Abstract
Coxsackievirus A7 (CAV7) is a rarely detected and poorly characterized serotype of the Enterovirus species Human enterovirus A (HEV-A) within the Picornaviridae family. The CAV7-USSR strain has caused polio-like epidemics and was originally thought to represent the fourth poliovirus type, but later evidence linked this strain to the CAV7-Parker prototype. Another isolate, CAV7-275/58, was also serologically similar to Parker but was noninfectious in a mouse model. Sequencing of the genomic region encoding the capsid proteins of the USSR and 275/58 strains and subsequent comparison with the corresponding amino acid sequences of the Parker strain revealed that the Parker and USSR strains are nearly identical, while the 275/58 strain is more distant. Using electron cryomicroscopy and three-dimensional image reconstruction, the structures of the CAV7-USSR virion and empty capsid were resolved to 8.2-Å and 6.1-Å resolutions, respectively. This is one of the first detailed structural analyses of the HEV-A species. Using homology modeling, reconstruction segmentation, and flexible fitting, we constructed a pseudoatomic T = 1 (pseudo T = 3) model incorporating the three major capsid proteins (VP1 to VP3), addressed the conformational changes of the capsid and its constituent viral proteins occurring during RNA release, and mapped the capsid proteins' variable regions to the structure. During uncoating, VP4 and RNA are released analogously to poliovirus 1, the interfaces of VP2 and VP3 are rearranged, and VP1 rotates. Variable regions in the capsid proteins were predicted to map mainly to the surface of VP1 and are thus likely to affect the tropism and pathogenicity of CAV7.
INTRODUCTION
The Picornaviridae family contains numerous enteroviruses, which collectively are the most prevalent, economically costly causes of human viral infections (20). Although most enterovirus infections are asymptomatic or result in only mild respiratory symptoms, clinical manifestations include rash, carditis, neonatal sepsis-like disease, and infections of the central nervous system such as acute flaccid paralysis, meningitis, and encephalitis (12, 32, 48, 80). Enteroviruses have also been linked to chronic diseases such as type 1 diabetes mellitus (51, 52, 59, 69).
There are over one hundred enteroviruses that infect humans, and they are currently classified into four species, Human enterovirus A (HEV-A) to HEV-D (73). The prevalence and clinical significance of the members of the HEV-A species have been underscored by severe outbreaks during the past decade, including significant epidemics of hand, foot, and mouth disease in Asia, mainly caused by enterovirus 71 (EV71). EV71 has been specifically associated with neurological disease causing high mortality in children (2, 40, 41, 92). Like EV71, coxsackievirus A7 also belongs to the HEV-A species (60). CAV7 (prototype CAV7-Parker) was originally identified in the United States based on its pathogenicity in suckling mice and its serological properties (16, 17). Another strain, CAV7-USSR, was isolated in the former Soviet Union from the feces of a boy suffering from acute flaccid paralysis and was originally designated as poliovirus 4 (15, 33, 60, 79). The CAV7-275/58 strain was recovered from a case of aseptic meningitis but was nonpathogenic in newborn mice and therefore originally classified as an echovirus type in the Enterovirus genus (66). CAV7 is one of the few picornaviruses besides poliovirus and enterovirus 71 (EV71) that has been associated with outbreaks of flaccid paralysis (13, 22, 28, 30, 31, 41, 79). Several isolates were detected in the 1950s and 1960s during paralytic and meningitis epidemics in the United States, the former Soviet Union, and Scotland, but the serotype has rarely been detected since (8, 32). Although the genomic sequences of all the HEV-A prototype strains have been determined previously, the reasons why EV71 is now very prevalent (60) and CAV7 is not are unknown. Three receptor candidates have recently been identified for EV71, which may improve our understanding of this clinically important enterovirus species (57, 87, 90). CAV7, unlike EV71, does not use SCARB2 as a receptor (87).
Picornaviruses consist of an ∼7.5-kb, single-stranded, positive-sense RNA genome enclosed by an icosahedrally symmetric, nonenveloped capsid composed of 60 copies of each of the four capsid proteins, VP1 to VP4 (VP1-VP4). The introduction of picornaviral genomes into the host cell cytoplasm is thought to be facilitated by conformational changes occurring in the viral capsid due to, for instance, receptor binding or exposure to the low pH of the endosome (29, 76, 84, 85). These changes allow the exposure of membrane-active peptides that can form a fusion pore in the endosomal membrane for release of the RNA. This phenomenon is particularly well studied for poliovirus, where genome release is accompanied by the exposure of the N-terminal region of VP1 and the release of the myristoylated VP4 (14, 24, 50). Several authors have reported that the release of the genome may lead to expansion of the particles, such as the 4% expansion seen in human rhinoviruses (HRVs) after uncoating using atomic models fitted into rhinovirus reconstructions (38, 39). In poliovirus, these conformational changes related to expansion were also first explained by comparing pseudoatomic models from reconstructions at ∼22-Å resolution and later at 10-Å resolution of 160S, 135S, and 80S particles with the atomic model of the 160S particle (7, 49). Significantly, domain movements of up to 9 Å were modeled. The first atomic models outlining the conformational changes in the HRV2 80S particle and between empty and mature EV71 particles were described as this work was under review (27, 64, 81). As in poliovirus, 4% expansions through large domain movements were seen, disrupting the protomer interfaces to open up a pore on the 2-fold and at the base of the canyon.
We determined the sequences coding for the capsid proteins of CAV7-Parker, CAV7-USSR, and CAV7-275/58 strains. As there were no atomic models available at the time for any members of the HEV-A species, we then investigated the three-dimensional (3D) structure of the CAV7-USSR strain and its response to heat treatment by electron cryomicroscopy (cryoEM), 3D image reconstruction, homology modeling, and flexible fitting. Major conformational changes occur at the interfaces of the capsid proteins VP1, VP2, and VP3. Sequence comparisons and modeling suggest potential pathogenicity and/or receptor binding sites that may explain the differential behavior of CAV7 strains.
MATERIALS AND METHODS
Purification and stability of CAV7.
Three CAV7 strains (Parker, USSR, and 275/58) were obtained from the ATCC (ATCC VR-166, VR-319, and VR-673 for Parker, USSR, and 275/58, respectively) (15, 33, 66). The viruses were passaged twice through B-Vero cells (Bio-Cult Laboratories Ltd., UK) prior to propagation on the same cell line for purification. The infected cells were collected, freeze-thawed three times, and cleared by centrifugation at 10,000 × g for 30 min at 4°C. The supernatant was precipitated with 7% polyethylene glycol, 300 mM NaCl, harvested by low-speed centrifugation, and dissolved in 20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM MgCl2 (TNM), 0.3% sodium deoxycholate, 0.6% NP-40 for 30 min at 4°C followed by a short spin (1). The virus was further purified in 10-to-40% CsCl gradients (150,000 × g, 17 h, 4°C) prepared in phosphate-buffered saline (PBS) buffer with 1 mM MgCl2 (46). Fractions containing virus were collected and dialyzed against TNM buffer and cleared by centrifugation for 2 min (10,000 × g, 4°C). The resulting virus preparations were concentrated to ∼3 mg/ml in 25 mM phosphate buffer, 0.5 mM MgCl2, pH 7.4, and stored at −80°C until required. The identity of the purified viruses was confirmed by neutralization using CAV7-specific World Health Organization reference antiserum (42) and sequencing of the VP1 gene (58). Empty particles were induced by heating the gradient-purified virus at 56°C for 30 min, while the control aliquot was kept on ice (38). The samples were then analyzed by cryoEM for the presence of empty particles and the specific infectivity was determined.
Sequencing of the capsid protein-encoding regions.
Viral RNA was extracted from purified viruses using a Qiagen, ViralAmp RNA purification kit. Reverse transcription-PCR (RT-PCR) was performed using ImProm RT polymerase (Promega), Phusion DNA polymerase (Finnzymes, Finland), with the 5′ primer TCCTCCGGCCCCCTGAAT (65) and the 3′ primer CCTTGGGCAGTAGTGGATGAGA. The 3-kb amplicon, covering the genes coding for the capsid proteins, was subjected to stepwise sequencing (ABI PRISM 3130xl; Turku Centre for Biotechnology Sequencing Core Facility, Turku, Finland) using primers that were generated according to the CAV7-Parker nucleotide sequence (GenBank accession number AY421765). The consensus DNA sequences covering the open reading frames were generated in BioEdit (v7.0.0). The sequence alignments against the prototypic Parker strain (AY421765) were generated using ClustalW 1.82 with the default values (DNA gap open penalty, 15.0; DNA gap extension penalty, 6.66) and manually adjusted to equal lengths using the SeaView editor (74). Phylogenetic relationships from aligned sequences were inferred using programs DNADIST and PROTDIST in the Phylip package (version 3.65) (19).
