Abstract
Influenza virus infection results in strong, mainly T-dependent, extrafollicular and germinal center B cell responses, which provide lifelong humoral immunity against the homotypic virus strain. Follicular T helper cells (TFH) are key regulators of humoral immunity. Questions remain regarding the presence, identity, and function of TFH subsets regulating early extrafollicular and later germinal center B cell responses. This study demonstrates that ICOS but not CXCR5 marks T cells with B helper activity induced by influenza virus infection and identifies germinal center T cells (TGC) as lymph node-resident CD4+ ICOS+ CXCR4+ CXCR5+ PSGL-1lo PD-1hi cells. The CXCR4 expression intensity further distinguished their germinal center light and dark zone locations. This population emerged strongly in regional lymph nodes and with kinetics similar to those of germinal center B cells and were the only TFH subsets missing in influenza virus-infected, germinal center-deficient SAP−/− mice, mice which were shown previously to lack protective memory responses after a secondary influenza virus challenge, thus indicting the nonredundant functions of CXCR4- and CXCR5-coexpressing CD4 helper cells in antiviral B cell immunity. CXCR4-single-positive T cells, present in B cell-mediated autoimmunity and regarded as “extrafollicular” helper T cells, were rare throughout the response, despite prominent extrafollicular B cell responses, revealing fundamental differences in autoimmune- and infection-induced T-dependent B cell responses. While all ICOS+ subsets induced similar antibody levels in vitro, CXCR5-single-positive T cells were superior in inducing B cell proliferation. The regulation of T cell localization, marked by the single and coexpression of CXCR4 and CXCR5, might be an important determinant of TFH function.
INTRODUCTION
Tcell-dependent B cell responses, hallmarks of adaptive immunity and protection from infections, such as infections with influenza virus, take place in distinct locations within secondary lymphoid tissues. Following activation, B cells move to medullary cords to establish extrafollicular foci or into B cell follicles to establish germinal centers (secondary follicles). Extrafollicular B cells quickly differentiate into antibody-secreting cells, a process that can involve T-dependent B cell activation but does not require continuous T cell help (28, 43). Germinal center B cells, sources of high-affinity antibodies and humoral memory, require ongoing CD4 T cell help. Follicular dendritic cells (FDC) and T cells regulate the selection of high-affinity B cells and differentiation into long-lived plasma cells and memory cells (45). Follicular T helper cells (TFH), found throughout B cell follicles, are a major CD4 T cell subset regulating humoral immunity to model antigens (3, 6, 24, 41) and influenza virus infection (22, 42). Many questions remain regarding the role of helper T cell subsets in initiating, maintaining, and regulating extrafollicular and germinal center responses.
As recently highlighted (25), current studies underscore the need for a more complete understanding of the role of TFH functions during influenza virus infection, because of their importance in shaping protective B cell responses during infection and vaccination. Moreover, T cells deficient in SLAM adaptor protein (SAP), a signaling molecule required for their ability to migrate into germinal centers, have been shown to be critical for the generation of a protective humoral memory response to influenza virus infection (21). However, functional studies of TFH populations have been hampered by the lack of markers distinguishing T cells supporting different B cell fate decisions (reviewed in reference 49).
The existence of CXCR4+ CXCR5− helper T cells that support extrafollicular B cell responses in autoimmune-prone mice has been indicated (32). This is consistent with previous studies demonstrating that CXCR4 directs the migration and retention of extrafollicular plasmablasts within medullary regions of lymph nodes (16). Whether such cells are hallmarks of an aberrant autoimmune process or are part of the normal immune response following immunization or infection remains unclear. Interestingly, CXCR4 also regulates germinal center dark and light zone demarcation (1).
The acquisition of CXCR5 and the loss of CCR7 direct the migration of primed CD4 T cells into B cell follicles, where they acquire effector functions needed to support germinal centers (18). The precise T cell signals are not understood, but interleukin-21 (IL-21) secretion was recently found to be necessary for the optimal affinity maturation of B cells and sustained germinal center responses (7, 51). The coexpression of CXCR5 and ICOS identifies TFH. Interestingly, the transcriptional repressor Bcl-6, shown previously to direct TFH development (20, 31, 48), regulates the expression of CXCR5 and CXCR4 and thereby may regulate TFH migration (48). Other receptors associated with TFH are PD-1 and SAP (9, 18, 30, 44). Functionally, CXCR5 is required for homing to primary follicles; however, CXCR5 is neither required for CD4 T cell migration into germinal centers (secondary follicles) (18) nor sufficient for CD4 T cells to support germinal centers (34). Thus, although CXCR5 is a marker for T cells in the B cell follicle, it does not discriminate between locations in primary and secondary B cell follicles.
Germinal center TFH are currently identified as a fraction of CXCR5+ TFH with the highest level of PD-1 expression (18); however, the PD-1 expression level on TFH varies, and thus, a PD-1hi population is not clearly distinguished. SAP, an adaptor protein involved in cell-cell contacts, facilitates the migration of TFH into germinal centers (8, 34). The lack of SAP expression by CD4 T cells, one underlying cause of human X-linked lymphoproliferative disease (10, 29, 40), strongly diminishes germinal center sizes and numbers (12). Mice lacking SAP also lack PD-1hi TFH (34) yet still form ICOS+ CXCR5+ CD4+ TFH that home to B cell follicles but not to germinal centers (12, 34), indicating that germinal center CD4 T cells (TGC) differ from other TFH. Recently, GL-7 expression was shown to mark TGC (50). However, in wild-type mice, both GL-7 and PD-1 are induced transiently during early T cell-B cell interactions in the interfollicular bridge zones (23) and thus do not exclusively mark TGC. In humans, but not in mice, TGC might express CD57 (24, 35). Other markers described previously for TGC, such as B and T lymphocyte attenuator (BTLA), stain most CXCR5+ cells and thus seem of limited use for differentiating TFH (18, 30). Markers to discriminate CD4 T cells located in germinal center light and dark zones are also missing (2). Consequently, little is known about any functional differences of T helper cells that support B cells in their different lymph node niches.
Influenza virus infection results in strong, mainly T-dependent, extrafollicular and germinal center B cell responses, which provide lifelong humoral immunity against the homotypic virus strain. Here we investigated T cell subsets that regulate this strong humoral response. We demonstrate that ICOS expression, but not CXCR5 expression, defines the possession of B helper function among CD4 T cells in vitro, while the differential expression of two chemokine receptors, CXCR4 and CXCR5, identifies TFH subsets that reside in distinct compartments of the lymph node. We identify strongly induced germinal center CD4 T cells as being CXCR5/CXCR4 double positive and show that the CXCR4 intensity discriminates between TGC residing in germinal center light and dark zones. CXCR4-single-positive “extrafollicular” T cells were rare throughout the course of infection, despite strong extrafollicular B cell responses. Functionally, CXCR5-single-positive TFH were superior at inducing B cell proliferation, while all subsets provided similar support for antibody production in vitro. Collectively, our studies point to links between chemokine-receptor-mediated CD4 T cell positioning within lymph node niches and their helper cell function after infection.
MATERIALS AND METHODS
Mice.
Female BALB/c (Harlan Laboratories), C57BL/6, and MRL/MpJ-Faslpr mice were purchased (The Jackson Laboratory). SAP−/− mice (breeders kindly provided by P. Schwartzberg, NIH) and TS-1 mice (breeders kindly provided by A. Caton, The Wistar Institute) expressing a transgenic T cell receptor specific for influenza virus A/Puerto Rico/34/8 (H1N1) (A/PR8) hemagglutinin (HA) peptide residues 110 to 119 presented on I-Ed were bred and maintained at the University of California, Davis (26). All mice were used between 8 and 14 weeks of age and kept under conventional housing conditions in filter-top cages. The University of California, Davis, Institutional Animal Care and Use Committee approved all experimental protocols.
Infections and immunizations.
Intranasal influenza virus infections were conducted under isoflurane anesthesia with a sublethal dose of A/PR8 (H1N1) (10 PFU) or the reassortant influenza virus A/Mem71 (H3N1) (12,000 PFU) (5) in 40 μl of phosphate-buffered saline (PBS) per mouse. For immunizations, mice were injected subcutaneously on each side of the tail base with 100 μl of 1,000 hemagglutinating units (HAU) of A/PR8 or A/Mem71 in complete Freund's adjuvant. Virus was propagated in embryonated hen eggs, and infectious titers were established as previously described (13).
FACS staining and sorting.
