through modulation of RIG-I pathway components such as IRF-3, have potential applications as antiviral drugs or immune modulators. Here we report on the identification of novel compounds that modulate the activity of the RIG-I pathway, mediate translocation of IRF-3 to the nucleus, and have broad-spectrum antiviral activity.
MATERIALS AND METHODS
Reporter cell line synthesis and library selection.
Huh7 cells were stably transfected with a luciferase promoter construct driven by the consensus ISG54 promoter. The following sequence was inserted into the pGL4.17 construct (Promega) by the use of the restriction sites indicated (where underlined characters signify restriction sites used and boldface characters signify the TATA box): GAGCTCCTCCGGAGGAAAAAGAGTCCTCTAAAGTATAATAAAAAGAAAAAAAGAAAAAGAGTCCTGCCAATTTCACTTTCTAGTTTCACTTTCCCTTTTGTAACGTCAGCTGAAGGGAAACAAACAAAAAGGAACCAGAGGCCACTTGTATATATAGGTCTCTTCAGCATTTATTGGTGGCAGAAGAGGAAGATTTCTGAAGAGTGCAGCTGCGATATC.
Huh7 cells were transfected with plasmid DNA and grown under conditions of G418 selection until resistant clones were isolated and tested for luciferase expression. The B-actin cell line was created in a similar manner, and the following actin promoter sequence was inserted into the pGL4.76 vector (Promega): GAGCTCCCCAAGGCGGCCAACGCCAAAACTCTCCCTCCTCCTCTTCCTCAATCTCGCTCTCGCTCTTTTTTTTTTTCGCAAAAGGAGGGGAGAGGGGGTAAAAAAATGCTGCACTGTGCGGCGAAGCCGGTGAGTGAGCGGCGCGGGGCCAATCAGCGTGCGCCGTTCCGAAAGTTGCCTTTTATGGCTCGAGCGGCCGCGGCGGCGCCCTATAAAACCCAGCGGCGCGACGCGCCACCACCGCCGAGACCAAGCTT.
Huh7 cells were transfected with plasmid DNA and grown under conditions of hygromycin selection until resistant clones were isolated. The diversity library contains 20,000 small molecules from Life Chemicals that fulfill the requirements for drug-like properties (Lipinski's rule of five). Molecules were selected to maintain maximal structural diversity in the library.
High-throughput screening assay.
Reporter cell lines were cultured under conditions of antibiotic selection until seeding. Cells were seeded in white opaque-bottomed 96-well tissue culture plates at a density of 1 × 104cells/well and grown for 24 h without antibiotic selection. Each assay plate had wells A1, B1, C1, and D1 treated with dimethyl sulfoxide (DMSO) only to reach a final concentration of 0.5%; wells E1, F1, G1, and H1 were infected with 10 hemagglutinin activity (HA) Sendai virus as a positive control for ISG54-driven luciferase induction. The remaining wells were treated with compound diluted to a final concentration of 10 μM in media containing 0.5% DMSO. Cells were incubated with control or compound treatment for 24 h. Steady-Glo luciferase reagent (Promega) was added at 50 μl per well, mixed thoroughly, and incubated at room temperature for 20 min. Luminescence was quantitated per well using a luminometer (Berthold). Confirmation of initial hits was performed in triplicate.
MTS assay.
Huh7 cells were seeded in clear-bottomed 96-well tissue culture plates and grown for 24 h. Seeded cells were treated with increasing concentrations of compound (0, 5, 10, 20, and 50 μM in media containing 0.5% DMSO) for 24 h. Negative-control wells contained 0.5% DMSO only, and a positive control for cytotoxicity was examined using an encephalomyocarditis virus (EMCV) infection which causes a 100% cytopathic effect. The proportion of viable cells was calculated using a cell viability assay that measures conversion of a tetrazolium compound [MTS; 3-(4,5-dimethyl-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt] to a colored formazan compound in live cells. The conversion of MTS to formazan was detected by a 96-well microtiter plate reader, and the resulting optical densities (OD) were used directly to estimate cell viability. Cells were incubated for 3 h in the presence of one-step reagent (Cell Titer One; Promega) before OD reading was done. Experiments were performed in triplicate.
Immunofluorescent assay for nuclear translocation of IRF-3.
Huh7 cells were seeded in clear-bottomed 96-well tissue culture plates at a density of 5 × 103 cells per well and grown for 24 h. Cells were treated with compounds at final concentrations of 20 μM and 10 μM in media containing 0.5% DMSO. Negative-control cells contained 0.5% DMSO only, and positive-control cells were infected with 10 HA of Sendai virus. Cells were incubated for 24 h, and then monolayers were fixed in 4% paraformaldehyde, permeabilized, and stained for IRF-3 protein. Staining was performed as directed in the instructions provided with a Cellomics translocation kit (Thermo Scientific). Rabbit polyclonal antibody against IRF-3 (Kineta) was used (1:400). Secondary anti-rabbit antibody conjugated to Dylight 488 (fluorescein isothiocyanate [FITC] equivalent) and Hoescht dye (nuclear stain) were used for detection of cellular IRF-3 and cell nuclei. Following secondary antibody incubation, the monolayers were washed and left in 100 μl of wash buffer for imaging. IRF-3 staining (FITC) and nuclear staining (DAPI; 4′,6-diamidino-2-phenylindole) were viewed and quantitated by the use of an inverted microscope. Images were taken using MetaMorph software and saved as high-resolution image files. Compound treatments were done in triplicate.
HCV immunofluorescence-based antiviral assay.
Huh7 cells were seeded as described above for the IRF-3 immunofluorescence assay. Cells were treated with 10 μM compound in media containing 0.5% DMSO. After 24 h, the media solution with compound added was removed temporarily from wells. The cells were washed with phosphate-buffered saline (PBS) before HCV2a virus was added at the stated multiplicity of infection (MOI). Virus was incubated for 2 to 4 h and then removed. The cells were washed with PBS, and the compound solutions were replaced into each well. The cells were grown overnight, and then monolayers were fixed and stained for HCV proteins. Primary serum PHS no. 72 (1:3,000) was used. Secondary anti-human antibody conjugated to Dylight 488 or Alexa 488 (FITC equivalent) and Hoescht dye (nuclear stain) were used for detection of HCV protein and cell nuclei. Following secondary antibody incubation, the monolayers were washed and left in 100 μl of wash buffer for imaging as described above. The number of infected cells was quantitated based on positive staining. Compound treatments were done in triplicate.
Real-time qPCR to measure HCV RNA.
Huh7 cells were seeded and treated as described above for the HCV antiviral assay. Alternatively, the compound was added postinfection 6 h after HCV was added. The cells were infected as described above at an MOI greater than or equal to 1.0. At 72 h postinfection, the viral RNA was purified from culture media using a Qiagen Viral Amp kit as directed by the manufacturer. The numbers of genome copies of HCV2a in 5 μl of RNA purified from supernatants were determined by quantitative PCR (qPCR). To create a standard curve for HCV RNA, in vitro-transcribed HCV2a was serially diluted from 101 to 1012 in nuclease-free water and 5 μl was added to the master mixture of PCR reagents. HCV2a primers were optimized at Kineta, and the following primers were utilized: 5′-GCACCATGAGCACAAATCCTAA-3′ and 5′ GGAACTTAACGTCTTCTGGG 3′.
