Abstract
Viruses are known to use virally encoded envelope proteins for cell attachment, which is the very first step of virus infection. In the present study, we have obtained substantial evidence demonstrating that hepatitis C virus (HCV) uses the cellular protein apolipoprotein E (apoE) for its attachment to cells. An apoE-specific monoclonal antibody was able to efficiently block HCV attachment to the hepatoma cell line Huh-7.5 as well as primary human hepatocytes. After HCV bound to cells, however, anti-apoE antibody was unable to inhibit virus infection. Conversely, the HCV E2-specific monoclonal antibody CBH5 did not affect HCV attachment but potently inhibited HCV entry. Similarly, small interfering RNA-mediated knockdown of the key HCV receptor/coreceptor molecules CD81, claudin-1, low-density lipoprotein receptor (LDLr), occludin, and SR-BI did not affect HCV attachment but efficiently suppressed HCV infection, suggesting their important roles in HCV infection at postattachment steps. Strikingly, removal of heparan sulfate from the cell surface by treatment with heparinase blocked HCV attachment. Likewise, substitutions of the positively charged amino acids with neutral or negatively charged residues in the receptor-binding region of apoE resulted in a reduction of apoE-mediating HCV infection. More importantly, mutations of the arginine and lysine to alanine or glutamic acid in the receptor-binding region ablated the heparin-binding activity of apoE, as determined by an in vitro heparin pulldown assay. HCV attachment could also be inhibited by a synthetic peptide derived from the apoE receptor-binding region. Collectively, these findings demonstrate that apoE mediates HCV attachment through specific interactions with cell surface heparan sulfate.
INTRODUCTION
Hepatitis C virus (HCV) is a leading cause of liver diseases, chronically infecting an estimated 130 million to 170 million people worldwide (71, 82). HCV infection results in acute and chronic hepatitis, cirrhosis, and hepatocellular carcinoma (59), which ranks as the fifth most common cancer and the third most frequent cause of cancer death worldwide. Hepatitis C is also the most common indication for liver transplantation (15). Coinfection of HCV and HIV is very common, particularly among drug abusers (3). Thus, HCV infection poses a major global health problem. Current standard therapy with pegylated alpha interferon (peg-IFN-α) and ribavirin is less than 50% effective against HCV genotype 1, the dominant virus accounting for up to 70% of infections (27, 41, 54). Although two HCV NS3 protease-specific inhibitors, telaprevir and boceprevir, have recently been approved (33), their combination with peg-IFN-α and ribavirin has limitations such as severe side effects, long duration of treatment, and high cost. Discovery and development of more efficacious and safer anti-HCV drugs are urgently needed.
HCV is the prototype virus of the Hepacivirus genus in the Flaviviridae family (68). It is an enveloped RNA virus containing a single positive-strand RNA genome that encodes a long open reading frame (19). The translation initiation of HCV polyprotein is mediated by the highly structured internal ribosomal entry site (IRES) element within the 5′ untranslated region (5′UTR) of the HCV RNA genome (78). Upon translation, viral structural proteins (C, E1, and E2) and viral nonstructural (NS) proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) are produced from the viral polyprotein precursor by the action of cellular peptidases and viral NS2/NS3 metalloprotease and NS3/NS4A serine protease (45). Over the last decade, a number of genetic studies with subgenomic HCV RNA replicons and infectious HCV RNAs have determined the important roles of viral structural and NS proteins in the HCV life cycle. The structural proteins C, E1, and E2 together with p7 and NS2 are required for the production of infectious HCV (37, 38, 61, 72, 77). NS3, NS4A, NS4B, NS5A, and NS5B were found to be the minimal set of viral proteins essential for HCV RNA replication in the cell (14, 49). Interestingly, recent studies suggested that HCV NS proteins also play important roles in the production of infectious virus particles (6, 75). However, the underlying molecular mechanisms of viral NS proteins in HCV assembly and/or egression are unknown. Likewise, the importance of cellular proteins in the HCV life cycle has yet to be determined.
It is thought that HCV enters cells via receptor-mediated endocytosis and subsequent fusion between the viral and cellular membranes (13, 34, 56). A number of cell surface proteins were shown to interact with the viral envelope glycoproteins E1 and E2 (10, 67). Human CD81 was identified as the first HCV receptor/coreceptor by interacting with HCV E2 (23, 64). Subsequently, many other cell surface molecules were found to be important for HCV cell entry, including the low-density lipoprotein receptor (LDLr) (2, 58, 62), scavenger receptor class B type I (SR-BI) (8, 11, 70), claudin-1 (25), occludin (48, 65), dendritic cell-specific intercellular adhesion molecule 3 grabbing nonintegrin (DC-SIGN) and liver/lymph node-specific SIGN (L-SIGN) (22, 66), heparin sulfate proteoglycans (HSPGs) (9, 40, 60), and asialoglycoprotein receptor (69). However, it is not clear why HCV cell entry requires so many different cell surface receptors and/or coreceptors.
Recently, our studies have demonstrated that the cellular protein apolipoprotein E (apoE) is important for HCV infection (18, 36). The role of apoE in HCV infection was initially suggested by several interesting observations. It had long been thought that HCV was associated with lipoproteins in the plasma of hepatitis C patients (5). Several studies showed that the HCV RNA-containing particles could be precipitated by apoB- and apoE-specific antibodies (5). It was also found that the HCV RNA-containing particles isolated from the plasma of hepatitis C patients were rich in triglycerides and contained at least HCV RNA, core protein, and apoB. These findings led to the proposed model that HCV may be assembled as a lipoviroparticle (LVP) (4, 5). Apart from these studies, we had found that purified HCV particles grown in cell culture contained apoE and that the abundance of apoE correlated very well with HCV infectivity, as determined by HCV fractionation studies (18). More significantly, apoE-specific but not apoB-specific monoclonal or polyclonal antibodies were found to efficiently neutralize HCV infectivity. The complete blockade of apoB-containing lipoprotein secretion by microsomal transfer protein (MTP) inhibitors did not affect HCV production unless apoE expression and secretion were inhibited. Additionally, we found that apoE in purified HCV particles was sensitive to protease digestion like HCV E2, suggesting that apoE is a structural component located on the surface of the HCV envelope (36). The structural nature of apoE in HCV particles has recently been confirmed by two elegant studies with immunogold electron microscopy (EM) (29, 57). Both studies have shown that apoE and HCV E2 are colocalized on the surface of the HCV envelope. Collectively, these findings demonstrate that apoE is an integral part of HCV particles and plays an important role in HCV infection and assembly.
However, the underlying molecular mechanism of apoE in HCV infection has not been determined. In this study, we have defined the mechanism of action of apoE in HCV infection using a virus attachment assay in conjunction with site-directed mutagenesis analysis, small interfering RNAs (siRNAs), and a heparin-binding assay. Findings derived from our studies demonstrate that the cellular protein apoE mediates HCV attachment. That HCV uses a cellular protein for cell attachment may partly contribute to its persistent infection via the apoE-mediated evasion of host immunity.
MATERIALS AND METHODS
Cell culture.
The Huh-7.5 cell line and adenovirus packaging cell line AD293 (Agilent) were maintained in Dulbecco's modified Eagle's medium (DMEM; HyClone) supplemented with 10% fetal bovine serum (Atlantic Biologicals), 0.1 mM nonessential amino acids, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in a 5% CO2 incubator.
Antibodies and chemicals.
HCV NS3 monoclonal antibody and apoE-specific monoclonal antibodies 23 (MAb23) and WuE4 (ATCC) were all produced in the lab as previously described (16, 18). HCV NS5A monoclonal antibody (9E10) was provided by Charlie Rice (44). β-Actin monoclonal antibody (AC15) and normal mouse IgG1 were purchased from Sigma-Aldrich. Horseradish peroxidase-conjugated goat anti-mouse IgG was from Pierce. CD81 monoclonal antibodies were from Santa Cruz (clone 5A6). Rabbit anti-scavenger receptor BI (anti-SR-BI) antibodies were from Abcam (EP1556Y) and Invitrogen (catalog no. 486800). Claudin-1- and occludin-specific mouse monoclonal and rabbit polyclonal antibodies were purchased from Invitrogen. Mouse anti-LDLr (clone HL1) monoclonal antibodies were provided by Jin Ye (62). Heparinases I, II, and III were purchased from Sigma-Aldrich. Heparin-immobilized agarose beads were from Pierce.
Construction of DNA plasmids and preparation of recombinant adenoviruses.
The pCMV6-XL5-mApoE3 DNA contains an apoE3 variant (mApoE3) with three silent nucleotide mutations in the apoE siRNA-targeting region. Thus, the apoE-specific siRNA used in this study would only silence endogenous but not ectopic expression of apoE (24). Individual mutations in the apoE receptor-binding region (see Fig. 7A, amino acid residues 136 to 150) were introduced into the mApoE3 gene by PCR using synthetic oligonucleotide primers. Two sets of primers were used to perform overlapping PCR to create each apoE3 mutant as described previously (24). The DNA fragments containing specific mutations were cloned into the pShuttle-CMV vector (Stratagene) between the restriction enzyme sites KpnI and XbaI. The plasmid DNAs were confirmed by sequence analysis at the Northwestern University Biotechnology Laboratory (Chicago, IL). Recombinant adenoviruses expressing apoE mutant proteins were produced using AdEasy XL system (Stratagene). Briefly, pShuttle-CMV vectors were linearized by digestion with restriction enzyme PmeI and were then transformed into BJ5183-AD-1 competent cells, which contain the AdEasy-1 plasmid encoding the adenovirus-5 genome (Stratagene). The resulting adenoviral DNA constructs were amplified in XL-10 gold competent cells and were purified using a Qiagen DNA isolation kit. Purified adenoviral DNAs were transfected into AD293 cells to produce recombinant adenoviruses by following the manufacturer's instructions. Recombinant adenoviruses prepared in large quantity were purified using a kit from Virapur (San Diego, CA), aliquoted, and stored at −80°C in an ultra-low-temperature freezer. The titers of recombinant adenoviruses were determined by a plaque assay.