Single-cell suspensions of mediastinal lymph nodes (MedLN) from infected mice or inguinal lymph nodes from immunized mice were prepared as described previously (39). Cells were stained at 2.5 × 107 cells/ml in “staining medium” (39), blocked with an anti-Fc receptor monoclonal antibody (MAb) (2.4.G2 at 5 μg/ml) on ice for 15 min, and then washed and stained with antibodies against mouse CXCR5-biotin (BD Biosciences) and CXCR4-Alexa 647 (eBioscience) or CXCR4-allophycocyanin (BD Biosciences) at 37°C for 30 to 45 min. Cells were then stained for 20 min on ice with the following conjugates, generated in-house unless otherwise specified: streptavidin-QDot605 (Molecular Probes), ICOS-fluorescein isothiocyanate (FITC) or ICOS-Pacific Blue (7E.17G9), PSGL-1–phycoerythrin (PE) (BD Biosciences), CD62L-Cy5PE (eBioscience), CD4-Cy5.5PE, CD3-efluor780-allophycocyanin (eBioscience), CD11a-Cy7PE (BD Biosciences), CD44-allophycocyanin (eBioscience), PD-1–PE (BD Biosciences), OX-40–biotin (BD Biosciences), Bcl-6–PE (BD Biosciences), biotinylated peanut agglutinin (PNA; Vector Laboratories), anti-TS-1 T cell receptor (TCR)–biotin antibody (6.5-2), CD19-Cy5PE (eBioscience), CD4-Pacific Blue, CD8a-Pacific Blue, CD45R-allophycocyanin, CD38-FITC, CD24-Cy5.5PE, and CD138-biotin (BD Biosciences). Cells were stained with the Live/Dead fixable violet dead cell discriminator (Invitrogen) on ice for 30 min, washed, and resuspended in staining medium for fluorescence-activated cell sorter (FACS) analysis. For Bcl-6, cells were fixed (BD Cytofix/Cytoperm) for 20 min at 37°C, resuspended in BD Perm/Wash buffer for 15 min at room temperature, washed in BD Perm/Wash buffer, and stained with Bcl-6–PE (BD Biosciences) for 45 min at room temperature. For sorting, cells were stained with MAb conjugates against CXCR5-biotin, CD19-Cy5PE, CD8a-Cy5PE, CXCR4-Alexa 647, streptavidin (SA)-QDot605, ICOS-FITC, and CD4-Cy7-allophycocyanin and resuspended in staining medium containing propidium iodide. Samples were run on a FACSAria instrument (BD Biosciences) (39). Data were analyzed by using FlowJo software (Tree Star Inc.). Purities of sorted CD4 T cell subsets were >95% based on ICOS expression and >90% based on chemokine receptor expression.
Magnetic cell separation.
Some cell isolation was done by the depletion of unwanted labeled cells by magnetic cell separation. For B cell enrichments, spleen cells were stained, as described above, with biotinylated antibodies against CD4, CD8 (in-house generated), and TCRγ/δ and CD49b (eBioscience). For TS-1 CD4+ cell enrichments, spleen cells from naïve TS-1 mice were labeled with biotinylated antibodies against CD8, CD19, CD11b (in-house), and CD49b. Suspensions were then labeled with anti-biotin MicroBeads (Miltenyi Biotec) and separated by the use of an autoMACS instrument (Miltenyi Biotec). Purities were >93%, as determined by FACS analysis with anti-CD19- and anti-CD4-allophycocyanin antibodies.
CFSE labeling.
Cells were resuspended in PBS at 107 cells/ml, and an equal volume of 2 mM carboxyfluorescein succinimidyl ester (CFSE) in PBS was added. Cells were incubated at 37°C for 10 min and then washed twice with staining medium.
B cell proliferation.
Magnetically activated cell sorting (MACS)-enriched, CFSE-labeled B cells at 6.25 × 106 cells/ml were pulsed with 1,000 HAU/ml influenza virus A/Mem/71 in culture medium (36) for 3 h at 37°C. A total of 2.5 × 105 pulsed B cells were cocultured in 96-well round-bottom plates for 4 days with graded numbers of purified CD4 T cell subsets from MedLN of infected mice in triplicates.
Adoptive transfers.
A total of 1 × 106 MACS-enriched CD4 T cells from spleens of noninfected TS-1 mice were transferred intravenously (i.v.) in 100 μl PBS, followed by infection with A/PR8.
ELISA.
An enzyme-linked immunosorbent assay (ELISA) was performed as described previously (13). Briefly, Maxisorb plates (Thermo Scientific) were coated with 1,000 HAU of the relevant influenza virus (13). Plates were blocked, and tissue culture supernatants were prediluted 2.5-fold, added to the plate, 2-fold serially diluted in PBS, and incubated for 3 h. Antibody binding was revealed by using biotinylated goat anti-mouse Ig (H+L; Southern Biotech), streptavidin-horseradish peroxidase (Vector Labs), and TMB substrate (13). Influenza virus-specific hyperimmune serum served as the standard to determine the relative units/ml of virus-specific Ig, as described previously (13).
Histology.
Lymph nodes frozen in Tissue-Tek optimal cutting temperature compound (OCT; Sakura) on dry ice and stored at −80°C were cut into 5-μm-thick sections on a cryostat (Leica) and dried onto Superfrost/Plus slides (Fisher Scientific) for 2 h at room temperature (RT). Slides were fixed in ice-cold acetone for 10 min, dried for 1 to 2 h at RT, rehydrated in PBS plus 0.1% bovine serum albumin (BSA), and stained with PBS–0.1% BSA–1% normal horse serum containing anti-CD4-efluor450 antibody (RM4-5; eBiosciences) and either anti-CXCR4-biotin antibody (BD Biosciences), PNA-biotin (Vector Labs), or FDC-M2-biotin (ImmunoKontact) for 2 to 3 h at RT in humid chambers. Slides were washed twice in PBS and once in PBS plus 0.1% BSA, for 5 min each. Slides were then stained with anti-IgD-FITC antibody and streptavidin-Alexa Fluor 594 (Invitrogen) for 1 h, washed three times in PBS, and mounted with Fluoromount-G (SouthernBiotech). Images were collected on an Olympus BX61 microscope with an Olympus DP72 color camera and were processed with MetaMorph (Molecular Devices) and ImageJ (NIH) software. For the quantification of CD4+ cells expressing CXCR4 in each region of the lymph node, four 5-μm sections spaced 150 to 250 μm apart from 3 mice were analyzed. Lymph node regions were identified as CD4+ T zone, IgD+ 1° follicle, IgD− germinal center within an IgD+ follicle, and a germinal center/B follicle border zone, which are located near the edges of germinal centers and contain both IgD+ and IgD− cells. Within each region, the numbers of CD4+ CXCR4+ and CD4+ CXCR4− cells were tallied from 6 to 20 field views at a ×60 magnification per section, totaling an average of 43 field views at a ×60 magnification per mouse. Data points are the frequencies of CD4+ CXCR4+ cells among the total CD4+ cells counted in each region, per mouse. Confocal images were collected with an Olympus FV1000 laser scanning confocal microscope and were processed by using FluoView (Olympus) and ImageJ (NIH) software.
Quantitative reverse transcription-PCR.
RNA was isolated with RNeasy (Qiagen) and stored in THE RNA Storage (Ambion) at −80°C. cDNA was prepared by using random hexamers (Promega) with SuperScript II (Invitrogen). Amplification was performed with Clontech polymerase and the commercial primer probes Mm00477633_m1 for Bcl-6, Mm00517640_m1 for IL-21, Mm00445259_m1 for IL-4, and Mm00484668_m1 for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Applied Biosystems), using the following amplification cycles on a Prism 7700 instrument (Applied Biosystems): 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. Data were normalized for expression relative to the expression of GAPDH.
RESULTS
CD4 T cell helper activity is not restricted to ICOS+ CXCR5+ TFH.
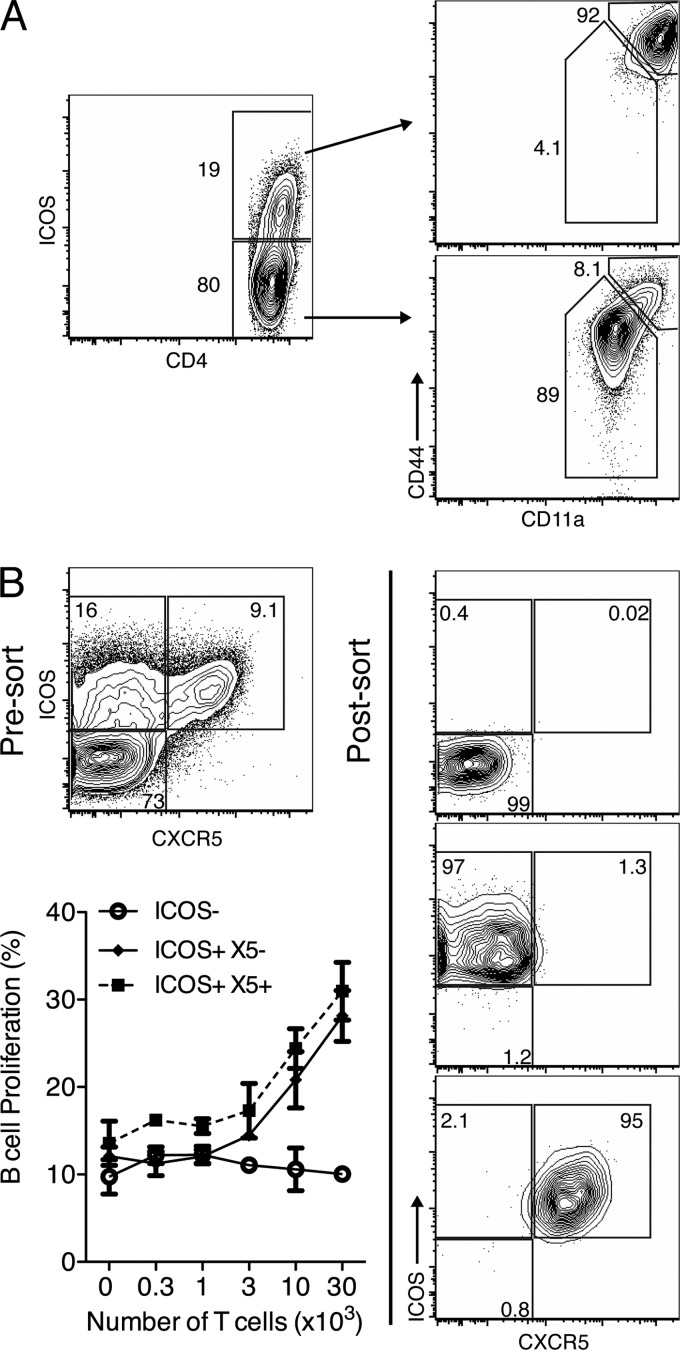
Infection of humans and mice with influenza virus results in antibody-mediated lifelong protection from reinfections with the same influenza virus strain. To identify the CD4 T cell subsets supporting T-dependent B cell responses, CD4 T cells from regional mediastinal lymph nodes (MedLN) of A/Mem71-infected BALB/c mice were analyzed by 9-color flow cytometry for the expressions of various costimulators and activation markers. Day 7 of infection was chosen for initial studies because extrafollicular B cell responses peak and germinal center B cell responses are being established (our unpublished data); thus, T helper cells should be abundant. Initially, we focused on CD4+ ICOS+ CXCR5+ TFH. Consistent with data from previous studies (46, 47), all ICOS+ cells were activated CD4+ CD11ahi CD44hi T cells (Fig. 1A), and all CXCR5+ cells were ICOS+ (Fig. 1B). However, CXCR5 was found for only a subset of all ICOS+ cells (Fig. 1B).
