Abstract
While pandemic 2009 H1N1 influenza A viruses were responsible for numerous severe infections in humans, these viruses do not typically cause corresponding severe disease in mammalian models. However, the generation of a virulent 2009 H1N1 virus following serial lung passage in mice has allowed for the modeling of human lung pathology in this species. Genetic determinants of mouse-adapted 2009 H1N1 viral pathogenicity have been identified, but the molecular and signaling characteristics of the host response following infection with this adapted virus have not been described. Here we compared the gene expression response following infection of mice with A/CA/04/2009 (CA/04) or the virulent mouse-adapted strain (MA-CA/04). Microarray analysis revealed that increased pathogenicity of MA-CA/04 was associated with the following: (i) an early and sustained inflammatory and interferon response that could be driven in part by interferon regulatory factors (IRFs) and increased NF-κB activation, as well as inhibition of the negative regulator TRIM24, (ii) early and persistent infiltration of immune cells, including inflammatory macrophages, and (iii) the absence of activation of lipid metabolism later in infection, which may be mediated by inhibition of nuclear receptors, including PPARG and HNF1A and -4A, with proinflammatory consequences. Further investigation of these signatures in the host response to other H1N1 viruses of various pathogenicities confirmed their general relevance for virulence of influenza virus and suggested that lung response to MA-CA/04 virus was similar to that following infection with lethal H1N1 r1918 influenza virus. This study links differential activation of IRFs, nuclear receptors, and macrophage infiltration with influenza virulence in vivo.
INTRODUCTION
The 2009 H1N1 influenza virus was responsible for the first pandemic of the 21st century and caused more than 18,000 deaths worldwide between April 2009 and August 2010 (50). People infected during the pandemic experienced mostly mild symptoms, but there were reports of severe primary viral pneumonia in young and healthy people, which is not typically seen during seasonal influenza epidemics. The 2009 H1N1 virus is highly transmissible among humans and is now circulating as a seasonal influenza virus strain (51), and thus there is a threat of emergence of more virulent strains in the future resulting from continuous mutation of the virus. Therefore, it is crucial to improve our understanding of 2009 H1N1 pathogenicity.
The mouse model has been used to study the pathogenic potential of 2009 H1N1 virus and characterize viral factors critical for virulence. Most 2009 H1N1 isolates replicate efficiently in the lungs of BALB/c mice but are almost avirulent for this animal (50% mouse lethal dose [MLD50] ≥ 106 PFU [18, 5, 30] for A/California/04/09 and A/Mexico/4482/09). However, they can gain virulence and become lethal at a lower inoculum after serial lung passages in mice (16, 34, 39, 53, 56). Virulence determinants of mouse-adapted H1N1 virus have been identified in the ribonucleoprotein (RNP) complex (16, 34, 39, 53, 56) and hemagglutinin (HA) (16, 34, 39, 53). Several mutations have been reported, with independent and synergistic effects on virulence (39, 53). Mutations in HA have been associated with a change in receptor binding specificity (16, 39, 53), and mutations in PA, PB1, PB2, and NP have been associated with enhanced polymerase activity (16, 39, 53, 56). These studies have characterized the mouse-adapted virus pathogenic determinants; however, the corresponding effects on host response following infection have not been described.
To gain a better understanding of virulence and pathogenic potential following adaptation of 2009 H1N1 virus, we performed a time course study in BALB/c mice to compare the prototypic 2009 H1N1 strain A/California/04/09 (CA/04) and a corresponding mouse-adapted variant, MA1-A/California/04/09 (MA-CA/04) (16). MA-CA/04 contains five genetic mutations (in HA, G155E, S183P, and D222G; in PB2, E158G; and in NP, D101G) and is associated with a dramatic increase in virulence in mice (16). Using global gene expression profiling in tandem with bioinformatics methods, we characterized the differences in the host response to both viruses and found that the increased pathogenicity of MA-CA/04 was associated with an early and sustained inflammatory response, infiltration of inflammatory macrophages, and perturbation of lipid metabolism. Our data suggest several transcription factors (TFs) could mediate these responses. To put our findings into a larger context of influenza virulence, we compared these signatures to gene expression in the lungs of mice infected with either lethal r1918 or other nonlethal H1N1 influenza A viruses. Similar expression pattern of these biological signatures among viruses with similar pathogenicities suggests their importance in differential responses to lethal and nonlethal influenza viruses.
MATERIALS AND METHODS
Viruses.
Viral stocks for A/California/04/2009, mouse-adapted MA1-A/California/04/2009 (generously provided by Richard Webby, St. Jude Children's Hospital, Memphis, TN [16]), A/Mexico/4482/09 (Mex/4482), and A/Brisbane/59/07 (Brisbane/59/07) were propagated in Madin-Darby canine kidney (MDCK) cells for 48 h at 37°C. A/New Jersey/8/76 (NJ/8/76) was propagated in the allantoic cavity of 10-day-old embryonated hens' eggs for stock preparation. The supernatants were clarified by centrifugation, aliquoted, and stored at −70°C. Virus titers were determined by standard plaque assay on MDCK cells (55). The identities of virus genes were confirmed by sequence analysis to verify that no inadvertent mutations were present during the generation of virus stocks.
Mouse experiments.
Six- to eight-week-old female BALB/c mice were purchased from Charles River Laboratories (Wilmington, MA). All 2009 H1N1 in vivo experiments were performed under biosafety level 3 enhanced (BSL3+) containment. Mice were anesthetized with 2,2,2-tribromoethanol in tert-amyl alcohol (Avertin; Sigma-Aldrich, St. Louis, MO) and intranasally (i.n.) inoculated with 50 μl of phosphate-buffered saline (PBS; mock) or with 106 PFU of virus in a 50-μl volume. Six animals in each group were monitored for morbidity and mortality as measured by weight loss and survival over a period of 16 days. Viral titers were determined in the lungs, spleen, and brain on days 1, 3, and 5 postinoculation in nine animals per group (three animals for each time point). An additional nine animals were used for transcriptional profiling, again three animals per time point. Histopathology was performed on another batch of nine mice for each virus, with three animals used per time point. Animal research was conducted under the guidance of the CDC's Institutional Animal Care and Use Committee in an animal facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International.
Histopathology and immunohistochemistry.
Lung sections were fixed by submersion in 10% neutral buffered formalin, routinely processed, and embedded in paraffin. Histopathological lesions were examined after staining with hematoxylin and eosin (HE) following standard protocols and scored as follows: 1, no lesion; 2, mild lesion; 3, moderate lesion; 4, severe lesion. Immunohistochemical viral antigen detection was performed using a mouse-derived monoclonal antibody (P13C11) specific for a type A influenza virus nucleoprotein as previously described (4), and the following scores were used: 0, no antigen; 1, rare positive cells; 2, infrequent positive cells; 3, common positive cells; and 4, widespread positive cells.
Microarray experiments.
Transcriptional profiles were determined by microarray analysis of RNA isolated from lung tissue from three individual animals per group at days 1, 3, and 5 postinfection (p.i.). Total RNA extraction from lung samples was performed as previously described (4). Probe labeling and microarray slide hybridization for each biological replicate was performed using a whole mouse genome microarray kit (G4122-60520; Agilent Technologies) according to the manufacturer's instructions.
Data analysis.
Extracted raw data were background corrected using the “norm-exp” method with an offset of 1 and quantile normalized using Agi4x44PreProcess (28) in the R software environment. Replicate probes were mean summarized, and all probes were required to pass Agilent QC flags for 75% replicates of at least one infected time point (30,827 probes passed).
Mean expression values were calculated by determining the arithmetic mean for the individual replicates. From this, ratios of values for the infected samples to the mean values of time-matched mock samples were calculated and logarithm base 2 transformed. These values are referred to as “log2 ratios.”
A Welch t test on log2-scaled intensity values was performed to determine the probes that were differentially expressed (DE) between each virus and time-matched mock and between two different viruses at each time point. Criteria for DE were an absolute log2 ratio of >1 and a Benjamini-Hochberg adjusted q value of <0.05.
Genes discriminating CA/04 and MA-CA/04 were grouped in three sets based on their expression levels. Set 1 (upregulated lethal signature) was defined as genes whose expression conformed, for at least 1 day p.i., to the following rules: log2 ratio(CA/04) > −1 and log2 ratio(MA-CA/04) > 0 and log2 ratio(MA-CA/04) > log2 ratio(CA/04); set 2 (survival/recovery signature) was defined as genes with log2 ratio(CA/04) >1 and log2 ratio(CA/04) > log2 ratio(MA-CA/04) for at least 1 day p.i.; and set 3 (downregulated lethal signature) was defined as genes with log2 ratio(CA/04) < 1 and log2 ratio(MA-CA/04) < −1 and log2 ratio(MA-CA/04) < log2 ratio(CA/04) for at least 1 day p.i.
Functional analysis of statistically significant gene expression changes was performed using the Ingenuity Pathways Knowledge Base (IPA; Ingenuity Systems) as previously described (4). All enrichment scores were calculated in IPA using all probes that passed our QC filter as the background data set.
TF analysis.
We used two different methods to identify potential transcriptional regulators that could control the gene expression response to infection. Transcription factor (TF) binding motif enrichment was performed with the PSCAN software program (54) using the JASPAR database and promoter definition from −450 to +50 nucleotides (relative to the transcription start site). PSCAN computes a z-test P value for each TF, which assesses whether its binding motif is significantly over- or underrepresented in promoters of the query genes. We also used a second complementary method, based on prior knowledge of expected effects between transcriptional regulators and their known target genes according to the IPA database, to predict regulators and infer their activation state. The IPA TF analytic tool calculates a z score that determines whether gene expression changes for known targets of each TF are consistent with what is expected from the literature (z > 0, TF predicted to be “activated”) or if the changes are anticorrelated with the literature (z < 0, TF predicted to be “inhibited”). z scores greater than 2 or smaller than −2 are considered significant. Using both JASPAR and IPA complementary approaches in our study, we may reduce the likelihood of missing significant enrichment of TFs that might be absent in one database or the other.