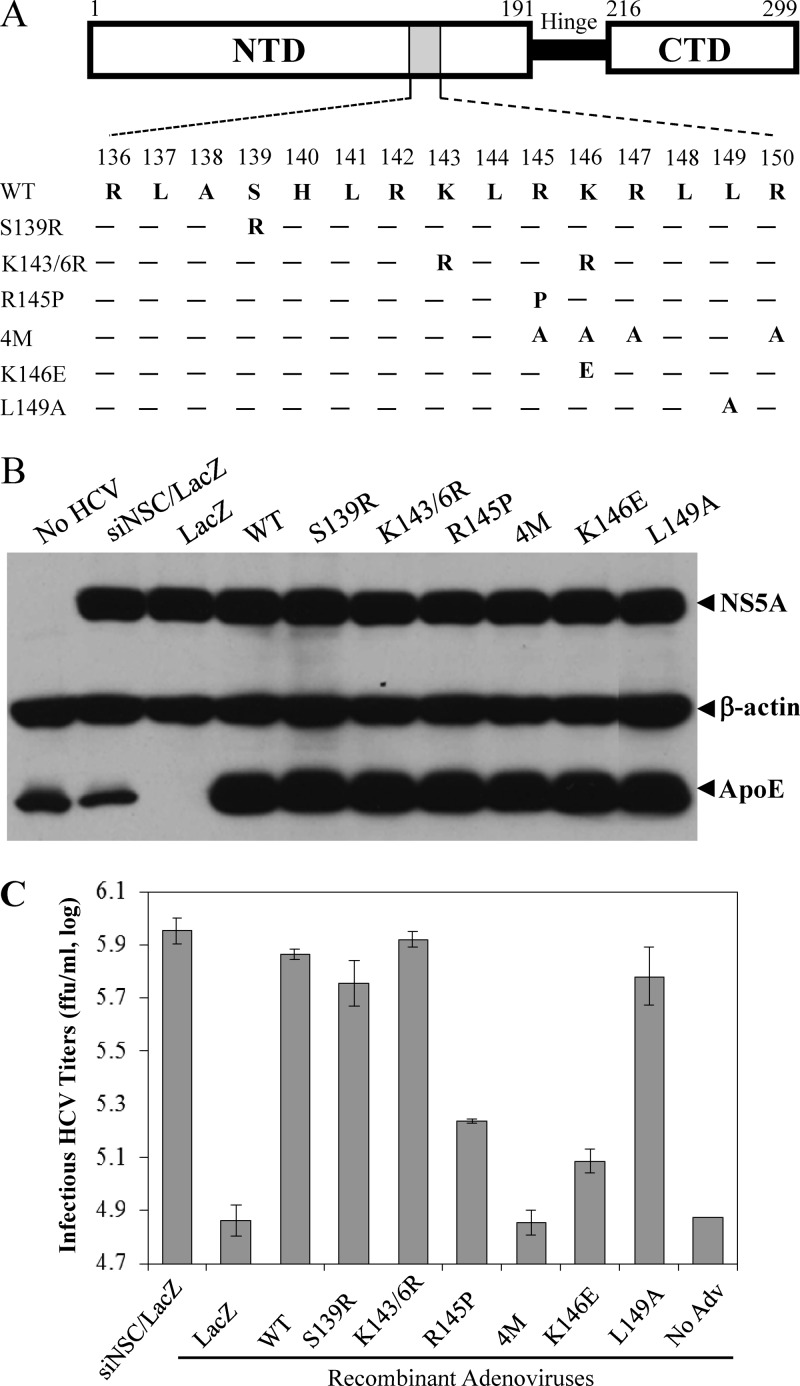
Fig 7.
Site-directed mutagenesis analysis of the apoE receptor-binding region. (A) Mutations of the N-terminal receptor-binding region. Three domains of apoE are schematically shown: the N-terminal domain (NTD) containing the receptor-binding region (gray box), the hinge region (Hinge), and the C-terminal domain (CTD) involved in interactions with phospholipids. Numbers indicate amino acid positions from the N terminus of mature apoE. Amino acid residues of the receptor-binding region (136 to 150) are shown underneath their positions. Each amino acid mutation is highlighted at its corresponding position. Wild-type and mutant apoE were ectopically expressed by recombinant adenoviruses (see Materials and Methods). (B) Effects of ectopic expression of wild-type and mutant apoE on HCV replication. Huh-7.5 cells were transfected with 50 nM apoE-specific siRNA. At 24 h p.t., Huh-7.5 cells were superinfected with HCV at an MOI of 5 at 37°C for 3 h, followed by adenovirus at an MOI of 6 at 37°C for 6 h. At 24 h p.i., cell lysates were harvested and used for the detection of apoE and NS5A by Western blotting with β-actin as a control. (C) Determination of infectious HCV titers in the supernatants by IHC. The supernatants of the experiments described in panel A were collected at 24 h p.i. The infectious HCV titers in the supernatants were determined by serial dilution.
HCV attachment assay.
Cell culture plates or dishes were coated with either 0.1 mg/ml of poly-l-lysine (Sigma-Aldrich) or 50 μg/ml of collagen (BD Biosciences). Huh-7.5 cells or primary human hepatocytes (Celsis In Vitro Technologies, Baltimore, MD) in 12-well cell culture plates or cell culture dishes 10 cm in diameter were incubated with HCV in the absence or presence of indicated antibodies or synthetic peptides derived from the apoE receptor-binding region (Biomatik, Wilmington, DE) at 4°C (on ice) for 2 h. The unbound HCV was removed by aspiration and by washing cells three times with phosphate-buffered saline (PBS). The virion RNA (vRNA) of the cell-bound HCV was isolated with an RNeasy purification kit (Qiagen). The levels of HCV vRNA were determined by RNase protection assay (RPA) and/or by quantitative reverse transcription-PCR (qRT-PCR).
Antibody neutralization of HCV infectivity.
Huh-7.5 cells in 6- or 12-well cell culture plates were infected with HCV at a multiplicity of infection (MOI) of 5 in the presence of antibodies specific to HCV E2 (MAb CBH5) and apoE (MAb23), respectively. Isotype-matched normal mouse and human IgGs were used as negative controls. After a 2-h incubation on ice or at 37°C, the unbound HCV was removed by aspiration and by washing cells with PBS, and then the HCV-infected cells were incubated with DMEM at 37°C. At 24 h postinfection (p.i.), the culture supernatants were collected for the measurement of infectious HCV titers, whereas the levels of HCV RNA and proteins in the HCV-infected Huh-7.5 cells were determined by RPA and Western blotting.
Knockdown of endogenous apoE expression and infection of recombinant adenoviruses for ectopic expression of apoE.
The endogenous expression of apoE was silenced by transfection with a specific siRNA. The apoE-specific siRNA and a nonspecific control (NSC) siRNA used in this study were previously described (24, 36). Huh-7.5 cells in 6-well cell culture plates were transfected with 0.05 nmol siRNA using RNAiMax transfection reagent (Invitrogen). At 24 h posttransfection (p.t.), cells were infected with HCV at an MOI of 5 at 37°C for 3 h and then superinfected with apoE-expressing recombinant adenoviruses at an MOI of 6. The unbound viruses were removed by aspiration and by washing cells with PBS. At 24 h p.i., cell culture supernatants were collected for determination of infectious HCV titers, vRNA, and the levels of ectopic apoE expression and secretion. The infected cells were either lysed in radioimmunoprecipitation assay (RIPA) buffer containing a cocktail of protease inhibitors (Roche) or used for isolation of total RNA with an RNeasy mini purification kit (Qiagen). The levels of apoE in the supernatants and apoE and NS5A in the HCV-infected cells were determined by Western blotting, whereas the levels of infectious HCV titer and vRNA were quantified by limiting dilution coupled with immunohistochemistry (IHC) and qRT-PCR.
Assay for dominant negative effect of apoE mutants.
Various amounts (0, 0.11, 0.33, and 1.0 μg) of the pCMV6-XL5 DNAs expressing wild-type apoE3 and the apoE3/4M (where 4M is four mutations) and apoE3/K146E mutants (see Fig. 7A), respectively, were transfected into Huh-7.5 cells in each well of a 12-well cell culture plate. At 24 h p.t., cells were infected with HCV at an MOI of 5 at 37°C for 2 h. After 24 h p.i., the supernatants were collected for virus titration and the HCV-infected cells were lysed for detection of HCV NS5A by Western blotting.
Synthetic siRNAs and silence of endogenous expression of HCV receptors/coreceptors.
Specific siRNAs targeting human CD81, claudin-1, LDLr, occludin, and SR-BI genes were synthesized by Dharmacon based on the previously reported target sequences (62). Their respective target sequences are UGAUGUUCGUUGGCUUCCU (CD81), UAACAUUAGGA CCUUAGAA (claudin-1), GGACAGAUAUCAUCAACGA (LDLr), GUGAAGAGUACAUGG CUGC (occludin), and GGUUGACUUCUGGCAUUCC (SR-BI). Huh-7.5 cells in each well of a 12-well cell culture plate were transfected with 0.2 nmol of each siRNA using RNAiMax transfection reagent (Invitrogen). At 48 h p.t., cells were either lysed for determination of gene silencing or used for analysis of the effects of gene silencing on HCV attachment and infection. HCV attachment and infection assays were carried out by incubation of siRNA-transfected cells with HCV at 37°C for 2 h. The unbound HCV was removed by extensive washing with PBS. The vRNA of the cell-bound HCV was extracted with TRI Reagent/RNAzol (Molecular Research Center) and quantified by qRT-PCR. The NS5A protein in the HCV-infected cells was detected by Western blotting.
Removal of heparan sulfate by heparinase treatment.
Huh-7.5 cells in a 12-well cell culture plate were washed with PBS and then incubated with various concentrations of heparinase I in a buffer containing 20 mM Tris-HCl (pH 6.8), 50 mM NaCl, 4 mM CaCl2 and 0.01% bovine serum albumin at 37°C for 1 h (40). The heparinase-containing buffer was removed, and cells were washed three times with PBS. The heparinase-treated Huh-7.5 cells were incubated with HCV on ice for 2 h. The unbound HCV was removed, and the cells were washed three times with PBS. The vRNA of the cell-bound HCV was extracted with TRI Reagent and quantified by qRT-PCR using the StepOnePlus real-time PCR system.
Western blot analysis.
The protein concentration of the cell lysate was determined using a protein assay reagent (Bio-Rad). An equal amount of total protein for each sample was analyzed by 10% SDS-PAGE. Separated proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane using a semidry blotter (Bio-Rad). After blocking with 5% dry milk, immunoblot analysis of NS5A, apoE, CD81, claudin-1, occludin, LDLr, and SR-BI was done using primary monoclonal or polyclonal antibody specific to each protein, secondary anti-mouse or anti-rabbit antibody, and an enhanced chemiluminescence kit (Pierce).
RNA extraction and quantification by RPA.
Total RNAs were extracted from the HCV-infected Huh-7.5 cells using a Qiagen RNeasy mini purification kit or TRI Reagent/RNAzol (Molecular Research Center). The levels of positive-strand HCV RNA in the cells were determined by RPA using a [32P]UTP-labeled probe containing the negative-strand HCV 3′UTR RNA as described previously (16). After digestion with RNase A/T1, RNA products were analyzed in a 6% polyacrylamide-urea (7.7 M) gel, visualized by autoradiography, and quantified using a PhosphorImager.
Titration of infectious HCV.