PCR was performed under the normal conditions for a 20-μl Power SYBR green (Applied Biosystems) reaction mixture with 125 nM each HCV primer.
Influenza A virus enzyme-linked immunosorbent assay (ELISA).
MRC5 cells were seeded in a clear-bottomed 96-well tissue culture plate at a density of 1 × 104 cells/well and grown for 16 h. Seeded cells were treated with increasing concentrations of compound (0, 5, 10, 20, and 50 μM in media containing 0.5% DMSO) for 6 h and then infected with 125 PFU of influenza virus strain WSN. Diluted virus was added directly to the well, and compound was not removed. Cells were grown for a total of 24 h post-compound treatment and fixed with methanol:acetone. Cells were blocked with horse serum and bovine serum albumin (BSA) in the presence of Triton X-100. Mouse anti-influenza A virus nucleoprotein monoclonal antibody (Chemicon) was used (1:3,000). Secondary anti-mouse IgG-horseradish peroxidase (IgG-HRP) (Pierce) was used (1:3,000). The reaction was developed using TMBK BioFX reagents as suggested by the manufacturer. Following reagent addition, the cells were incubated at room temperature for 2 to 5 min and 2 N HCl was used to stop the reaction. Plates were read at 450 nm on a microtiter plate reader.
Influenza A virus and IRF-3 Western blot analysis.
Cells were grown, treated, and infected as described above for the influenza virus WSN ELISA. Alternatively, infected or drug-treated cells were harvested for protein lysates 24 h postinfection/treatment by washing the cells once with PBS and lysing the cells in radioimmunoprecipitation (RIPA) buffer. IRF-3 lysates contained a phosphatase inhibitor cocktail in the RIPA buffer. Lysates were harvested and analyzed for proteins by Western blot analysis with the same anti-influenza virus antibody as described above. The IRF-3 lysates were probed with the monoclonal rabbit anti-IRF-3 antibody that is specific to phosphorylated protein on serine 396 (Cell Signaling Technology catalog no. 4947) at a 1:1,000 dilution in TBST buffer (Tris-buffered saline with 0.1% Tween). HRP-conjugated anti-rabbit secondary antibody was used for detection (1:5,000). Both Western blots were probed with hsp90 as a loading control.
Influenza A virus focus-forming antiviral assay.
MRC5 cells were seeded in a clear-bottomed 96-well tissue culture plate at a density of 1 × 104 cells/well and grown for 16 h. Seeded cells were treated with increasing concentrations of compound (0, 5, 10, 20, and 50 μM in media containing 0.5% DMSO) for 6 h and then infected with 125 PFU of influenza virus strain WSN. Diluted virus was added directly to the well, and compound was not removed. Cells were grown for a total of 14 h post-compound treatment and fixed with methanol:acetone. Cells were blocked with horse serum and BSA in the presence of Triton X-100. Mouse anti-influenza A virus nucleoprotein monoclonal antibody (Chemicon) was used (1:3,000). Dylight 488 anti-mouse secondary antibody and Hoescht stain were used for fluorescence detection (1:5,000). Images were visualized and quantitated using an Array Scan VTI instrument (Thermo Fisher Scientific).
Expression microarray analysis and bioinformatics.
Total RNA isolation and mRNA amplification were performed on equal masses of total RNA from MRC5 cells treated with DMSO, negative control, or 10 μM KIN 101 at 20 h posttreatment and from MRC5 cells infected with Sendai virus at 20 h postinfection.
Probe labeling and microarray slide hybridization were performed with Agilent Whole Human Genome microarrays. Each microarray contains 44,000 probes, corresponding to approximately 41,000 human genes. Slides were scanned with an Agilent DNA microarray scanner, and the resulting images were analyzed using Agilent Feature Extractor (AFE) version 9.5.3.1. All data were entered into a custom-designed Oracle 9i-backed relational database (Labkey, Inc., Seattle, WA) and uploaded into Rosetta Resolver System 7.2 (Rosetta Biosoftware) and Spotfire Decision Site 9.1.1 (Spotfire AB) for analysis and visualization. All primary expression microarray data are available at http://viromics.washington.edu.
One-way textbook analysis of variance (ANOVA; Benjamini-Hochberg multiple-test corrected P value < 0.01) was performed to determine gene expression differences across treatments (KIN 101, negative control, and DMSO). Probes identified by ANOVA were further filtered to include only those genes found to have significantly different expression compared to that seen with DMSO treatment (post hoc test of ANOVA results; Scheffe P value < 0.1) that also differed >2-fold from the expression from DMSO-treated cells.
Functional analysis of statistically significant gene expression changes was performed with Ingenuity pathways analysis (Ingenuity Systems). This software analyzes RNA expression data in the context of known biological response and regulatory networks as well as other higher-order response pathways. Ingenuity functional analysis identified the corresponding biological functions and/or diseases that were most significant among differentially regulated genes. In the functional networks, genes are represented as nodes, and the biological relationship between 2 nodes is represented as an edge (line). All edge determinations are supported by at least 1 published reference or by canonical information stored in the Ingenuity Pathways knowledge base.
Quantitative real-time PCR was used to validate certain transcriptional changes found by microarray analysis. Primer and probe sets for each of the target sequences were chosen from the Applied Biosystems Assays-on-Demand product list. Assays were performed using an ABI 7500 Real-Time PCR system and TaqMan chemistry.
RESULTS
Identification of small-molecule inducers of the ISG54 promoter.
To identify agonists of the RIG-I pathway, we developed a screening platform consisting of Huh7 cells harboring a luciferase reporter gene under the control of the ISG54 promoter. Since Huh7 cells do not express TLRs, their use as a screening platform biased the screen toward selection of compounds that activate RIG-I-like receptor (RLR)-signaling programs (13). Moreover, the ISG54 promoter encodes tandem IRF elements that bind activated IRF-3 and an IFN-stimulated response element that confers promoter induction by alpha/beta IFN (IFN-α/β). The screening of compounds was performed using noninfected cells to avoid identifying compounds with direct antiviral activity against viral proteins. Our assay conditions were optimized to yield low background under nonstimulated conditions (the absence of virus infection or IFN treatment) and reproducibly high levels of dose-dependent induction in response to Sendai virus infection, which strongly induces the ISG54 promoter (26).
We used the luciferase reporter assay to screen a small-molecule diversity library of 20,000 compounds that was selected to contain maximally diverse and drug-like molecules. Compounds that activated luciferase activity to 4 standard deviations above the mean over the entire diversity library were selected for further validation (Fig. 1A). This resulted in an initial hit rate of 0.49%. In addition, we used computational docking studies to identify small molecules that were predicted to bind to the ligand binding domain of RIG-I. This “targeted set” was also subsequently evaluated using the ISG54-luciferase screening assay and yielded a 4% hit rate, a significant enrichment over the diversity screen rate.