Fig 1.
B helper function is restricted to ICOS+ CD4 helper T cells. (A) BALB/c mice (n = 2) were infected with influenza virus A/Mem71 for 5 days and examined by flow cytometry. Shown are 5% contour plots with outliers gated on live CD3+ CD4+ lymphocytes, with percentages of CD3+ CD4+ cells or ICOS+ cells indicated. Plots are representative of data from 3 similar experiments using BALB/c, BALB/cByJ, or C57BL/6 mice. (B) BALB/c mice (n = 12) were infected with influenza virus A/Mem71 for 12 days, and CD4+ T cells were sorted from pooled MedLN based on ICOS and CXCR5 expressions. Shown are 5% contour plots with outliers of presort and postsort samples. Graded numbers of sorted T cells were cultured for 4 days with 2.5 × 105 CFSE-labeled B cells from pooled inguinal lymph nodes (IngLN) of BALB/c mice (n = 4) at 12 days post-influenza virus immunization. Shown are mean frequencies ± standard deviations (SD) for live CD45R+ B cells which had proliferated (CFSElo) (left), from one representative experiment of two performed.
To determine whether T helper functions were restricted to ICOS+ CXCR5+ TFH, we cocultured FACS-purified ICOS−, ICOS+ CXCR5−, and ICOS+ CXCR5+ CD4 T cells from MedLN of A/Mem71-infected mice at day 12 of infection with MACS-enriched, CFSE-labeled B cells from A/Mem71-immunized mice. Immunization was used to increase B cell yields. Measurements of B cell proliferation demonstrated that among the ICOS+ cells, both CXCR5− and CXCR5+ CD4 T cells induced comparable levels of B cell proliferation (Fig. 1B). Thus, helper function was not restricted to CXCR5-expressing CD4 TFH in vitro.
Low frequencies of CXCR5− CXCR4+ CD4+ T cells following influenza virus infection.
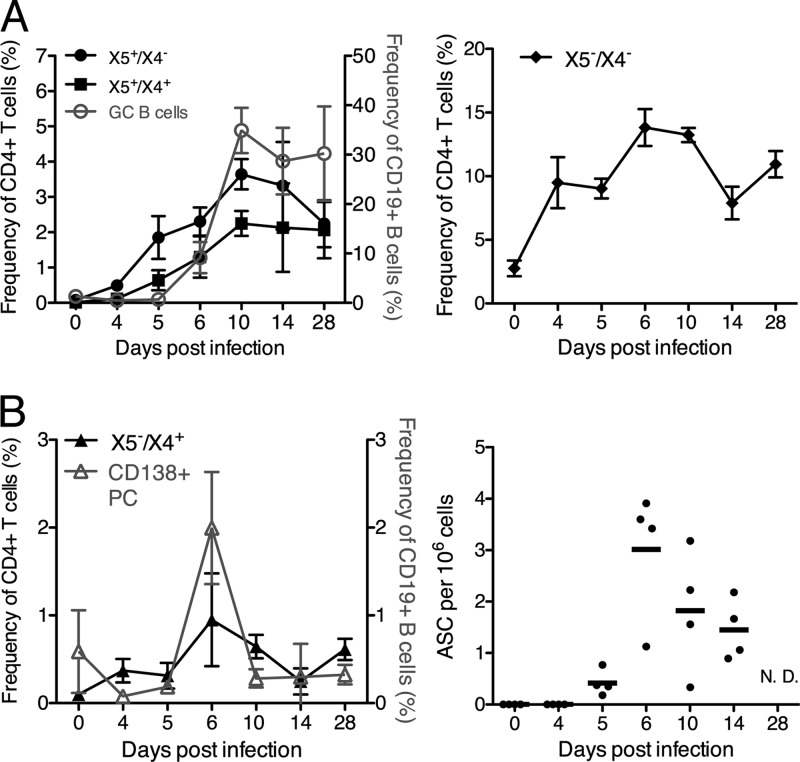
Recently, ICOS+ CXCR5− CXCR4+ T cells associated with extrafollicular focus responses in multiple mouse models of autoimmunity were described (32). We asked whether the helper function of ICOS+ CXCR5− cells in vitro was due to the presence of ICOS+ CXCR5− CXCR4+ “extrafollicular” T cells. Overall, our data do not support that conclusion. Despite the presence of strong extrafollicular B cell responses at day 10 of influenza virus infection (38) (see Fig. 3B), only 3.6% ± 0.17% (n = 4) of CD4+ ICOS+ cells were CXCR5− CXCR4+ (X5−/X4+) (Fig. 2A and B). These cells expressed low levels of CXCR4 and were poorly demarcated from the CXCR4− cells (Fig. 2A). This is in contrast to the high frequencies of ICOS+ CXCR4+ cells present in mouse models of autoimmune disease (32) (see Fig. S1 in the supplemental material). A time course study confirmed the presence of only low frequencies of X5−/X4+ CD4 T cells in the regional lymph nodes throughout the response (Fig. 3B), although a slight rise did occur on day 6 of infection, coinciding with the peak of antibody secretion and CD138+ plasma cell accumulation (Fig. 3B).
Fig 3.
Kinetics of ICOS+ CXCR5+ CXCR4+ T cells correlate with germinal center B cell responses during influenza virus infection. MedLN of influenza virus A/Mem71-infected BALB/c mice (n = 4 per time point) were analyzed by 8-color flow cytometry as described in the legend of Fig. 2A. (A and B) Shown are mean frequencies ± SD of X5+/X4−, X5+/X4+ (left), and X5−/X4− (right) CD4 T cells compared to germinal center B cell frequencies (GC B cells) (live, CD19+ CD38low CD24high) (A) and frequencies of X5−/X4+ cells and plasma cells (CD138+) (left) and antibody-secreting foci, as assessed by an enzyme-linked immunosorbent spot (ELISPOT) analysis (right) (B). ASC, antibody-secreting cells.
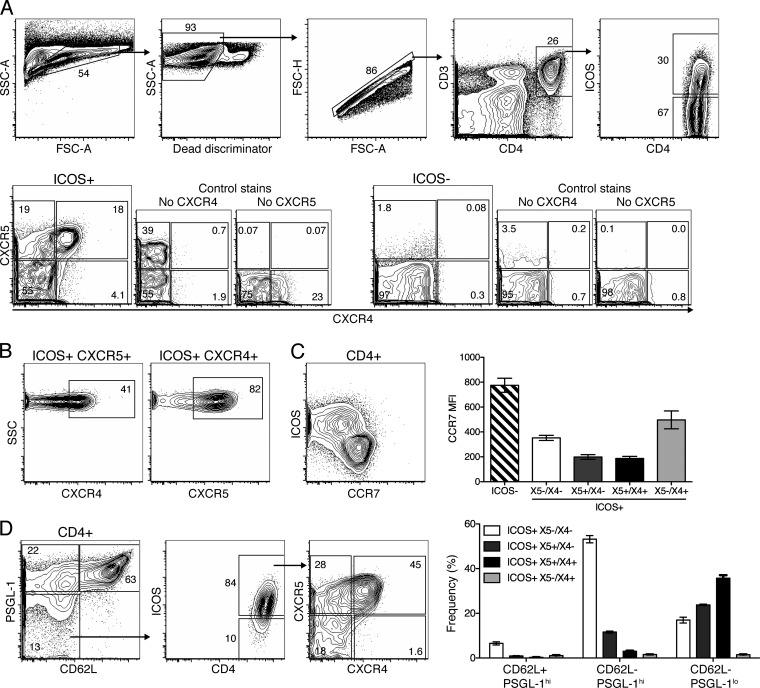
Fig 2.