Meta-analysis of immune cells.
A meta-analysis was performed using publicly available microarray data from Mouse GeneAtlas v3 of major types of immune cells and lung tissue in mice. Raw CEL files were downloaded from the NCBI GEO website (GSE10246) (24, 42), preprocessed in the R program using the affy package, and normalized using the robust multiarray analysis (RMA) method (17). For comparison with our data set, Unigene ID was chosen as the common gene identifier. A gene was defined as highly expressed in a cell subset if its expression was 20-fold greater in this subset than in the lung samples (from Mouse GeneAtlas v3). Fisher's exact test was used to determine whether the proportion of highly expressed immune genes in each viral signature was significantly greater than the proportion of immune genes in the background (30,827 probes passing quality control [QC] assessment). Enrichment scores were calculated as −log10(P value).
Meta-analysis of other H1N1 influenza A viruses.
Data for meta-analysis of the host transcriptional response to other H1N1 viruses was obtained from mouse infection experiments using Mex/4482, Brisbane/59/07, and NJ/8/76, performed as described above. We also used microarray data derived from stored mRNA isolated from lungs of BALB/c mice infected with r1918 virus in the study by Kash et al. (21). For comparison with CA/04 and MA-CA/04 expression profiles, all data were quantile normalized together and the COMBAT algorithm in R was used to correct batch effects (19).
To visualize transcriptional dissimilarity between samples in a 2-dimensional (2D) space, Euclidian distances were calculated for transcriptomic data expressed in intensity scale, and nonmetric multidimensional scaling (MDS) was performed with the MASS package in the Bioconductor software program (48) to map the distances into the 2-dimensional space with minimal loss of information (evaluated by Kruskal's stress).
Microarray data accession number.
Raw microarray data have been deposited in NCBI's Gene Expression Omnibus under GEO Series accession number GSE36328 and are also accessible through the Systems Virology website (http://www.systemsvirology.org).
RESULTS
Mouse adaptation results in increased virulence and lung pathology.
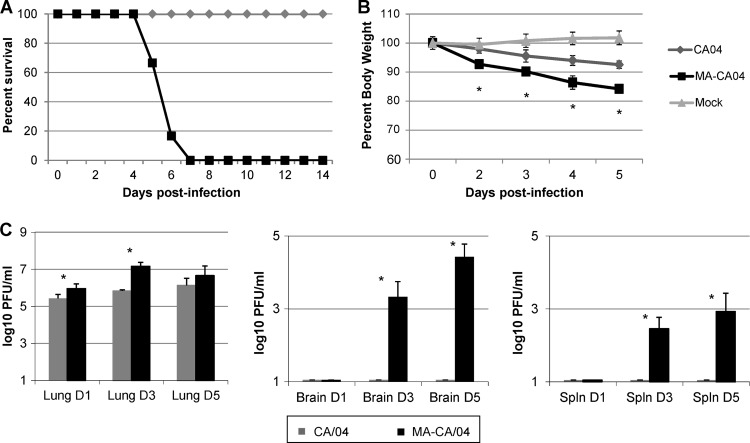
BALB/c mice were infected with 106 PFU of either CA/04 or MA-CA/04 virus; weight loss, survival, and viral titers were determined at different days p.i. While mice infected with CA/04 lost minimal weight and recovered by day 8 p.i., infection with MA-CA/04 resulted in continuous weight loss and 100% lethality by day 7 p.i. (Fig. 1A and B). This increase in virulence was accompanied by increased replication of MA-CA/04 virus in lungs, as quantified by plaque assay (Fig. 1C). Since extrapulmonary replication was previously reported for MA-CA/04 (16) but not for other 2009 H1N1 mouse-adapted viruses, we verified that under our experimental conditions, MA-CA/04 disseminated to the brain and spleen on days 3 and 5 p.i. but CA/04 did not.
Fig 1.
Characterization of mortality, weight loss, and viral titers in MA-CA/04- and CA/04-infected mice. Mice were inoculated with 106 PFU of each virus or PBS as a mock control. (A) Mortality data from mice inoculated with influenza virus or mock inoculated (6 mice/group). (B) Average weight of surviving animals at each time point (expressed as mean ± standard deviation) from a total of 18 animals (6 mice/group). (C) Influenza virus titers measured in lungs, brain, and spleen at 1, 3, and 5 day p.i. (3 mice/virus/time point). Differences between MA-CA/04 and CA/04 for weight and titer were determined by a Student t test (P < 0.05). Statistical significance is denoted with an asterisk.
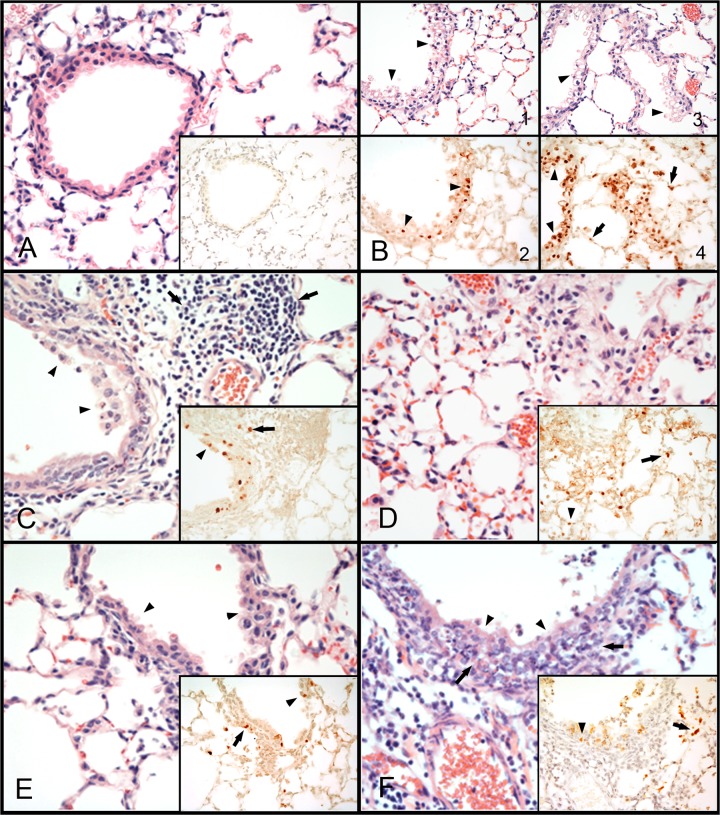
Microscopic lesions were examined by hematoxylin-and-eosin staining of lung tissue sections from infected mice, and viral antigen was detected by immunohistochemistry. Infection with either CA/04 or MA-CA/04 virus produced lesions typical of influenza A infections: bronchiolitis and bronchitis with accompanying necrosis of respiratory epithelium and associated histiocytic alveolitis (Fig. 2; see also Table S1 in the supplemental material). In general, the lesions produced by MA-CA/04 were more severe than the lesions observed with CA/04. At 1 day p.i., both viruses produced mild to moderate bronchiolitis and bronchitis with degenerative changes, including vacuolation, of the respiratory epithelium. Viral staining was commonly present in bronchioles and bronchial respiratory epithelium (Fig. 2B). The MA-CA/04 virus also produced mild histiocytic alveolitis at this time point, and infrequent viral staining was present in alveolar epithelium and macrophages. At 3 days p.i., mild to moderate bronchiolitis and bronchitis with necrosis and loss of the respiratory epithelium, accompanied by mild to moderate peribronchiolar histiocytic to lymphocytic alveolitis, was observed with both viruses (Fig. 2C and D). Alveolar necrosis with congestion and edema was more evident in mice infected with the MA-CA/04 virus. Infrequent viral staining in bronchioles and bronchial respiratory epithelium was seen with both viruses, but viral staining in alveolar epithelium and macrophages was more common in tissues from mice infected with MA-CA/04 (Fig. 2C and D, inserts). At 5 days p.i., mice infected with CA/04 had mild lymphocytic to histiocytic peribronchiolar to diffuse alveolitis, as well as bronchiolitis and bronchitis with necrosis and loss of the respiratory epithelium. In some cases neutrophils were also present (Fig. 2E). Rare to common viral staining was present in alveolar epithelium, in macrophages, and in bronchioles and bronchial respiratory epithelium. At this same time point, mice infected with MA-CA/04 had moderate to severe peribronchiolar to diffuse alveolitis, with histiocytes, lymphocytes and uncommon neutrophils present (Fig. 2F). Moderate to severe bronchiolitis and bronchitis with necrosis and loss of the respiratory epithelium were also observed. Viral staining was infrequent in bronchioles and bronchial respiratory epithelium but common in alveolar epithelium and macrophages.
Fig 2.