Preparation of vitrified samples and cryoEM.
Vitrified samples of CAV7-USSR were prepared from 3-μl aliquots of purified virus or heat-treated virus on freshly glow-discharged Quantifoil R2/2 (Quantifoil Micro Tools GmbH, Germany) and C-Flat 224 (Electron Microscopy Sciences) grids as described previously (5). The grids were held in a GATAN 626 cryoholder maintained at −180°C in an FEI Tecnai F20 microscope (EM Unit, Institute of Biotechnology, University of Helsinki) operated at 200 kV. Images were recorded on Kodak SO163 film at a magnification of ×62,000 and scanned at 7 μm per pixel as described previously (70).
Image processing.
Micrographs were discarded if they had noticeable drift or astigmatism or indicated the presence of crystalline ice. The defocus level of each micrograph was determined using CTFFIND3 (55). Particles were automatically picked using ETHAN (44) and then manually screened, separated into filled- and empty-particle data sets, and windowed in EMAN (53). Starting models for both data sets were generated using a random model computation procedure (88, 89) operating on 150 images selected from the furthest from focus micrographs and resulted in initial reconstructions at ∼30 Å. These reconstructions then served as starting models for full orientation and origin determination of the entire data set using AUTO3DEM (89). Initially, the full and empty capsids were resolved to 8.5-Å and 6.8-Å resolutions, respectively. Subsequent refinement of magnification factors and defocus levels together with application of a temperature factor of 1/200 Å2 during the orientation search to emphasize contributions at higher spatial frequencies led to further improvement. The resolution of the empty capsid was estimated to be reliable to 6.1 Å and that of the filled capsid to 8.2 Å by Fourier shell correlation analysis (threshold criterion of 0.5) (77). The final reconstructions were calculated out to Nyquist frequency, the B-factors were estimated and corrected with EM-Bfactor, and then the data were truncated to the resolution indicated by the Fourier shell correlation analysis (21, 67). The segmentation of capsid proteins VP1-VP3 was done manually in Chimera, rendering the density at 3 standard deviations above the mean to allow sufficient separation of the proteins.
Modeling of CAV7 capsid proteins.
Predictions of the structure of CAV7-USSR VP1-VP4 capsid proteins were obtained by multiple-template comparative modeling using the I-TASSER server (68, 91). For VP1, the template structures were bovine enterovirus Protein Data Bank Identification (PDB ID):1bev (72), P1/Mahoney strain of poliovirus PDB ID:1hxs (54), human rhinovirus 16 PDB ID:1aym (34), human rhinovirus serotype 1A PDB ID:1r1a (43), coxsackievirus A9 PDB ID:1d4m (37), and coxsackievirus A21 PDB ID:1z7s (84). For VP2, the template structures were coxsackievirus A9 PDB ID:1d4m (37), Seneca Valley virus-001 PDB ID:3cji (78), coxsackievirus B3 PDB ID:1cov (56), Mahoney poliovirus PDB ID:1pov (6), human rhinovirus 14 PDB ID:4rhv (4), and bovine enterovirus PDB ID:1bev (72). For VP3, the template structures used were Mahoney poliovirus PDB ID:1pov (6), echovirus 1 PDB ID:1ev1 (23), equine rhinitis A virus PDB ID:2xbo (26), Seneca Valley virus-001 PDB ID:3cji (78), coxsackievirus A9 PDB ID:1d4m (37), echovirus 1 PDB ID:1ev1 (23), human rhinovirus serotype 1A PDB ID:1r1a (43), and bovine enterovirus PDB ID:1bev (72). For VP4, the template structures were coxsackievirus A21 PDB ID:1z7s (84), human rhinovirus 16 PDB ID:1aym (34), swine vesicular disease virus PDB ID:1oop (25), Mahoney poliovirus PDB ID:1al2 (82), Holliday junction DNA helicase ruvA PDB ID:1ixs (86), and human rhinovirus 16 PDB ID:1c8m. Initially, each of the models was treated as a rigid body and fitted manually into the respective segmented densities of the CAV7 VP1-VP3 proteins from both full and empty capsids. The fits were further improved using the fit-in-map tool for automated rigid fitting in Chimera (63). Flexible fitting was then applied, first to each model individually and then to the whole asymmetric unit. The flexible-fitting procedure included the identification of coarse rigid bodies within the individual models using RIBFIND (62) and a multistage refinement protocol (62) using Flex-EM (75). First, a refinement was performed based on the RIBFIND rigid bodies. In the second stage of refinement, each of the secondary structure elements (β-sheets and α-helices) was treated as a separate rigid body. The refined models for individual capsid proteins were then merged together into a single asymmetric unit and further refinement was applied, in the context of the density of the whole unit, to relieve any clashes among the different proteins (possibly occurring due to segmentation errors). Using the refined asymmetric unit, a pseudoatomic model for the whole capsid was generated by the oligomer generator utility in VIPERdb (71). However, clashes were observed in the interfaces between the asymmetric units. The unresolved loops causing clashes at the interface between the neighboring asymmetric units were removed, and a further refinement was performed. The amount of conformational change occurring in the proteins as a result of movement from the filled particle to the empty particle was estimated by calculating the Cα root mean square deviation (RMSD) upon the superposition of the refined models of each corresponding pair of capsid proteins, using MODELLER-9v7 (18). In addition, the component placement scores (62, 75) for corresponding proteins and individual secondary structure elements between empty and full were calculated. Pairwise three-dimensional alignment of the CAV7-USSR strain capsid proteins VP1-VP3 (filled capsid) against other picornaviruses was calculated using the PDBeFold server (47).
The CAV7 homology model of VP4 from I-TASSER was aligned with the VP4 atomic models from CBV3 (PDB: 1cov), CAV21 (red; PDB: 1z7s), PV1 (cyan; PDB: 1hxs), HRV16 (magenta; PDB: 1ayn), and HRV3 (yellow; PDB: 1rhi) using the matchmaker feature in Chimera (54, 56, 61, 63, 84, 93).
Accession numbers.
The reconstructions have been deposited in the EMDB with the accession numbers EMD-2027 and EMD-2028. The pseudoatomic models have been deposited in the PDBe and assigned wwPDB ID codes 4agx and 4agy. The gene sequences coding for the four structural proteins (VP1-VP4) of the CAV7 strains were deposited in the GenBank database with the following accession numbers: JN100649 (USSR VP1), JN100650 (Parker VP1), JN100651 (275/58 VP1), JN100652 (USSR VP2), JN100653 (Parker VP2), JN100654 (275/58 VP2), JN100655 (USSR VP3), JN100656 (Parker VP3), JN100657 (275/58 VP3), JN100658 (USSR VP4), JN100659 (Parker VP4), and JN100660 (275/58 VP4).
RESULTS
Sequence comparisons.
Genomic RNA from CAV7-Parker, USSR, and 275/58 strains was used in RT-PCR to generate cDNA for cloning and sequence analysis. The resulting nucleic acid sequences of the genes coding for the structural proteins were compared to each other (Table 1) and to those of CAV7-Parker (60). Surprisingly, the USSR and Parker strain nucleotide sequences were nearly identical despite the fact that the former originated from the former Soviet Union and the latter from the United States (Table 1). There were minor differences among the sequences of USSR (GenBank accession numbers JN100649, JN100652, JN100655, and JN100658), our resequenced Parker (GenBank accession numbers JN100650, JN100653, JN100656, and JN100659), and CAV7-Parker (GenBank accession number AY421765), which may be due to different RT-PCR/sequencing approaches. In contrast, the CAV7-275/58 sequences (GenBank accession numbers JN100651, JN100654, JN100657, and JN100660) were clearly more distant from these two strains (Table 1). The nucleotide differences among CAV7-275/58, CAV7-USSR, and CAV7-Parker strains in comparison to other enterovirus types are still within the limits of the typing criteria for enteroviruses (58), i.e., CAV7-275/58 is an isolate of CAV7 within the HEV-A species (58). CAV7-275/58 was also neutralized by CAV7-specific WHO antiserum (data not shown), thereby indicating that it has similar neutralizing antigenic sites to CAV7-Parker and CAV7-USSR strains. Interestingly, the differences in the capsid protein amino acid sequences among the three CAV7 strains were not concentrated in specific linear regions but were scattered throughout the sequences, which implies rapid evolution of the strain (8, 32).
Table 1.