HCV in the culture supernatants was 10-fold serially diluted. Fifty microliters of diluted HCV was used to infect Huh-7.5 cells in each well of 96-well plates. After an 8-h incubation at 37°C, 100 μl of fresh DMEM containing 1% methylcellulose was added. At 48 h p.i., Huh-7.5 cells were fixed with 10% neutralized formalin. Focus-forming units (FFU) were determined by immunofluorescence assay (IFA) or immunohistochemistry using an NS3-specific monoclonal antibody and peroxidase-conjugated secondary anti-mouse antibody and 3,3′diaminobenzidine (DBA) substrate (Vector Laboratories). The viral titer was determined by the average number of NS3-positive focus-forming units (FFU/ml) in triplicate assays.
Heparin-binding assay.
Heparin-immobilized beads (Pierce) were preequilibrated with PBS and then incubated with 200 μl of cell culture medium containing the wild type or mutant apoE from adenovirus-infected Huh-7.5 cells. After a 1-h incubation at room temperature, apoE-bound beads were spun down, while the supernatant was collected and used for the detection of unbound apoE protein. The apoE-bound beads in the pellet were washed with 500 μl of PBS. Heparin-bound, unbound, and input apoE proteins were detected by Western blotting using the WuE4 monoclonal antibody.
HCV vRNA extraction and quantification by qRT-PCR.
HCV vRNA in the medium was extracted with TRI Reagent (Molecular Research Center). Total cellular RNA was extracted using an RNeasy minikit (Qiagen). The level of HCV vRNA was quantified by real-time RT-PCR using a QuantiFast probe RT-PCR kit (Qiagen). The oligonucleotide primers 2aF (5′-AGCCATGGCGTTAGTATGAGTGTC-3′) and 2aR (5′-ACAAGGCCTTTCGCAACCCAA-3′) are complementary to the HCV 5′UTR. The double-quenched probe (5′-AAACCCACTCTATGCCCGGCCATTT-3′) contains 5′-FAM (where FAM is 6-carboxyfluorescein), 3′-Iowa black FQ (IBFQ), and an internal ZEN quencher (IDT). Reactions were run in an ABI 7700 sequence detection system or StepOnePlus real-time PCR system (Applied Biosystems) as follows: 10 min at 50°C, 5 min at 95°C, and then cycled 40 times at 95°C for 10 s and 60°C for 30 s. The in vitro T7 transcript of the HCV JFH1 RNA genome was used as a standard.
RESULTS
Importance of apoE in HCV infection.
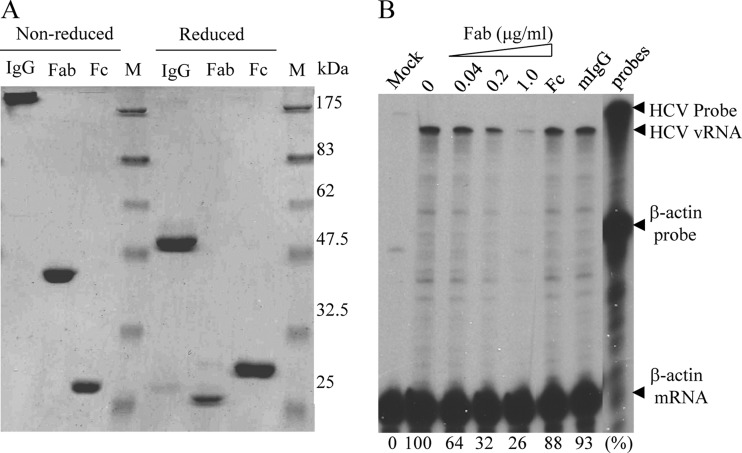
Our previous studies have demonstrated that apoE is an important determinant of HCV infection and assembly (12, 18, 24, 36). Consistent with its dual functions in the HCV life cycle, apoE was found to be a structural component of HCV particles, as demonstrated by studies with protease digestion and immunogold EM (29, 36, 57). The importance of apoE in HCV infection was suggested by our previous findings that apoE-specific monoclonal antibodies could efficiently neutralize HCV infectivity (18, 36). However, it remains unclear whether the blockade of HCV infection by apoE-specific monoclonal antibodies was due to antibody-mediated HCV aggregation. To exclude this possibility, we prepared the Fab fragments of an apoE-specific monoclonal antibody (MAb23) by papain digestion using a Pierce Fab preparation kit. The undigested IgG and the Fc fragment were separated from the Fab fragments using protein A-conjugated agarose beads (Fig. 1A). A cell culture-adapted JFH1 HCV variant, which grows to a titer more than 1,000-fold higher than that of wild-type virus, was used in this study (J. J. Jiang and G. L. Luo, unpublished data). Various amounts of purified Fab fragments were added during HCV infection on ice for 2 h. The HCV-infected cells were incubated at 37°C for 24 h (single-cycle virus growth). Infectious HCV titers in the supernatants were determined by serial dilution and staining for NS3-positive foci and were calculated as focus-forming units per milliliter (FFU/ml). The levels of positive-stranded HCV RNA were determined by RNase protection assay (RPA) as described previously (16). Similar to the whole IgG, purified Fab fragments potently blocked HCV infectivity in a dose-dependent manner, resulting in a reduction of positive-stranded HCV RNA by more than 70% at a concentration of 1 μg/ml (Fig. 1B and data not shown). Likewise, the Fab fragments reduced HCV yields similarly to MAb23 (data not shown). In contrast, the Fc fragment and normal mouse IgG1 had no significant effect on HCV infectivity and production of infectious virus (Fig. 1). These results confirmed our previous finding demonstrating that apoE plays an important role in HCV infection.
Fig 1.
Inhibition of HCV infection by the Fab fragments of MAb23. (A) Analysis of MAb23 and Fab and Fc fragments. The MAb23 whole IgG (IgG) and Fab and Fc fragments (1 μg each) were analyzed by 10% SDS-PAGE and stained with Coomassie blue. The sizes of protein markers are indicated on the right. (B) Inhibition of HCV infection by the MAb23 Fab fragments. Huh-7.5 cells were incubated with HCV on ice for 2 h in the presence of various amounts (0.04 to 1 μg/ml) of the Fab fragments (Fab), 1 μg/ml of the Fc fragment (Fc), or 1 μg/ml of normal mouse IgG (mIgG). Upon removal of the unbound HCV and washing of cells with PBS, the HCV-infected cells were incubated with medium at 37°C. At 24 h p.i., total cellular RNAs were extracted with TRI Reagent/RNAzol (Molecular Research Center, Inc.) and the levels of positive-stranded HCV RNA were determined by RPA as previously described (16). The β-actin mRNA was used as an internal control. The protected HCV RNA products migrated about 40 nucleotides faster due to a mismatched sequence derived from the vector DNA.
Blockade of HCV attachment by apoE-specific but not HCV E2-specific monoclonal antibody.
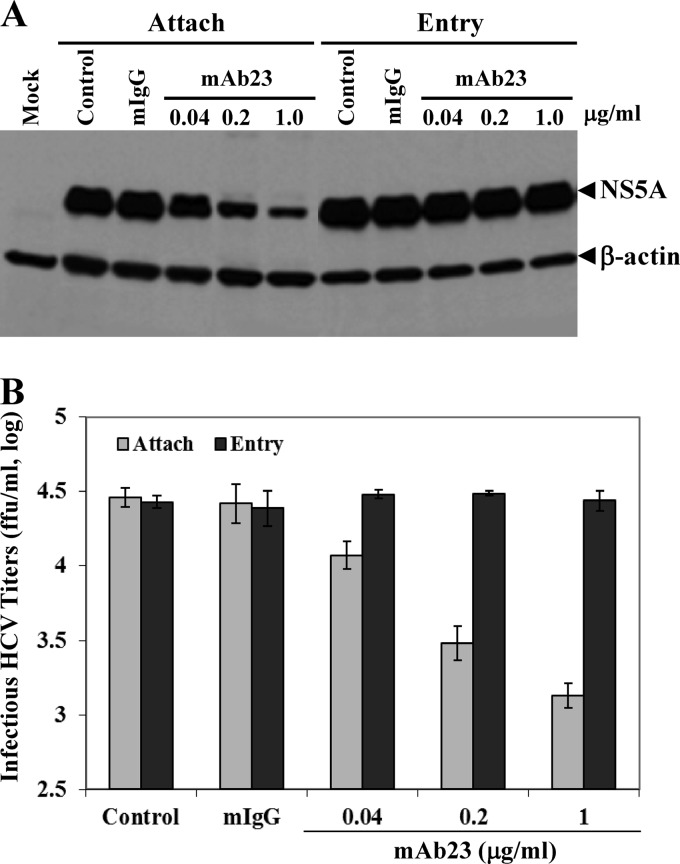
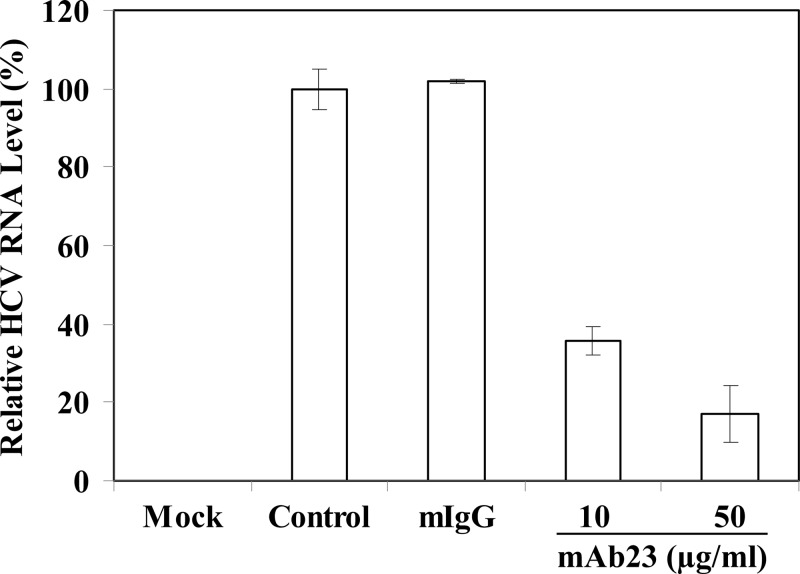
To illustrate the mechanism of apoE in mediating HCV infection, experiments were carried out to determine whether apoE MAb23 would block HCV attachment and/or cell entry. Initially, Huh-7.5 cells were incubated with HCV (on ice) at 4°C, which allows virus attachment but not cell entry, in the absence or presence of apoE MAb23 and normal mouse IgG. After a 2-h incubation on ice, the unbound HCV was removed by extensive washing with PBS. The HCV-bound cells were incubated with fresh medium for an additional 24 h at 37°C prior to lysis. The NS5A in the HCV-infected cells was detected by Western blotting (Fig. 2A), whereas the infectious HCV in the supernatants were titrated by a serial dilution method (Fig. 2B). Interestingly, apoE MAb23 blocked HCV infection in a dose-dependent manner, resulting in a nearly 80% reduction of NS5A protein, when added during virus attachment. Similar to normal mouse IgG, MAb23 failed to inhibit HCV infection when added after HCV attachment, as shown by comparable levels of NS5A in the HCV-infected cells (Fig. 2A) and infectious HCV in the supernatants (Fig. 2B) in the presence of MAb23 or with no antibody or normal mouse IgG controls. These results suggest that apoE is important for HCV attachment but not cell entry. We also determined the inhibitory activity of MAb23 against HCV infection in primary human hepatocytes (PHHs). As shown in Fig. 3, MAb23 inhibited HCV infection of PHHs in a dose-dependent manner, resulting in a reduction of HCV RNA by more than 80%. More significantly, MAb23 could inhibit the attachment and infection of clinical HCV of genotype 1b (J. Jiang, X. Wu, H. Tang, and G. Luo, unpublished data), demonstrating the importance of apoE in HCV attachment in vivo.