Histopathologic changes and immunostaining of tissues from H1N1 virus-inoculated mice. Photomicrographs of lung tissue sections stained with hematoxylin and eosin and for the presence of viral antigen (×400). (A) Lung tissue from mock-infected mouse; lack of viral antigen staining in lung (insert). (B to E) Lung tissues from mice infected with the H1N1 viruses. (B) Moderate bronchiolitis with vacuolation of the respiratory epithelium (arrowheads) in mice infected with CA/04 (B-1 and B-2) or MA-CA/04 (B-3 and B-4) at 1 day p.i.. Viral antigen staining (in brown) found in cells of the bronchiole respiratory epithelium (arrowheads) (B-2 and -4) and in adjacent alveolar epithelial cells (arrows) (B4). (C) Moderate bronchiolitis with necrosis and loss of the respiratory epithelium (arrowheads) and associated moderate peribronchiolar lymphocytic alveolitis (arrows) in mouse infected with CA/04 at 3 days p.i. Viral antigen (in brown) found in bronchiolar epithelial cells (arrowheads) and macrophages (arrows) (insert). (D) Moderate interstitial pneumonia in mouse infected with MA-CA/04 at 3 days p.i. Viral antigen (in brown) found in alveolar epithelium (arrowheads) and macrophages (arrows) (insert). (E) Mild bronchiolitis (arrowheads) with mild peribronchial alveolitis in mouse infected with CA/04 at 5 days p.i. Viral staining (in brown) in bronchiole respiratory epithelium (arrowheads) and alveolar epithelium and macrophages (arrows) (insert). (F) Severe bronchiolitis (arrowheads) and peribronchiolar alveolitis (arrows), mainly histiocytic but also with lymphocytes and some neutrophils, in mouse infected with MA-CA/04 at 5 days p.i. Viral staining (in brown) found in bronchiole respiratory epithelium (arrowheads), desquamated cells, and alveolar epithelium and macrophages (arrows) (insert).
In summary, increased virulence of MA-CA/04 was associated with more severe lung lesions, more evident infection of alveolar epithelium and macrophages, and indications of a greater number of histiocytes, lymphocytes, and neutrophils infiltrating the lung than with wild-type CA/04 virus.
MA-CA/04 induces a strong host transcriptional response after infection.
To characterize the impact of mouse adaptation on host responses, pulmonary gene expression was compared between time matched mock-infected mice and mice infected with either CA/04 or MA-CA/04 virus at 1, 3, and 5 days p.i. We observed that MA-CA/04 infection resulted in more differentially expressed (DE) genes at each time point than did infection with CA/04 (Fig. 3A). The number of DE genes was greatest on day 3 p.i. for both viruses; however, importantly, transcriptional responses to CA/04 infection decreased dramatically on day 5, whereas for MA-CA/04, the number of DE genes decreased only marginally. Functional analysis of upregulated genes showed that MA-CA/04 infection resulted in a sustained increase in the expression of genes associated with the inflammatory response, immune cell trafficking, cell death, and cellular growth and proliferation (see Fig. S1 in the supplemental material). After CA/04 infection, only 195 transcripts were significantly upregulated on day 1 p.i., and they were not strongly associated with any biological function (see Fig. S1). In contrast, on day 3 p.i., 1,374 genes were upregulated after CA/04 infection, and their functional enrichment profile was very similar to that of genes induced by MA-CA/04 at the same time. At day 5 p.i., lipid and amino acid metabolism processes were upregulated in CA/04-infected mice but not in mice infected with MA-CA/04. Genes downregulated following MA-CA/04 and CA/04 virus infection were enriched to a limited extent in different general biological functions, such as tissue development, genetic disorder, cellular function, and maintenance (see Fig. S1). On day 3 p.i., 60% of the functions associated with downregulated genes after CA/04 or MA-CA/04 infection were in common (see Fig. S1).
Fig 3.
Number of genes differentially expressed (DE) after infection for each virus compared to results for time-matched mocks (A) or overlap between genes differentially regulated between MA-CA/04 and CA/04 at each time point (B). (A) Numbers of upregulated (red) and downregulated (green) DE probes after MA-CA/04 and CA/04 infections on days 1, 3, and 5. DE genes were defined with a Welch t test between the time-matched mock and each infected group (q value < 0.05; absolute log2 ratio > 1). (B) Venn diagram representing relative overlap between genes differentially regulated between MA-CA/04 and CA/04 on day 1 p.i. (blue), day 3 p.i. (gray), and day 5 p.i. (red). The total number of genes differentiating MA-CA/04 and CA/04 infection is indicated for each time point along, with the percentage of those genes unique to the time point. These genes were defined with a Welch t test between MA-CA/04 and CA/04 for each time point (q value < 0.05; absolute log2 ratio [CA/04 to MA-CA/04] >1).
The enhanced virulence of MA-CA/04 is therefore characterized by an increased host response, with upregulated genes strongly associated with immune/inflammatory processes across all time points. In contrast, whereas CA/04 infection also induced a strong immune response, the response was limited at day 3, with metabolism functions being induced later in infection.
MA-CA/04 and CA/04 elicit different transcriptional programs.
To directly examine differences in the host response to CA/04 and MA-CA/04, we determined the genes that were DE between CA/04 and MA-CA/04 at each day p.i. (Welch t test, absolute log2 ratio > 1 and q value < 0.05). A total of 4,202 unique DE transcripts were identified, with an overlap between time points of 36%. At day 3, only 25% of the transcripts discriminating CA/04 and MA-CA/04 were not DE at the other time points, whereas at days 1 and 5 p.i., there were 41% and 53% unique genes, respectively (Fig. 3B).
Genes that were DE between MA-CA/04 and CA/04 were categorized into three sets based on their general expression patterns (Fig. 4; see also Data Set S1 in the supplemental material). Genes in set 1 (upregulated lethal signature; 2,107 genes) were more highly expressed during MA-CA/04 infection at at least one time point, and many of these genes remained highly expressed throughout infection. These genes were only transiently upregulated, and to a lesser extent, on day 3 p.i. in CA/04-infected animals. Genes in set 2 (survival/recovery signature; 968 genes) were upregulated in CA/04-infected animals at at least one time point while unchanged or downregulated during MA-CA/04 infection. The largest differential within this group was found on day 5 p.i. Finally, genes in set 3 (downregulated lethal signature; 1,126 genes) were more downregulated in MA-CA/04-infected mice at at least one time point and showed a trend opposite from that of genes in set 1. Many of the genes in set 3 were highly downregulated early after MA-CA/04 infection and remained so throughout infection, while they were only transiently downregulated, and to a lesser extent, on day 3 p.i. in CA/04-infected animals.
Fig 4.
Genes DE between MA-CA/04 and CA/04 infection formed 3 main transcriptional programs. The heat map represents expression levels (in log2 ratios) of 4,202 transcripts that were DE between MA-CA/04 and CA/04 infection at at least one time point. Genes were clustered based on their expression values across samples using Pearson correlation and average linkage function. Row colors on the left correspond to the Venn diagram of Fig. 3B and indicate on which day p.i. each gene was found DE between MA-CA/04 and CA/04 infection. Three sets of genes were determined based on gene expression levels: set 1 represents the upregulated lethal signature (2,107 probes), set 2 is the survival/recovery signature (968 probes), and set 3 is the downregulated lethal signature (1,126 probes). There was a minimal overlap between each set. Only 2 genes (Ear5 and Chi3l4) were found more downregulated for CA/04 than for MA-CA/04 infection at at least one time point, and set 4 is therefore not depicted.
Genes highly induced after MA-CA/04 infection are associated with the immune response and enriched with interferon regulatory factor (IRF) binding motifs.
Functional analysis was performed using IPA to determine the canonical pathways and biological functions (Table 1) that were most significantly associated with differences in gene expression between the two viruses (Fig. 4). Set 1 was strongly enriched in pathways related to innate immunity and relationships between innate and adaptive immunity (Table 1). Approximately 14% of the transcripts (291 probes) in set 1 encoded factors involved in immune cell trafficking (Table 1; see also Data Set S1 in the supplemental material), including a large number of CCL and CXCL cytokines and their receptors. We also noted an increase in the expression of genes coding for inflammatory molecules, such as the classical proinflammatory cytokines IL-1b, IL-6, and TNF-α, and molecules involved in tissue destruction and cell death, including matrix metallopeptidases, those encoded by FAS, TNF, and the TNF receptor, TNFRSF1B, and several receptors of Fc fragments. We also compared the list of genes in set 1 with the list of genes that were DE following intranasal treatment of mice with alpha/beta interferon (IFN-α/β) (see Data Set S2) (9). Among the 2,107 transcripts in set 1, 39% (825 probes) were thus defined as IFN-regulated genes (Fisher's exact P value < 2.2e−16), suggesting that a large part of the set 1 transcriptional program was driven by an upregulation of the innate immune response.
Table 1.
IPA canonical pathways and biological functions enriched in each transcriptional program discriminating MA-CA/04 and CA/04a
| Canonical pathway | Canonical Pathway q value | Category | Function annotation | Function q value |
|---|---|---|---|---|
| Set 1: upregulated genes associated with lethality | ||||
| Communication between innate and adaptive immune cells | 3.E−17 | Inflammatory response | Immune response | 2.E−80 |
| Role of pattern recognition receptors in recognition of bacteria and viruses | 5.E−13 | Cell-to-cell signaling and interaction | Activation of cells | 3.E−49 |
| Dendritic cell maturation | 3.E−12 | Immune cell trafficking | Leukocyte migration | 6.E−47 |
| Role of hypercytokinemia/hyperchemokinemia in the pathogenesis of influenza | 3.E−12 | Cell-to-cell signaling and interaction | Activation of blood cells | 8.E−46 |
| Cross talk between dendritic cells and natural killer cells | 4.E−11 | Inflammatory response | Inflammatory response | 4.E−45 |
| Set 2: transcriptional program associated with survival/recovery | ||||
| FXR/RXR activation | 2.E−28 | Lipid metabolism | Metabolism of terpenoid | 4.E−30 |
| Metabolism of xenobiotics by cytochrome P450 | 4.E−28 | Lipid metabolism | Steroid metabolism | 2.E−27 |
| LPS/IL-1-mediated inhibition of RXR function | 4.E−27 | Lipid metabolism | Fatty acid metabolism | 7.E−25 |
| LXR/RXR activation | 1.E−25 | Amino acid metabolism | Metabolism of amino acids | 1.E−21 |
| Fatty acid metabolism | 1.E−21 | Lipid metabolism | Transport of lipid | 7.E−21 |
| Set 3: downregulated genes associated with lethality | ||||
| LXR/RXR activation | 4.E−02 | Cardiovascular disease | Venous thrombosis | 4.E−03 |
| Metabolism of xenobiotics by cytochrome P450 | 4.E−02 | Cancer | Metastatic colorectal cancer | 7.E−03 |
| Glycolysis/gluconeogenesis | 4.E−02 | Lipid metabolism | Release of cholesterol | 8.E−03 |
| Lipid metabolism | Transport of lipid | 1.E−02 | ||
| Lipid metabolism | Oxidation of lipid | 1.E−02 |
The top-ranked significant pathways and functions are represented (cutoff for significance, q < 0.05).