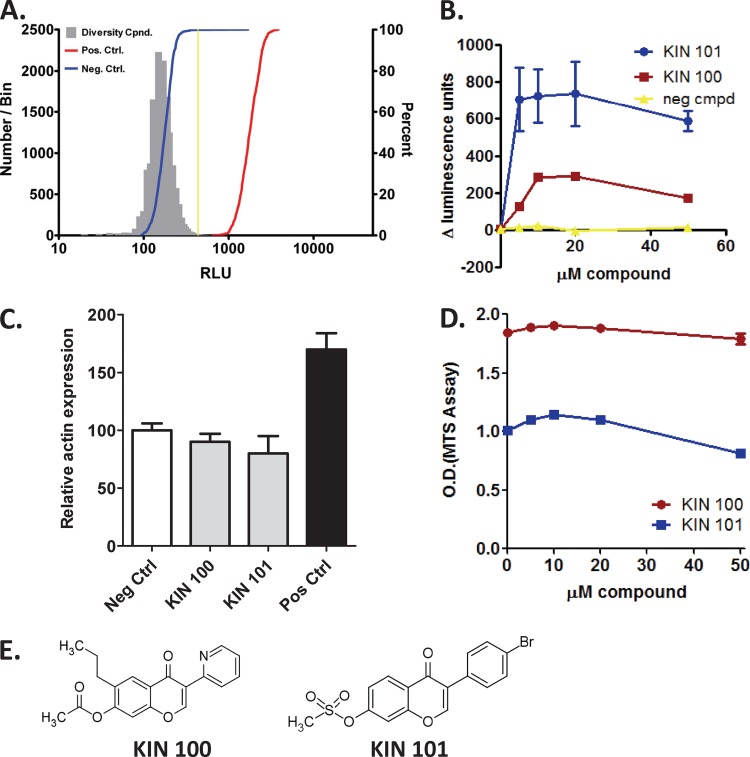
Fig 1.
Identification of small-molecule inducers of the ISG54 promoter. (A) A 20,000-member diversity library was screened at 10 μM to identify compounds (Cpnd.) that induced ISG54 luciferase reporter activity. Uninfected cells (negative control [Neg. Ctrl.]) and Sendai virus-infected cells (positive control [Pos. Ctrl.]) are represented on the histogram along with the sample population. The threshold (4 standard deviations) that was used to identify positive hits is indicated by the vertical yellow line. (B) Dose-dependent increase in ISG54 promoter activity. Huh7 cells stably expressing a luciferase reporter gene driven by the ISG54 promoter were treated with compound for 18 h and then analyzed for luciferase production. A negative compound (neg cmpd) was selected as a library member with no activity. Data were normalized by setting the DMSO control in each data set to 1. (C) Actin promoter induction. Huh7 cells expressing a luciferase construct driven by the actin promoter were treated with 10 μM compound and then analyzed for luciferase production. Negative-control cells were treated with vehicle alone (0.5% DMSO). Positive-control cells were treated with a compound from the diversity library that showed reproducible activation of the actin promoter. (D) Effects on cellular metabolism. Huh7 cells were grown for 48 h with increasing doses of compound and analyzed using an MTS assay to measure cellular metabolism (the higher the OD, the higher the metabolic activity). (E) Structures of isoflavone molecules KIN 100 and 101 are shown.
Initial compound hits were then validated to select molecules that activated the ISG54 promoter in a dose-dependent and -specific manner. This resulted in the identification of two isoflavone compounds, KIN 100 and KIN 101. Both compounds caused significant dose-dependent activation of the ISG54 promoter compared to inactive compounds within the library (Fig. 1B) without causing induction of β-actin promoter activity (Fig. 1C). KIN 101 had more potent activity in the ISG54 promoter assay, causing about a 5-fold increase in luciferase production over background. An MTS assay was used to analyze the in vitro cytotoxicity of these compounds in multiple cell types (Huh7, human liver; MRC5, lung fibroblast; and 293, human embryonic kidney cells). Neither KIN 100 nor KIN 101 had a significant effect on cellular metabolism or cytotoxicity at 50 μM (data from Huh7 cells are shown in Fig. 1D).
KIN 101 has broad-spectrum activity against RNA viruses.
Because KIN 100 and 101 were selected for their ability to act as RIG-I pathway agonists, they were predicted to have antiviral properties consistent with their function as innate immune modulators. To assay for antiviral activity, we first used a synthetic clone of HCV2a based upon the published JFH1 sequence (13a, 27a) in an HCV2a focus-forming assay to examine KIN 101 for its ability to inhibit HCV infection. Huh7 cells were treated for 18 h with KIN 101 at concentrations between 1 and 20 μM and subsequently infected with HCV2a at an MOI of 1. Under these conditions, KIN 101 had a significant and dose-dependent effect on the formation of foci (Fig. 2A). The 50% inhibitory concentration (IC50) for KIN 101 was calculated to be 0.2 μM, indicating potent inhibition of HCV infection in vitro for this library hit.
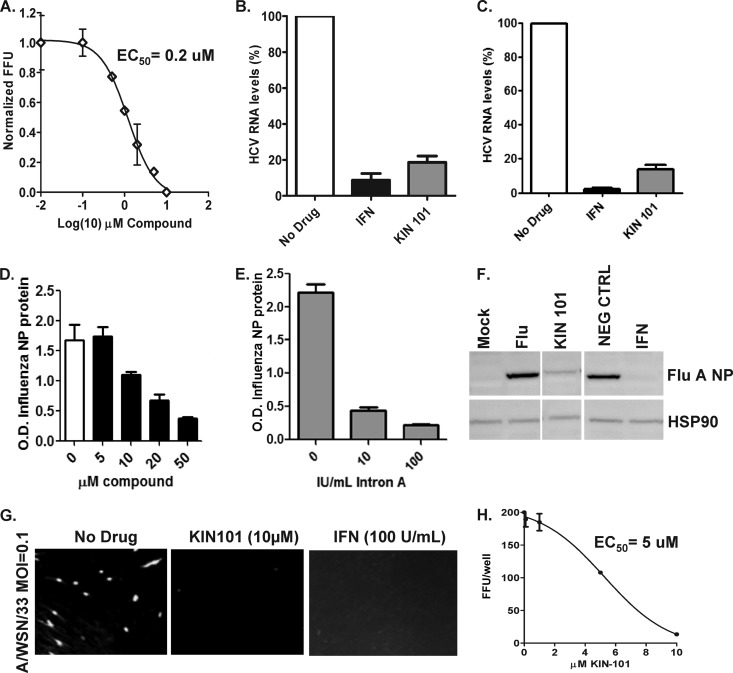
Fig 2.