Fig 2.
Inhibition of HCV attachment by MAb23. The effects of MAb23 on HCV attachment were determined by incubating Huh-7.5 cells with HCV on ice for 2 h in the absence (control) or presence of various concentrations (0.04, 0.2, and 1 μg/ml) of MAb23 or a normal mouse IgG (mIgG). The unbound HCV was removed, and the HCV-bound cells were washed with PBS and then incubated at 37°C for 24 h. To determine the effects of MAb23 on HCV entry, HCV was allowed to bind to Huh-7.5 cells on ice for 2 h prior to the addition of MAb23 or mIgG. After HCV attachment on ice for 2 h, the unbound HCV was aspirated and the cells were washed with PBS. Subsequently, the HCV-bound cells were incubated with cell culture medium containing 0.04, 0.2, and 1 μg/ml of MAb23 or mIgG at 37°C for 6 h. Cell culture medium was replaced with fresh medium without antibody. At 24 h p.i. at 37°C, the levels of NS5A in the HCV-infected cells were detected by Western blotting (A), while the infectious HCV titers in the supernatants were determined by limiting dilution and IFA staining for HCV NS3-positive cells (B). Mean values and standard deviations of three experiments are shown in panel B. Attach, HCV attachment assay; Entry, HCV entry assay.
Fig 3.
Inhibition of HCV infection of primary human hepatocytes by apoE MAb23. Freshly isolated primary human hepatocytes (PHHs) in 12-well plates were purchased from Celsis In Vitro Technologies (Baltimore, MD) and maintained according to the manufacturer's instructions. PHHs were infected with HCV in the absence (control) or presence of mIgG (50 μg/ml) or apoE MAb23 (10 and 50 μg/ml) at 37°C for 8 h. At 48 h p.i., HCV RNAs in the infected PHHs were isolated with TRI reagent (Invitrogen). The levels of positive-strand HCV RNA were determined by qRT-PCR as described in Materials and Methods. The average levels of relative HCV RNA to the control (100%), from triplicate experiments, are shown.
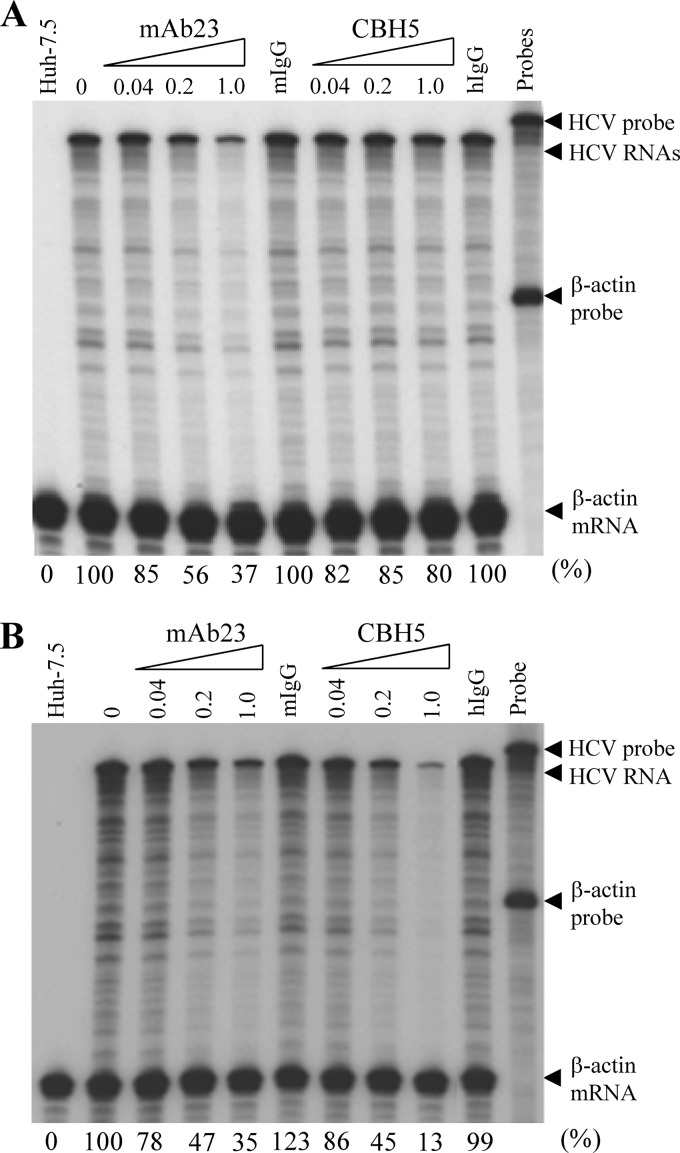
To further confirm the importance of apoE in HCV attachment, we determined the effects of MAb23 on HCV binding and/or cell entry in a comparison with an HCV E2-specific monoclonal antibody, CBH5, in a parallel experiment. Our previous studies showed that CBH5 is a highly potent inhibitor of JFH1 HCV infection with a 50% effective concentration (EC50) of 0.056 μg/ml (39). For the HCV attachment assay, Huh-7.5 cells were incubated with HCV on ice for 2 h in the presence of increasing amounts of MAb23 and CBH5. The unbound HCV was removed by extensive washing. The level of HCV attachment was determined by quantifying the levels of the cell-bound HCV vRNA by RPA. Strikingly, MAb23 inhibited HCV attachment in a dose-dependent manner. It suppressed HCV binding by more than 60% at a concentration of 1 μg/ml, whereas CBH5 did not significantly affect HCV attachment at concentrations up to 25 μg/ml (Fig. 4A and data not shown). Next, we sought to determine whether MAb23 and CBH5 bound to HCV at 4°C (after attachment) would still inhibit HCV infection. Again, the HCV attachment assay was done by an incubation of Huh-7.5 cells with HCV on ice for 2 h in the presence of various amounts of MAb23 and CBH5. Upon removal of unbound HCV by extensive washing with PBS, the HCV-bound cells were incubated at 37°C for 24 h. The levels of positive-stranded HCV RNA in the cell were determined by RPA. Both MAb23 and CBH5 similarly inhibited HCV infection in dose-dependent manners (Fig. 4B), suggesting that CBH5 did bind to HCV at 4°C and subsequently suppressed HCV entry into cells even though it did not affect HCV attachment (Fig. 4A). These findings are consistent with previous findings reported by others that HCV E2 acts at a postattachment step(s) via interactions with known receptors and/or coreceptors (25, 26, 30, 46, 60). More importantly, our findings indicate that HCV cell attachment is mediated by apoE but not HCV E2, suggesting for the first time that a cellular protein can mediate HCV attachment.
Fig 4.
Effects of MAb23 and HCV E2-specific MAb (CBH5) on HCV attachment and infection. (A) Inhibition of HCV attachment by apoE but not E2 MAb. Huh-7.5 cells were incubated with HCV on ice in the presence of various amounts of MAb23 or CBH5. Normal mouse IgG (mIgG) and human IgG (hIgG) were used as negative controls. After 2 h incubation on ice, the unbound HCV was removed and cells were washed three times with PBS. The vRNA of the cell-bound HCV was extracted with RNeasy minikit (Qiagen) and quantified by RPA as previously described (24, 36). (B) Inhibition of HCV infection by MAb23 and CBH5. HCV infection was the same as in panel A. After washing with PBS, the HCV-infected cells were incubated at 37°C for 24 h. Total RNAs were extracted from cells, and the levels of positive-stranded HCV RNA were determined by RPA. The β-actin mRNA was used as an internal control. Numbers on the top indicate the amounts of MAb (μg/ml). The levels of HCV RNA relative to the control (%) are shown at the bottom with 100% representing the RNA level in the absence of antibodies.
HCV attachment is mediated by apoE binding to cell surface heparan sulfate.