To identify potential transcriptional regulators of the genes within set 1, we used PSCAN to analyze the promoter regions of these genes for binding motifs of 130 transcription factors (TFs) (Table 2; see also Data Set S3 in the supplemental material). We found a strong enrichment for IRF1 and -2 binding sites, as well as NF-κB and STAT3. IRF3 and NF-κB have been previously described as being highly activated during pathogenic influenza virus infection of cell lines (36, 49), and STAT TFs are extensively involved in the antiviral response in vivo (26). It may be interesting to note that among ISGF3 components (STAT1-STAT2-IRF9), only STAT1 is present in PSCAN. STAT1 binding motif was enriched for set 1 (P value = 3.6e−4) (see Data Set S3) but not for the other sets. The sequence of the ISGF3 binding site partially overlaps the IRF binding element, and we found significant enrichment for IRF binding element in set 1. This may suggest that ISGF3 also plays a role in regulating the transcriptional program associated with lethality. Besides, since IRF1 and -2 were the only IRFs in the JASPAR database (see Data Set S3), and since IRFs bind similar DNA motifs (though with some specificities [38]), it is likely that other IRFs could also control the expression of genes that were upregulated after MA-CA/04 infection. Other novel TFs potentially regulating the set 1 transcriptional response based on binding motif enrichment (Table 2; see also Data Set S3) were myeloid zinc finger protein 1 (MZF-1) and SPI1 (also known as PU-1), TFs that are both involved in myeloid cell development.
Table 2.
TF binding motifs enriched in each transcriptional program discriminating MA-CA/04 and CA/04a
| TF | P value (PSCAN) |
|---|---|
| Set 1: upregulated genes associated with lethality | |
| NF-κB | 1.38E−18 |
| NFKB1 | 1.27E−15 |
| IRF2 | 2.82E−15 |
| IRF1 | 1.24E−14 |
| RELA | 2.19E−12 |
| MZF1_1-4 | 8.38E−12 |
| Set 2: transcriptional programs associated with recovery | |
| HNF4A | 9.34E−46 |
| NR2F1 | 6.05E−31 |
| HNF1A | 2.05E−24 |
| NR1H2::RXRA | 1.42E−21 |
| HNF1B | 4.90E−21 |
| PPARG::RXRA | 8.20E−17 |
| Set 3: downregulated genes associated with lethality | |
| ZEB1 | 4.73E−08 |
| INSM1 | 1.08E−07 |
| SP1 | 1.57E−07 |
| Myf | 1.34E−06 |
| PLAG1 | 1.24E−05 |
| PPARG::RXRA | 2.86E−05 |
Results for all 130 TFs analyzed in PSCAN are provided in Data Set S3 in the supplemental material.
Together, these results suggest that MA-CA/04 may trigger a greater activation of pattern recognition receptors (PRRs) resulting in IRF and NF-κB activation, leading subsequently to a high IFN and proinflammatory response, mediated by STAT TFs. This cytokine environment could affect the functions of infiltrating innate (dendritic cells and macrophages) and adaptive immune cells and also be responsible for lung lesions. In contrast, in CA/04 virus-infected animals, this activation was more limited and restrained to day 3 p.i.
Survival is associated with increased expression of lipid metabolism genes and genes enriched with nuclear receptor binding sites.
Survival of infection with CA/04 was associated with the induction of 968 genes that were significantly more upregulated in CA/04-infected mice than in MA-CA/04 infected mice (Fig. 4). Functional analysis of these genes found that 268 transcripts (28%) encoded molecules involved in lipid metabolism (see Data Set S1 in the supplemental material), including several ATP-binding cassette (ABC) transporters, apolipoprotein, diverse enzymes involved in fatty acid metabolism, vitamin D, arachidonic acid, lipid synthesis and uptake, several solute carrier family members (SLC), cytochrome P450 members, and surfactant proteins (SP-D and A1). Induction of these genes in CA/04-infected mice could be involved in viral clearance, as with SP-D (25), and in limiting the inflammatory response, as with CYP2C and -2J synthesizing anti-inflammatory metabolites (44), and enzymes involved in arachidonic acid metabolism. In addition, functional analysis showed a significant enrichment in genes related to amino acid metabolism. The upregulation of amino acid and lipid metabolism genes may be required to build cellular components essential for recovery from lung tissue damage.
To find potential regulators of the survival/recovery signature, we looked for TF binding motifs in promoters of genes from set 2. The top-ranked TFs were nuclear receptors acting in a heterodimer with RXR (NR1H2::RXRA and PPARG::RXRA) or as a homodimer (NR2F1 and several hepatocyte nuclear factors). These results were consistent with canonical pathway analysis, because three out of the five most enriched pathways involved RXR and one pathway (FXR/RXR activation) involved hepatocyte nuclear factor TFs (Table 1). Overall, these data indicate that nuclear receptors may be involved in lipid and amino acid metabolism induction potentially required for recovery from influenza infection.
Lethal infection is associated with suppression of coagulation genes.
The last set of genes DE between MA-CA/04 and CA/04 included genes that were more downregulated in MA-CA/04 mice at at least one time point (Fig. 4). Most of these genes were consistently more downregulated throughout infection with MA-CA/04 while being unchanged in CA/04-infected animals, but a small fraction were upregulated in these animals on days 1 and 5 p.i. and therefore also belonged to set 2. Overall enrichment scores were less significant for this set than for the other two sets (Table 1). However, we found a significant representation of lipid metabolism and coagulation-related genes in this set (Table 1; see also Data Set S1 in the supplemental material). Lipid enrichment was only partially explained by overlap with set 2, since several genes belonging only to set 3 were also associated with lipid metabolism. Procoagulant (F2, fibrinogen) and anticoagulant (Serpin C1 = Antithrombin III) factors, as well as fibrinolytic (Plasminogen) and antifibrinolytic (Serpin F2) factors, were more downregulated in MA-CA/04 mice on day 3 p.i. On day 5 p.i., several anticoagulants (encoded by Protein C, Serpin D1, C1, and A1) and profibrinolytic factors (encoded by Plasminogen, F12 and Kallikrein B) were more upregulated in CA/04-infected than in MA-CA/04-infected mice (see Data Set S1) and could limit fibrin deposition involved in acute lung injury.
Transcription factor analysis using PSCAN found significant enrichment for PPARG::RXRA, which could therefore also be implicated in downregulation of set 3 genes, as well as several transcriptional repressors (ZEB1, INSM1, SP1, and PLAG1). In conclusion, MA-CA/04 infection induced a consistent downregulation of lipid metabolism and coagulation-associated genes, which could be due to regulation of PPARG and/or transcriptional repressors.
Prediction of TF activation state supports IRF and nuclear receptor regulation of transcription.
Because we found several TF binding motifs that were highly enriched in each set of genes discriminating CA/04 and MA-CA/04, we were interested in determining their activation state following infection. For this purpose, we used the IPA TF analytic tool, which can predict the activation of TFs based on the expression levels of their known targets. We performed this analysis using log2 ratio expression values for the 4,203 genes discriminating CA/04- and MA-CA/04-infected animals. Figure 5A represents z scores for 16 TFs that were found to be very likely (|z| > 4) to be involved in the transcriptional response to infection at at least one time point and in one group. Based on the annotation of their target genes, we found that these TFs were involved either in lipid metabolism control or in the inflammatory response.
Fig 5.
TF activation state prediction in MA-CA/04- and CA/04-infected mice. (A) z scores predicting activation state of TFs (|z| > 4) based on expression values of genes discriminating CA/04 and MA-CA/04. Dashed lines depict |z| = 2, which is considered to be the limit of significance. z > 2 predicts an activation of the regulator. z < −2 predicts an inhibition. “*” denotes TFs that were found among the top 5 ranked TFs in PSCAN analysis (Table 2). (B) Expression levels (in log2 ratio) for 353 targets of the 3 most significant TFs controlling inflammatory response. Genes that belong to the “inflammatory response” category in IPA are indicated in purple. (C) Expression levels (in log2 ratio) for 597 targets of the 3 most significant TFs controlling lipid metabolism. Genes that belong to the “lipid metabolism” category in IPA are indicated in yellow.
Binding motifs for several TFs likely to be involved in inflammatory response control were also found in our previous PSCAN analysis to be present in promoters of genes from the upregulated lethal signature (Table 2; see also Data Set S3 in the supplemental material). These TFs were predicted to be activated throughout infection with MA-CA/04 virus, while in CA/04-infected mice they would be activated only by day 3 p.i., with a possible decrease in their activation on day 5 p.i. (Fig. 5A). Consistently, most of their target genes belonged to the upregulated lethal signature (set 1) and were more expressed in MA-CA/04-infected mice than in CA/04-infected mice (Fig. 5B). Two additional IRFs (IRF7 and -8) were also predicted to be more activated in MA-CA/04-infected mice, which supports the general implication of IRFs in controlling the transcriptional program associated with lethality. Additionally, two negative regulators, GFI1 and TRIM24, were found to be constantly inactivated in MA-CA/04 samples, and we verified that their targets were also more upregulated in MA-CA/04-infected mice (Fig. 5B).