KIN 101 exhibits activity against HCV2a and influenza virus in vitro. (A) Huh7 cells were treated with KIN 101 for 24 h and then infected with HCV2a at an MOI of 1.0. Cells were grown for an additional 48 h and then stained for HCV proteins. The number of infected cells versus concentration of drug added is shown. FFU, focus-forming units. (B) Huh7 cells were pretreated with KIN 101 (10 μM) and grown for 18 h; cells were then infected with HCV2a at an MOI of 1.0 and grown for an additional 72 h. Supernatants from infected cells were harvested, and HCV RNA was purified and quantified using real-time PCR with HCV-specific primers. Cells infected with HCV that did not receive KIN 101 (No Drug) were used as a negative control and values set to 100% infection. IFN-β (intron A; 100 IU/ml) was used as a positive control for HCV inhibition. (C) Huh7 cells were infected with HCV2a at an MOI of 1.0, and 4 h following infection, compounds or IFN was added to infected cells. The remainder of the experiment was done as described for panel B. (D) MRC5 cells were pretreated with increasing doses of KIN 101 for 4 h, infected with influenza A virus WSN at an MOI of 1.0, and grown for 24 h. Infected cells were detected using an ELISA method against the influenza virus NP protein. Infected cells with no drug treatment were used as a positive control for infection. (E) IFN-β was used as a positive control for antiviral activity. Cells were treated with indicated doses of IFN-β and infected with influenza virus, and infected cells were detected using the ELISA method. (F) MRC5 cells were treated and infected as described for panel D. Cells were harvested 24 h after infection and lysed, and whole-cell protein lysates were analyzed by Western blotting for the influenza virus NP protein. Mock-infected cells were used to determine nonspecific background, and influenza virus-infected cells with no drug treatment (Flu) served as a positive control for infection and detection. IFN was used a positive control for inhibition of influenza virus infection. The bottom panel shows staining with Hsp90 as a loading control. NEG CTRL, negative control. (G) MRC5 cells were treated and infected as described for panel D. At 16 h postinfection, the cells were fixed and the influenza virus NP protein was detected using specific and fluorescence-tagged antibodies. Negative-control cells that did not contain drug are shown in the left panel, cells treated with KIN 101 (10 μM) are shown in the middle, and cells treated with IFN-β are shown on the right. The infected cells were quantitated using the Array Scan instrument. (H) Influenza virus-infected cells were measured on the Array Scan instrument, and the relative number of infected cells following drug treatment is shown.
To determine the effect of KIN 101 on the production of HCV viral RNA, we used quantitative PCR. Huh7 cells were pretreated with KIN 101 for 18 h and subsequently infected with HCV2a at an MOI of 1.0. Cells were then grown for an additional 72 h, and viral RNA in the supernatants was quantified. Cells that were pretreated with KIN 101 showed a >1 log decrease in HCV RNA levels (Fig. 2B). The decrease in HCV RNA detected in supernatant following KIN 101 treatment was similar to the effect seen following treatment with IFN-β (intron A; 100 IU/ml) (Fig. 2B). In addition, we performed a drug challenge experiment in which KIN 101 was added 4 h after HCV infection was established. Consistent with potent antiviral function, KIN 101 decreased the production and release of HCV2a RNA in the supernatants of infected cells when added following infection (Fig. 2C). Notably, the antiviral effects of KIN 101 were similar when cells were pretreated with the compound for several hours prior to infection and when KIN 101 was added following HCV infection. This suggests that KIN 101 is fast acting in inducing innate immune pathways required for an antiviral response. Similar results were observed with KIN 100 (data not shown).
To determine whether KIN 100 and 101 have broad-spectrum antiviral activity, we also tested these compounds for antiviral effects against influenza virus. MRC5 cells were pretreated with KIN 101 for 4 h and infected with the WSN strain of H1N1 influenza A virus (125 focus-forming units per well or an MOI = 0.1). Using an ELISA method that measures levels of viral NP protein, we found that KIN 101 caused a dose-dependent decrease in influenza virus infection similar to that observed in cells treated with IFN-β (intron A; 100 IU/ml) (Fig. 2D and E). This decrease in influenza virus infection shown by the ELISA was confirmed by Western blot analysis. We found that treatment with KIN 101 caused a significant decrease in the NP protein abundance (Fig. 2F, lane 3) compared with that observed in untreated cells (lane 2) or cells treated with a negative-control compound (lane 4) and infected with the same amount of virus. The negative-control compound was selected from the diversity library and had no activity in screening or validation assays for ISG induction. IFN-treated cells were used as a positive control and showed a decrease in NP similar to that caused by KIN 101 (lane 5). To normalize for differences in protein amounts between samples, Hsp90 was used as a loading control (bottom panel), demonstrating that amounts of total protein in the lysates analyzed were equal. Additionally, a focus-forming assay was utilized to measure the inhibition of influenza virus infection. MRC5 cells were pretreated with compound for 4 h, and influenza WSN strain virus was used to infect cells at an MOI = 0.1. In the presence of vehicle alone (DMSO, no drug), influenza virus-infected cells or foci can be detected by staining with an NP-specific antibody and visualized by fluorescence microscopy. In the influenza virus-infected cells, approximately 10 to 20% of the cell monolayer was infected 24 h postinfection (Fig. 2G, left panel). In contrast, when compound was added to the cells, there was a complete inhibition of focus formation similar to that seen with IFN-treated cells and no influenza virus-infected cells were detected (Fig. 2G, center and right panels). A KIN 101 dose-response assay was performed to calculate the 50% effective concentration (EC50) of this compound in the influenza virus focus-forming assay, and treatments with compound at concentrations between 1 and 10 uM were utilized to create a drug regression curve (Fig. 2H). The EC50 of KIN 101 in the influenza virus infection assay was approximately 5 uM.
In contrast to KIN 101, KIN 100 had no significant activity against the WSN strain of influenza A virus despite testing using several cell types that are amenable to influenza virus infection (data not shown). This finding suggests a difference between the two isoflavone molecules in the mechanism of action. Further analyses are necessary to identify the structural components of KIN 101 that mediate broad-spectrum antiviral activity.
KIN 100 and 101 cause IRF-3 translocation that is not inhibited by viral countermeasures.
The transcription factor IRF-3 is a key component of the innate immune response that mediates the clearance of many RNA viruses. Following RNA virus infection, RIG-I signals through its essential adaptor protein, IPS-1, leading to recruitment of factors that direct the phosphorylation and activation of IRF-3. Once IRF-3 is in the phosphorylated state, it forms a dimer, translocates into the nucleus, and induces the expression of ISGs and cytokines through promoters containing IRF-3 binding sites (15). To examine the direct involvement of IRF-3 in the activity of KIN 100 and 101, we performed nuclear translocation assays using Huh7 cells. Huh7 cells express RIG-I but lack other pathogen-recognition receptors (8). Therefore, nuclear accumulation of IRF-3 is a specific indicator of RIG-I pathway activation (23).
In untreated Huh7 cells, IRF-3 was detected in both the cytoplasm and the nucleus, resulting in diffuse cellular staining (Fig. 3A). Upon activation of the RIG-I pathway by Sendai virus, IRF-3 translocated to and accumulated in the nucleus. KIN 100 and 101 both stimulated IRF-3 translocation to an extent similar to that induced by Sendai virus but did not alter the distribution of poly(A) binding protein. Poly(A) binding protein was used as a control to confirm that compounds were not causing a nonspecific deregulation of protein shuttling within the cell or a detrimental effect on cellular membranes that would cause a change in the subcellular distribution of IRF-3. Additionally, compounds that caused no ISG54 induction were selected from the diversity screen and used as an additional negative control. Negative-control compounds did not alter IRF-3 localization, demonstrating that this was a specific effect of KIN 100 and 101.