ICOS+ CXCR5+ TFH coexpressing CXCR4 are induced in response to infection. MedLN isolated from BALB/c mice (n = 4) at 10 days postinfection with influenza virus A/Mem71 were analyzed by 8- to 10-color flow cytometry. Shown are 5% contour plots with outliers of a representative sample from one of at least two repeat experiments (n = 4 each). (A) Gates used to identify CD4+ ICOS+ T cells expressing CXCR5 and/or CXCR4. SSC, side scatter; FSC, forward scatter. (B) CD4+ T cells were further analyzed to display the frequency of ICOS+ CXCR5+ cells coexpressing CXCR4 (left) and ICOS+ CXCR4+ cells coexpressing CXCR5 (right). (C) Expression levels of ICOS and CCR7 were examined on CD4+ T cells (left). The bar chart indicates mean fluorescent intensities (MFI) ± SD for CCR7 on the indicated CD4+ T cell populations. (D) ICOS, CXCR5, and CXCR4 expression levels in CD4+ CD62L− PSGL-1low cells were analyzed. The bar chart represents the mean frequencies ± SD for the four ICOS+ populations identified by CXCR5 and CXCR4 expression among cells indicated on the x axis.
The downregulation of CD62L and PSGL-1 has also been associated with CXCR4+ extrafollicular helper T cells (32), although this might also occur in other TFH (33). Our data indicate that the loss of CD62L and PSGL-1 is associated with CD4 T cell activation, as CD62Lneg PSGL-1low CD4 cells contained mostly ICOS+ and CCR7low cells, while the CD62L+ PSGL-1hi population was mostly ICOS− CCR7hi (Fig. 2C and D). However, the CD62Lneg PSGL-1low ICOS+ CD4+ T cells were not enriched for extrafollicular X4+/X5− cells but instead contained many X4+/X5+ cells. Thus, following influenza virus infection, the downregulation of PSGL-1 is not restricted to X5−/X4+ helper T cells.
The frequencies of extrafollicular CD4+ ICOS+ PSGL-1low CD62Llow CXCR4+ CXCR5− T cells were very low during influenza virus infection (38) (Fig. 3B). Whether this small population is sufficient for the T cell help of extrafollicular focus responses to influenza virus infection, responses that might not require ongoing T cell help once the foci are initiated (28), requires further study. Importantly, it seemed unlikely that the strong in vitro T helper activity observed among the CXCR5− ICOS+ cells (Fig. 1) was due to the presence of these few cells.
Many ICOS+ CXCR5+ TFH coexpress CXCR4.
Nearly half (40% ± 1.1%; n = 4 [representative of data from 2 experiments]) of the ICOS+ CXCR5+ T cells coexpressed CXCR4 (X5+/X4+) at day 10 after infection and expressed low levels of CCR7 (Fig. 2A to C). A significant population of X5+/X4+ cells was also observed for autoimmune MRL/MpJ-Faslpr mice (see Fig. S1 in the supplemental material). Both X5+/X4− and X5+/X4+ populations steadily increased in frequencies (Fig. 3A, left) and absolute numbers (data not shown), which peaked at around day 14 after influenza virus infection of BALB/c mice. These increases correlated with CD19+ CD24hi CD38low germinal center B cell kinetics (Fig. 3A, left). Over 50% of ICOS+ cells expressed neither CXCR5 nor CXCR4 (X5−/X4−) and were CCR7intermediate (Fig. 2A and C). This population rapidly increased after infection and then remained high for >28 days (Fig. 3A, right). Thus, X5−/X4− and X5+/X4+ CD4 T cells are major subpopulations of activated, CD4+ ICOS+ helper T cells. The kinetics of the X5+/X4+ CD4 T cells was most closely correlated with the appearance and maintenance of germinal center B cells after influenza virus infection.
Virus-specific TFH coexpress CXCR4.
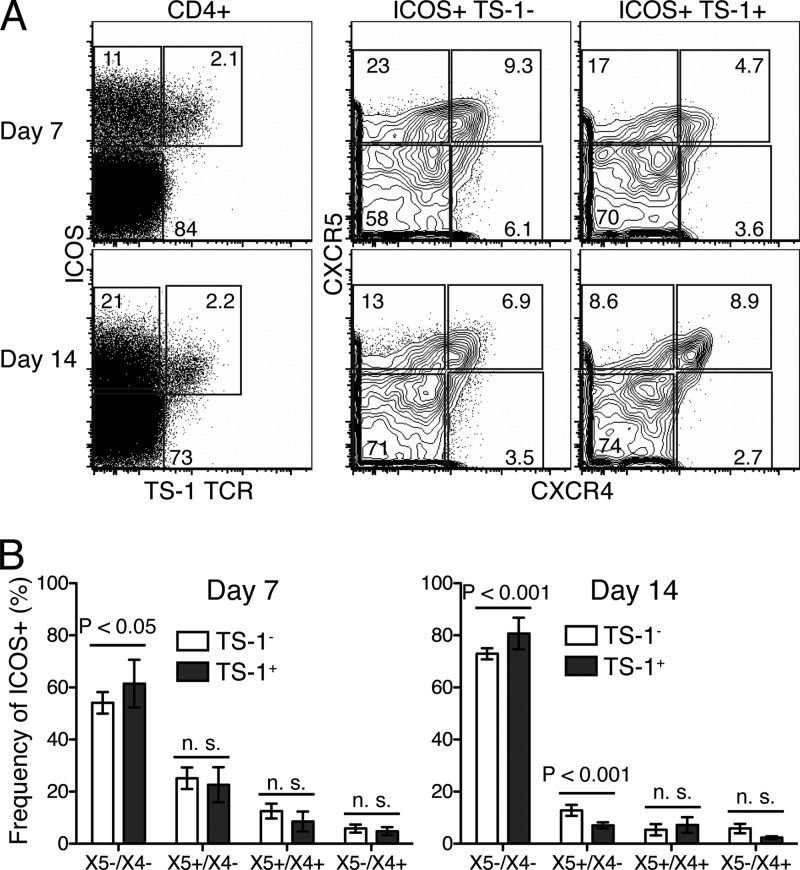
We next asked whether the observed phenotypic heterogeneity among ICOS+ CD4+ T cells is the result of antigen specificity differences. Thus, we compared chemokine receptor expressions of transgenic, influenza virus A/PR8 hemagglutinin-specific T cells (TS-1) following their adoptive transfer into wild-type mice with those of endogenous T cells. At days 7 and 14 postinfection, virus-specific TS-1 T cells were identified by using a biotinylated transgene-specific MAb (26). This required the use of CXCR5-PE instead of CXCR5-biotin, which, in our hands, gives superior staining. Despite the somewhat suboptimal staining, virus-specific TS-1 and endogenous ICOS+ CD4+ T cells showed the same profile of chemokine receptor expression and were present at similar frequencies (Fig. 4). One exception was the X5+/X4− population, which was reduced among the transgenic T cells at 14 days postinfection. The reasons for this finding are unclear but could be related to the transfer of relatively large numbers of transgenic T cells, which was shown previously to result in the faster contraction of transferred cells (17). The data demonstrate that CD4+ T cells of the same antigen specificity exhibit similar degrees of heterogeneity with regard to chemokine receptor expression as polyclonal T cells. Therefore, differences in antigen specificity do not underlie the heterogeneity observed among activated ICOS+ CD4+ T cells.
Fig 4.
Influenza virus-specific T cells coexpress CXCR5 and CXCR4. A total of 1 × 106 MACS-enriched CD4+ T cells from TCR transgenic TS-1 mice, specific for HA of influenza virus A/PR8, were transferred into BALB/c mice (n = 4 per time point). Mice were infected immediately, and MedLN were analyzed by 9-color flow cytometry at 7 and 14 days postinfection. (A) Transgenic CD4+ T cells were identified by using a clonotype-specific MAb and analyzed for chemokine receptor expression as described in the legend of Fig. 2A. Shown are dot plots and 5% contour FACS plots with outliers of a representative sample for each time point. (B) Bar charts indicating mean frequencies ± SD of each CD4 T cell subset among TS-1 transgenic and endogenous cells. Data were pooled from two replicate experiments.
ICOS+ CXCR5+ CXCR4+ TFH are located within germinal centers.