Among the four TFs that could control lipid metabolism during infection, three were previously found in PSCAN (HNF1A, HNF4A, and PPARG). The hepatocyte nuclear factors (HNFs) were predicted to be activated in CA/04-infected lungs on days 1 and 5 p.i., while they would be repressed or unchanged at the same time points in MA-CA/04-infected animals (Fig. 5A). Consistently, most of their target genes were more expressed in CA/04 animals than in MA-CA/04 animals on days 1 and 5 p.i. (Fig. 5C). PPARG was another nuclear receptor found by the two complementary TF analyses. It was predicted to be inhibited throughout infection with MA-CA/04 virus, while not with infection with CA/04, on days 1 and 5 p.i. (Fig. 5A). Most of its targets were more downregulated in MA-CA/04 samples and were associated with lipid metabolism. Notably, the small subset of PPARG targets that were found to be more upregulated in MA-CA/04-infected mice are genes that are known to be repressed by PPARG, including those encoding several proinflammatory cytokines (IL-1, IL-6, IFNG, and TNF) and several chemokines (CXCL-1, CXCL-3, CXCL-6, and CXCL-14 and CCL-13, CCL-17, and CCL-22). This was consistent with an inhibition of PPARG activity in MA-CA/04-infected animals and indicates that this regulation could be involved in both the absence of lipid metabolism upregulation and increased inflammatory response observed in MA-CA/04-infected animals.
These results overall support the implication of several nuclear receptors (HNF1A, HNF4A, and PPARG), IRFs, and the NF-κB complex in the differential host response to MA-CA/04 or CA/04 infection. We also found novel transcription regulators not present in the JASPAR database, notably TRIM24, whose repression could play a role in increasing the inflammation in MA-CA/04 virus-infected animals.
Genes associated with inflammatory macrophages and granulocytes are highly induced during MA-CA/04 infection.
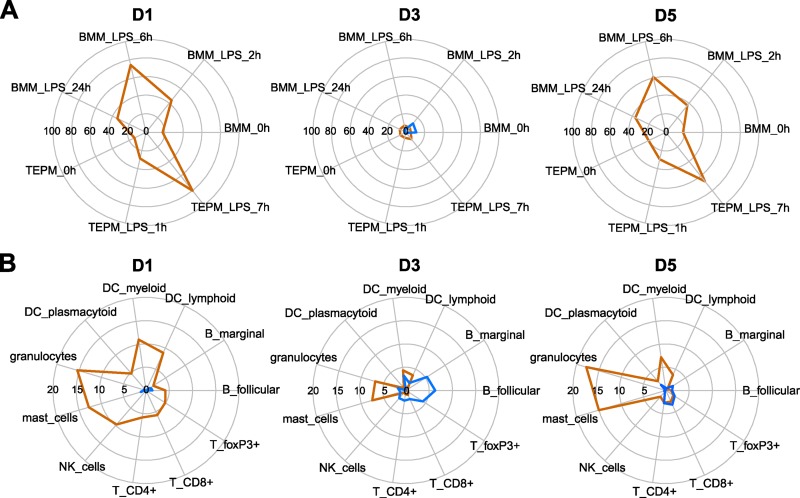
Since several pathways differentially induced between MA-CA/04 and CA/04 were related to specific immune cells or included chemoattractants, we hypothesized that observed transcriptional differences elicited by MA-CA/04 and CA/04 infection could be partially due to differences in immune cell populations and/or their activation state in the lungs. To further explore this possibility, we compared our gene expression profiles to microarray data for each immune cell population present in the GNF Mouse Gene Atlas v3 database (42). We defined, for each immune cell population present in the database, a list of genes that were expressed 20-fold more in a given cell subset than in whole lung. We then analyzed whether there was significant enrichment in genes that were more upregulated in response to infection with either MA-CA/04 (Fig. 6, orange lines) or CA/04 (Fig. 6, blue lines). We could therefore evaluate whether differences in gene expression profiles between MA-CA/04 and CA/04 infection were associated with immune cell infiltration.
Fig 6.
Enrichment in immune cell highly expressed genes for genes that were differentially changed between MA-CA/04 and CA/04 infection. Genes specific to each immune cell population were determined using the Gene Atlas v3 data set to be genes that were 20-fold more increased in each immune population than for whole lung tissue. Genes upregulated in MA-CA/04 infection are shown in orange, and genes upregulated in CA/04 infection are shown by blue lines. On each day p.i., we calculated the proportion of immune cell-related genes in 2 lists of genes DE between MA-CA/04 and CA/04 infection: (i) genes with a log2 ratio in MA-CA/04 infection > that in CA/04 infection > 0 (orange); (ii) genes with a log2 ratio in CA/04 infection > that in MA-CA/04 infection > 0 (blue). Enrichment scores (ES) were calculated as a −log10 P value, using a right-tailed Fisher exact test. Because of differences of scale between ES in macrophages and in other immune cells, they were depicted in 2 different radial plots for each day p.i. (A) ES for bone marrow-derived macrophages (BMM) and thioglycolate-elicited peritoneal macrophages (TEPM). (B) ES for other immune cells: lymphoid (CD8a+) dendritic cells (DC), myeloid (CD8a−) DC, plasmacytoid (B220+) DC, B cells from follicular and marginal zones, different T cell populations (CD4+, CD8+, and FoxP3+), natural killer (NK) cells, mast cells, and granulocytes (mac1+ gr1+).
The list of genes that exhibited no change in expression after CA/04 infection but which were strongly induced by MA-CA/04 at days 1 and 5 p.i. was highly enriched in genes associated with lipopolysaccharide (LPS)-stimulated macrophages (from both bone marrow-derived macrophages [BMM] and thioglycolate-elicited peritoneal macrophages [TEPM]) (Fig. 6A). LPS was used as an in vitro stimulus in the Mouse Gene Atlas study to trigger proinflammatory pathways activating macrophages (24). Higher infiltration of inflammatory macrophages in MA-CA/04-infected lungs may therefore contribute to the transcriptional program associated with lethality. Similarly, analysis of the other immune cell-associated genes (Fig. 6B) suggested a higher infiltration of NK cells at day 1 p.i. and of granulocytes and mast cells for all time points after MA-CA/04 infection. In contrast, on day 3 p.i., there were fewer genes more highly expressed in MA-CA/04-infected mice, and there were fewer associated with macrophages, suggesting a similar proportion of macrophages in CA/04- and MA-CA/04-infected lungs on day 3 p.i. Macrophage infiltration after CA/04 infection may therefore be transient, while such infiltration is sustained in MA-CA/04-infected animals.
Closer examination of enrichment scores at day 3 p.i. revealed that genes highly induced by CA/04 included genes that were highly expressed in unstimulated bone marrow-derived macrophages (Fig. 6A, “BMM_0h,” enrichment score of 11.1,), B cells, and FoxP3+ T cells (Fig. 6B). However, most of the genes from the transcriptional program associated with survival/recovery (set 2) that were highly expressed on day 5 p.i. with CA/04 infection were not associated with immune cells (Fig. 6).
Overall, these results suggest that macrophages present in lungs after infection with MA-CA/04 or CA/04 could be functionally different, with noninflammatory macrophages contributing to the gene expression response to CA/04, while inflammatory macrophage, granulocyte, and mast cell infiltration contributes to the transcriptional response measured in lung samples from MA-CA/04-infected animals.
Transcriptional programs associated with lethality and recovery in the context of other H1N1 influenza virus infections.
We next examined whether the transcriptional signatures associated with lethality and recovery that were derived from the comparison of CA/04- and MA-CA/04-infected mice corresponded to the expression patterns elicited by other lethal and nonlethal H1N1 influenza viruses. Several nonlethal H1N1 viruses were selected for comparison. These included Mex/4482 and NJ/8/76, both of which were isolated from patients with severe respiratory illness, and Brisbane/59/07, a seasonal H1N1 virus. Intranasal infection by these viruses in BALB/c mice resulted in a range of virulence, in agreement with a previous report of H1N1 murine virulence (5). Brisbane/59/07 and NJ/8/76 caused minimal weight loss (see Fig. S2A in the supplemental material) and were classified as low pathogenicity. Mex/4482 caused significant weight loss, which was intermediate between those of CA/04 and MA-CA/04 infection (Student's t test, P < 0.05 from days 4 and 2 p.i., respectively) (see Fig. S2A). Viral titers in the lungs were much lower in Brisbane/59/07-infected animals than in all other groups (P < 0.05), while titers were similar in mice infected with Mex/4482, NJ/8/76, or CA/04 (see Fig. S2B). Stored RNA isolated from the lungs of mice infected with the reconstructed 1918 pandemic influenza virus (r1918) was used as an additional lethal H1N1 comparator (see Materials and Methods). As described previously, mice infected with r1918 had 75% weight loss at day 4 p.i., and there was 100% lethality by day 6 p.i. (21). Viral titers in the spleen and brain of animals infected with these lethal and nonlethal viruses indicated that none of these viruses disseminated systemically in this mouse model.