Comparison of the percentage sequence similarities among resequenced CAV7 typesa
| Comparison strain | Similarity to CAV7-Parker in indicated genome region |
|||||||
|---|---|---|---|---|---|---|---|---|
| VP1 |
VP2 |
VP3 |
VP4 |
|||||
| nt | aa | nt | aa | nt | aa | nt | aa | |
| CAV7–USSR | 99.5 | 98.6 | 100.0 | 100.0 | 99.4 | 98.3 | 99.5 | 100.0 |
| CAV7–275/58 | 83.5 | 94.9 | 80.0 | 97.6 | 84.0 | 97.1 | 82.0 | 94.2 |
Comparisons are between resequenced CAV7- Parker and CAV7-USSR and between resequenced CAV7-Parker and CAV7-275/58 at the nucleotide (nt) and amino acid (aa) levels for the coding sequence corresponding to the major structural proteins VP1-VP4.
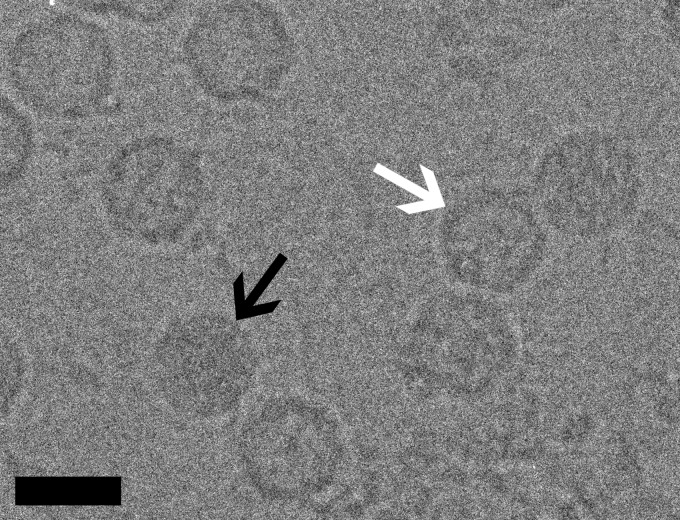
CryoEM images of CAV7.
The purified virus preparation contained both filled and empty particles of approximately 28 nm in diameter. The images of these particles were thus manually segregated during the image preprocessing (Fig. 1). To significantly increase the data set size for the structure determination of the empty capsid, we generated homogeneous samples of empty particles by incubating CAV7 preparations at 56°C for 30 min. The heat treatment of the virus preparation led to a significant drop in the infectivity of the preparation from 4.3 × 107 50% tissue culture infective doses (TCID50)/ml to 6.7 × 103 TCID50/ml, and the fraction of empty particles seen in cryoEM images increased from 68% to 100%.
Fig 1.
Micrographs of CAV7 particles suspended in a layer of vitreous water over holes in a carbon support film were recorded at a nominal magnification of ×62,000 in an FEI Tecnai F20 microscope operated at an accelerating voltage of 200 kV. Black and white arrows indicate filled particles and empty capsids, respectively. This image was recorded at a 4.3-μm underfocus. Bar, 30 nm.
3D image reconstruction and modeling of CAV7 capsid proteins.
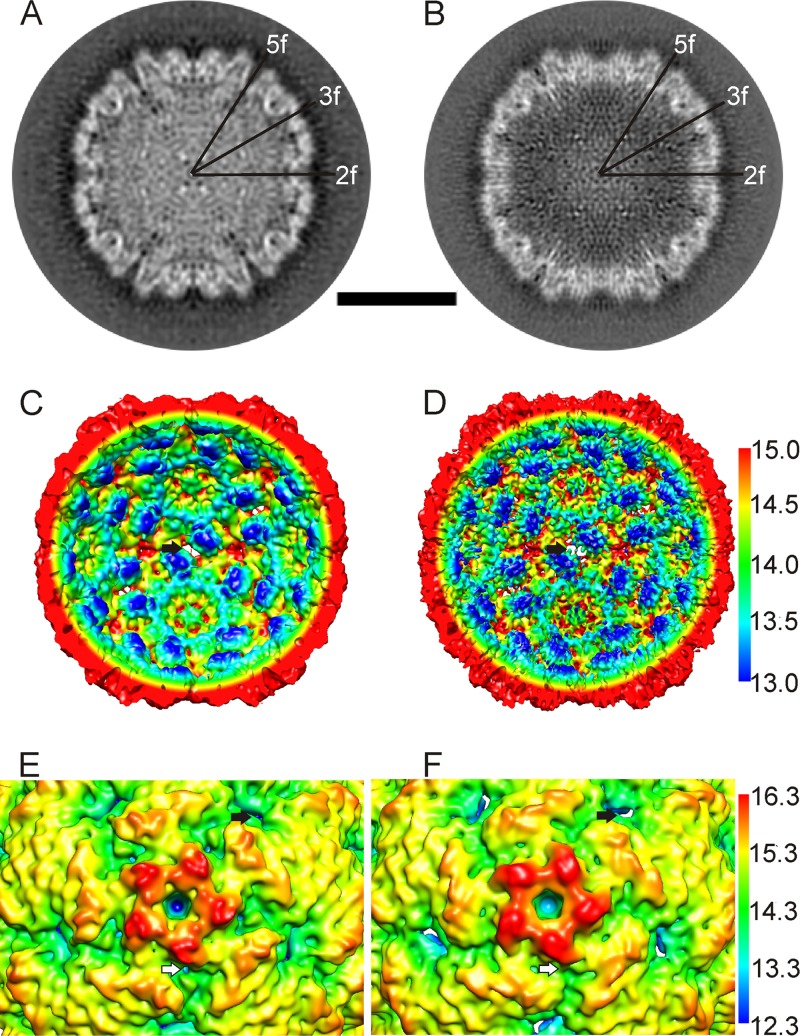
Using cryoEM and 3D image reconstruction, we solved the structure of the full CAV7 capsid to 8.2-Å resolution and that of the empty CAV7 capsid to 6.1-Å resolution (Fig. 2 and Table 2). The reconstructions are very similar in size: a radial profile calculated from the reconstructions showed only a 1-pixel (0.113-nm) radial expansion in the empty particle (radius, 13.9 nm) compared to the filled (radius, 13.8 nm). The resolutions of both the full- and empty-capsid reconstructions were high enough to enable segmentation of the capsid proteins. The intertwining strands of neighboring VPs could not be identified, but the folding cores of individual VPs were easily detected. Although the empty-capsid proteins show more detail, the densities attributed to proteins VP2 and VP3 were found to be nearly identical in the full and empty capsids. In contrast, the density attributed to protein VP1 in the vicinity of the vertices shows obvious changes after genome release. An open channel with a diameter of roughly 12 Å was observed in the 5-fold vertex of the full-capsid reconstruction (Fig. 2A and E). The channel is open all the way through the protein layer even at 1 standard deviation above the mean. In the empty-capsid reconstruction, the channel is blocked off at the inner surface of the capsid and remains blocked even at 3 standard deviations above the mean (Fig. 2B and F). In addition, holes were observed in the capsid at the 2-fold axes in both empty and filled capsids (black arrows in Fig. 2C to F). Deep canyons circling the 5-fold vertices were also observed in both empty and filled capsids (white arrows in Fig. 2E and F; also see Movie S1 in the supplemental material).
Fig 2.
Subtle changes in the capsid occur on release of RNA and VP4. (A and B) Comparison of central cross-sections through the filled particle reconstruction (A) and the empty particle reconstruction (B) shows that in the empty particle, the channel on the vertex closes, and the subunits around the 2-fold symmetry axes move. Symmetry axes are marked 5-fold (5f), 2-fold (2f), and 3-fold (3f). Bar, 15 nm. (C and D) Radially depth-cued, isosurface representations of the interior of the CAV7 full capsid with the RNA density removed (C) and the CAV7 empty capsid (D), shown at 2.5 standard deviations above the mean. The nm scale for the radial depth cueing in D is similar to that in C. The black arrow indicates an opening on a 2-fold axis of symmetry. (E and F) Radially depth-cued, isosurface representations of a closeup of a 5-fold vertex (filtered to 8.23-Å resolution) from the filled particle (E) and the empty particle (F). The nm scale for the radial depth cueing in F is the same as that in E. White arrows indicate the canyon around the vertices. Black arrows indicate an opening on a 2-fold axis of symmetry.
Table 2.