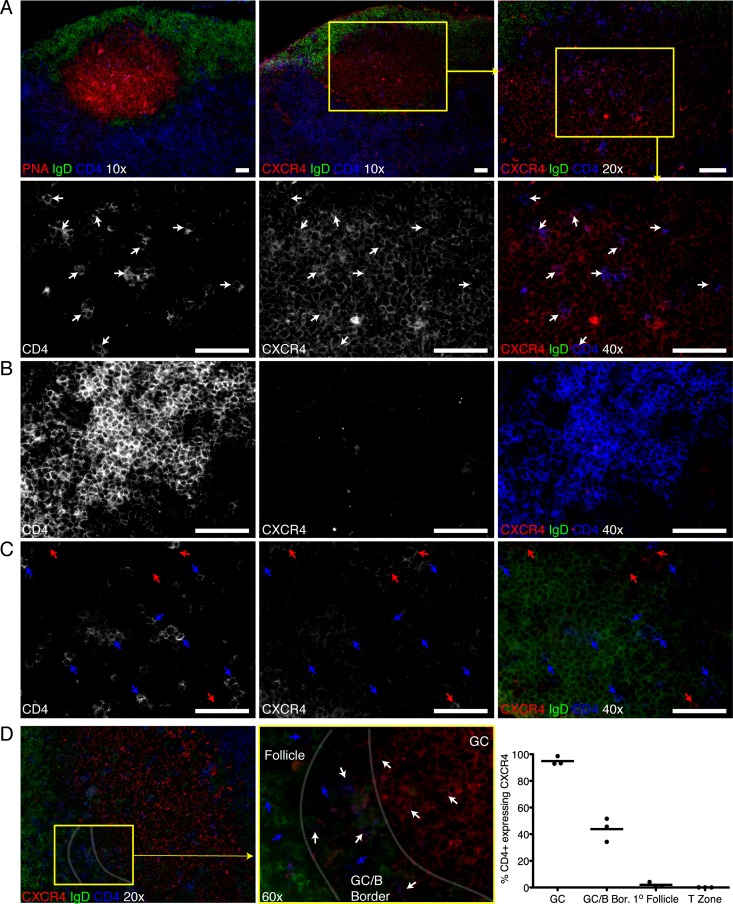
CXCR4 expression among CXCR5+ B cells has been linked to their positioning within germinal centers (1), and thus, the coexpression of CXCR4 and CXCR5 on TFH might indicate a germinal center localization. We therefore performed immunofluorescence microscopy on MedLN from BALB/c mice 10 days after influenza virus infection. Serial sections were stained for (i) IgD, to visualize both B cell follicles and germinal centers, as the latter appear as IgD− zones within the IgD+ primary follicle; (ii) CD4; and (iii) either PNA to stain germinal centers, FDC-M2 to identify germinal center light zones, or CXCR4. As expected, CD4+ T cells were found in both light and dark zones of germinal centers and were more prevalent in the (T-zone-distal) light zone. Importantly, only CD4+ T cells in germinal centers (CD4+ CXCR4+) (Fig. 5A and D, white arrows) expressed CXCR4 but not in T cell zones (Fig. 5B and D) or primary follicles (CD4+ CXCR4−) (Fig. 5C and D, blue arrows). Since germinal center B cells express CXCR4, it was possible that the apparent CD4/CXCR4 dual staining represented an artifact of bleed-over staining from the CXCR4 expressed on the B cells surrounding the CD4 T cells in the germinal centers. However, high-resolution laser scanning confocal microscopy, imaging only a fraction of the tissue plane, confirmed the colocalization of CXCR4 on CD4+ germinal center T cells (see Fig. S2 in the supplemental material) but not in other parts of the lymph node (data not shown). Moreover, the quantification of CD4+ cells coexpressing CXCR4 in each region of the lymph node showed that within germinal centers, 95% ± 3.2% of the CD4+ T cells coexpressed CXCR4, while in B cell follicles and T cell zones, only 2.0% ± 1.9% and 0.04% ± 0.04% coexpressed CXCR4, respectively (Fig. 5D). At the germinal center-B cell follicle border, a region which has scattered PNA staining (not shown) and a mixed population of both IgD− germinal center and IgD+ naïve B cells also had a more mixed population of T cells, 44% ± 8.8% of which coexpressed CXCR4 (Fig. 5D).
Fig 5.
Germinal center T cells express CXCR4. Serial sections from MedLN of influenza virus A/Mem71-infected mice at day 10 stained with IgD (green), CD4 (blue), and either PNA or CXCR4 (red). At a magnification of ×40, CD4 and CXCR4 are shown as black and white images of their respective channels, while merged images include IgD. Yellow boxes indicate the area within the next-higher magnification. Shown are representative images from one mouse of three individually sectioned and stained mice. White bars indicate 50-mm scale bars. (A to C) PNA staining confirms an IgD− germinal center, and CD4/CXCR4 staining on the serial section shows that germinal center T cells express CXCR4 (white arrows) (A), while T zone (B)- and primary B cell follicle (C)-localized T cells lack CXCR4 expression. Red arrows indicate CXCR4+ but CD4− cells. (D) Images taken at a ×60 magnification show the frequency of CD4+ T cells coexpressing CXCR4, germinal center T cells coexpressing CXCR4 (white arrows), and follicle T cells lacking CXCR4 (blue arrows), whereas there is a mixed population of CD4 T cells coexpressing CXCR4 in the border between the germinal center and follicle. CD4+ CXCR4+ and CD4+ CXCR4− cells were counted in germinal centers, follicles, the germinal center-follicle border, and T zones from 4 sections per mouse (n = 3) and added, and the frequency of CD4+ CXCR4+ cells among CD4+ cells is shown in the chart. Bars indicate average values.
Consistent with dark/light zone B cells expressing high/low levels of CXCR4 (1), dark zone CD4+ T cells strongly expressed CXCR4, whereas light zone T cells were more dimly stained (Fig. 5A). Occasionally, small numbers of CXCR4+ CD4+ T cells were found in small clusters of CXCR4+ IgD− CD4− cells, a phenotype consistent with plasma cells. These cells seemed to represent extrafollicular focus T cells (data not shown). Thus, CXCR4+ CXCR5+ ICOS+ CD4+ expression identifies germinal center TFH (TGC), and CXCR4+ expression level identifies their sublocation within germinal center light and dark zones.
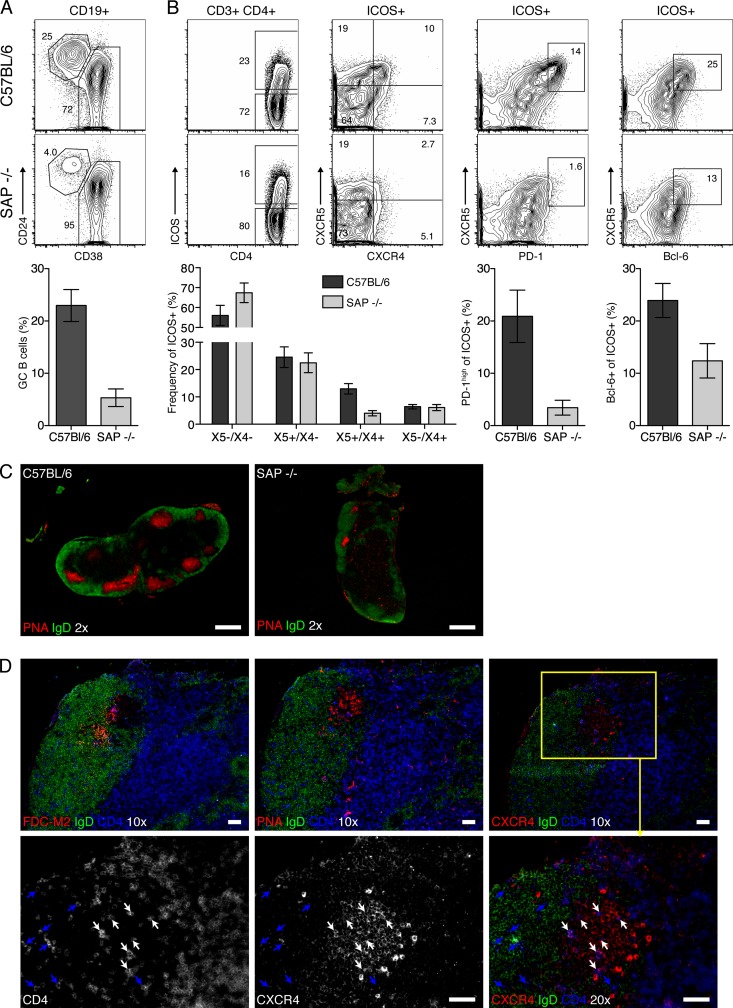
SAP−/− mice lack X5+/X4+ TFH.
SAP-deficient mice have a CD4 T cell-intrinsic defect in germinal center formation, which was correlated with the inability of SAP−/− T cells to efficiently migrate into germinal centers following T cell-B cell interactions (8, 34). These mice do, however, generate ICOS+ CXCR5+ TFH in primary follicles following antigen challenge (12, 27). To determine the functional relevance of the TGC subset identified above, we asked whether the strongly reduced germinal center formation in SAP−/− mice was associated with a lack of TGC. Consistent with data from previous studies (12), SAP−/− mice had few germinal center B cells and lower frequencies of ICOS+ CXCR5+ T cells (Fig. 6A) and completely lacked CXCR5+ PD-1high TFH 10 days after infection (Fig. 6B). Strikingly, there was a selective loss of CXCR4 coexpressers among ICOS+ CXCR5+ T cells in the SAP−/− mice (P < 0.001), while the other T cell subsets were present at frequencies comparable to those of the wild type (Fig. 6B). Histology confirmed the reduced numbers and sizes of germinal centers in SAP−/− mice compared to those in wild-type mice (Fig. 6C), which appeared morphologically intact based on PNA and FDC-M2 staining and the presence of light/dark zones and CD4+ T cells (Fig. 6D, top). The remaining CD4+ T cells present showed low expression levels of CXCR4 (Fig. 6D, bottom, blue arrows), explaining the lack of discernible CXCR4+ CD4+ T cells in SAP−/− mice by FACS analysis. These data further show that the coexpression of CXCR4 and CXCR5 identifies TGC and suggest that germinal centers require X5+/X4+ TGC.