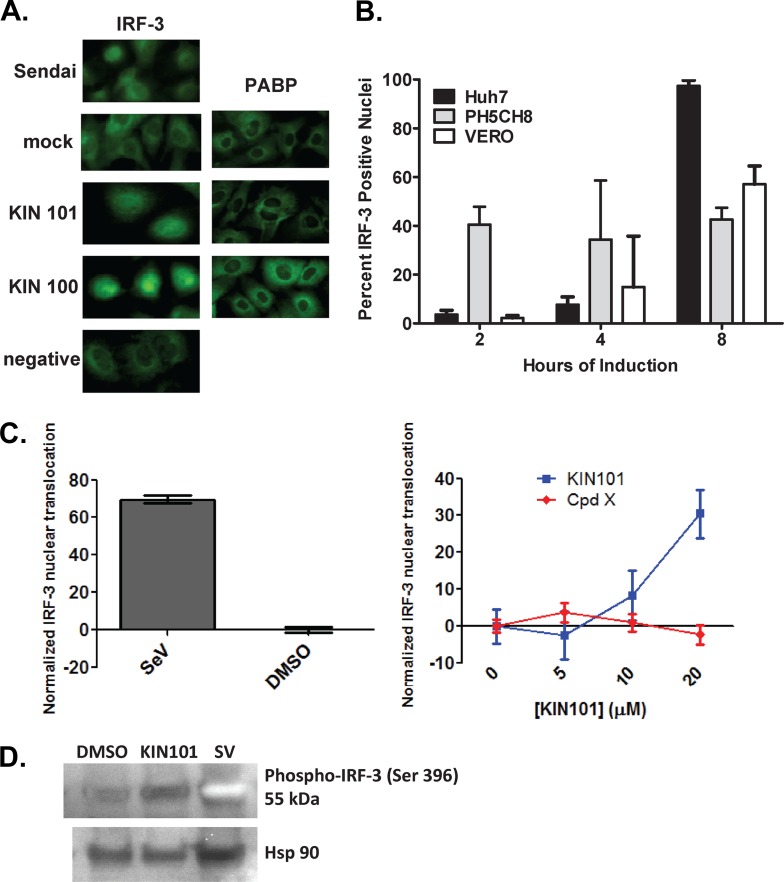
Fig 3.
KIN 100 and 101 induce IRF-3 nuclear translocation. (A) Huh7 cells (human liver carcinoma) were treated with drug or controls for 18 h, fixed, and stained with IRF-3 polyclonal rabbit serum and an FITC secondary antibody (green; left panel). Mock-infected cells were treated with vehicle alone (0.5% DMSO media), and Sendai virus infection served as a positive control for causing IRF-3 nuclear translocation. KIN 101 and KIN 100 were added to cells in media containing 0.5% DMSO at a concentration of 10 μM as well as the negative samples that were treated with a drug that showed no activity in screening or validation assays for ISG induction. Cells shown in the right panel were treated in a similar fashion and stained for poly(A) binding protein (PABP) as a control for a cytoplasmic protein whose cellular distribution should not be altered by drug treatment. (B) Quantitation of IRF-3 translocation in various cell types at different treatment time points. Various cell types were treated with negative controls or KIN 101 as described for Fig. 2A for the amounts of time indicated. Following IRF-3 staining, the cells were analyzed using an Array Scan VTI instrument to quantitate the IRF-3 in the defined nucleus and cytoplasmic regions of the cells. Cells that were positive for IRF-3 nuclear staining were quantified by measuring the IRF-3-specific stain intensity, and the quantitation represents the average results determined for triplicate wells, with 1,000 cells per well analyzed. Cells positive for IRF-3 nuclear localization had 3 standard deviations more IRF-3 staining in the nucleus than the negative-control cells. (C) PH5CH8 cells were treated with different concentrations of KIN101 or a negative-control compound, and IRF-3 translocation was measured with the Array Scan instrument (right panel). The IRF-3 translocation was normalized by setting the value for the no-drug conditions to 1 in each data set. Sendai virus (SeV) and DMSO served as positive and negative controls, respectively (left panel). (D) Phosphorylated IRF-3 was detected by immunodetection with a rabbit monoclonal antibody that specifically recognizes IRF-3 protein that is phosphorylated on serine 396 (Cell Signaling Technology catalog no. 4947). HSP90 was detected as a loading control.
In addition to analysis of IRF-3 localization through immunofluorescence microscopy, we also used an Array Scan VTI instrument. The Array Scan technology uses immunofluorescence assays and mathematic algorithms to quantify the fluorescent signal in defined compartments within the cell and allows more assay conditions to be accurately compared within an experiment. We used the Array Scan VTI to perform an in-depth analysis of KIN 101 in the IRF-3 assay over multiple time points in three different cell lines (Fig. 3B). To quantify the effect of KIN 101 on IRF-3 translocation, the intensity of IRF-3 in the nucleus was measured and compared to that seen with control cells that were treated with a negative-control compound. Cells that showed nuclear IRF-3 levels that were increased by 3 standard deviations above negative-control cell levels were considered positive for IRF-3 nuclear translocation. KIN 101 induced the greatest degree of IRF-3 nuclear translocation in Huh7 cells but also induced a significant response in another liver cell line (PH5CH8) and in kidney epithelial cells (Vero cells) derived from African green monkeys. In all cell lines tested, the effect of KIN 101 on IRF-3 nuclear translocation occurred rapidly. IRF-3 activation was detected as soon as 4 h posttreatment of PH5CH8 and Vero cells and 8 h posttreatment of Huh7 cells. To measure dose-dependent IRF-3 translocation, PH5CH8 cells were treated with KIN 101 in a concentration range of 5 to 20 uM for 18 h. IRF-3 translocation was measured using the Array Scan instrument, compared to that of a negative-control compound, and normalized by setting the amount of IRF-3 translocation to zero in each compound set (Fig. 3C). At the highest concentration of KIN 101 (20 uM), a 30-fold increase in IRF-3 translocation was observed, in contrast to the level seen with Sendai virus-infected cells (positive control; right panel), which caused a 60-fold increase. An alternative method to measure IRF-3 activation is examining IRF-3 phosphorylation by immunodetection of the phosphorylated IRF-3 protein. PH5CH8 cells were treated with vehicle, KIN101, or Sendai virus as a control for 18 h (Fig. 3D). Whole-cell lysates were harvested, and Western blot analysis was used to detect phosphorylated IRF-3 with an antibody that specifically detects IRF-3 phosphorylated on serine 396 (Cell Signaling Technology catalog no. 4947). Although the phosphospecific antibody for IRF-3 showed weak detection, the cells treated with KIN101 showed an increased amount of phosphorylated IRF-3 compared to the DMSO control (Fig. 3D). Sendai virus infection causes a strong increase in phosphorylated IRF-3 as expected. Together, these data provide strong evidence that ISG activation and the resulting antiviral properties are caused through activation and translocation of IRF-3.