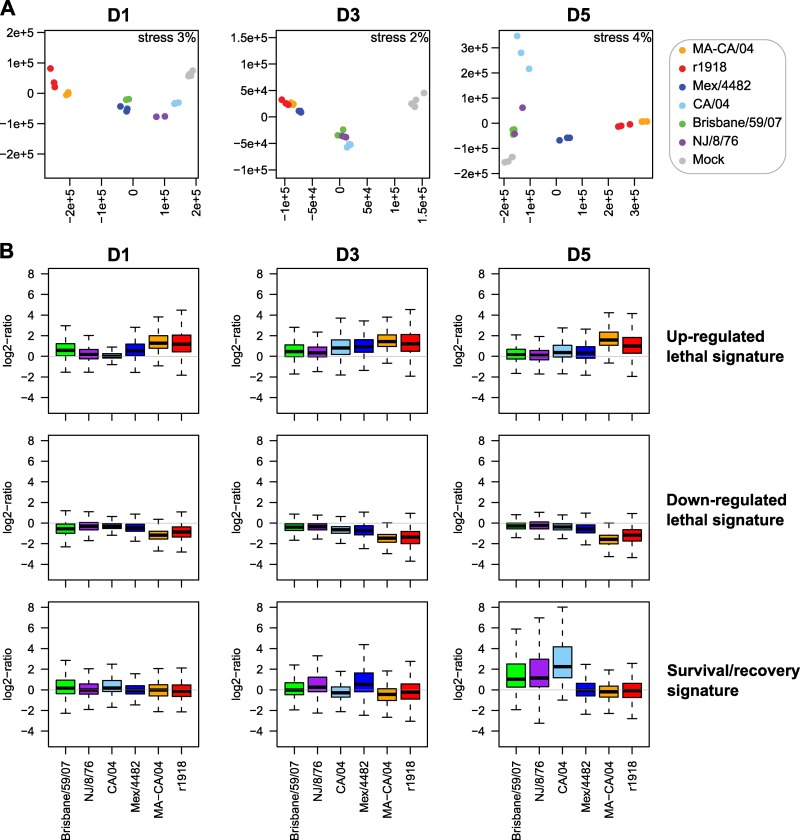
To determine whether genes discriminating the transcriptional response to CA/04 and MA-CA/04 on each day p.i. were also able to discriminate low-pathogenicity H1N1 (Brisbane/59/07 and NJ/8/76) from the intermediate-pathogenicity Mex/4482 and from the high-pathogenicity r1918, we used nonparametric multidimensional scaling (MDS) to visualize global gene expression concordance between all samples and for each signature (Fig. 7A). Kruskal's stress was used to measure the information loss during dimensionality reduction. We had very small stress values (<5%), indicating a good fit of the MDS representations. On day 1 p.i., and using the genes that were DE between mice infected with CA/04 and those infected with MA-CA/04 on that time point (1,868 genes), MA-CA/04- and r1918-infected samples were located in the same 2D space, indicating that these genes were expressed at similar levels for both lethal viruses. Low and medium pathogens were clustered closer to the mock inoculation. This confirmed that, as with CA/04, less pathogenic viruses induced limited changes in the expression of these genes on day 1 p.i., compared to results for the highly pathogenic viruses MA-CA/04 and r1918.
Fig 7.
Comparison of MA-CA/04 and CA/04 transcriptional profiles with those of r1918, Mex/4482, Brisbane/59/07, and NJ/8/76. (A) Nonparametric multidimensional scaling (MDS) representing the Euclidian distances between samples on each day p.i. The matrix distance was calculated using genes that were found DE between MA-CA/04 and CA/04 on each day p.i. Kruskal's stress evaluates the loss of information in the representation. (B) Box plot of each transcriptional signature discriminating MA-CA/04 and CA/04 for days 1, 3, and 5 p.i. data.
Likewise, MDS analysis showed that genes DE between mice infected with CA/04 or MA-CA/04 on day 3 p.i. (1,041 genes) also differentiated highly pathogenic (r1918 and MA-CA/04) from less pathogenic viruses (CA/04, Brisbane/59/07, and NJ/8/76). Interestingly, Mex/4482, which induced significant weight loss at this time point, elicited a host response similar to that for the lethal viruses on day 3 p.i. Finally, on day 5 p.i., we observed three different clusters of samples based on expression of 3,123 genes DE between mice infected with MA-CA/04 or CA/04. Mice infected with MA-CA/04 or r1918 expressed these genes similarly, since Mex/4482 elicited a response intermediate between those for low- and high-pathogenicity viruses, and a third cluster was formed by the less pathogenic viruses, with results for Brisbane/59/07 and NJ/8/76 being more similar to those for the mock samples than those for CA/04. These results indicate that genes DE between mice infected with CA/04 and MA-CA/04 discriminate different levels of pathogenicity of other H1N1 viruses.
We next compared these DE gene signatures across viruses by examining the distribution and correlation of gene expression across groups (Fig. 7B). We observed that the upregulated lethal signature was expressed to similar levels between MA-CA/04 and r1918 (Pearson correlation, r = 0.81), with a higher median of expression than that for the other viruses on each day p.i. In addition, these genes were overall slightly more expressed in response to Mex/4482 than to CA/04. Downregulated genes from the lethal transcriptional signatures were also more downregulated by the lethal viruses on each day p.i. (Pearson correlation between MA-CA/04 and r1918, r = 0.51). Finally, the recovery signature on day 5 p.i. was upregulated only by less pathogenic viruses, with the highest upregulation for CA/04-infected mice but good correlation with results for Brisbane/59/07 (r = 0.89) and NJ/8/76 (r = 0.9). The intermediate-pathogenicity virus, Mex/4482, which induced weight loss continuously until day 8 p.i., did not upregulate the recovery-associated genes on day 5 p.i., confirming that these genes were associated with recovery and that their expression might be delayed during Mex/4482 virus infection. This analysis confirmed the general relevance of the transcriptional programs associated with lethality and survival/recovery for H1N1 pathogenicity.
Overall, using global gene expression profiling, we found that the increased virulence of the mouse-adapted CA/04 virus was associated with higher and earlier expression of inflammatory and immune cell-related genes following activation of IRF, NB-κB, and STAT TFs. Comparison of our data to metadata collected in other studies from multiple immune cell populations suggested that these differences may be due in part to increases in inflammatory macrophage infiltration in the lungs of mice infected with MA-CA/04. Lethality was also associated with the lack of upregulation of lipid and amino acid metabolism genes, which were upregulated in CA/04-infected mice. Transcription factor analysis suggested that this absence of signaling during lethal infection may be mediated in part by nuclear receptors. Finally, comparison of the gene expression patterns associated with lethality and recovery suggests similar patterns in the expression of these genes between CA/04 and other nonlethal H1N1 influenza viruses and between the lethal viruses MA-CA/04 and r1918.
DISCUSSION
The 2009 pandemic H1N1 virus requires only a few mutations following limited passage in mice to increase its virulence extraordinarily in this animal model (16, 34, 39, 53, 56). As 2009 H1N1 continued circulating in humans in 2010-2011 and in the current influenza season, concern remains about its pathogenic potential. The mouse-adapted virus therefore provides an important opportunity to identify host response correlates to a more virulent form of the 2009 H1N1 virus.
In the current study, we compared the host response to the wild-type prototypic 2009 H1N1 viral CA/04 and its mouse-adapted variant, MA-CA/04. These strains have only 5 amino acid differences, but while the wild-type strain was a mild pathogen in BALB/c mice, MA-CA/04 induced 100% lethality by day 7. Histopathological examination revealed differences in lung lesions associated with lethality. Additionally, for MA-CA/04-infected mice, some virus was detected in brain and spleen at later time points of infection. CA/04 and MA-CA/04 viruses both lack viral determinants that were previously reported to be responsible for systemic spread (HA polybasic site [13] and lysine at position 627 of PB2 [10]). Moreover, only one mutation in MA-CA/04, HA G155E, has not been found in the other nondisseminating mouse-adapted 2009 H1N1 virus. This specific mutation could be involved in systemic spread, but dissemination could also result from the unique combination of mutations present in MA-CA/04. Extrapulmonary dissemination could also be explained by the high efficiency of viral replication overwhelming the host response, as was shown for H5N1 HK483 (41).
To our knowledge, this is the first use of global gene expression profiling to characterize the host response to a lethal 2009 H1N1 virus. Several transcriptomic studies have compared the host response to 2009 H1N1 viruses and other seasonal H1N1 viruses in macaques (33), mice (22), and pigs (29). These studies described a higher upregulation of genes related to immune and inflammatory responses following infection with pandemic strains compared than with seasonal virus. Our microarray analysis also revealed a strong immune response in CA/04-infected mice at day 3, but this response was delayed and transient compared to that elicited by MA-CA/04. These findings are consistent with those of previous studies (10, 21) showing that early and sustained activation of the inflammatory response is associated with lethal influenza virus infection. In addition to higher expression of inflammatory and immune cell-related genes, lethality following MA-CA/04 infection was also associated with the lack of upregulation of lipid metabolism genes. Genes discriminating MA-CA/04 and CA/04 were differentiating other lethal and nonlethal H1N1 viruses, confirming their general relevance for virulence of influenza virus. Distribution of expression for signatures associated with lethality and recovery was not contingent on differences in virus levels. Therefore, although we cannot exclude the possibility that the level of viral replication impacts pathogenesis and the host response, our data suggest that identified signatures were more related to virulence than viral replication.