Fig 6.
SAP−/− mice lack ICOS+ X5+/X4+ TFH. (A and B) MedLN from influenza virus A/Mem71-infected C57BL/6 (top) and SAP−/− mice (bottom) (n = 4 to 5/group) at day 10 were analyzed by 8-color flow cytometry. Shown are 5% contour plots with outliers of CD19+ germinal center B cells (CD24hi CD38lo gate) (A) and ICOS expression on live CD4+ T cells (B, left) and the expression of the indicated markers on CD4+ ICOS+ T cells (B, right). Bar charts indicate mean frequencies ± SD of gated cell populations in SAP−/− (light bars) and C57BL/6 (dark bars) mice, respectively. Data are pooled from two experiments (n = 4 and n = 5). (C and D) Serial sections from MedLN of influenza virus-infected mice at 10 days stained for IgD (green), CD4 (blue), and either PNA, FDC-M2, or CXCR4 (red), as indicated. Shown are representative images of MedLN from one mouse of two individually sectioned and stained mice. White bars indicate 500-mm (×2 magnification) or 50-mm (×10 and ×20 magnifications) scale bars. At a ×20 magnification, CD4 and CXCR4 are shown as black and white images of their respective channels, while merged images include IgD. The yellow box indicates the area within the next-higher magnification. White arrows indicate CD4+ CXCR4+ cells, and blue indicates CD4+ CXCR4− cells.
The adoptive transfer of wild-type X5+/X4+ TFH from MedLN back into infected mice demonstrated their inability to enter lymph nodes (not shown), likely because they lack CD62L and PSGL-1, molecules that facilitate T cell migration through high endothelial venules (HEV) (4, 15) (Fig. 2), further indicating their residence and function within secondary lymphoid tissues. This finding prevented us from testing directly whether the transfer of wild-type X5+/X4+ TGC reconstitutes germinal centers in SAP−/− mice. Since CXCR4 is required for thymocyte maturation (19), CXCR4−/− mice lack normal numbers of properly matured T cells, making results from studies of germinal center formation in these mice ambiguous. Systems to deplete CXCR4 expression upon T cell activation are not currently available. However, CXCR4 might not be solely responsible for the germinal center homing of TGC, as CXCR4 expression by B cells is important for light/dark zone organization but not germinal center formation (1). Collectively, the data suggest that the regulation of X5/X4 coexpression directs the germinal center sublocalization of TFH. The low expression level of CXCR4, determined by immunofluorescence, among the few TGC cells present in the SAP−/− mice further supports this conclusion.
X5+/X4+ TFH have characteristics associated with germinal center T cells.
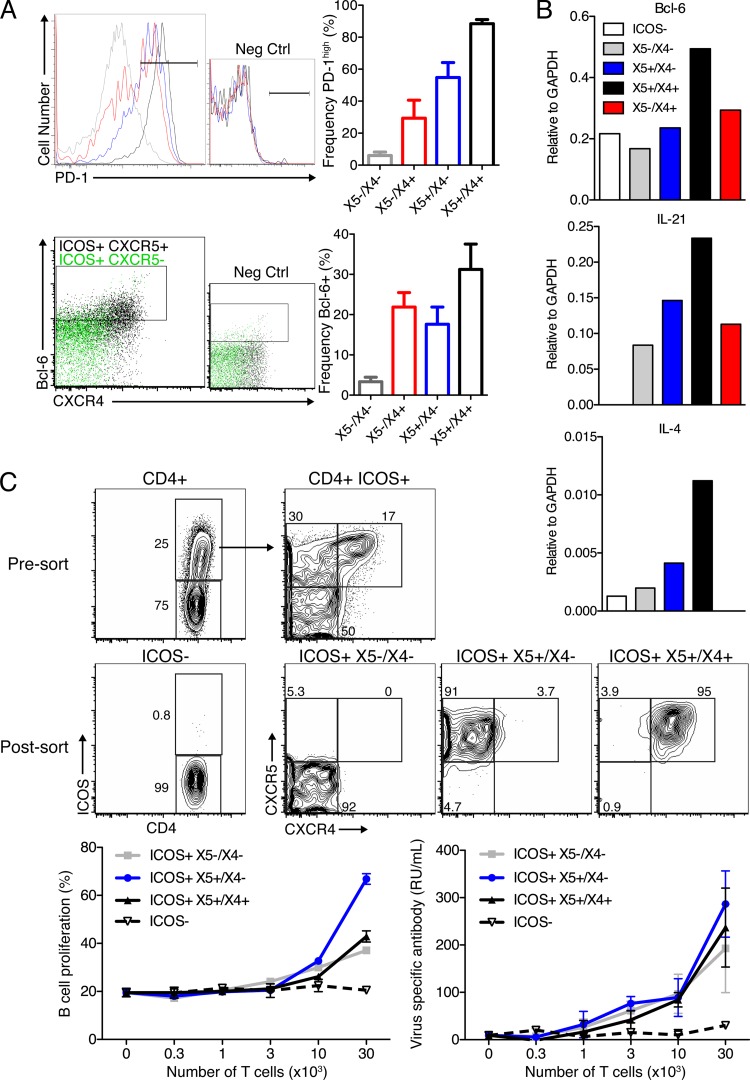
Having identified markers to distinguish TGC from TFH, we next tested their phenotypic and functional attributes. Previous studies suggested that high levels of PD-1 are associated with germinal center T cells (18). Strikingly, the ICOS+ X5+/X4+ CD4+ T cells were nearly uniformly PD-1high (Fig. 7A). Notably, PD-1high was not an exclusive phenotype of TGC, as both X5+/X4− and X5−/X4+ but not X5−/X4− ICOS+ T cells contained PD-1high cells (Fig. 7A), further demonstrating that PD-1hi expression does not fully discriminate between TFH and TGC. It is conceivable, however, that PD-1hi TFH are TGC that have recently left the germinal centers without having decreased PD-1 expression levels (11).
Fig 7.
Chemokine receptor expression is not associated with large differences in B helper function. (A, top) Representative overlay histogram plots of PD-1 expression among ICOS+ CD4+ T cells (gray line, X5−/X4−; red line, X5−/X4+; blue line, X5+/X4−; black line, X5+/X4+). (Bottom) Colored dot plots for the expression of Bcl-6 and CXCR4 among ICOS+ CXCR5+ (black) and ICOS+ CXCR5− (green) cells. CXCR4+ cells (i.e., largely ICOS+ X5+/X4+) are enriched for Bcl-6 expression in both frequency and intensity. Bar graphs indicate mean frequencies ± SD of expression of the indicated surface marker within the given ICOS+ population. Data were pooled from two experiments analyzing individual mice (n = 4 each). (B) Gene expression analysis of FACS-purified MedLN CD4 T cell subsets isolated from influenza virus A/Mem71-infected mice at day 10 (n = 4). Shown are transcript levels relative to GAPDH expression levels. Data are representative of two replicate experiments (one with BALB/c mice and one with C57BL/6 mice). (C, top) Shown are 5% contour plots of CD4 T cells pre- and post-FACS purification. T cells were cocultured with MACS-enriched CFSE-labeled B cells from lymph nodes of influenza virus-immunized mice, as outlined in the legend of Fig. 1. Cells were cultured in triplicate, and mean frequencies ± SD of proliferating CFSElo B cells (left) and relative antibody concentrations in supernatants (right), as assessed by ELISA, are displayed. RU, relative units. Results are from one of two experiments that yielded comparable results.
Bcl-6 is a transcriptional repressor that is highly expressed by germinal center B cells and is thought to be required for TFH cell differentiation (20, 31, 48). Bcl-6 expression frequencies were highest among TGC (about 35%) (Fig. 7A), and TGC expressed the highest levels of Bcl-6 mRNA (Fig. 7B). The reasons for the apparent lack of uniform Bcl-6 expression by TGC are unclear. Also, even in those CD4 T cells that expressed Bcl-6, the expression levels were well below those of the uniformly Bcl-6+ germinal center B cells (not shown). As expected, ICOS− CD4+ T cells lacked Bcl-6 expression (not shown), while a smaller fraction of ICOS+ X5−/X4− (Fig. 7A) cells was Bcl-6 positive.
TGC are thought to secrete IL-21, which acts directly on B cells to control germinal center B cell fate and Bcl-6 expression (51). Furthermore, others previously reported the selective expression of IL-4 by TGC (37, 50). Cytokine production by the T helper subsets was assessed at the mRNA expression level following their FACS separation from MedLN 10 days after influenza virus infection. X5+/X4+ TGC expressed the highest levels of IL-21 and IL-4 mRNAs (Fig. 7B). However, IL-4 expression levels overall were very low, consistent with a predominant switch of B cells to IgG2a/c but not IgG1 after influenza virus infection (36). Together, the data show X5+/X4+-expressing TGC to be most closely associated with the functions and phenotypes thought to characterize their functions. However, no characteristic was exclusive to TGC.