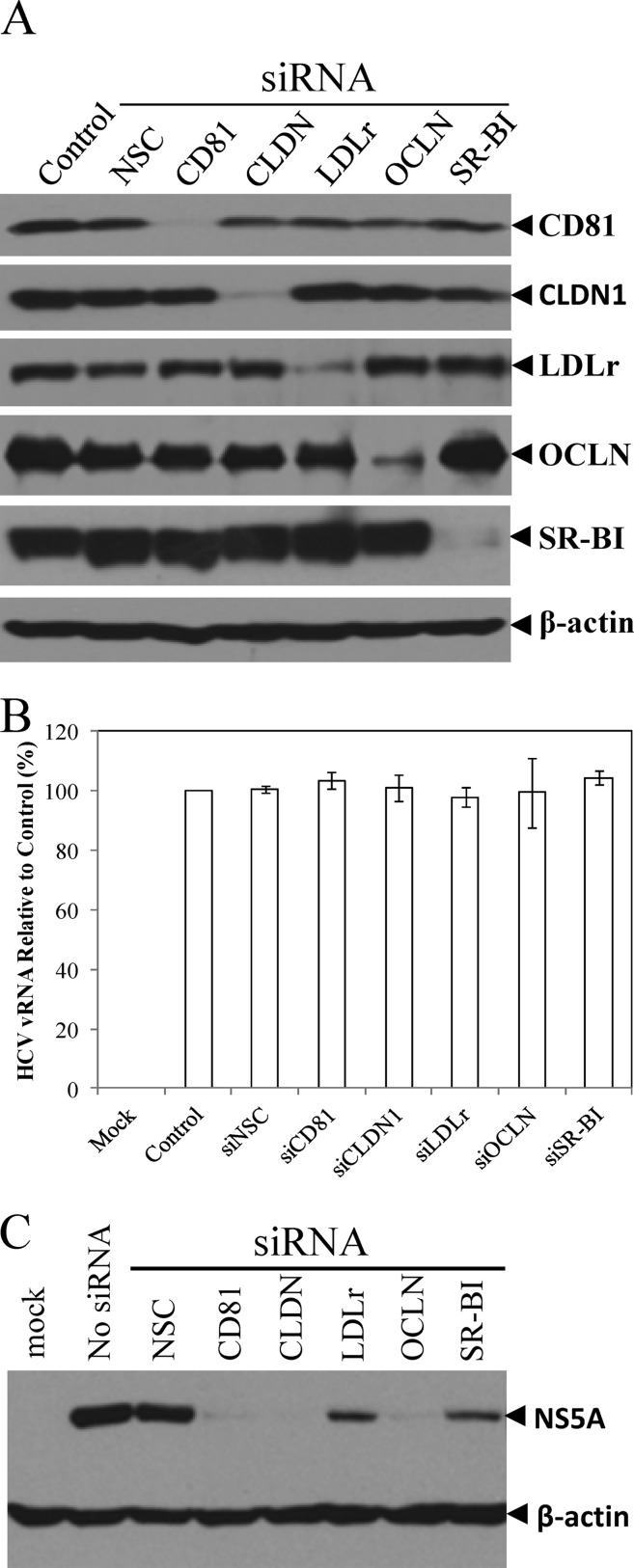
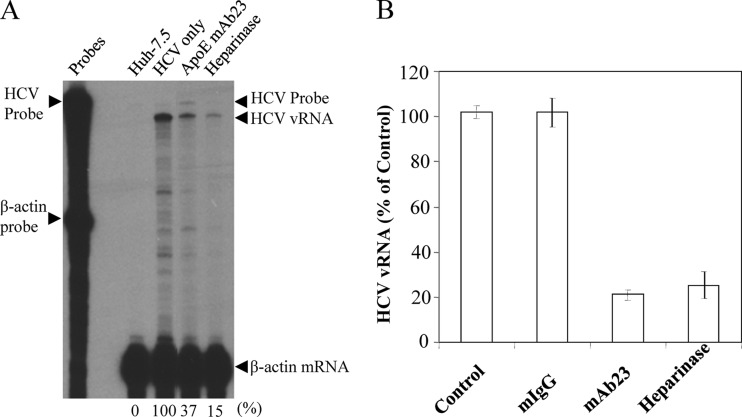
To define the underlying molecular mechanism of apoE in HCV attachment, we sought to determine the cell surface receptor(s) important for apoE binding and HCV entry. Many apoE receptors have been described in the literature, including HSPGs, LDLr, very-low-density lipoprotein receptor (VLDLr), SR-BI, and various types of LDLr-related proteins (LRPs). First, we sought to determine the roles of several known HCV receptors/coreceptors in HCV attachment and/or infection by silencing the endogenous expression of CD81, claudin-1, occludin, SR-BI, and LDLr. Specific siRNAs targeting the mRNAs of CD81, claudin-1, occludin, SR-BI, and LDLr were individually transfected into Huh-7.5 cells. Consistent with previous studies by others (62), CD81-, claudin-1-, occludin-, and SR-BI-specific siRNAs knocked down their endogenous expression by more than 90%, as determined by Western blot analysis (Fig. 5A). The LDLr-specific siRNA silenced the endogenous expression of LDLr by about 78% (Fig. 5A), similar to that observed by others (62). Strikingly, the siRNA-mediated knockdown of CD81, claudin-1, LDLr, occludin, and SR-BI expression did not significantly affect HCV attachment (Fig. 5B), suggesting that none of these known HCV receptors/coreceptors plays any significant role in HCV attachment. Next, we determined the effects of the siRNA-mediated knockdown of known receptors on HCV entry and infection. As shown in Fig. 5C, the siRNA-mediated silence of CD81, claudin-1, and occludin expression suppressed HCV infection by more than 95%, while knockdown of LDLr and SR-BI expression caused about 80% reduction of HCV infection (Fig. 5C). These results are in line with the findings reported by others that CD81, claudin-1, occludin, SR-BI, and LDLr are important for HCV infection (2, 8, 11, 23, 25, 48, 55, 58, 62, 65, 70, 89). Taken together, these findings suggest that the HCV receptors/coreceptors examined here play important roles in HCV infection at postattachment steps rather than at an attachment step. More significantly, we found that the removal of HSPGs by heparinase treatment resulted in a remarkable inhibition of HCV attachment by 85% and 75%, as determined by RPA and qRT-PCR, respectively, which was similar to MAb23 (Fig. 6). These results were reproduced by different experiments, as determined by both RPA (Fig. 6A) and qRT-PCR assays (Fig. 6B). Collectively, the findings derived from these studies demonstrate that apoE mediates HCV attachment through a specific interaction with cell surface heparan sulfate but not via other known HCV receptors and/or coreceptors such as CD81, claudin-1, LDLr, occludin, and SR-BI.
Fig 5.
Effects of siRNA-mediated knockdown of key HCV receptors on HCV attachment and infection. Huh-7.5 cells in 12-well plates were transfected with 0.2 nmol of each siRNA specific to CD81, claudin-1, LDLr, occludin, and SR-BI using RNAiMax reagent (Invitrogen) as described in Materials and Methods. (A) At 48 h p.t., the levels of CD81, claudin-1, LDLr, occludin, and SR-BI expression were determined by Western blotting using specific antibodies to each protein as indicated on the right. (B) Effects of silencing CD81, claudin-1, LDLr, occludin, and SR-BI expression on HCV attachment. At 48 h after siRNA transfection, Huh-7.5 cells were incubated with HCV at 37°C for 2 h. Upon extensive washing, total cellular RNA was extracted with RNAzol reagent. The levels of HCV vRNA were quantified by qRT-PCR and were converted to a percentage of the control with 100% representing the level of HCV vRNA without antibody treatment. Average levels of HCV vRNA and deviations from three independent experiments are shown. (C) Effects of the siRNA-mediated knockdown of the above receptors on HCV infection. At 48 h after siRNA transfection, Huh-7.5 cells were incubated with HCV on ice for 2 h, followed by extensive washing. The HCV-attached Huh-7.5 cells were incubated at 37°C for 24 h. HCV NS5A in the cell lysates was detected by Western blotting using β-actin as a control.
Fig 6.
(A) Effects of heparinase treatment on HCV attachment. Heparan sulfate on the cell surface was removed by treatment of Huh-7.5 cells with 1 unit/ml of heparinase I (Sigma) at 37°C for 1 h as described previously (38). The heparinase-treated Huh-7.5 cells were subsequently incubated with HCV on ice for 2 h. The unattached HCV was aspirated and the HCV-attached Huh-7.5 cells were washed three times with PBS. Total cellular RNA was prepared using an RNeasy mini purification kit (Qiagen). The apoE-specific MAb23 was used as a positive control. The HCV vRNA in 50 μg of total RNA was quantified by RPA as previously described (16, 34). The β-actin mRNA was used as an internal control to normalize the amounts of total RNA used between different samples. (B) Quantification of HCV vRNA by qRT-PCR. HCV attachment inhibition experiments were carried out the same way as in panel A except for the use of a 12-well plate of Huh-7.5 cells. Upon HCV attachment and extensive washing of the heparinase-treated Huh-7.5 cells, total cellular RNA was extracted with RNAzol reagent (Molecular Research Center, Inc.). The HCV vRNA was quantified by qRT-PCR using the StepOnePlus PCR system (Applied Biosystems). Percentage of average HCV vRNA levels and deviations derived from three independent experiments are shown. The HCV vRNA level relative to that of the control (100%) is shown.
Mutagenesis analysis of the apoE receptor-binding region.
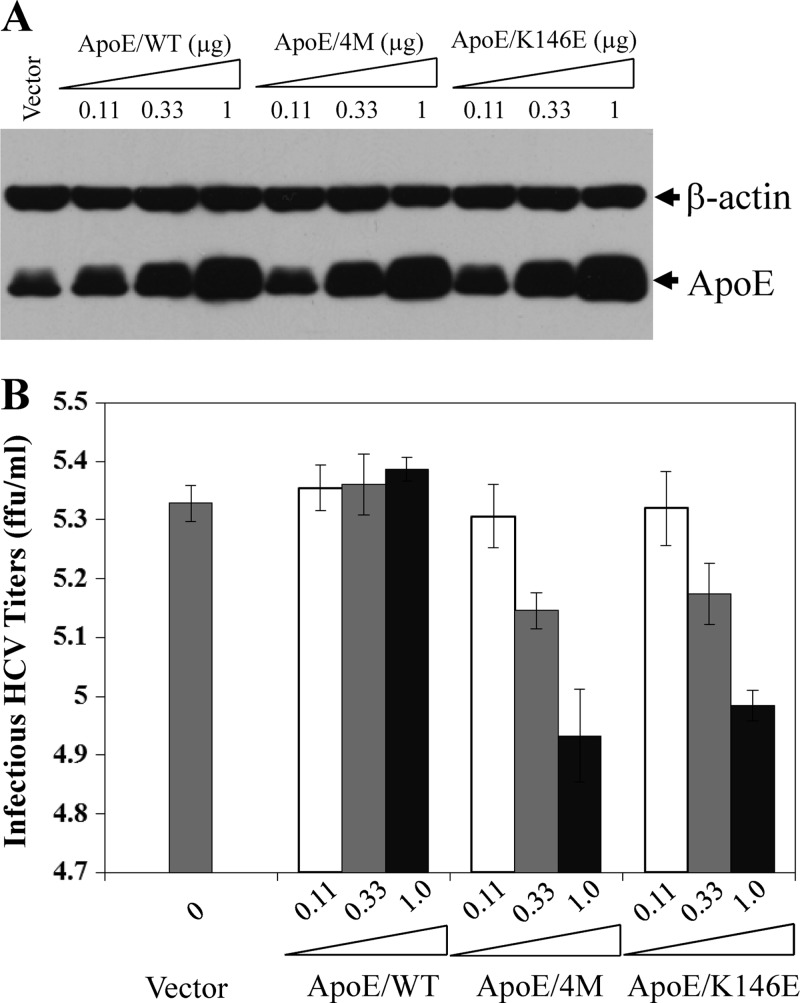
To better understand the mechanism of action of apoE in HCV attachment, we carried out a mutagenesis analysis of the apoE receptor-binding region. ApoE is a 34-kDa (299 amino acids) apoprotein containing a 22-kDa N-terminal domain (residues 1 to 191) that is recognized by the receptor(s) and a 10-kDa C-terminal domain (residues 222 to 299) that interacts with phospholipids (31, 35, 51). It is conceivable that the receptor-binding domain of apoE mediates HCV attachment. A number of previous studies have demonstrated that the amino acid residues 136 to 150 of apoE are the core receptor-binding region (43, 53, 79, 83). Therefore, we mutated several key residues of this region (Fig. 7A) based on their effects on the binding of apoE to the LDLr family and HSPG, as determined by numerous previous studies (31, 43, 50–53, 79, 84). The endogenous expression of apoE was efficiently silenced by transfection with an apoE-specific siRNA as described in our previous work (18, 24, 36). Recombinant adenoviruses were constructed to ectopically express the wild type and each apoE mutant. Huh-7.5 cells were first transfected with an apoE-specific siRNA, followed by superinfection with individual recombinant adenoviruses and HCV. A nonspecific control siRNA (siNSC) and an adenovirus expressing a lacZ gene (LacZ) were used as negative controls (Fig. 7). As expected, the endogenous expression of apoE was silenced to undetectable levels (Fig. 7B, lane 3). Upon superinfection with adenovirus and HCV, nearly all of the Huh-7.5 cells stained positive for both β-galactosidase (β-Gal) and HCV NS3, suggesting that Huh-7.5 cells were coinfected with adenovirus and HCV in the same cell (data not shown). The wild type and each apoE mutant were ectopically expressed to comparable levels (Fig. 7B). Consistent with our previous findings, HCV replication in the cell was not affected by ectopic overexpression of apoE, as shown by similar levels of NS5A (Fig. 7B). However, the knockdown of endogenous apoE expression resulted in more than 10-fold reduction of HCV infectivity in the supernatants (Fig. 7C, LacZ and No Adv). As in our previous studies, ectopic expression of a wild-type apoE restored the majority of HCV infectivity (Fig. 7C, WT) compared to the siNSC control (Fig. 7C, siNSC/LacZ). Similarly, the defect of HCV infectivity resulting from the knockdown of endogenous apoE expression was restored by ectopic expression of apoE variants with a serine-to-arginine mutation at residue 139 (S139R), lysine-to-arginine mutations at both amino acid residues 143 and 146 (K143/146R), and a leucine-to-alanine mutation at residue 149 (L149A) (Fig. 7C). In contrast, apoE mutations from arginine to proline at residue 145 (R145P), arginine at residues 145, 147 and 150, lysine to alanine at residue 146 (4M), and lysine to glutamic acid at residue 146 (K146E) were unable to restore the reduced HCV infectivity (Fig. 7C). The reduced HCV infectivity was not due to the effect of mutations on HCV assembly since the infectious titers and vRNA levels of intracellular HCV particles were comparable among the wild type and all apoE mutant proteins (data not shown). To further confirm these findings, we sought to determine whether the 4M and K146E apoE mutants would have dominant negative effects on endogenous apoE. To test this, Huh-7.5 cells were infected with HCV and then transfected with various amounts (0, 0.11, 0.33, and 1.0 μg) of DNA vectors expressing the wild type and 4M and K146E apoE mutants, respectively. The levels of apoE expression in the cell were detected by Western blotting (Fig. 8A), while infectious HCV titers in the supernatants were determined by serial dilution (Fig. 8B). The levels of apoE expression were proportional to increasing amounts of DNAs transfected into cells (Fig. 8A). Strikingly, the increasing levels of apoE mutant expression correlated very nicely with decreasing HCV infectivity in the supernatants (Fig. 8B), suggesting that ectopic expression of apoE mutants (4M and K146E) had a dominant negative effect on HCV infectivity by competing with endogenous apoE. Taken together, these results are consistent with previous findings that the positively charged residues of the apoE receptor-binding region are critical to the recognition by apoE receptors.