Previous studies have shown that immune cell infiltration in the lungs is an important pathological feature of lethal influenza viruses. Patients who died from 2009 H1N1 virus infection showed profound inflammatory infiltration in the lungs and diffuse alveolar damage similar to that seen in past pandemics (11). Enrichment analysis leveraging immune cell transcription information accessed through the Mouse Gene Atlas suggested differences in kinetics and in activation of immune cell subsets infiltrating the lungs of virus-infected mice. While CA/04-induced genes were transiently associated with noninflammatory macrophage and B and T-regulatory cell (FoxP3+) infiltration on day 3 p.i., genes that were more upregulated in MA-CA/04 mice were associated with inflammatory macrophages, granulocytes, and mast cells for all time points. The relevance of these findings is supported by histopathological examination of infected lungs, pointing out histiocyte and lymphocyte infiltration in the peribronchiolar area of CA/04-infected mice from day 3 only. In contrast, these cells were present in the lungs of MA-CA/04-infected mice from day 1 and increased during infection in the peribronchiolar and alveolar compartments.
Infiltrating macrophages and neutrophils are essential to control viral replication at early time points (46), but excessive inflammatory cell recruitment contributes to lung injury (1, 12, 27, 35). Similarly, several studies suggest that CD4 and CD8 T cells also contribute to both influenza virus clearance and immunopathology (reviewed in reference 32). Our results suggest that a transient and strong immune infiltration could be important for the protective response to CA/04, with certain cell types being associated with survival. For example, regulatory T cells could play a role in moderating the inflammatory response on day 3 p.i. in CA/04-infected mice. In contrast, persistent and strong lung infiltration with inflammatory macrophages, granulocytes, and mast cells in response to lethal viruses was associated with hypercytokinemia (Table 1) and was likely to induce airway epithelium destruction (12, 14). Beyond immune cell types, our results suggest that specific signaling pathways activated in infiltrating cells, such as the LPS-stimulated pathway in macrophages, were associated with influenza pathogenesis. It is possible that MA-CA/04 infection of macrophages plays a role in this pathway induction. Indeed, we detected viral antigen more frequently in macrophages for MA-CA/04-infected animals, which suggests a change in viral tropism that could be mediated by HA mutations. In addition, LPS activation and antiviral response result in activation of common signaling pathways, such as the activation of IRF3.
Our data support the role for the IFN regulatory factor (IRF) family in influenza virulence. Binding motifs for IRF1 and IRF2 were found to be enriched in the promoters of genes that were induced more highly by MA-CA/04 than by CA/04. IRF1 was also predicted to be activated early after infection with MA-CA/04 but not CA/04 based on the expression of its target genes. Since the upregulated lethal signature was also strongly associated with immune cells, especially macrophages, it is possible that IRFs were activated in immune cells infiltrating the lung during infection, with possible consequences for their functions (reviewed in reference 2). In particular, IRF1 and IRF8 are involved in macrophage activation and inhibit generation of regulatory T cells, which could have an important impact in influenza pathogenesis. IRFs may be activated in immune and epithelial cells by cytokines secreted during infection and by direct infection (47). A recent in vitro study concluded that IRFs were equally involved in the response of primary endothelial cells to infection with low- or high-pathogenicity influenza viruses (49). In contrast, our results suggest that IRFs were more activated in mice infected with lethal viruses, being in part responsible for a sustained inflammatory response and macrophage activation.
In addition to IRFs, sustained activation of NF-κB and STATs was implicated in the inflammatory response following MA-CA/04 infection. These TFs have been described previously as activated in response to highly pathogenic influenza virus infection (26, 36). Since NF-κB and STAT signaling pathways are involved in immune cell activation following different stimuli, their higher activation in the lungs of mice infected with MA-CA/04 may also reflect a difference in the activation state of infiltrating immune cells. Finally, we report that some TFs, which have not previously been associated with influenza pathogenesis to our knowledge, could potentially regulate the transcriptional program associated with lethal infection. These transcription regulators include MZF1, whose binding motif was found to be enriched in these genes (Table 2), and TRIM24 and GFI1, whose regulation was predicted to be significant based on the expression of their known targets (Fig. 5A). TRIM24 and GFI1 are two transcriptional repressors that were predicted to be inhibited during MA-CA/04 infection. Interestingly, GFI1 suppression in macrophages results in an exaggerated production of inflammatory cytokines following LPS stimulation (20). In addition, TRIM24 knockout (KO) mice have an overactivation of the IFN/STAT pathway (45). Inhibition of both TRIM24 and GFI1 could therefore have important inflammatory consequences in MA-CA/04-infected mice.
Transcription factors involved in the survival/transcriptional program associated with recovery include members of the nuclear receptor ligand-dependent family. HNF1A and -B, HNF4A, and PPARG were predicted to be activated in CA/04-infected mice and/or repressed in MA-CA/04-infected mice based on expression values of their target genes. To our knowledge, this is the first report associating HNF TFs with influenza virus pathogenesis. HNF1A and -4A are two major liver TFs that control diverse metabolic functions. These TFs are also expressed to a lesser extent in pathological or normal lung (3, 23) and were predicted to be activated in CA/04-infected mice on days 1 and 5 p.i. and either unchanged or repressed for HNF1A in MA-CA/04-infected animals. Known targets of HNF1A and -4A include many genes involved in lipid and amino acid metabolism, as well as several blood coagulation factors (15, 40). Pulmonary and systemic activation of coagulation has been implicated in a fatal human case of 2009 H1N1 infection (11) and in lethal H1N1 influenza virus infection in mice (37). We found different changes in coagulation factor synthesis in the lungs of mice infected with MA-CA/04 or CA/04 viruses. Absence of fibrinolytic and anticoagulant factor induction on day 5, in combination with plasma protein leakage and induction of tissue factor and plasminogen activator inhibitor (PAI)-1 by activated macrophages (52), could contribute to intra-alveolar fibrin deposition in the lungs of mice infected with lethal influenza viruses. In addition to coagulation, activation of lipid and amino acid metabolism may be required to build cellular membrane components for cell proliferation during recovery from influenza virus infection. Similar to our results, microarray analysis of lungs from pigs infected with CA/04 also showed that recovery from infection is associated with an overall increase in lipid metabolism at day 5 (29). The NS1 segment of r1918 has been previously implicated in lipid metabolism and CYP repression in cell culture (7). However, there was no difference in NS1 sequence between MA-CA/04 and CA/04, suggesting that other viral properties could be implicated in this regulation.
Our results suggest that lipid metabolism inhibition after MA-CA/04 infection could also involve PPARG repression. This nuclear receptor is active as a heterodimer with retinoid X receptor (RXR). Repression of PPARG activity could involve suppression of RXRα-dependent gene expression due to Jun N-terminal protein kinase (JNK) phosphorylation, itself activated by tumor necrosis factor alpha (TNF-α) and interleukin 1 (IL-1) signaling (57), or RXRα repression mediated by IRF3 in hepatocytes and BMMs (8). Known targets of PPARG that were DE between MA-CA/04 and CA/04 infection were associated with lipid metabolism and the inflammatory response. This TF has been extensively studied for its role in bridging lipid metabolism and inflammation in several diseases, including atherosclerosis (reviewed in references 6 and 43). Considering their functions in transrepressing inflammatory TF target genes, such as those encoding NF-κB, NFAT, STAT, and AP-1, PPARG activation could be crucial to limiting inflammation in CA/04-infected mice. Besides, PPARG activation potentiates polarization of circulating monocytes into macrophages of the M2 phenotype, which generate anti-inflammatory products and mediate tissue repair. This suggests that macrophages present in CA/04-infected lung may have a protective M2 phenotype, compared to the sustained infiltration with inflammatory M1 macrophages in MA-CA/04-infected mice. Based on their anti-inflammatory role, PPARG agonists have been used as immune modulators, providing protection in mice infected with highly pathogenic and pandemic strains of influenza virus (1, 31). We provide for the first time evidence that PPARG, together with other nuclear receptors, is involved in inducing genes associated with recovery from influenza virus infection.
In conclusion, the virulence of CA/04 was dramatically increased following host adaptation. Enhanced virulence was associated with an increased and sustained inflammatory response, immune cell infiltration, and no increase in lipid metabolism later in infection. Our results suggest that regulation of several key transcription factors may be involved in these processes, including induction of IRF1 and -2 and lack of activation of HNF4A, PPARG, and LXR-β. These TFs may also regulate the function of infiltrating immune cells, including macrophages. Identifying specific genes involved in deleterious infiltrating cell subsets may point to new therapies to prevent severe disease associated with influenza virus infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Richard Webby of St. Jude Children's Research Hospital for use of the MA1-CA/04 virus. We also thank Lynn Law, Sean Proll, and Marcus Korth for valuable feedback on the manuscript.
This project was funded in part by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract HHSN272200800060C (to M.G.K.).
The findings and conclusions in this report are those of the authors and do not necessarily reflect the views of the funding agency.