Statistics for the reconstructions
| Parameter | Full-capsid reconstruction | Empty-capsid reconstruction |
|---|---|---|
| Electron dose (e−/Å2) | 16–17 | 16–17 |
| No. of micrographs | 223 | 264 |
| No. of particles boxed | 2,152 | 7,849 |
| No. of particles used in the reconstruction | 2,152 | 7,849 |
| Defocus range (μm) | 0.62–4.81 | 0.62–4.81 |
| Resolution (Å) | 8.2 | 6.1 |
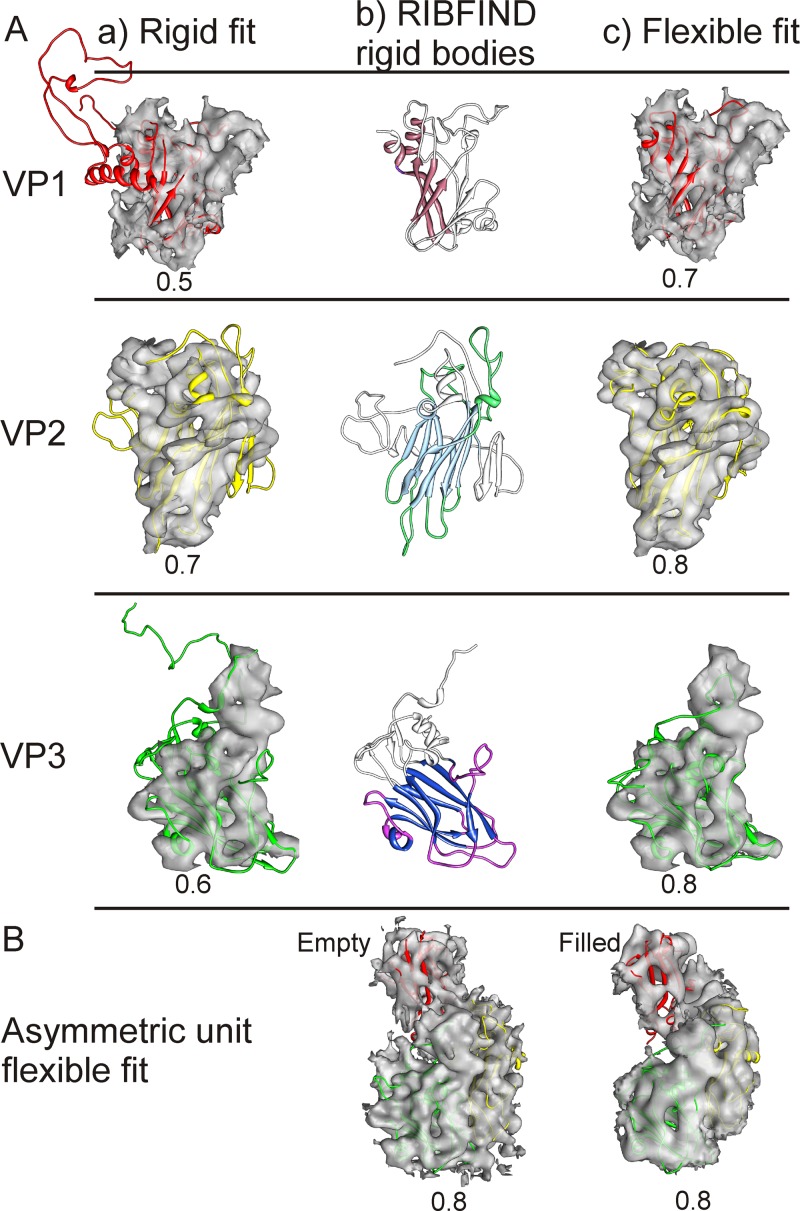
In order to model the changes occurring in the capsid after RNA release and to investigate protein-RNA interactions, we generated homology models of the CAV7-USSR and 275/58 capsid proteins (VP1-VP4) using the I-TASSER server (68, 91). All of the models were reliably predicted (I-TASSER C-scores: VP1, 0.60; VP2, 1.08; VP3, 1.33; VP4, −0.93). As expected, the viral proteins VP1-VP3 possess the typical β-barrel structures commonly found in other picornaviruses. The homology models were further refined by fitting them into the corresponding density segmented from both the full and the empty-particle reconstructions in a conservative, stepwise manner illustrated in Fig. 3 (63, 75). Initially, the position of the β-barrels was found manually and the fit of the entire model was locally optimized by an automated rigid fitting (Fig. 3Aa). The β-barrels are very large, distinctive features, where the walls of the barrel and the hollow in the middle are obvious at this resolution, even if the individual β-strands are not. Then, the termini from the models which had very low sequence identity and did not fit into the segmented density were trimmed away, leaving the conserved core (e.g., removal of VP1 N terminus) (Table 2; Fig. 3Ab). At this stage, even if most of the model fits well within the electron density, often additional refinement can improve the fit of the homology model (75). Here, to improve the fit further, a two-stage flexible-fitting procedure was employed, using RIBFIND (62) and Flex-EM (75) (see Materials and Methods), resulting in an optimal fit around the position defined in step 1 (Fig. 3Ac). Note that the use of the RIBFIND program helps to avoid overfitting (62). Finally, the asymmetric unit was refined to remove clashes at the interface, and the whole capsids were generated. These additional rounds of refinement led to further trimming of loops, especially in VP1 (Table 2). The final models are shown fitted into the asymmetric unit of both the empty and filled capsids in Fig. 3B. A good fit was found between the EM and the homology models with a cross-correlation score for the asymmetric unit of 0.8 (calculated in the Chimera tool “fit in map”) for both reconstructions. As a consequence of this approach, even though the typical picornaviral annulus formed by the N terminus of VP3 is present in both reconstructions, the model does not extend that far due to the uncertainty of tracing an extended chain at this resolution (see Movie S2 in the supplemental material).
Fig 3.
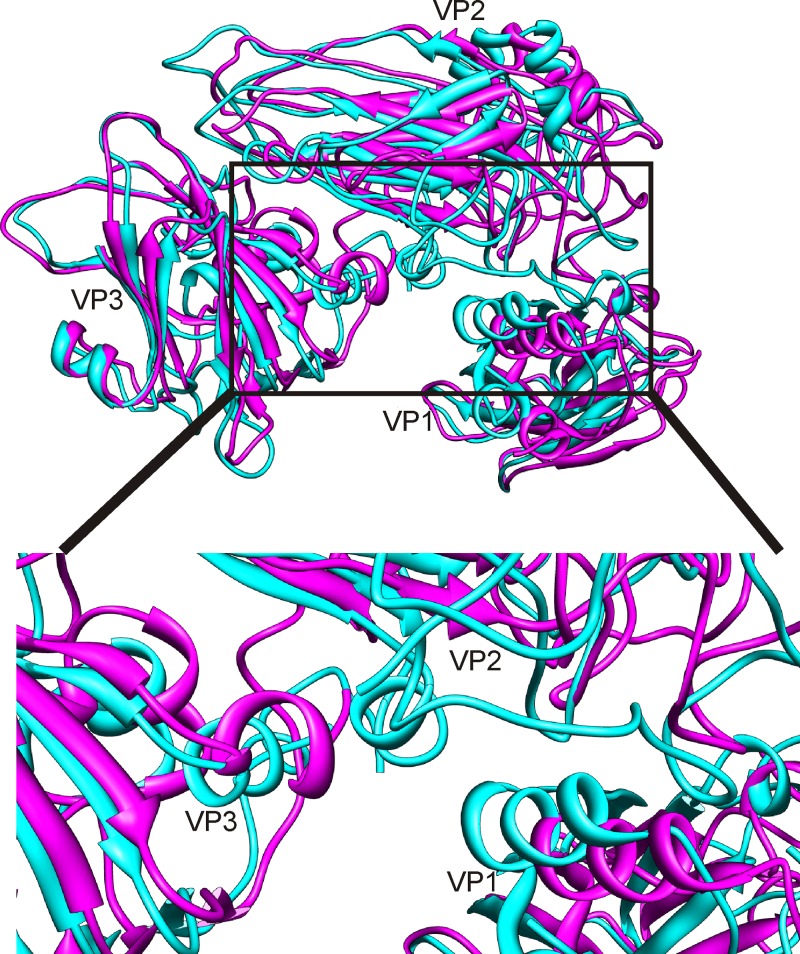
Refinement of the homology models of VP1 (red), VP2 (yellow), and VP3 (green). (A) Progress from the initial rigid fit of the homology model in Chimera (a) through RIBFIND (b) to flexible fitting within the segmented protein density (c) of the CAV7 empty-capsid proteins VP1, VP2, and VP3 (63). (b) Clustered rigid bodies were identified using RIBFIND (62). The clustered secondary structure elements and loops used in the rigid-body fitting are in color rather than in white. (c) Flexible fitting carried out to improve the fit using Flex-EM (75). (B) Final fit of the homology models within an asymmetric unit of the CAV7 empty and filled capsids. (Aa, Ac, and B) The correlation value for each fit is displayed below each model.