Fig 8.
Dominant negative effects of the 4M and K146E apoE mutants. Huh-7.5 cells in 12-well cell culture plates were transfected with increasing amounts (0.11, 0.33, and 1 μg) of pCMV6-XL5 vectors expressing wild-type apoE, the apoE mutant with arginine-to-alanine mutations at residues 145, 146, 147 and 149 (4M), and the apoE mutant with a lysine to glutamic acid mutation at amino acid residue 146 (K146E), respectively. The total amount of DNA was kept constant at 1 μg using vector DNA. DNA was transfected into Huh-7.5 cell using DMRIE-C reagent (Invitrogen) by following the manufacturer's instructions. At 6 h p.t., the medium was replaced with DMEM containing 10% FBS. At 24 h p.t., Huh-7.5 cells were infected with HCV at an MOI of 5. At 24 h p.i., cell lysates and the culture media were collected for detection of apoE by Western blotting (A) and for determination of infectious HCV titers by IHC (B). Mean values and standard deviations of three experiments are shown in panel B.
Effects of apoE mutations on heparin binding.
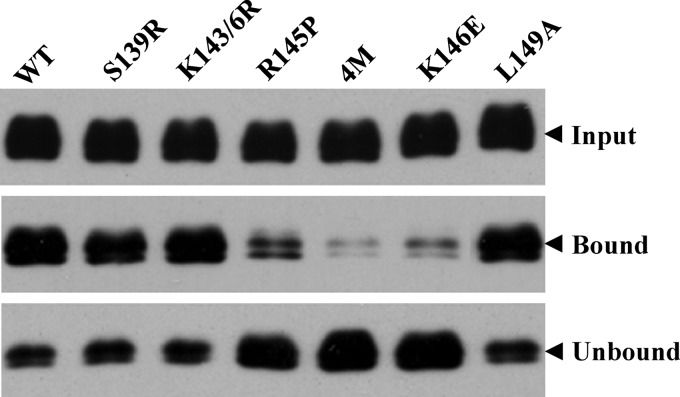
To provide direct evidence to demonstrate specific interactions between apoE and HSPGs, we performed an in vitro heparin-binding assay, which reflects the apoE and HSPG interactions. The endogenous apoE expression in Huh-7.5 cells was silenced to undetectable levels (Fig. 7B). The wild type and apoE mutant were ectopically expressed by recombinant adenoviruses, as described in Fig. 7. The supernatants containing both secreted apoE and apoE in HCV virions were collected and subject to a pulldown assay with heparin-immobilized beads. The heparin-bound (pulldown) and unbound apoE proteins were determined by Western blotting. As shown in Fig. 9, mutations of Arg to Pro at amino acid residue 145 (R145P), Lys to Glu at amino acid residue 146 (K146E), Arg to Ala at amino acid residues 145, 147, and 150, and Lys to Ala at position 146 (4M) all remarkably decreased the heparin-binding activity of apoE. However, mutations from Ser to Arg at residue 139, Lys to Arg at residues 143 and 146, and Leu to Ala at residue 149 had no effect on the heparin-binding activity of apoE, which was similar to that of wild-type apoE (Fig. 9). These results are in line with the findings derived from the experiments described in Fig. 7 and 8 that mutations of the positively charged amino acids to neutral or negatively charged residues in the receptor-binding domain inactivated the ability of apoE to bind to the cell surface heparin sulfate. Collectively, the findings derived from our studies demonstrate that the ability of apoE to mediate HCV attachment and infection is closely correlated with its heparin-binding activity, suggesting that apoE mediates HCV attachment through specific interaction with cell surface heparan sulfate.
Fig 9.
Effects of mutations in the receptor-binding region on the heparin binding of apoE. The endogenous apoE expression was silenced to undetectable levels by a synthetic apoE-specific siRNA as described previously (24). The wild type and individual apoE mutants (indicated on the top) were ectopically expressed by recombinant adenoviruses. The supernatants were collected and subjected to a pulldown assay with heparin-conjugated beads. The heparin-bound (pulldown) and unbound apoE proteins were determined by Western blotting using apoE-specific WuE4 monoclonal antibody and an ECL kit (Pierce).
Inhibition of HCV infection by a synthetic peptide derived from the apoE receptor-binding region.
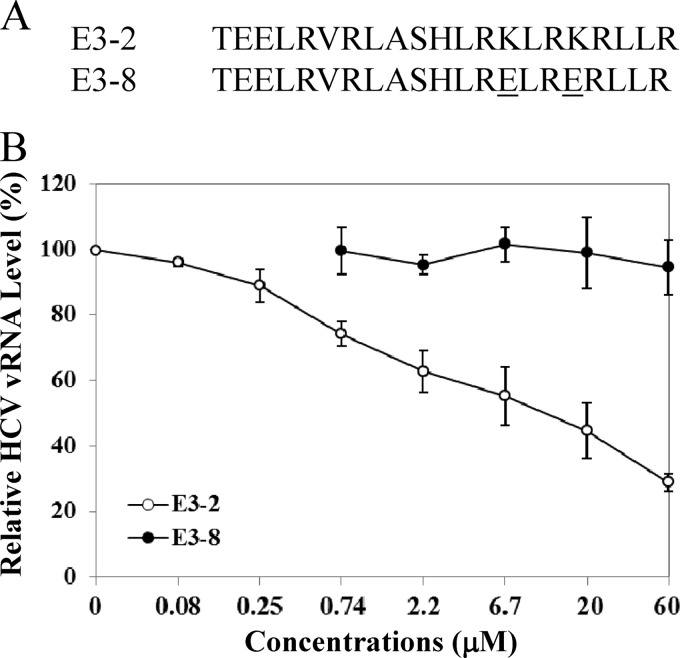
To validate the importance of the receptor-binding domain of apoE in HCV attachment, we determined the inhibition of HCV attachment by a synthetic peptide of 21 amino acids derived from the apoE receptor-binding region (Fig. 10A, E3-2). As a control, two lysine residues at amino acid residues 143 and 146 were mutated to glutamic acids (Fig. 10A, E3-8). Huh-7.5 cells were incubated with HCV on ice for 2 h in the presence of various concentrations of each peptide. Upon removal of HCV and extensive washing of cells with PBS, the vRNA of the cell-bound HCV was extracted with RNAzol reagent and was quantified by qRT-PCR. Results show that HCV attachment was inhibited by the wild-type peptide but not the mutant peptide (Fig. 10B). Again, these results corroborate the findings derived from mutagenesis analysis of apoE (Fig. 7 and 8).
Fig 10.
Inhibition of HCV attachment by a synthetic peptide derived from the apoE receptor-binding region. (A) Huh-7.5 cells in 12-well cell culture plates were incubated with HCV on ice for 2 h in the absence or presence of various concentrations of wild-type (E3-2) and mutant (E3-8) peptides. The unbound HCV was removed by extensive washing with PBS. The vRNA of the cell-bound HCV was isolated with RNAzol reagent (Molecular Research Center). (B) The average levels of HCV vRNA, from triplicate experiments, were determined by qRT-PCR as described in Materials and Methods.