Finally, we determined whether the chemokine receptor expression profile of CD4+ ICOS+ T cells was associated with specific helper activities. For this, we cocultured MACS-purified, CFSE-labeled, antigen-pulsed B cells with FACS-purified ICOS+ CD4+ T cell subsets isolated from infected MedLN at day 8, as shown in Fig. 1. Due to their low frequencies, the ICOS+ X5−/X4+ cells were tested only in single instead of triplicate cultures, with 1 × 104 cells or fewer (not shown). Consistent with the data shown in Fig. 1, ICOS expression identified cells capable of in vitro T cell help (Fig. 7C). Among the ICOS+ T cells, the levels of induction of virus-specific antibody secretion were similar (Fig. 7C) (data not shown for X5−/X4+). However, X5+/X4− helpers consistently showed the greatest ability to induce B cell proliferation. When the antigen was omitted, few B cells survived and proliferated over the 4-day culture period (not shown), indicating that helper activity was antigen dependent. The higher ability of X5+/X4− helpers than that of X5+/X4+ TGC was not explained by the differing levels of FoxP3+ regulatory cells, as the frequencies were similar 10 days after infection (3.4 and 3.0%, respectively) (data not shown). The data demonstrate that ICOS expression identifies cells with B cell helper functions in vitro. Overall, remarkably little functional heterogeneity exists among ICOS+ cells; however, X5+/X4− helpers consistently induced more B cell proliferation in vitro.
Collectively, the study demonstrates that influenza virus infection induces ICOS+ CD4+ T helper cells with differing chemokine receptor expression profiles, which determines their location within (X5+/X4+) and outside (X5+/X4−) germinal centers and primary follicles (X5−/X4−) and between light/dark zones of germinal centers (CXCR4lo/high). While ICOS but not CXCR5 expression was correlated with B cell help in vitro, some functional differences among the defined T cell subsets were apparent. However, while TGC were associated with the highest levels of expression of Bcl-6, IL-21, IL-4, and PD-1, none were exclusive attributes of TGC. The extent to which T cell-intrinsic and -extrinsic factors regulate the outcome of T cell-B cell interactions can now be delineated, based on this study.
DISCUSSION
CD4+ ICOS+ CXCR5+ “follicular” T helper cells are thought to control intrafollicular T-dependent B cell responses during infection and immunization, with ICOS expression providing the necessary costimulatory signals and CXCR5 directing the follicular localization. However, while CXCR5 expression is required for CD4 T cell migration into primary B cell follicles, CXCR5 expression is neither necessary nor sufficient for the germinal center localization of TFH (18, 34). Moreover, mice deficient in SAP still generate ICOS+ CXCR5+ T cells in response to influenza virus infection (12, 22) (Fig. 6), even though they are unable to maintain germinal centers due to a CD4 T cell-intrinsic migration defect, suggesting the presence of additional signals that regulate the positioning of CD4 T cells to and within germinal centers. Here we demonstrate that the coexpression of CXCR4 and CXCR5 among CD4+ ICOS+ T cells identifies germinal center T helper cells and that CXCR4 expression levels were correlated with their sublocalization to light and dark zones. This TGC subset appears to be necessary for germinal center maintenance, as we show that X4+/X5+ TFH are the only subset missing in germinal center-deficient, influenza virus-infected SAP−/− mice. Importantly, these mice were previously shown to have a T cell-intrinsic defect in the ability to form a protective memory response to influenza virus infection (21). While the expression of ICOS was indispensable for CD4 T cell helper activity in vitro, chemokine receptor expression, including CXCR5, defined their location but not their in vitro helper activity.
Influenza virus infection induced robust CXCR5+ CXCR4+ germinal center-resident TGC responses in wild-type but not SAP−/− mice (Fig. 5 to 7). Remarkably, while their phenotypes and localizations were distinct, TGC provided similar in vitro help to induce the proliferation and differentiation of B cells as the other ICOS+ T cell subsets, except for a slightly better ability of X5+/X4− TFH to induce B cell proliferation (Fig. 7). This finding is in agreement with a previously reported human study showing that the expression of CD57, a marker associated with human TGC, did not correlate with B cell helper function (35). While our studies show that mouse TGC expressed Bcl-6 and PD-1 to a greater extent than the other TFH cells and expressed higher levels of IL-4 and IL-21 (Fig. 7), none of those attributes were specific for TGC. Such largely quantitative differences might play more important roles in the provision of B cell help in vivo. Alternatively, they could point to currently unknown qualitative differences between TGC and other helper cells. Another attractive possibility for future study is that their location within distinct lymph node niches and with B cells that also differ by location is a primary determinant of their helper quality. The developed marker set can now be employed to further study potential functional differences between TFH subsets within the various compartments of the lymph node.
Germinal centers require CXCR5 and CXCR4 expression by B cells for their proper location within the B cell follicle and for the organization of light and dark zones, respectively (1). Thus, it is not surprising that TGC express the same chemokine receptors. Compared to germinal center B cells, however, the level of Bcl-6 protein expression by TGC was low and only roughly one-third of the measurable levels of TGC-expressed Bcl-6 protein 10 days after influenza virus infection. It is possible that TGC only transiently express Bcl-6 following their activation or at levels below the detection limit of our FACS-based assay. Alternatively, since Bcl-6 regulates both CXCR4 and CXCR5 expressions, alterations in Bcl-6 expression could facilitate the movement of CD4 T cells within germinal center dark and light zones and/or between germinal centers and primary follicles (11).
The migration of TFH into germinal centers seems to require extensive T cell-B cell interactions facilitated by the cell adhesion SLAM family members CD84 and Ly108 and the SLAM adaptor protein (SAP) (8, 34). Consistent with the lack of robust germinal center formation in SAP−/− mice, TFH from SAP−/− mice have strongly reduced frequencies of CXCR4+ TGC (Fig. 6). As discussed above, the data shown here (Fig. 6) and by others (1) indicate that strong CXCR4 expression might be a consequence of, rather than a mechanism for, the location of TGC within germinal centers. Further proof of this concept will require the inducible depletion of CXCR4 expression upon CD4 T cell activation, as CXCR4 is required for the proper maturation of T cells within the thymus (19). An induction or strong enhancement of the expression of CXCR4 by CD4 T cells after their entry into germinal centers would also be consistent with data from previous studies demonstrating that human FDC modulate CXCR4 expression on T cells in vitro (14) and our studies showing low but detectable levels of CXCR4 in the few TGC present in SAP−/− mice by immunofluorescent microscopy (Fig. 6). The finding that the brightest CXCR4-staining TGC cells were present in the dark zones, while FDC are more abundant in the light zone (1) (Fig. 7, and data not shown), however, may indicate additional mechanisms that mediate CXCR4 induction within germinal centers. From an experimental standpoint, the differences in CXCR4 expression levels of light and dark zone TGC identified here for the first time enable studies of potential functional differences between TGC in these compartments.
The expression of CXCR4 on CXCR5− ICOS+ CD4 T cells has been linked to a population of CD4 T cells providing help for extrafollicular plasmablast responses in autoimmune-prone strains of mice (see Fig. S1 in the supplemental material) (32), consistent with the role of CXCR4 in the extrafollicular accumulation of plasmablasts in medullary cord areas of the secondary lymphoid tissues (16). Somewhat surprisingly, our studies showed only a very small and poorly demarcated population of such T cells in lymph nodes of influenza virus-infected mice, although robust extrafollicular responses were induced (Fig. 3). Such differences between infection- and autoimmune-induced T helper subsets may indicate that the continued presence of high frequencies of X4+/X5− T helper cells maintains the long-term extrafollicular foci in autoimmune disorders, while the short-lived (38) extrafollicular responses induced following influenza virus infection might not require their continued presence, consistent with previous work by others (28, 43). This conclusion is also consistent with our findings that plasmablast peak responses coincided with the peak of the X4+/X5− CD4 T cell numbers in lymph nodes of influenza virus-infected mice (Fig. 3) and that small clusters of CXCR4+ T cells were present in extrafollicular foci (data not shown).
Collectively, this study indicates that the orchestration of T cell-B cell movement and interactions within secondary lymphoid tissues could fine-tune T cell help by signals acquired at least in part within the local niche. The identification of CXCR4 and CXCR5 as markers to distinguish TGC from non-germinal-center-resident TFH can now be used to examine this and other hypotheses to define the mechanisms underlying the distinct outcomes of intra- and extrafollicular T cell-B cell interactions. Such studies may aid in the development of improved vaccines that can achieve both short- and long-term immunity to influenza virus infection.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grant T32-AI-060555 (R.A.E.) and NIAID grants AI073911 and AI051354 (N.B.).
We thank Gabe McKenzie for initial studies; Abigail Spinner (California National Primate Research Center [CNPRC]) for expert help with flow cytometry; Frank Ventimiglia (CNPRC), Bob Munn (University of California, Davis), and Jason Cyster and his laboratory member Lisa Kelly (University of California, San Francisco) for help with immunofluorescent microscopy; Pamela Schwartzberg (NIH) and Shane Crotty (La Jolla Institute for Allergy and Immunology) for providing SAP−/− mice; and Adam Treister (Treestar Inc.) for FlowJo software.