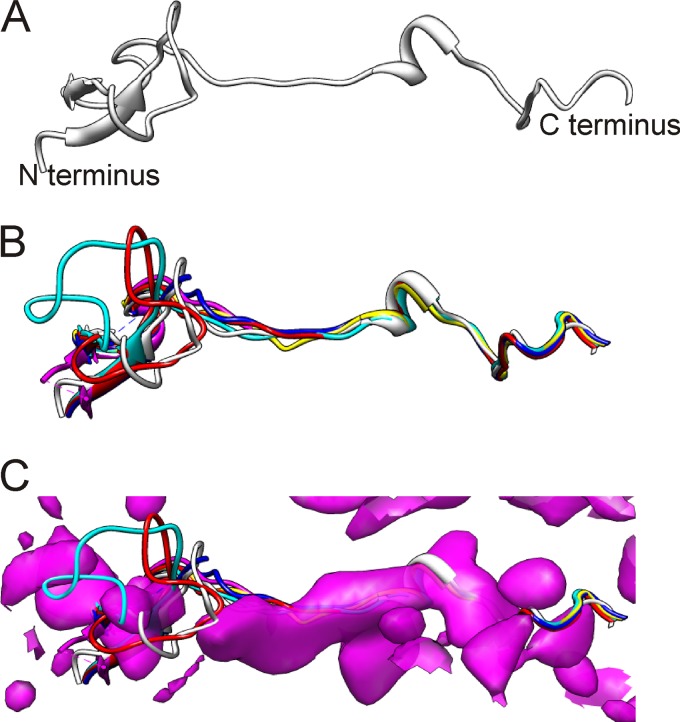
The same approach did not work for VP4. This protein could not be reliably identified in the presence of RNA inside the filled capsid, as it has very little secondary structure and thus was not clearly evident at 8.2-Å resolution for segmentation. However, it was possible to superpose the CAV7 VP4 homology model (Fig. 4A) onto atomic models of 5 picornavirus capsids (Fig. 4B). This gives an estimate of the probable position of VP4 in CAV7. There is some density at this exact position in the filled-particle reconstruction (Fig. 4C; also see Movie S2 in the supplemental material), and it corresponds best to the C-terminal half of the model. However, we did not include VP4 in the final homology model of the full capsid (Fig. 3B), as some of the density could belong to VP1, VP3, or RNA. The density attributed to VP4 is absent from the empty-capsid reconstruction, as is the RNA. Noticeably, the EV71 VP4 structure that has just been reported is extended in the N-terminal 14 to 31 residues compared to what is seen for other enteroviruses (81), but the C-terminal half is conserved in structure and position, which adds some weight to our prediction for CAV7.
Fig 4.
Modeling of VP4. (A) VP4 model generated from I-TASSER. (B) VP4 model (white) for CAV7 superimposed on VP4 structures of CBV3 (blue; PDB: 1cov), CAV21 (red; PDB: 1z7s), PV1 (cyan; PDB: 1hxs), HRV16 (magenta; PDB: 1ayn), and HRV3 (yellow; PDB: 1rhi) (54, 56, 61, 84, 93). (C) VP4 models from B shown with CAV7 filled-capsid density in the background. The density was zoned to a radius of 12 Å in Chimera (63) by selecting the VP4 model of HRV3.
The fits of the homology models into the segmented densities of the full and empty capsids revealed considerable changes in protein position measured as both the RMSD of the whole proteins and the component placement score of individual secondary structure elements (Fig. 3B; Tables 2, 3, and 4). The largest change in the orientation and position of the capsid proteins in the full versus the empty capsid is observed for VP1, with an overall translation score of 3.5 Å and a rotational score of 12° (Table 3; Fig. 5). VP3 also moved 2.5 Å and 7.8°. These movements are responsible for the changes seen on the 5-fold axis (Fig. 2). Noticeably, such large domain movements are transmitted throughout the structure, resulting in considerable changes at protein-protein interfaces. Some of the most noticeable interface changes occur in the canyon with the helix at residues 116 to 125 (hereafter referred to as helix 116–125) (VP1), helix 172–177 (VP2), and helix 44-78 (VP3) all moving significantly (Table 4; Fig. 5, inset). Helix 91-98 (VP2) has a symmetry-related copy on the opposite side of the 2-fold axis of symmetry, among protomers. The movement of this helix away from the axis is responsible for the opening up of the pore on the 2-fold axis of symmetry from the full to the empty capsid (Fig. 2).
Table 3.
Structure comparison among final models of the filled and empty capsids
| Protein name (residue range) | No. of chains & regions modeled | C-alpha RMSD of empty to full (Å) |
|---|---|---|
| VP1 (1–296) | 77–200, 223–257 | 4.7 |
| VP2 (1–254) | 1–254 | 6.0 |
| VP3 (1–242) | 41–198, 211–242 | 4.7 |
| VP4 (1–69) | 1–69 | Not applicable |
| VP1–VP3 | 5.3 |
Table 4.
Component placement scores for the individual proteins between empty and filled capsids (75)
| Protein name (residue range) | Translation (Å) | Rotation (°) |
|---|---|---|
| VP1 (77–200, 223–257) | 3.5 | 12.1 |
| VP2 (1–254) | 2.5 | 2.7 |
| VP3 (41–198, 211–242) | 2.5 | 7.8 |
Fig 5.
Movements associated with the transition from the filled to the empty particle. Superposition of the modeled asymmetric units shows major conformational changes in the capsid proteins. The modeled asymmetric units from the filled (magenta) and empty (cyan) particles are shown as ribbon models. The largest change is seen in VP1 where the whole protein undergoes an ∼12° rotation and a translation of 3.5 Å The inset shows the conformational changes detected at the interface of VP1, VP2, and VP3 (75). The helices with the greatest movement at the interfaces are labeled in the inset helix 116–125 (VP1), helix 172–177 (VP2), and helix 44–78 (VP3).
We identified mutations on the capsid surface that might explain the differential pathogenic properties of the strains by mapping the amino acid differences of the 275/58 or the resequenced Parker strain relative to the USSR strain homology models in order to identify surface-exposed residues. Three mutations are apparently exposed on the capsid outer surface in the Parker strain (VP1, E142G; VP3, C83G, A84P), compared to 10 275/58 mutations (VP1, V91F, K103E, G109E, T163S, R164K, C224W; VP2, T156A; VP3, G83C, P84A, T236S, A238S). Apparently, no mutations are exposed on the capsid inner surface in the Parker strain, in contrast to what is seen for two 275/58 mutations (VP2, H141L, N143D).
Comparison of the CAV7 to other picornaviruses.
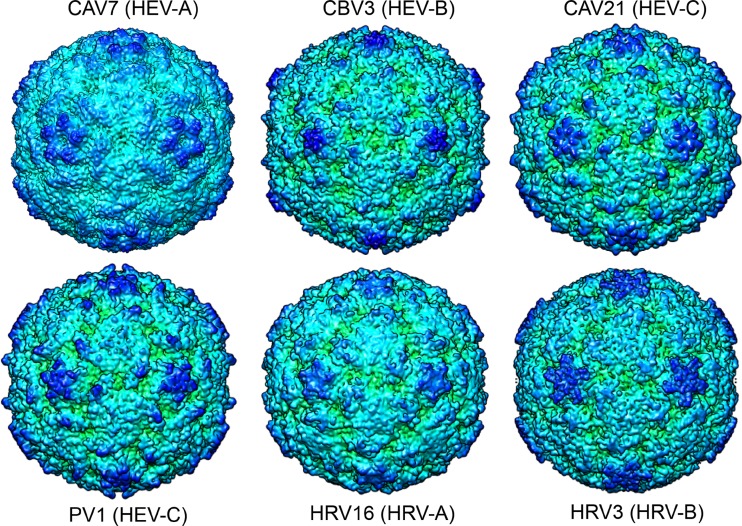
Comparison of the structure of CAV7 (VP1-VP3) to that of other picornaviruses was quantified using structural alignment with the PDBeFold server (47), which takes into account both the number of residues aligned and the Cα RMSD. The CAV7 quaternary structure was found to closely resemble those of the coxsackieviruses and rhinoviruses such as CBV3 and HRV16 (Fig. 6; also see Table S1 in the supplemental material). We could also compare the conformational changes that occur on genome release to those reported for poliovirus, HRV2, and EV71 (7, 27, 81). The expansion that we see is smaller than the ∼4% increase in the size reported for poliovirus, EV71, and HRV2 going from the mature virus to the empty particle, where the shells become thinner. In HRV2, this thinning is mainly explained by a hinge movement in VP1, where the C-terminal domain moves away from the β-barrel. In EV71, the 5-fold proximal end of the VP1 β-barrel moves upwards and rotates. In poliovirus, the thinning has been explained by rigid-body rotation of the individual capsid proteins, likened to tectonic plate movement. The helix movements that we see at the interfaces were not reported for poliovirus, where rigid-body fitting has been carried out. In HRV2 and EV71, the observed pore that opens on the 2-fold axis of symmetry is due to the separation of the αA helices in adjacent VP2 molecules. We observed a similar movement in the CAV7 VP2 helix 91–98 (Table 5).