DISCUSSION
Virus attachment to target cells is the very first step of the viral life cycle. It has been the theory in virology that virus attachment is mediated by the binding of viral envelope proteins to the cell surface receptor(s). In contrast to this dogma, we have obtained several lines of evidence demonstrating that the cellular protein apoE mediates HCV attachment. First of all, an apoE-blocking monoclonal antibody and its Fab fragments efficiently inhibited HCV infection (Fig. 1 and data not shown). When examined under the virus attachment conditions, the apoE antibody was able to specifically block the binding of HCV to Huh-7.5 cells and primary human hepatocytes (Fig. 2 and 3). After HCV bound to cells, however, the apoE antibody failed to inhibit HCV cell entry (Fig. 2). Unlike apoE, the HCV E2 acts at a postattachment step(s) of HCV infection. The E2-specific monoclonal antibody CBH5, which was previously shown to be a highly potent inhibitor of HCV infection (39), did not affect HCV attachment, although it inhibited HCV cell entry even after virus attachment (Fig. 4). Another study recently found that a synthetic peptide of 16 amino acids derived from the E2 transmembrane domain inhibited HCV infection at a postbinding step (47). It has been thought that E2 mediates HCV attachment through binding to cell surface receptors such as CD81, claudin-1, LDLr, occludin, SR-BI, and HSPGs (7, 10, 21, 63, 87). Now, increasing evidence suggests that the E2-binding receptors CD81, claudin-1, occludin, and SR-BI may act at postattachment steps during HCV infection (25, 26, 30, 60). HCV E1 may also mediate HCV entry at a postbinding step(s) because E1-specific antibodies were shown to inhibit HCV infection at a postattachment step(s) (30). Additionally, the important role of apoE in HCV attachment is supported by findings derived from the mutagenesis analysis of the apoE receptor-binding region (Fig. 7). Numerous previous studies have demonstrated that the apoE-binding receptors share the same binding site located in helix 4, consisting of amino acid residues 136 to 150. Substitutions of the positively charged arginine and lysine with neutral or negatively charged residues in the receptor-binding site impaired the apoE's ability to mediate HCV infection (Fig. 7, 8, and 10). The role of apoE in HCV attachment is also consistent with the findings that apoE is an integral component localized to the HCV envelope (18, 29, 36, 57). Our previous studies found that apoE was detected in purified HCV particles and that HCV infectivity was proportional to the abundance of apoE in infectious HCV (18). ApoE in purified HCV was found to be sensitive to protease digestion, suggesting its localization on the surface of the HCV envelope (36). The C-terminal α-helical domain of apoE is important for interaction with NS5A, and the apoE-NS5A interaction is critical to HCV assembly and production (12, 24). The structural nature of apoE has recently been confirmed by EM studies (29, 57). Both apoE and HCV E2 were visualized on the envelope of the singular HCV particles by immunogold labeling. However, either apoE or E2, but not both, was detected in a small fraction of HCV particles (57). We believe that apoE plays a major role in mediating HCV attachment. It is also possible that apoE-deficient HCV may use HCV E1/E2 proteins for attachment. It was estimated that each HCV particle contained up to 300 molecules of apoE, which outnumbered E2 protein in the HCV particle (57). This may explain the dominant role of apoE in HCV attachment. Nevertheless, compelling evidence derived from our studies demonstrates that apoE is a structural component mediating HCV attachment.
Several previous studies suggested that HSPGs play an important role in HCV infection (7, 40, 60). Removal of heparan sulfate from cell surface by heparinase treatment reduced the susceptibility of cells to HCV infection (9, 40, 60). In this study, we further found that HCV attachment was blocked by pretreatment of cells with heparinase (Fig. 6). More significantly, we have determined the underlying mechanism of HSPGs in HCV attachment by binding to apoE on the surface of the HCV envelope. Mutations of basic amino acids to neutral or acidic residues in the receptor-binding region ablated the heparin-binding activity of apoE (Fig. 9). The other known HCV receptors and/or coreceptors such as CD81, claudin-1, LDLr, occludin, and SR-BI do not play significant roles in mediating HCV attachment, as demonstrated by specific blocking antibodies and siRNAs (Fig. 5 and data not shown). Although apoE mediates HCV attachment through interactions with the cell surface heparan sulfate, there are many questions waiting to be answered. Why does HCV attachment use a cellular protein unlike most other viruses using a virally encoded envelope protein(s)? Why are so many receptors/co-receptors required for HCV entry to cells? In the case of HIV, cellular proteins facilitate HIV attachment (17, 73, 76). The unique advantage for viruses that use cellular proteins for cell attachment is that this makes it easy to get around the host immune system. The host proteins on virions can evade the host immune response and therefore help viruses to establish persistent infections. The requirement of apoE for HCV attachment may partially contribute to the establishment of a persistent HCV infection. Similar to HIV, HCV uses multiple cellular receptors for its cell entry. The underlying molecular mechanisms of cellular receptors/coreceptors in HCV entry have not been determined. The current model is that each receptor/coreceptor plays a distinct role at a specific stage or step of HCV infection in a highly orchestrated manner through sequential interactions with HCV E1 and E2 proteins (63). It is also possible that multiple receptors are used preferentially by different virus variants containing different mutations in viral envelope proteins because of the quasispecies nature associated with both HCV and HIV. Defining the role and mechanism of action of each receptor in HCV entry will be very challenging.
A number of studies have demonstrated that apoE receptor binding is mediated by a highly basic, α-helical sequence (∼15 residues) within residues 136 to 150 in the receptor-binding domain which interacts with the LDLr family members, including LDLr, LRPs, and HSPGs (43, 51, 53, 79, 83). SR-BI is also a known receptor for apoE binding and HCV infection (1, 8, 11, 70). Like other key HCV receptors, SR-BI functions at a postbinding step(s), as determined by SR-BI-specific small molecule inhibitors and antibodies (74, 88). A previous study suggested that apoE on HCV virions facilitated HCV infection through interaction with LDLr (62). However, findings derived from our studies clearly show that only HSPGs, but not LDLr or SR-BI, are important for HCV attachment. The siRNA-mediated knockdown of both LDLr and SR-BI reduced HCV infection by 80% but did not significantly inhibit HCV attachment. Interestingly, the Lys-to-Arg mutations at residues 143 and 146 (K143/146R) in the receptor-binding region, which were previously shown to decrease the LDLr-binding activity to ∼30% (28, 85), did not affect the heparin-binding activity of apoE (Fig. 9). In fact, the K143/146R apoE mutant slightly increased the HCV infectivity (Fig. 7). These findings are consistent with those reported by others that basic residue specificity is not required for the effective binding of apoE to heparin, unlike its binding to LDLr (28). Furthermore, our recent studies demonstrated that apoE2, which is defective in LDLR binding, had no effect on HCV infectivity and production (24) unlike the findings reported by others (32). Taken together, this compelling evidence suggests that HSPG but not LDLr serves as a receptor for the apoE-mediated HCV attachment. It is not clear why the apoE-binding receptor SR-BI is not important for HCV attachment. The preferential binding of apoE to different receptors on the cell surface may also be influenced by the abundance of receptors expressed among different cell types. Protein modification, oligomerization, and structure configuration of apoE may also modulate the receptor binding. ApoE is a glycosylated protein with at least two glycosylation sites and also forms different oligomers (20, 42, 80, 81, 84, 86). ApoE on the HCV virions appears to be hyperglycosylated compared to apoE expressed in the cell and secreted to supernatant (data not shown). Future studies are warranted to determine the importance of glycosylation and oligomerization of apoE in HCV attachment and the underlying molecular mechanism of apoE in HSPG binding and HCV attachment.
ACKNOWLEDGMENTS
We thank Charlie Rice (Rockefeller University) for kindly providing the Huh-7.5 cell line and NS5A monoclonal antibody 9E10, Takaji Wakita (Japan NIH) for the JFH1 cDNA clone, Steven Foung (Stanford University) for E2 monoclonal antibody CBH5, Jin Ye (UT Southwestern Medical Center) for LDLr-specific monoclonal antibody (clone HL1), and Theodore Mazzone (University of Illinois at Chicago) for the pcDNA3.1/hApoE3 plasmid DNA.
This work is supported by NIH grants (AI091953, AI097318, and AI092074) and in part by a grant from the Kentucky Science and Engineering Foundation (KSEF-148-502-10-272) and by grant AI079150 to H.T.
Footnotes
Published ahead of print 24 April 2012
REFERENCES
- 1. Acharya P, et al. 2002. Comparison of the stabilities and unfolding pathways of human apolipoprotein E isoforms by differential scanning calorimetry and circular dichroism. Biochim. Biophys. Acta 1584:9–19 [DOI] [PubMed] [Google Scholar]
- 2. Agnello V, Abel G, Elfahal M, Knight GB, Zhang QX. 1999. Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. U. S. A. 96:12766–12771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alter MJ. 2006. Epidemiology of viral hepatitis and HIV co-infection. J. Hepatol. 44:S6–S9 [DOI] [PubMed] [Google Scholar]
- 4. André P, et al. 2002. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J. Virol. 76:6919–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. André P, Perlemuter G, Budkowska A, Brechot C, Lotteau V. 2005. Hepatitis C virus particles and lipoprotein metabolism. Semin. Liver Dis. 25:93–104 [DOI] [PubMed] [Google Scholar]
- 6. Appel N, et al. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4:e1000035 doi:10.1371/journal.ppat.1000035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barth H, et al. 2003. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 278:41003–41012 [DOI] [PubMed] [Google Scholar]
- 8. Barth H, et al. 2008. Scavenger receptor class B is required for hepatitis C virus uptake and cross-presentation by human dendritic cells. J. Virol. 82:3466–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barth H, et al. 2006. Viral and cellular determinants of the hepatitis C virus envelope-heparan sulfate interaction. J. Virol. 80:10579–10590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bartosch B, Dubuisson J. 2010. Recent advances in hepatitis C virus cell entry. Viruses 2:692–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bartosch B, et al. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624–41630 [DOI] [PubMed] [Google Scholar]
- 12. Benga WJ, et al. 2010. Apolipoprotein E interacts with hepatitis C virus nonstructural protein 5A and determines assembly of infectious particles. Hepatology 51:43–53 [DOI] [PubMed] [Google Scholar]
- 13. Blanchard E, et al. 2006. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol. 80:6964–6972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blight KJ, Kolykhalov AA, Rice CM. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972–1974 [DOI] [PubMed] [Google Scholar]
- 15. Brown RS. 2005. Hepatitis C and liver transplantation. Nature 436:973–978 [DOI] [PubMed] [Google Scholar]
- 16. Cai Z, et al. 2005. Robust production of infectious hepatitis C virus (HCV) from stably HCV cDNA-transfected human hepatoma cells. J. Virol. 79:13963–13973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chaipan C, et al. 2010. Incorporation of podoplanin into HIV released from HEK-293T cells, but not PBMC, is required for efficient binding to the attachment factor CLEC-2. Retrovirology 7:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang KS, Jiang J, Cai Z, Luo G. 2007. Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J. Virol. 81:13783–13793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Choo QL, et al. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359–362 [DOI] [PubMed] [Google Scholar]