R.A.E. and N.B. declare no conflict of interest. D.N.E. is an employee of BD Biosciences.
Footnotes
Published ahead of print 24 April 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Allen CD, et al. 2004. Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nat. Immunol. 5:943–952 [DOI] [PubMed] [Google Scholar]
- 2. Allen CD, Okada T, Cyster JG. 2007. Germinal-center organization and cellular dynamics. Immunity 27:190–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ansel KM, McHeyzer-Williams LJ, Ngo VN, McHeyzer-Williams MG, Cyster JG. 1999. In vivo-activated CD4 T cells upregulate CXC chemokine receptor 5 and reprogram their response to lymphoid chemokines. J. Exp. Med. 190:1123–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arbones ML, et al. 1994. Lymphocyte homing and leukocyte rolling and migration are impaired in L-selectin-deficient mice. Immunity 1:247–260 [DOI] [PubMed] [Google Scholar]
- 5. Baumgarth N, Brown L, Jackson D, Kelso A. 1994. Novel features of the respiratory tract T-cell response to influenza virus infection: lung T cells increase expression of gamma interferon mRNA in vivo and maintain high levels of mRNA expression for interleukin-5 (IL-5) and IL-10. J. Virol. 68:7575–7581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Breitfeld D, et al. 2000. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J. Exp. Med. 192:1545–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bryant VL, et al. 2007. Cytokine-mediated regulation of human B cell differentiation into Ig-secreting cells: predominant role of IL-21 produced by CXCR5+ T follicular helper cells. J. Immunol. 179:8180–8190 [DOI] [PubMed] [Google Scholar]
- 8. Cannons JL, et al. 2010. Optimal germinal center responses require a multistage T cell:B cell adhesion process involving integrins, SLAM-associated protein, and CD84. Immunity 32:253–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chtanova T, et al. 2004. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J. Immunol. 173:68–78 [DOI] [PubMed] [Google Scholar]
- 10. Coffey AJ, et al. 1998. Host response to EBV infection in X-linked lymphoproliferative disease results from mutations in an SH2-domain encoding gene. Nat. Genet. 20:129–135 [DOI] [PubMed] [Google Scholar]
- 11. Crotty S. 2011. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29:621–663 [DOI] [PubMed] [Google Scholar]
- 12. Crotty S, Kersh EN, Cannons J, Schwartzberg PL, Ahmed R. 2003. SAP is required for generating long-term humoral immunity. Nature 421:282–287 [DOI] [PubMed] [Google Scholar]
- 13. Doucett VP, et al. 2005. Enumeration and characterization of virus-specific B cells by multicolor flow cytometry. J. Immunol. Methods 303:40–52 [DOI] [PubMed] [Google Scholar]
- 14. Estes JD, et al. 2002. Follicular dendritic cell-mediated up-regulation of CXCR4 expression on CD4 T cells and HIV pathogenesis. J. Immunol. 169:2313–2322 [DOI] [PubMed] [Google Scholar]
- 15. Harakawa N, et al. 2007. P-selectin glycoprotein ligand-1 mediates L-selectin-independent leukocyte rolling in high endothelial venules of peripheral lymph nodes. Int. Immunol. 19:321–329 [DOI] [PubMed] [Google Scholar]
- 16. Hargreaves DC, et al. 2001. A coordinated change in chemokine responsiveness guides plasma cell movements. J. Exp. Med. 194:45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hataye J, Moon JJ, Khoruts A, Reilly C, Jenkins MK. 2006. Naive and memory CD4+ T cell survival controlled by clonal abundance. Science 312:114–116 [DOI] [PubMed] [Google Scholar]
- 18. Haynes NM, et al. 2007. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J. Immunol. 179:5099–5108 [DOI] [PubMed] [Google Scholar]
- 19. Janas ML, Turner M. 2010. Stromal cell-derived factor 1alpha and CXCR4: newly defined requirements for efficient thymic beta-selection. Trends Immunol. 31:370–376 [DOI] [PubMed] [Google Scholar]
- 20. Johnston RJ, et al. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 325:1006–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kamperschroer C, Dibble JP, Meents DL, Schwartzberg PL, Swain SL. 2006. SAP is required for Th cell function and for immunity to influenza. J. Immunol. 177:5317–5327 [DOI] [PubMed] [Google Scholar]
- 22. Kamperschroer C, Roberts DM, Zhang Y, Weng NP, Swain SL. 2008. SAP enables T cells to help B cells by a mechanism distinct from Th cell programming or CD40 ligand regulation. J. Immunol. 181:3994–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kerfoot SM, et al. 2011. Germinal center B cell and T follicular helper cell development initiates in the interfollicular zone. Immunity 34:947–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim CH, et al. 2001. Subspecialization of CXCR5+ T cells: B helper activity is focused in a germinal center-localized subset of CXCR5+ T cells. J. Exp. Med. 193:1373–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim TS, Sun J, Braciale TJ. 2011. T cell responses during influenza infection: getting and keeping control. Trends Immunol. 32:225–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kirberg J, et al. 1994. Thymic selection of CD8+ single positive cells with a class II major histocompatibility complex-restricted receptor. J. Exp. Med. 180:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Linterman MA, et al. 2009. Follicular helper T cells are required for systemic autoimmunity. J. Exp. Med. 206:561–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. MacLennan IC, et al. 2003. Extrafollicular antibody responses. Immunol. Rev. 194:8–18 [DOI] [PubMed] [Google Scholar]
- 29. Nichols KE, et al. 1998. Inactivating mutations in an SH2 domain-encoding gene in X-linked lymphoproliferative syndrome. Proc. Natl. Acad. Sci. U. S. A. 95:13765–13770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nurieva RI, et al. 2008. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity 29:138–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nurieva RI, et al. 2009. Bcl6 mediates the development of T follicular helper cells. Science 325:1001–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Odegard JM, et al. 2008. ICOS-dependent extrafollicular helper T cells elicit IgG production via IL-21 in systemic autoimmunity. J. Exp. Med. 205:2873–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Poholek AC, et al. 2010. In vivo regulation of Bcl6 and T follicular helper cell development. J. Immunol. 185:313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qi H, Cannons JL, Klauschen F, Schwartzberg PL, Germain RN. 2008. SAP-controlled T-B cell interactions underlie germinal centre formation. Nature 455:764–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rasheed AU, Rahn HP, Sallusto F, Lipp M, Muller G. 2006. Follicular B helper T cell activity is confined to CXCR5(hi)ICOS(hi) CD4 T cells and is independent of CD57 expression. Eur. J. Immunol. 36:1892–1903 [DOI] [PubMed] [Google Scholar]
- 36. Rau FC, Dieter J, Luo Z, Priest SO, Baumgarth N. 2009. B7-1/2 (CD80/CD86) direct signaling to B cells enhances IgG secretion. J. Immunol. 183:7661–7671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reinhardt RL, Liang HE, Locksley RM. 2009. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat. Immunol. 10:385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rothaeusler K, Baumgarth N. 2010. B-cell fate decisions following influenza virus infection. Eur. J. Immunol. 40:366–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rothaeusler K, Baumgarth N. 2006. Evaluation of intranuclear BrdU detection procedures for use in multicolor flow cytometry. Cytometry A 69:249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sayos J, et al. 1998. The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM. Nature 395:462–469 [DOI] [PubMed] [Google Scholar]
- 41. Schaerli P, et al. 2000. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. J. Exp. Med. 192:1553–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stevenson PG, Doherty PC. 1999. Non-antigen-specific B-cell activation following murine gammaherpesvirus infection is CD4 independent in vitro but CD4 dependent in vivo. J. Virol. 73:1075–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Toellner KM, Gulbranson-Judge A, Taylor DR, Sze DM, MacLennan IC. 1996. Immunoglobulin switch transcript production in vivo related to the site and time of antigen-specific B cell activation. J. Exp. Med. 183:2303–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Veillette A, et al. 2008. SAP expression in T cells, not in B cells, is required for humoral immunity. Proc. Natl. Acad. Sci. U. S. A. 105:1273–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vinuesa CG, Linterman MA, Goodnow CC, Randall KL. 2010. T cells and follicular dendritic cells in germinal center B-cell formation and selection. Immunol. Rev. 237:72–89 [DOI] [PubMed] [Google Scholar]
- 46. Watanabe M, Hara Y, Tanabe K, Toma H, Abe R. 2005. A distinct role for ICOS-mediated co-stimulatory signaling in CD4+ and CD8+ T cell subsets. Int. Immunol. 17:269–278 [DOI] [PubMed] [Google Scholar]
- 47. Yoshinaga SK, et al. 1999. T-cell co-stimulation through B7RP-1 and ICOS. Nature 402:827–832 [DOI] [PubMed] [Google Scholar]
- 48. Yu D, et al. 2009. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity 31:457–468 [DOI] [PubMed] [Google Scholar]
- 49. Yu D, Vinuesa CG. 2010. The elusive identity of T follicular helper cells. Trends Immunol. 31:377–383 [DOI] [PubMed] [Google Scholar]
- 50. Yusuf I, et al. 2010. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150). J. Immunol. 185:190–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zotos D, et al. 2010. IL-21 regulates germinal center B cell differentiation and proliferation through a B cell-intrinsic mechanism. J. Exp. Med. 207:365–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.