Footnotes
Published ahead of print 24 April 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Aldridge JR, Jr, et al. 2009. TNF/iNOS-producing dendritic cells are the necessary evil of lethal influenza virus infection. Proc. Natl. Acad. Sci. U. S. A. 106:5306–5311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Battistini A. 2009. Interferon regulatory factors in hematopoietic cell differentiation and immune regulation. J. Interferon Cytokine Res. 29:765–780 [DOI] [PubMed] [Google Scholar]
- 3. Baumhueter S, Courtois G, Crabtree GR. 1988. A variant nuclear protein in dedifferentiated hepatoma cells binds to the same functional sequences in the beta fibrinogen gene promoter as HNF-1. EMBO J. 7:2485–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Belisle SE, et al. 2010. Genomic profiling of tumor necrosis factor alpha (TNF-alpha) receptor and interleukin-1 receptor knockout mice reveals a link between TNF-alpha signaling and increased severity of 1918 pandemic influenza virus infection. J. Virol. 84:12576–12588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Belser JA, et al. 2010. Pathogenesis of pandemic influenza A (H1N1) and triple-reassortant swine influenza A (H1) viruses in mice. J. Virol. 84:4194–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bensinger SJ, Tontonoz P. 2008. Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature 454:470–477 [DOI] [PubMed] [Google Scholar]
- 7. Billharz R, et al. 2009. The NS1 protein of the 1918 pandemic influenza virus blocks host interferon and lipid metabolism pathways. J. Virol. 83:10557–10570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chow EK, et al. 2006. A role for IRF3-dependent RXRalpha repression in hepatotoxicity associated with viral infections. J. Exp. Med. 203:2589–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cilloniz C, et al. 2012. Molecular signatures associated with Mx1-mediated resistance to highly pathogenic influenza virus infection: mechanisms of survival. J. Virol. 86:2437–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fornek JL, et al. 2009. A single-amino-acid substitution in a polymerase protein of an H5N1 influenza virus is associated with systemic infection and impaired T-cell activation in mice. J. Virol. 83:11102–11115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gill JR, et al. 2010. Pulmonary pathologic findings of fatal 2009 pandemic influenza A/H1N1 viral infections. Arch. Pathol. Lab. Med. 134:235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Herold S, et al. 2008. Lung epithelial apoptosis in influenza virus pneumonia: the role of macrophage-expressed TNF-related apoptosis-inducing ligand. J. Exp. Med. 205:3065–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Horimoto T, Kawaoka Y. 1994. Reverse genetics provides direct evidence for a correlation of hemagglutinin cleavability and virulence of an avian influenza A virus. J. Virol. 68:3120–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hu Y, et al. 2012. Mast cell-induced lung injury in mice infected with H5N1 influenza virus. J. Virol. 86:3347–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hwang-Verslues WW, Sladek FM. 2010. HNF4alpha—role in drug metabolism and potential drug target? Curr. Opin. Pharmacol. 10:698–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ilyushina NA, et al. 2010. Adaptation of pandemic H1N1 influenza viruses in mice. J. Virol. 84:8607–8616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Irizarry RA, et al. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Itoh Y, et al. 2009. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 460:1021–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johnson WE, Li C, Rabinovic A. 2007. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8:118–127 [DOI] [PubMed] [Google Scholar]
- 20. Karsunky H, et al. 2002. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat. Genet. 30:295–300 [DOI] [PubMed] [Google Scholar]
- 21. Kash JC, et al. 2006. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 443:578–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kash JC, et al. 2011. Lethal synergism of 2009 pandemic H1N1 influenza virus and Streptococcus pneumoniae coinfection is associated with loss of murine lung repair responses. mBio 2(5):e00172–11 doi:10.1128/mBio.00172-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kunii R, et al. 2011. The predominant expression of hepatocyte nuclear factor 4alpha (HNF4alpha) in thyroid transcription factor-1 (TTF-1)-negative pulmonary adenocarcinoma. Histopathology 58:467–476 [DOI] [PubMed] [Google Scholar]
- 24. Lattin JE, et al. 2008. Expression analysis of G protein-coupled receptors in mouse macrophages. Immunome Res. 4:5 doi:10.1186/1745-7580-4-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. LeVine AM, Whitsett JA, Hartshorn KL, Crouch EC, Korfhagen TR. 2001. Surfactant protein D enhances clearance of influenza A virus from the lung in vivo. J. Immunol. 167:5868–5873 [DOI] [PubMed] [Google Scholar]
- 26. Levy DE, Garcia-Sastre A. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:143–156 [DOI] [PubMed] [Google Scholar]
- 27. Lin KL, Suzuki Y, Nakano H, Ramsburg E, Gunn MD. 2008. CCR2+ monocyte-derived dendritic cells and exudate macrophages produce influenza-induced pulmonary immune pathology and mortality. J. Immunol. 180:2562–2572 [DOI] [PubMed] [Google Scholar]
- 28. Lopez-Romero P. Agi4x44PreProcess: PreProcessing of Agilent 4x44 array data. 2012 R package version 1.8.0. ed. [Google Scholar]
- 29. Ma W, et al. 2011. 2009 pandemic H1N1 influenza virus causes disease and upregulation of genes related to inflammatory and immune responses, cell death, and lipid metabolism in pigs. J. Virol. 85:11626–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maines TR, et al. 2009. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science 325:484–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moseley CE, Webster RG, Aldridge JR. 2010. Peroxisome proliferator-activated receptor and AMP-activated protein kinase agonists protect against lethal influenza virus challenge in mice. Influenza Other Respi. Viruses 4:307–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peiris JS, Hui KP, Yen HL. 2010. Host response to influenza virus: protection versus immunopathology. Curr. Opin. Immunol. 22:475–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Safronetz D, et al. 2011. Pandemic swine-origin H1N1 influenza A virus isolates show heterogeneous virulence in macaques. J. Virol. 85:1214–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sakabe S, Ozawa M, Takano R, Iwastuki-Horimoto K, Kawaoka Y. 2011. Mutations in PA, NP, and HA of a pandemic (H1N1) 2009 influenza virus contribute to its adaptation to mice. Virus Res. 158:124–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sakai S, et al. 2000. Therapeutic effect of anti-macrophage inflammatory protein 2 antibody on influenza virus-induced pneumonia in mice. J. Virol. 74:2472–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schmolke M, Viemann D, Roth J, Ludwig S. 2009. Essential impact of NF-kappaB signaling on the H5N1 influenza A virus-induced transcriptome. J. Immunol. 183:5180–5189 [DOI] [PubMed] [Google Scholar]
- 37. Schouten M, et al. 2010. Activated protein C ameliorates coagulopathy but does not influence outcome in lethal H1N1 influenza: a controlled laboratory study. Crit. Care 14:R65 doi:10.1186/cc8964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Servant MJ, Tenoever B, Lin R. 2002. Overlapping and distinct mechanisms regulating IRF-3 and IRF-7 function. J. Interferon Cytokine Res. 22:49–58 [DOI] [PubMed] [Google Scholar]
- 39. Seyer R, et al. 2012. Synergistic adaptive mutations in the hemagglutinin and polymerase acidic protein lead to increased virulence of pandemic 2009 H1N1 influenza A virus in mice. J. Infect. Dis. 205:262–271 [DOI] [PubMed] [Google Scholar]
- 40. Shih DQ, et al. 2001. Hepatocyte nuclear factor-1alpha is an essential regulator of bile acid and plasma cholesterol metabolism. Nat. Genet. 27:375–382 [DOI] [PubMed] [Google Scholar]
- 41. Shinya K, et al. 2004. PB2 amino acid at position 627 affects replicative efficiency, but not cell tropism, of Hong Kong H5N1 influenza A viruses in mice. Virology 320:258–266 [DOI] [PubMed] [Google Scholar]
- 42. Su AI, et al. 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. U. S. A. 101:6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Szanto A, Roszer T. 2008. Nuclear receptors in macrophages: a link between metabolism and inflammation. FEBS Lett. 582:106–116 [DOI] [PubMed] [Google Scholar]
- 44. Theken KN, et al. 2011. Activation of the acute inflammatory response alters cytochrome P450 expression and eicosanoid metabolism. Drug Metab. Dispos. 39:22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tisserand J, et al. 2011. Tripartite motif 24 (Trim24/Tif1alpha) tumor suppressor protein is a novel negative regulator of interferon (IFN)/signal transducers and activators of transcription (STAT) signaling pathway acting through retinoic acid receptor alpha (Raralpha) inhibition. J. Biol. Chem. 286:33369–33379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tumpey TM, et al. 2005. Pathogenicity of influenza viruses with genes from the 1918 pandemic virus: functional roles of alveolar macrophages and neutrophils in limiting virus replication and mortality in mice. J. Virol. 79:14933–14944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Veckman V, et al. 2006. TNF-alpha and IFN-alpha enhance influenza-A-virus-induced chemokine gene expression in human A549 lung epithelial cells. Virology 345:96–104 [DOI] [PubMed] [Google Scholar]
- 48. Venables WN, Ripley BD. (ed). 2002. Modern applied statistics with S, 4th ed Springer, Berlin, Germany [Google Scholar]
- 49. Viemann D, et al. 2011. H5N1 virus activates signaling pathways in human endothelial cells resulting in a specific imbalanced inflammatory response. J. Immunol. 186:164–173 [DOI] [PubMed] [Google Scholar]
- 50. World Health Organization 2010. Pandemic (H1N1) 2009, update 112. World Health Organization, Geneva, Switzerland [Google Scholar]
- 51. World Health Organization 2011. Review of the 2011 winter influenza season, southern hemisphere. Wkly. Epidemiol. Rec. 86:480–496 [PubMed] [Google Scholar]
- 52. Wygrecka M, et al. 2007. Cellular origin of pro-coagulant and (anti)-fibrinolytic factors in bleomycin-injured lungs. Eur. Respir. J. 29:1105–1114 [DOI] [PubMed] [Google Scholar]
- 53. Ye J, et al. 2010. Variations in the hemagglutinin of the 2009 H1N1 pandemic virus: potential for strains with altered virulence phenotype? PLoS Pathog. 6:e1001145 doi:10.1371/journal.ppat.1001145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zambelli F, Pesole G, Pavesi G. 2009. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 37:W247–W252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zeng H, et al. 2007. Highly pathogenic avian influenza H5N1 viruses elicit an attenuated type i interferon response in polarized human bronchial epithelial cells. J. Virol. 81:12439–12449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhou B, et al. 2011. PB2 residue 158 is a pathogenic determinant of pandemic H1N1 and H5 influenza A viruses in mice. J. Virol. 85:357–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zimmerman TL, Thevananther S, Ghose R, Burns AR, Karpen SJ. 2006. Nuclear export of retinoid X receptor alpha in response to interleukin-1beta-mediated cell signaling: roles for JNK and SER260. J. Biol. Chem. 281:15434–15440 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.