Fig 6.
Comparison of the empty CAV7 capsid cryoreconstruction with representative enterovirus atomic models from coxsackievirus B3 (CBV3; PDB 1cov), coxsackievirus A21 (CAV21; PDB 1z7s), poliovirus 1 (PV1; 1hxs), human rhinovirus 16 (HRV16; PDB 1ayn), and human rhinovirus 3 (HRV3; PDB 1rhi) (54, 56, 61, 84, 93). The models were calculated to 6.1-Å resolution in CCP4 with a temperature factor of 100 Å2 (83), displayed as isosurface presentations 2 standard deviations above the mean and radially depth cued in Chimera (63). The view is down an icosahedral 2-fold axis of symmetry.
Table 5.
Component placement scores for individual secondary structure elements between filled and empty capsids (75)
| Protein name | Residue range | Secondary structure element type | Translation (Å) | Rotation (°) |
|---|---|---|---|---|
| VP1 | 79–84 | α–Helix | 5.6 | 6.9 |
| 100–103 | α–Helix | 2.4 | 4.7 | |
| 116–125 | α–Helix | 5.9 | 7.2 | |
| 87–93, 132–139, 188–191, 244–254 | β–Sheet | 3.4 | 8.2 | |
| 105–110, 149–155, 177–181, 230–236 | β–Sheet | 2.8 | 13.5 | |
| VP2 | 57–60 | α–Helix | 2.9 | 17.6 |
| 82–89 | α–Helix | 2.8 | 13.5 | |
| 91–98 | α–Helix | 4 | 56.2 | |
| 172–177 | α–Helix | 12.0 | 103.8 | |
| 16–18, 21–23 | β–Sheet | 2.4 | 22.9 | |
| 32–33, 64–65, 105–111, 190–194, 235–241 | β–Sheet | 2.2 | 7.3 | |
| 78–81, 121–128, 180–184, 212–218 | β–Sheet | 2.1 | 3.2 | |
| VP3 | 44–48 | α–Helix | 4.2 | 34.4 |
| 60–63 | α–Helix | 0.6 | 46.1 | |
| 94–97 | α–Helix | 4.5 | 33.5 | |
| 100–105 | α–Helix | 2.7 | 8.2 | |
| 146–151 | α–Helix | 0.9 | 16.2 | |
| 51–53, 115–121, 164–169, 214–220 | β–Sheet | 1.8 | 6.0 | |
| 82–87, 130–135, 154–158, 193–197 | β–Sheet | 1.8 | 14.6 | |
| 110–112, 178–179, 225–227 | β–Sheet | 3.4 | 11.2 |
DISCUSSION
In this study, we investigated the sequences and structure of an enterovirus within the HEV-A species in order to investigate its relationship to other picornaviruses, the release of RNA, and possible strain-specific sequence differences that affect cell tropism. This is one of the first structural models of a virus from the HEV-A species, which includes many major enteroviral pathogens. The cryoEM structures of empty and filled CAV7 capsids revealed features very typical for picornaviruses. CAV7 follows the T = 1 (pseudo T = 3) symmetry, which is common to the Picornaviridae. Segmentation of the CAV7 reconstruction was possible due to the subnanometer resolution of the reconstructions and revealed the position of the major capsid proteins and provided further evidence of the typical picornavirus structure. The closest structural similarity based on the alignment of the Cα backbones of VP1-VP3 (see Table S1 in the supplemental material) is to CBV3 (HEV-B) and rhinoviruses (HRV-A). However, not unexpectedly, poliovirus, CAV21 and HRV3 from the other species can also be aligned. As this paper was in review, two groups published structures of EV71, showing that it is closest in structure to bovine enterovirus, which was also used as a template in our modeling of all four capsid proteins (64, 81). The three-dimensional structure of CAV7 is thus more conserved than the genome or the epitopes used commonly in serotyping and classification.
In polioviruses, VP4 release is linked to the penetration of the endosome and the release of RNA (3). The role of VP4 in CAV7 is unknown, but we show that its removal coincided with the release of RNA after heat treatment. Hence, VP4 could well be involved in RNA release in CAV7, and this should be further explored. Currently, it is not known if receptor binding plays a role in this process in vivo.
The conformational changes in the CAV7 capsid that are seen after the release of RNA are the result of an orchestrated series of conformational changes occurring at the interfaces of the three major capsid proteins. The comparison raises the question of how the RNA actually escapes the capsid. Previous studies on poliovirus indicated that release could take place through the 5-fold vertices, and indeed we see major conformational changes in VP1, which result in the closing of a 12-Å-diameter channel at the CAV7 vertices (10, 11, 38, 45). However, recently, PV1 in the act of RNA uncoating has been revisited using electron tomography, and the footprint of RNA on the capsid outer surface was located about 20 Å from a 2-fold axis of symmetry (9, 49). This would suggest that conformational changes in VP2 and VP3 should occur, along with the obvious loss of VP4. Interestingly, not only does the CAV7 structure have holes on the 2-fold axis of symmetry, but the helices responsible for the opening of that pore in the empty capsid are similar to those reported in HRV2 80S particles and EV71 expanded particles (27, 81). We also recognized conformational changes in the interfaces among VP1, VP2, and VP3 that affect the canyon structure (Fig. 5, inset) around the N terminus of VP1 (although the VP1 model starts only at residue 77) (Table 3). Similar, more detailed observations of changes in the canyon have also been made for EV71 and noticeably were linked to the shrinking of the lipid-binding pocket in VP1 and the initiation of uncoating (81). Hence, although the scale of the conformational changes that occur on RNA release in CAV7 do not result in as large a capsid expansion as those seen in HRV2 and EV71, the movements seen are still in line with externalizing the VP1 N terminus through the canyon floor and RNA release from a hole in the capsid on a 2-fold axis of symmetry. Thus, in terms of virus entry into the cell, similar dissociation steps are predicted for HRV2, poliovirus, EV71, and CAV7.
In this study, we also analyzed the sequences of three CAV7 strains: Parker, USSR, and 275/58. While the Parker and USSR strains are practically identical, 275/58 is more distant. The Parker and USSR strains cause flaccid paralysis in a mouse model, typical for members of the HEV-A species, while 275/58 is nonpathogenic. Despite this difference in pathogenicity, all three strains can be typed by CAV7-specific neutralizing antibodies. Mapping of the point mutations of the Parker and 275/58 capsid proteins onto the USSR homology models revealed that a significant fraction of these mutations in VP1 are probably surface exposed, close to reported receptor binding sites of PV1, CAV21, and CBV3 (35, 36, 84). Hence, it is possible that the point mutations in 275/58, predicted to be on the surface, could explain the observed differences in CAV7 275/58 strain pathogenesis and tropism by affecting receptor selection and binding. Thus, we now have a set of potential sites to mutate to see directly whether or not they affect cell tropism.
In conclusion, we show that CAV7 has the typical structure of a picornavirus in which the interfaces of the protomer (VP1, VP2, VP3) undergo significant conformational changes in order to progress from a filled form of the capsid to an empty one. A number of surface residues that could be responsible for observed differences in pathogenicity were identified by comparison of different strains, which could shed light on the progression of disease caused by species A enteroviruses.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ritva Kajander and Carl-Mikael Suomivuori for excellent technical assistance and the Biocenter Finland National Cryo Electron Microscopy Unit, Institute of Biotechnology, Helsinki University and the CSC-IT Center for Science Ltd. for providing facilities. R.S.S. thanks Timothy S. Baker for encouraging this collaboration.