In order for innate immune agonists to function as effective antivirals, they must be capable of pathway stimulation in the presence of viral countermeasures. RNA viruses have adapted methods to suppress the RIG-I pathway by targeting and disrupting the function of various proteins involved in IRF-3 activation and ISG expression (3, 5). For example, the HCV protease NS3-4A cleaves IPS-1, which results in inhibition of IRF-3 phosphorylation and translocation into the nucleus (16, 18). To test the activity of KIN 101 in the presence of viral countermeasures, we used a cell line that expresses HCV NS3-4A under the control of a tetracycline-regulated promoter. Cells were grown in the absence of tetracycline to induce expression of the HCV NS3-4A protein (Fig. 4A), treated with 10 μM KIN 101 for 18 h, and analyzed for IRF-3 translocation using immunofluorescence microscopy and the Array Scan instrument. Our hypothesis is that KIN 101 can activate the RIG-I pathway through IRF-3 translocation in the presence of viral countermeasures such as HCV protease expression, resulting in the significant and broad-spectrum antiviral activity observed.
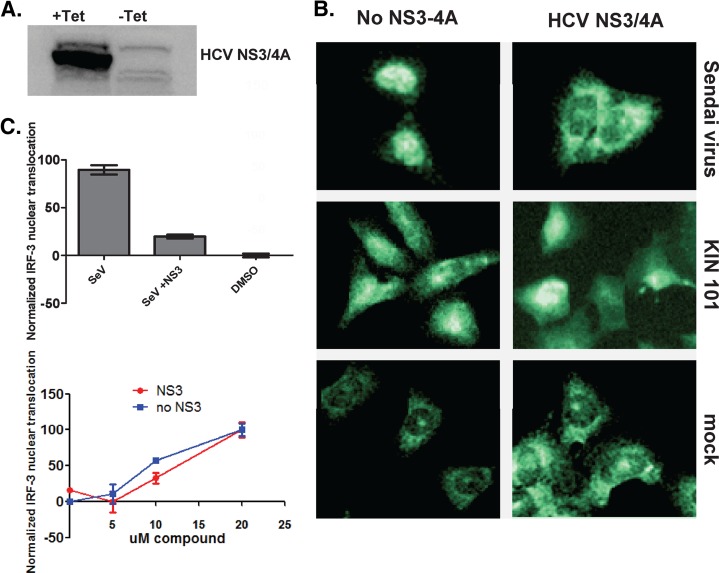
Fig 4.
KIN 101 induces IRF-3 nuclear translocation in cells expressing the HCV NS3 protease. (A) Human osteosarcoma cells were stably transformed with the HCV NS3-4A protease coding sequence under the control of a tetracycline-repressible promoter (+Tet). Cells were grown in the absence of tetracycline to induce high levels of HCV protease (−Tet). (B) Cells are grown with or without tetracycline and treated with KIN 101 (10 μM) or controls for 18 h. Cells were stained for IRF-3 as described for Fig. 3, and representative images are shown. Sendai virus infection was used as a control for IRF-3 translocation that is inhibited by HCV NS3-4A protease. Mock-treated cells were grown in media containing 0.5% DMSO. (C) Images were analyzed on the Array Scan VTI as described for Fig. 3. Values shown demonstrate IRF-3 intensity in the nucleus and represent the average results determined with triplicate wells, with 1,000 cells per well analyzed. Data were normalized by setting the value for the no-treatment DMSO control to 1 for each data set.
In the presence of NS3-4A, mock-infected cells exhibited diffuse IRF-3 staining that was detected predominantly in the cytoplasm. However, the translocation of IRF-3 that is normally induced by Sendai virus was blocked in the presence of NS3-4A (Fig. 4B). In contrast, KIN 101 induced significant IRF-3 translocation in the absence and presence of NS3-4A, suggesting that the agonist can function to upregulate the pathway in the presence of this viral countermeasure. Quantitation performed using the Array Scan instrument confirmed that conventional pathway activators such as Sendai virus infection are blocked by the HCV protease (Fig. 4C, upper panel). Sendai virus activates IRF-3 to almost 100-fold above the DMSO control level, but in the presence of NS3 protease, this activity is decreased by almost 90%. In contrast, the agonist compound KIN 101 caused significant IRF-3 nuclear translocation that was not inhibited by the expression of HCV protease NS3-4A (Fig. 4C, lower panel). KIN 101 causes dose-dependent IRF-3 translocation in cells without NS3 (blue line) and in cells expressing NS3 (red line). This initial pathway analysis suggests that KIN 101 is functioning to activate IRF-3 translocation downstream of IPS-1, since it is not inhibited by the HCV protease and IPS-1 cleavage. More detailed pathway mapping studies would be necessary to determine the exact targets of these isoflavone agonists.
KIN 101 causes a discrete induction of innate immune gene expression.
Since KIN101 exhibited broad antiviral activity, we used gene expression profiling to identify the genes and pathways that are affected by KIN 101 and determine whether there might be broader nonspecific gene expression changes that would be undesirable for an antiviral drug. MRC5 cells were treated with KIN 101 (10 μM) or a negative-control compound (selected from the diversity library for having no activity in screening assays) for 20 h. Sendai virus infection was again used as a positive control for the stimulation of innate immune pathways. Total RNA was purified from triplicate samples for microarray analysis.
We found that KIN 101 treatment resulted in the differential expression of 75 genes (ANOVA P value < 0.01; >2-fold change between groups [64 upregulated, 11 downregulated]) compared to DMSO treatment alone. This response was markedly constrained compared to the number of genes perturbed by either IFN treatment or virus infection in this or previous studies (6). Table 1 shows the top 20 genes that were upregulated following treatment with KIN 101 compared with the response seen in control cells treated with DMSO alone. (The complete list of 75 genes differentially expressed between KIN 101 and DMSO treatment is available in Table S1 in the supplemental material.) Gene expression patterns in cells that were treated with the negative-control compound showed no significant difference from those observed in DMSO-treated cells, and cells infected with Sendai virus as a positive control demonstrated a significant upregulation of innate immune pathways (see Fig. S1 in the supplemental material).
Table 1.