- 20. Choy N, Raussens V, Narayanaswami V. 2003. Inter-molecular coiled-coil formation in human apolipoprotein E C-terminal domain. J. Mol. Biol. 334:527–539 [DOI] [PubMed] [Google Scholar]
- 21. Cocquerel L, Voisset C, Dubuisson J. 2006. Hepatitis C virus entry: potential receptors and their biological functions. J. Gen. Virol. 87:1075–1084 [DOI] [PubMed] [Google Scholar]
- 22. Cormier EG, et al. 2004. L-SIGN (CD209L) and DC-SIGN (CD209) mediate transinfection of liver cells by hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 101:14067–14072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cormier EG, et al. 2004. CD81 is an entry coreceptor for hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 101:7270–7274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cun W, Jiang J, Luo G. 2010. The C-terminal alpha-helix domain of apolipoprotein E is required for interaction with nonstructural protein 5A and assembly of hepatitis C virus. J. Virol. 84:11532–11541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Evans MJ, et al. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446:801–805 [DOI] [PubMed] [Google Scholar]
- 26. Fofana I, et al. 2010. Monoclonal anti-claudin 1 antibodies prevent hepatitis C virus infection of primary human hepatocytes. Gastroenterology 139:953–964 doi:10.1053/j.gastro.2010.05.073 [DOI] [PubMed] [Google Scholar]
- 27. Fried MW, et al. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975–982 [DOI] [PubMed] [Google Scholar]
- 28. Futamura M, et al. 2005. Two-step mechanism of binding of apolipoprotein E to heparin: implications for the kinetics of apolipoprotein E-heparan sulfate proteoglycan complex formation on cell surfaces. J. Biol. Chem. 280:5414–5422 [DOI] [PubMed] [Google Scholar]
- 29. Gastaminza P, et al. 2010. Ultrastructural and biophysical characterization of hepatitis C virus particles produced in cell culture. J. Virol. 84:10999–11009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haberstroh A, et al. 2008. Neutralizing host responses in hepatitis C virus infection target viral entry at postbinding steps and membrane fusion. Gastroenterology 135:1719–1728 [DOI] [PubMed] [Google Scholar]
- 31. Hatters DM, Peters-Libeu CA, Weisgraber KH. 2006. Apolipoprotein E structure: insights into function. Trends Biochem. Sci. 31:445–454 [DOI] [PubMed] [Google Scholar]
- 32. Hishiki T, et al. 2010. Infectivity of hepatitis C virus is influenced by association with apolipoprotein E isoforms. J. Virol. 84:12048–12057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hofmann WP, Zeuzem S. 2011. A new standard of care for the treatment of chronic HCV infection. Nat. Rev. Gastroenterol. Hepatol. 8:257–264 [DOI] [PubMed] [Google Scholar]
- 34. Hsu M, et al. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. U. S. A. 100:7271–7276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang Y, Weisgraber KH, Mucke L, Mahley RW. 2004. Apolipoprotein E: diversity of cellular origins, structural and biophysical properties, and effects in Alzheimer's disease. J. Mol. Neurosci. 23:189–204 [DOI] [PubMed] [Google Scholar]
- 36. Jiang J, Luo G. 2009. Apolipoprotein E but not B is required for the formation of infectious hepatitis C virus particles. J. Virol. 83:12680–12691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jirasko V, et al. 2008. Structural and functional characterization of nonstructural protein 2 for its role in hepatitis C virus assembly. J. Biol. Chem. 283:28546–28562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jones CT, Murray CL, Eastman DK, Tassello J, Rice CM. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 81:8374–8383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Keck ZY, et al. 2007. Immunogenic and functional organization of hepatitis C virus (HCV) glycoprotein E2 on infectious HCV virions. J. Virol. 81:1043–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Koutsoudakis G, et al. 2006. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 80:5308–5320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lau GK, et al. 2005. Peginterferon Alfa-2a, lamivudine, and the combination for HBeAg-positive chronic hepatitis B. N. Engl. J. Med. 352:2682–2695 [DOI] [PubMed] [Google Scholar]
- 42. Lee Y, et al. 2010. Glycosylation and sialylation of macrophage-derived human apolipoprotein E analyzed by SDS-PAGE and mass spectrometry: evidence for a novel site of glycosylation on Ser290. Mol. Cell. Proteomics 9:1968–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Libeu CP, et al. 2001. New insights into the heparan sulfate proteoglycan-binding activity of apolipoprotein E. J. Biol. Chem. 276:39138–39144 [DOI] [PubMed] [Google Scholar]
- 44. Lindenbach BD, et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 45. Lindenbach BD, Rice CM. 2005. Unravelling hepatitis C virus replication from genome to function. Nature 436:933–938 [DOI] [PubMed] [Google Scholar]
- 46. Liu BM, et al. 2010. Characterization of potential antiviral resistance mutations in hepatitis B virus reverse transcriptase sequences in treatment-naive Chinese patients. Antiviral Res. 85:512–519 [DOI] [PubMed] [Google Scholar]
- 47. Liu R, et al. 2010. A peptide derived from hepatitis C virus E2 envelope protein inhibits a post-binding step in HCV entry. Antiviral Res. 86:172–179 [DOI] [PubMed] [Google Scholar]
- 48. Liu S, et al. 2009. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J. Virol. 83:2011–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lohmann V, et al. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113 [DOI] [PubMed] [Google Scholar]
- 50. Mahley RW. 1988. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science 240:622–630 [DOI] [PubMed] [Google Scholar]
- 51. Mahley RW. 1996. Heparan sulfate proteoglycan/low density lipoprotein receptor-related protein pathway involved in type III hyperlipoproteinemia and Alzheimer's disease. Isr. J. Med. Sci. 32:414–429 [PubMed] [Google Scholar]
- 52. Mahley RW, Rall SC., Jr 2000. Apolipoprotein E: far more than a lipid transport protein. Annu. Rev. Genomics Hum. Genet. 1:507–537 [DOI] [PubMed] [Google Scholar]
- 53. Mahley RW, Weisgraber KH, Huang Y. 2009. Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer's disease to AIDS. J. Lipid Res. 50(Suppl):S183–S188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Manns MP, et al. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958–965 [DOI] [PubMed] [Google Scholar]
- 55. Meertens L, et al. 2008. The tight junction proteins claudin-1, -6, and -9 are entry cofactors for hepatitis C virus. J. Virol. 82:3555–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Meertens L, Bertaux C, Dragic T. 2006. Hepatitis C virus entry requires a critical postinternalization step and delivery to early endosomes via clathrin-coated vesicles. J. Virol. 80:11571–11578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Merz A, et al. 2011. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J. Biol. Chem. 286:3018–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Molina S, et al. 2007. The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J. Hepatol. 46:411–419 [DOI] [PubMed] [Google Scholar]
- 59. Montalto G, et al. 2002. Epidemiology, risk factors, and natural history of hepatocellular carcinoma. Ann. N. Y. Acad. Sci. 963:13–20 [DOI] [PubMed] [Google Scholar]
- 60. Morikawa K, et al. 2007. The roles of CD81 and glycosaminoglycans in the adsorption and uptake of infectious HCV particles. J. Med. Virol. 79:714–723 [DOI] [PubMed] [Google Scholar]
- 61. Murray CL, Jones CT, Tassello J, Rice CM. 2007. Alanine scanning of the hepatitis C virus core protein reveals numerous residues essential for production of infectious virus. J. Virol. 81:10220–10231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Owen DM, Huang H, Ye J, Gale M., Jr 2009. Apolipoprotein E on hepatitis C virion facilitates infection through interaction with low-density lipoprotein receptor. Virology 394:99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pietschmann T. 2009. Virology: final entry key for hepatitis C. Nature 457:797–798 [DOI] [PubMed] [Google Scholar]
- 64. Pileri P, et al. 1998. Binding of hepatitis C virus to CD81. Science 282:938–941 [DOI] [PubMed] [Google Scholar]
- 65. Ploss A, et al. 2009. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457:882–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pöhlmann S, et al. 2003. Hepatitis C virus glycoproteins interact with DC-SIGN and DC-SIGNR. J. Virol. 77:4070–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Popescu CI, Dubuisson J. 2010. Role of lipid metabolism in hepatitis C virus assembly and entry. Biol. Cell 102:63–74 [DOI] [PubMed] [Google Scholar]
- 68. Robertson B, et al. 1998. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. International Committee on Virus Taxonomy. Arch. Virol. 143:2493–2503 [DOI] [PubMed] [Google Scholar]
- 69. Saunier B, et al. 2003. Role of the asialoglycoprotein receptor in binding and entry of hepatitis C virus structural proteins in cultured human hepatocytes. J. Virol. 77:546–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Scarselli E, et al. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shepard CW, Finelli L, Alter MJ. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5:558–567 [DOI] [PubMed] [Google Scholar]
- 72. Steinmann E, et al. 2007. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 3:e103 doi:10.1371/journal.ppat.0030103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. St-Pierre C, Ouellet M, Tremblay MJ, Sato S. 2010. Galectin-1 and HIV-1 infection. Methods Enzymol. 480:267–294 [DOI] [PubMed] [Google Scholar]
- 74. Syder AJ, et al. 2011. Small molecule scavenger receptor BI antagonists are potent HCV entry inhibitors. J. Hepatol. 54:48–55 [DOI] [PubMed] [Google Scholar]
- 75. Tellinghuisen TL, Foss KL, Treadaway J. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4:e1000032 doi:10.1371/journal.ppat.1000032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tsegaye TS, Pohlmann S. 2010. The multiple facets of HIV attachment to dendritic cell lectins. Cell Microbiol. 12:1553–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wakita T, et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang C, Sarnow P, Siddiqui A. 1993. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 67:3338–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Weisgraber KH. 1994. Apolipoprotein E: structure-function relationships. Adv. Protein Chem. 45:249–302 [DOI] [PubMed] [Google Scholar]
- 80. Wernette-Hammond ME, et al. 1989. Glycosylation of human apolipoprotein E. The carbohydrate attachment site is threonine 194. J. Biol. Chem. 264:9094–9101 [PubMed] [Google Scholar]
- 81. Westerlund JA, Weisgraber KH. 1993. Discrete carboxyl-terminal segments of apolipoprotein E mediate lipoprotein association and protein oligomerization. J. Biol. Chem. 268:15745–15750 [PubMed] [Google Scholar]
- 82. WHO 1998. WHO concerns hepatitis C. Lancet 351:1415 [Google Scholar]
- 83. Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA. 1991. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science 252:1817–1822 [DOI] [PubMed] [Google Scholar]
- 84. Yamauchi Y, et al. 2008. Role of the N- and C-terminal domains in binding of apolipoprotein E isoforms to heparan sulfate and dermatan sulfate: a surface plasmon resonance study. Biochemistry 47:6702–6710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zaiou M, et al. 2000. Apolipoprotein E–low density lipoprotein receptor interaction: influences of basic residue and amphipathic alpha-helix organization in the ligand. J. Lipid Res. 41:1087–1095 [PubMed] [Google Scholar]
- 86. Zanni EE, Kouvatsi A, Hadzopoulou-Cladaras M, Krieger M, Zannis VI. 1989. Expression of ApoE gene in Chinese hamster cells with a reversible defect in O-glycosylation. Glycosylation is not required for apoE secretion. J. Biol. Chem. 264:9137–9140 [PubMed] [Google Scholar]
- 87. Zeisel MB, Fofana I, Fafi-Kremer S, Baumert TF. 2011. Hepatitis C virus entry into hepatocytes: molecular mechanisms and targets for antiviral therapies. J. Hepatol. 54:566–576 [DOI] [PubMed] [Google Scholar]
- 88. Zeisel MB, et al. 2007. Scavenger receptor class B type I is a key host factor for hepatitis C virus infection required for an entry step closely linked to CD81. Hepatology 46:1722–1731 [DOI] [PubMed] [Google Scholar]
- 89. Zhang J, et al. 2004. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 78:1448–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]