R.S.S. thanks Timothy S. Baker for supporting the work through his NIH grants R37 GM-033050 and R01 AI-079095 and gift from the Agouron Foundation. This work was also supported by the Academy of Finland Centre of Excellence Programme in Virus Research (2006–2011; 129684 to S.J.B.), the Academy of Finland (139178 to S.J.B.; 128539 to P.S.; and 122215 to T.H.), the Sigrid Juselius Foundation (S.J.B. and T.H.), the National Graduate School in Informational and Structural Biology (J.J.T.S.), the Helsinki Graduate Program in Biotechnology and Molecular Biology (S.S.), the Medical Research Council Career Development Award (G0600084 to M.T.) and the Human Frontier Science Program (RGY0079/2009-C to M.T.).
Footnotes
Published ahead of print 18 April 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Abraham G, Colonno RJ. 1984. Many rhinovirus serotypes share the same cellular receptor. J. Virol. 51:340–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ang LW, et al. 2009. Epidemiology and control of hand, foot and mouth disease in Singapore, 2001–2007. Ann. Acad. Med. Singap. 38:106–112 [PubMed] [Google Scholar]
- 3. Arita M, Koike S, Aoki J, Horie H, Nomoto A. 1998. Interaction of poliovirus with its purified receptor and conformational alteration in the virion. J. Virol. 72:3578–3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arnold E, Rossmann MG. 1988. The use of molecular-replacement phases for the refinement of the human rhinovirus 14 structure. Acta Crystallogr. A 44:270–282 [DOI] [PubMed] [Google Scholar]
- 5. Baker TS, Olson NH, Fuller SD. 1999. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev. 63:862–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Basavappa R, et al. 1994. Role and mechanism of the maturation cleavage of VP0 in poliovirus assembly: structure of the empty capsid assembly intermediate at 2.9 A resolution. Protein Sci. 3:1651–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Belnap DM, et al. 2000. Molecular tectonic model of virus structural transitions: the putative cell entry states of poliovirus. J. Virol. 74:1342–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blomqvist S, Paananen A, Savolainen-Kopra C, Hovi T, Roivainen M. 2008. Eight years of experience with molecular identification of human enteroviruses. J. Clin. Microbiol. 46:2410–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bostina M, Levy H, Filman DJ, Hogle JM. 2011. Poliovirus RNA is released from the capsid near a twofold symmetry axis. J. Virol. 85:776–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bubeck D, et al. 2005. The structure of the poliovirus 135S cell entry intermediate at 10-Å resolution reveals the location of an externalized polypeptide that binds to membranes. J. Virol. 79:7745–7755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bubeck D, Filman DJ, Hogle JM. 2005. Cryo-electron microscopy reconstruction of a poliovirus-receptor-membrane complex. Nat. Struct. Mol. Biol. 12:615–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chambon M, et al. 2001. Circulation of enteroviruses and persistence of meningitis cases in the winter of 1999–2000. J. Med. Virol. 65:340–347 [DOI] [PubMed] [Google Scholar]
- 13. Chaves SS, Lobo S, Kennett M, Black J. 2001. Coxsackie virus A24 infection presenting as acute flaccid paralysis. Lancet 357:605. [DOI] [PubMed] [Google Scholar]
- 14. Chow M, et al. 1987. Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature 327:482–486 [DOI] [PubMed] [Google Scholar]
- 15. Chumakov MP, et al. 1956. Isolation and investigation of the IV immunological type of poliomyelitis virus. Vopr. Virusol. 1(1):16–19 (In Russian.) [PubMed] [Google Scholar]
- 16. Dalldorf G, Sickles GM. 1948. An unidentified, filtrable agent isolated from the feces of children with paralysis. Science 108:61–62 [DOI] [PubMed] [Google Scholar]
- 17. Dalldorf G, et al. 1949. A virus recovered from the feces of poliomyelitis patients pathogenic for suckling mice. J. Exp. Med. 89:567–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eswar N, et al. 2006. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics Chapter 5:Unit 5.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Felsenstein J. 1989. PHYLIP - phylogeny inference package (version 3.2). Cladistics 5:164–166 [Google Scholar]
- 20. Fendrick AM, Monto AS, Nightengale B, Sarnes M. 2003. The economic burden of non-influenza-related viral respiratory tract infection in the United States. Arch. Intern. Med. 163:487–494 [DOI] [PubMed] [Google Scholar]
- 21. Fernandez JJ. 2008. High performance computing in structural determination by electron cryomicroscopy. J. Struct. Biol. 164:1–6 [DOI] [PubMed] [Google Scholar]
- 22. Figueroa JP, Ashley D, King D, Hull B. 1989. An outbreak of acute flaccid paralysis in Jamaica associated with echovirus type 22. J. Med. Virol. 29:315–319 [DOI] [PubMed] [Google Scholar]
- 23. Filman DJ, Wien MW, Cunningham JA, Bergelson JM, Hogle JM. 1998. Structure determination of echovirus 1. Acta Crystallogr. D Biol. Crystallogr. 54:1261–1272 [DOI] [PubMed] [Google Scholar]
- 24. Fricks CE, Hogle JM. 1990. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J. Virol. 64:1934–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fry EE, et al. 2003. Crystal structure of swine vesicular disease virus and implications for host adaptation. J. Virol. 77:5475–5486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fry EE, et al. 2010. Crystal structure of equine rhinitis A virus in complex with its sialic acid receptor. J. Gen. Virol. 91:1971–1977 [DOI] [PubMed] [Google Scholar]
- 27. Garriga D, et al. 2012. Insights into minor group rhinovirus uncoating: the X-ray structure of the HRV2 empty capsid. PLoS Pathog. 8:e1002473 doi:10.1371/journal.ppat.1002473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gear JH. 1984. Nonpolio causes of polio-like paralytic syndromes. Rev. Infect. Dis. 6(Suppl. 2):S379–S384 [DOI] [PubMed] [Google Scholar]
- 29. Goodfellow IG, et al. 2005. Inhibition of coxsackie B virus infection by soluble forms of its receptors: binding affinities, altered particle formation, and competition with cellular receptors. J. Virol. 79:12016–12024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Grimwood K, et al. 2003. Acute flaccid paralysis from echovirus type 33 infection. J. Clin. Microbiol. 41:2230–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grist NR. 1962. Type A7 Coxsackie (type 4 poliomyelitis) virus infection in Scotland. J. Hyg. (Lond.) 60:323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grist NR, Bell EJ, Assaad F. 1978. Enteroviruses in human disease. Prog. Med. Virol. 24:114–157 [PubMed] [Google Scholar]
- 33. Habel K, Loomis LN. 1957. Coxsackie A7 virus and the Russian poliovirus Type 4. Proc. Soc. Exp. Biol. Med. 95:597–605 [DOI] [PubMed] [Google Scholar]
- 34. Hadfield AT, et al. 1997. The refined structure of human rhinovirus 16 at 2.15 A resolution: implications for the viral life cycle. Structure 5:427–441 [DOI] [PubMed] [Google Scholar]
- 35. He Y, et al. 2000. Interaction of the poliovirus receptor with poliovirus. Proc. Natl. Acad. Sci. U. S. A. 97:79–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. He Y, et al. 2001. Interaction of coxsackievirus B3 with the full length coxsackievirus-adenovirus receptor. Nat. Struct. Biol. 8:874–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hendry E, et al. 1999. The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses. Structure 7:1527–1538 [DOI] [PubMed] [Google Scholar]
- 38. Hewat EA, Blaas D. 2004. Cryoelectron microscopy analysis of the structural changes associated with human rhinovirus type 14 uncoating. J. Virol. 78:2935–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hewat EA, Neumann E, Blaas D. 2002. The concerted conformational changes during human rhinovirus 2 uncoating. Mol. Cell 10:317–326 [DOI] [PubMed] [Google Scholar]
- 40. Ho M, et al. 1999. An epidemic of enterovirus 71 infection in Taiwan. Taiwan Enterovirus Epidemic Working Group. N. Engl. J. Med. 341:929–935 [DOI] [PubMed] [Google Scholar]
- 41. Huang CC, et al. 1999. Neurologic complications in children with enterovirus 71 infection. N. Engl. J. Med. 341:936–942 [DOI] [PubMed] [Google Scholar]
- 42. Kamitsuka PS, Lou TY, Fabiyi A, Wenner HA. 1965. Preparation and standardization of coxsackievirus reference antisera. I. For twenty-four group A viruses. Am. J. Epidemiol. 81:283–306 [DOI] [PubMed] [Google Scholar]
- 43. Kim SS, et al. 1989. Crystal structure of human rhinovirus serotype 1A (HRV1A). J. Mol. Biol. 210:91–111 [DOI] [PubMed] [Google Scholar]