Top 20 genes upregulated in MRC5 cells following treatment with KIN 101
| Probe designation | Gene designation | Fold upregulation |
ANOVA P value | ||
|---|---|---|---|---|---|
| Negative vs DMSO | KIN 101 vs DMSO | Sendai vs DMSO | |||
| A_23_P17663 | MX1 | 1.04 | 93.64 | 100 | 0.00203 |
| A_23_P139786 | OASL | 1.41 | 86.52 | 100 | 0.00036 |
| A_24_P28722 | RSAD2 | 1.02 | 81.78 | 100 | 0.00095 |
| A_23_P45871 | IFI44L | −1.21 | 62.62 | 100 | 0.00095 |
| A_23_P52266 | IFIT1 | 1.01 | 28.15 | 100 | 0.00036 |
| A_23_P110196 | HERC5 | −1.03 | 28.09 | 100 | 0.00203 |
| A_24_P304071 | IFIT2 | 1.50 | 26.47 | 100 | 0.00538 |
| A_23_P64828 | OAS1 | −1.01 | 26.24 | 100 | 0.00446 |
| A_23_P24004 | IFIT2 | 1.02 | 23.73 | 100 | 0.00163 |
| A_23_P250358 | HERC6 | 1.96 | 18.86 | 98.83 | 0.00203 |
| A_23_P23074 | IFI44 | 1.21 | 18.79 | 99.91 | 0.00491 |
| A_23_P250353 | HERC6 | 1.31 | 17.26 | 100 | 0.00325 |
| A_24_P343929 | OAS2 | −1.02 | 17.09 | 100 | 0.00163 |
| A_23_P6263 | MX2 | −1.56 | 15.98 | 100 | 0.00016 |
| A_23_P384355 | LOC129607 | −1.69 | 15.87 | 100 | 0.0001 |
| A_24_P20607 | CXCL11 | 1.09 | 15.72 | 100 | 0.00915 |
| A_23_P68155 | IFIH1 | 1.13 | 14.02 | 100 | 0.00446 |
| A_23_P4283 | XAF1 | 1.09 | 13.35 | 97.17 | 0.00709 |
| A_23_P35412 | IFIT3 | −1.02 | 12.33 | 100 | 0.00003 |
| A_23_P48513 | IFI27 | 1.01 | 11.45 | 100 | 0.00543 |
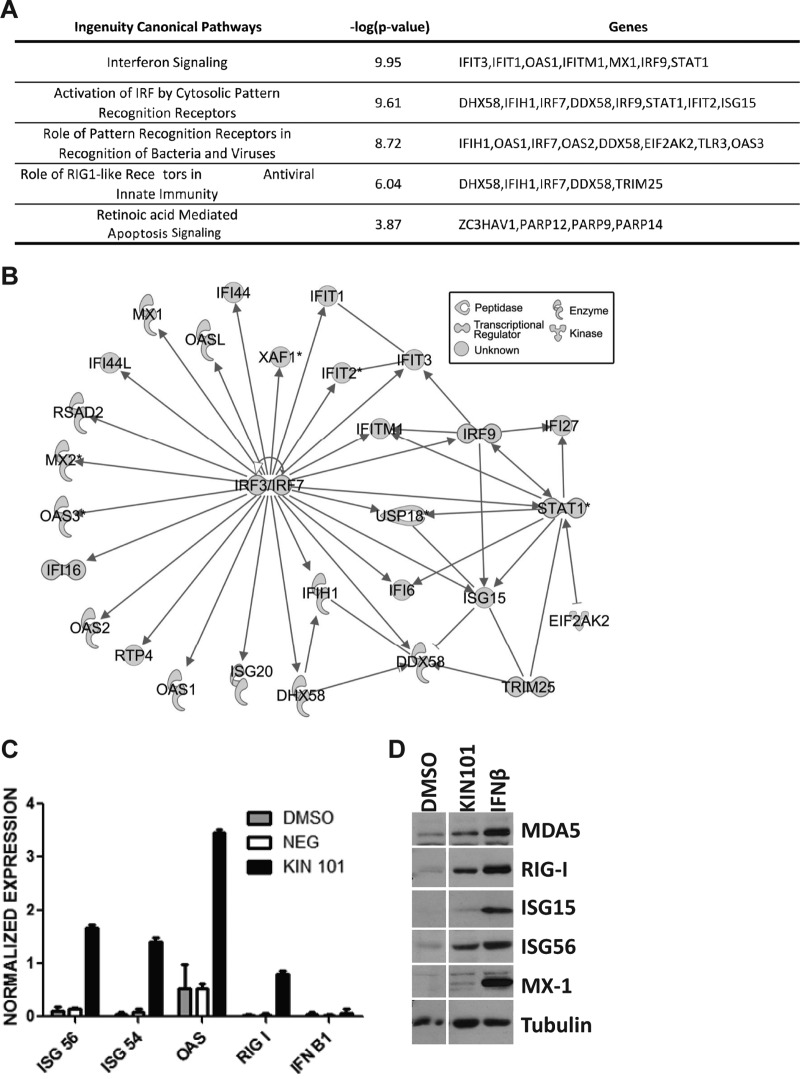
We used Ingenuity pathway analysis to identify the signaling pathways most affected by KIN 101 treatment. Figure 5A lists the top 5 Ingenuity canonical pathways that were identified as being most significantly perturbed, the genes involved, and their corresponding log P values. We also used Ingenuity pathway analysis to identify an interconnected network of genes stimulated by KIN 101 treatment. This network contained an abundance of downstream IRF effectors (IRF-3 and/or IRF-7) such as ISGs and other genes encoding innate immune proteins (Fig. 5B).
Fig 5.
KIN 101 induces the expression of discrete innate immune pathways centered on IRF signaling. (A) Top signaling pathways upregulated in MRC5 cells following treatment with KIN 101. (B) Network diagram generated using Ingenuity pathway analysis, showing an interconnected network of genes encoding innate immune proteins upregulated by KIN 101 treatment. (C) TaqMan confirmation of selected genes that were shown by microarray to be upregulated by KIN 101 treatment. Normalized expression is shown on a logarithmic scale. (D) Western blot analysis showing the increased abundance of proteins downstream of IRF activation following KIN 101 treatment. IFN-β treatment was used as a positive control.
To confirm gene expression changes identified by microarray, we performed TaqMan analysis on select genes that were induced following KIN 101 treatment. RNA quantitation by TaqMan confirmed the upregulation of key genes identified in the microarray, including those encoding ISG54, ISG56, OAS, and RIG-I (Fig. 5C). Additionally, genes that showed no change following KIN 101 treatment, including IFN-β, were not significantly upregulated in the PCR analysis. Levels of protein abundance were also analyzed by Western blotting to detect changes in the levels of proteins encoded by genes responsive to IRF-3. Treatment with KIN 101 resulted in a significant increase in the levels of ISGs as well as other proteins downstream of IRF activation such as RIG-I and MDA5 (Fig. 5D). Together, our analyses of host cell pathways responsive to KIN 101 revealed the significant induction of a distinct set of genes involved in host-mediated antiviral response pathways centered on IRF signaling.
DISCUSSION
Our results identify a novel class of innate immune pathway agonists that belong to the isoflavone family. These agonists caused specific and dose-dependent induction of the ISG54 promoter and had no significant toxicity or effect on cellular metabolism. Both agonists caused IRF-3 nuclear translocation even in the presence of the HCV NS3-4A protease, which is known to inhibit RIG-I signaling (16, 18). Additionally, KIN 101 was shown by microarray analysis to cause only discrete changes in gene expression profiles, with gene expression changes centering on IRF signaling. Both KIN 100 and 101 inhibited the replication of HCV, and KIN 101 also had activity against influenza virus.
Several isoflavones and related flavonoid compounds have previously been demonstrated to have antiviral properties (1). Although their mechanism of antiviral action is not well understood, isoflavones are thought to act both through direct-acting antiviral properties and through host-mediated functions. Genistein is the most studied isoflavone from an antiviral perspective, and it has been shown to inhibit the infectivity of a variety of viruses, including adenovirus (12), herpes simplex virus (17, 31), human immunodeficiency virus (22), and rotavirus (2). Isoflavones have been reported to affect virus binding, entry, replication, viral protein translation, and the formation of certain virus envelope glycoprotein complexes, suggesting broad mechanisms of action. Isoflavones also affect a variety of host cell signaling processes, including the induction of transcription factors and the secretion of cytokines (26). Isoflavone molecules have shown activity against several viruses that appears to be specific to their chemical structure. It has also been shown that isoflavone molecules, when added in combination, can exhibit synergistic activity (7), which provides additional evidence for unique mechanisms of action among isoflavone family members.
One of the isoflavone compounds we identified, KIN 100, was part of a targeted library that was chosen based upon predicted binding to the repressor domain of the RIG-I receptor. The repressor domain of RIG-I is known to be the binding site of the viral RNA ligand that results in activation of the IRF-3-dependent pathway and a resulting innate immune response. Actual binding studies to determine if KIN 100 binds to the RIG-I receptor have not been done and would be necessary to determine the exact target of the isoflavone molecules. Both isoflavone compounds described here activated ISG54 reporter gene expression and induced IRF-3 nuclear translocation. However, KIN 100 had activity against HCV but not influenza virus. This suggests that KIN 100 and KIN 101 could be acting at different points along the IRF-3 activation pathway.
Our finding that KIN 101 induced a limited and highly specific set of gene expression changes suggests that RIG-I pathway agonists, in comparison to IFN and other cytokines previously reported, have a narrower mechanism of action (i.e., limited stimulation of one or a small number of pathways that are critical to the innate immune response rather than global stimulation of the entire response) and better downstream homeostatic control. In addition, cellular inhibitors of RIG-I, including the LGP2 protein (19, 27), serve to suppress RIG-I signaling during the late stages of the innate immune response, further limiting the potential for long-acting inflammatory toxicities. The discrepancy between the RIG-I agonists described and a conventional interferon-mediated immune response is very interesting. Interferon-β induction following treatment with the isoflavone agonists has not been observed in various cell types at either the RNA or protein level. This suggests that the mechanism of antiviral activity is not dependent on induction of type I interferon. Ongoing studies are directly testing the role of secreted type I interferon and the signaling through the interferon receptor. Determining the innate immune target of these molecules would bring insight into the mechanism of action and unique antiviral properties of this novel drug class.
Further studies to map the specific targets of each isoflavone molecule are in progress. Additionally, in preliminary studies we found that other small-molecule agonists of the RIG-I pathway identified through our high-throughput screen, including additional isoflavones and other structural classes, have activity against West Nile virus, respiratory syncytial virus, dengue virus, and human immunodeficiency virus. Characterizing the full antiviral activity spectrum of isoflavones and additional structural classes of innate immune agonists should provide important information for therapeutic development as well as provide insight into their mechanism of action.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded in part by Public Health Service grant R43AI081335 and under contract HSSN 272200900035C from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services.
We kindly acknowledge the scientific team at Kineta, Inc., for critical input and the members of the Gale and Katze laboratories at the University of Washington for their scientific engagement. We also acknowledge Marcus Korth for assistance in preparing the manuscript for publication.
Footnotes
Published ahead of print 24 April 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Andres A, Donovan SM, Kuhlenschmidt MS. 2009. Soy isoflavones and virus infections. J. Nutr. Biochem. 20:563–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andres A, Donovan SM, Kuhlenschmidt TB, Kuhlenschmidt MS. 2007. Isoflavones at concentrations present in soy infant formula inhibit rotavirus infection in vitro. J. Nutr. 137:2068–2073 [DOI] [PubMed] [Google Scholar]
- 3. Barral PM, et al. 2009. Functions of the cytoplasmic RNA sensors RIG-I and MDA-5: key regulators of innate immunity. Pharmacol. Ther. 124:219–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baum A, Sachidanandam R, Garcia-Sastre A. 2010. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc. Natl. Acad. Sci. U. S. A. 107:16303–16308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Billharz R, et al. 2009. The NS1 protein of the 1918 pandemic influenza virus blocks host interferon and lipid metabolism pathways. J. Virol. 83:10557–10570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cilloniz C, et al. 2010. Lethal dissemination of H5N1 influenza virus is associated with dysregulation of inflammation and lipoxin signaling in a mouse model of infection. J. Virol. 84:7613–7624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cushnie TP, Lamb AJ. 2005. Antimicrobial activity of flavonoids. Int. J. Antimicrob. Agents 26:343–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Foy E, et al. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145–1148 [DOI] [PubMed] [Google Scholar]
- 9. Jiang F, et al. 25 September 2011, posting date Structural basis of RNA recognition and activation by innate immune receptor RIG-I. 479:423–427 doi:10.1038/nature10537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kato H, et al. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205:1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kawai T, et al. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981–988 [DOI] [PubMed] [Google Scholar]
- 12. Li E, et al. 2000. Association of p130CAS with phosphatidylinositol-3-OH kinase mediates adenovirus cell entry. J. Biol. Chem. 275:14729–14735 [DOI] [PubMed] [Google Scholar]
- 13. Li K, Chen Z, Kato N, Gale M, Jr, Lemon SM. 2005. Distinct poly(I-C) and virus-activated signaling pathways leading to interferon-beta production in hepatocytes. J. Biol. Chem. 280:16739–16747 [DOI] [PubMed] [Google Scholar]
- 13a. Lindenbach BD, et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 14. Loo YM, et al. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82:335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Loo YM, Gale M., Jr 2011. Immune signaling by RIG-I-like receptors. Immunity 34:680–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Loo YM, et al. 2006. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc. Natl. Acad. Sci. U. S. A. 103:6001–6006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lyu SY, Rhim JY, Park WB. 2005. Antiherpetic activities of flavonoids against herpes simplex virus type 1 (HSV-1) and type 2 (HSV-2) in vitro. Arch. Pharm. Res. 28:1293–1301 [DOI] [PubMed] [Google Scholar]
- 18. Meylan E, et al. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172 [DOI] [PubMed] [Google Scholar]
- 19. Saito T, et al. 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. U. S. A. 104:582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr 2008. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 454:523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669–682 [DOI] [PubMed] [Google Scholar]
- 22. Stantchev TS, Markovic I, Telford WG, Clouse KA, Broder CC. 2007. The tyrosine kinase inhibitor genistein blocks HIV-1 infection in primary human macrophages. Virus Res. 123:178–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sumpter R, Jr, et al. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79:2689–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Takahasi K, et al. 2008. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol. Cell 29:428–440 [DOI] [PubMed] [Google Scholar]
- 25. Tan SL, Ganji G, Paeper B, Proll S, Katze MG. 2007. Systems biology and the host response to viral infection. Nat. Biotechnol. 25:1383–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Terenzi F, Hui DJ, Merrick WC, Sen GC. 2006. Distinct induction patterns and functions of two closely related interferon-inducible human genes, ISG54 and ISG56. J. Biol. Chem. 281:34064–34071 [DOI] [PubMed] [Google Scholar]
- 27. Venkataraman T, et al. 2007. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J. Immunol. 178:6444–6455 [DOI] [PubMed] [Google Scholar]
- 27a. Wakita T, et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wilkins C, Gale M., Jr 2010. Recognition of viruses by cytoplasmic sensors. Curr. Opin. Immunol. 22:41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu LG, et al. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727–740 [DOI] [PubMed] [Google Scholar]
- 30. Yoneyama M, et al. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737 [DOI] [PubMed] [Google Scholar]
- 31. Yura Y, Yoshida H, Sato M. 1993. Inhibition of herpes simplex virus replication by genistein, an inhibitor of protein-tyrosine kinase. Arch. Virol. 132:451–461 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.