- 44. Kivioja T, Ravantti J, Verkhovsky A, Ukkonen E, Bamford D. 2000. Local average intensity-based method for identifying spherical particles in electron micrographs. J. Struct. Biol. 131:126–134 [DOI] [PubMed] [Google Scholar]
- 45. Kolatkar PR, et al. 1999. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J. 18:6249–6259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Korant BD, Lonberg-Holm K, Noble J, Stasny JT. 1972. Naturally occurring and artificially produced components of three rhinoviruses. Virology 48:71–86 [DOI] [PubMed] [Google Scholar]
- 47. Krissinel E, Henrick K. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60:2256–2268 [DOI] [PubMed] [Google Scholar]
- 48. Lee BE, Davies HD. 2007. Aseptic meningitis. Curr. Opin. Infect. Dis. 20:272–277 [DOI] [PubMed] [Google Scholar]
- 49. Levy HC, Bostina M, Filman DJ, Hogle JM. 2010. Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy. J. Virol. 84:4426–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lin J, et al. 2011. An externalized polypeptide partitions between two distinct sites on genome-released poliovirus particles. J. Virol. 85:9974–9983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lönnrot M, et al. 2000. Enterovirus infection as a risk factor for beta-cell autoimmunity in a prospectively observed birth cohort: the Finnish Diabetes Prediction and Prevention Study. Diabetes 49:1314–1318 [DOI] [PubMed] [Google Scholar]
- 52. Lönnrot M, et al. 2000. Enterovirus RNA in serum is a risk factor for beta-cell autoimmunity and clinical type 1 diabetes: a prospective study. Childhood Diabetes in Finland (DiMe) Study Group. J. Med. Virol. 61:214–220 [PubMed] [Google Scholar]
- 53. Ludtke SJ, Baldwin PR, Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82–97 [DOI] [PubMed] [Google Scholar]
- 54. Miller ST, Hogle JM, Filman DJ. 2001. Ab initio phasing of high-symmetry macromolecular complexes: successful phasing of authentic poliovirus data to 3.0 A resolution. J. Mol. Biol. 307:499–512 [DOI] [PubMed] [Google Scholar]
- 55. Mindell JA, Grigorieff N. 2003. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142:334–347 [DOI] [PubMed] [Google Scholar]
- 56. Muckelbauer JK, et al. 1995. Structure determination of coxsackievirus B3 to 3.5 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 51:871–887 [DOI] [PubMed] [Google Scholar]
- 57. Nishimura Y, et al. 2009. Human P-selectin glycoprotein ligand-1 is a functional receptor for enterovirus 71. Nat. Med. 15:794–797 [DOI] [PubMed] [Google Scholar]
- 58. Oberste MS, et al. 1999. Typing of human enteroviruses by partial sequencing of VP1. J. Clin. Microbiol. 37:1288–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Oberste MS, Pallansch MA. 2003. Establishing evidence for enterovirus infection in chronic disease. Ann. N. Y. Acad. Sci. 1005:23–31 [DOI] [PubMed] [Google Scholar]
- 60. Oberste MS, Penaranda S, Maher K, Pallansch MA. 2004. Complete genome sequences of all members of the species human enterovirus A. J. Gen. Virol. 85:1597–1607 [DOI] [PubMed] [Google Scholar]
- 61. Oliveira MA, et al. 1993. The structure of human rhinovirus 16. Structure 1:51–68 [DOI] [PubMed] [Google Scholar]
- 62. Pandurangan AP, Topf M. 2012. Finding rigid bodies in protein structures: application to flexible fitting into cryoEM maps. J. Struct. Biol. 177:520–531 [DOI] [PubMed] [Google Scholar]
- 63. Pettersen EF, et al. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 64. Plevka P, Perera R, Cardosa J, Kuhn RJ, Rossmann MG. 1 March 2012, posting date Crystal structure of human enterovirus 71. Science. doi:10.1126/science.1218713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pulli T, Koskimies P, Hyypiä T. 1995. Molecular comparison of coxsackie A virus serotypes. Virology 212:30–38 [DOI] [PubMed] [Google Scholar]
- 66. Richter FA, Rhodes AJ, Macpherson LW, Labzoffsky NA. 1971. A possible new enterovirus serotype isolated in Ontario. Arch. Gesamte Virusforsch. 35:218–222 [DOI] [PubMed] [Google Scholar]
- 67. Rosenthal PB, Henderson R. 2003. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333:721–745 [DOI] [PubMed] [Google Scholar]
- 68. Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Salminen K, et al. 2003. Enterovirus infections are associated with the induction of beta-cell autoimmunity in a prospective birth cohort study. J. Med. Virol. 69:91–98 [DOI] [PubMed] [Google Scholar]
- 70. Seitsonen JJ, Susi P, Lemmetty A, Butcher SJ. 2008. Structure of the mite-transmitted Blackcurrant reversion nepovirus using electron cryo-microscopy. Virology 378:162–168 [DOI] [PubMed] [Google Scholar]
- 71. Shepherd CM, et al. 2006. VIPERdb: a relational database for structural virology. Nucleic Acids Res. 34:D386–D389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Smyth M, et al. 1995. Implications for viral uncoating from the structure of bovine enterovirus. Nat. Struct. Biol. 2:224–231 [DOI] [PubMed] [Google Scholar]
- 73. Stanway G, et al. 2005. Family Picornaviridae, p 757–778 In Fauquet C, Mayo MA, Maniloff J, Desselberger U, Ball LA. (ed), Virus taxonomy. VIIIth Report of the International Committee on Taxonomy of Viruses Elsevier, London, United Kingdom [Google Scholar]
- 74. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Topf M, et al. 2008. Protein structure fitting and refinement guided by cryo-EM density. Structure 16:295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tuthill TJ, et al. 2009. Equine rhinitis A virus and its low pH empty particle: clues towards an aphthovirus entry mechanism? PLoS Pathog. 5:e1000620 doi:10.1371/journal.ppat.1000620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. van Heel M, Harauz G. 1986. Resolution criteria for three dimensional reconstruction. Optik 73:119–122 [Google Scholar]
- 78. Venkataraman S, et al. 2008. Crystallization and preliminary X-ray diffraction studies of Seneca Valley virus-001, a new member of the Picornaviridae family. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 64:293–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Voroshilova MK, Chumakov MP. 1959. Poliomyelitis-like properties of AB-IV-coxsackie A7 group of viruses. Prog. Med. Virol. 2:106–170 [Google Scholar]
- 80. Vuorinen T, Vainionpaa R, Hyypiä T. 2003. Five years' experience of reverse-transcriptase polymerase chain reaction in daily diagnosis of enterovirus and rhinovirus infections. Clin. Infect. Dis. 37:452–455 [DOI] [PubMed] [Google Scholar]
- 81. Wang X, et al. 2012. A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nat. Struct. Mol. Biology. 19:424–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wien MW, Curry S, Filman DJ, Hogle JM. 1997. Structural studies of poliovirus mutants that overcome receptor defects. Nat. Struct. Biol. 4:666–674 [DOI] [PubMed] [Google Scholar]
- 83. Winn MD, et al. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Xiao C, et al. 2005. The crystal structure of coxsackievirus A21 and its interaction with ICAM-1. Structure 13:1019–1033 [DOI] [PubMed] [Google Scholar]
- 85. Xing L, Casasnovas JM, Cheng RH. 2003. Structural analysis of human rhinovirus complexed with ICAM-1 reveals the dynamics of receptor-mediated virus uncoating. J. Virol. 77:6101–6107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yamada K, et al. 2002. Crystal structure of the RuvA-RuvB complex: a structural basis for the Holliday junction migrating motor machinery. Mol. Cell 10:671–681 [DOI] [PubMed] [Google Scholar]
- 87. Yamayoshi S, et al. 2009. Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat. Med. 15:798–801 [DOI] [PubMed] [Google Scholar]
- 88. Yan X, Dryden KA, Tang J, Baker TS. 2007. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J. Struct. Biol. 157:211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Yan X, Sinkovits RS, Baker TS. 2007. AUTO3DEM–an automated and high throughput program for image reconstruction of icosahedral particles. J. Struct. Biol. 157:73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yang B, Chuang H, Yang KD. 2009. Sialylated glycans as receptor and inhibitor of enterovirus 71 infection to DLD-1 intestinal cells. Virol. J. 6:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhang Y. 2007. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins 69:108–117 [DOI] [PubMed] [Google Scholar]
- 92. Zhang Y, et al. 2009. An outbreak of hand, foot, and mouth disease associated with subgenotype C4 of human enterovirus 71 in Shandong, China. J. Clin. Virol. 44:262–267 [DOI] [PubMed] [Google Scholar]
- 93. Zhao R, et al. 1996. Human rhinovirus 3 at 3.0 A resolution. Structure 4:1205–1220 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.