Abstract
The V protein of Sendai virus (SeV) suppresses innate immunity, resulting in enhancement of viral growth in mouse lungs and viral pathogenicity. The innate immunity restricted by the V protein is induced through activation of interferon regulatory factor 3 (IRF3). The V protein has been shown to interact with melanoma differentiation-associated gene 5 (MDA5) and to inhibit beta interferon production. In the present study, we infected MDA5-knockout mice with V-deficient SeV and found that MDA5 was largely unrelated to the innate immunity that the V protein suppresses in vivo. We therefore investigated the target of the SeV V protein. We previously reported interaction of the V protein with IRF3. Here we extended the observation and showed that the V protein appeared to inhibit translocation of IRF3 into the nucleus. We also found that the V protein inhibited IRF3 activation when induced by a constitutive active form of IRF3. The V proteins of measles virus and Newcastle disease virus inhibited IRF3 transcriptional activation, as did the V protein of SeV, while the V proteins of mumps virus and Nipah virus did not, and inhibition by these proteins correlated with interaction of each V protein with IRF3. These results indicate that IRF3 is important as an alternative target of paramyxovirus V proteins.
INTRODUCTION
The family Paramyxoviridae includes numerous human and animal pathogens such as measles virus (MeV), mumps virus (MuV), respiratory syncytial virus, canine distemper virus, Newcastle disease virus (NDV), Nipah virus (NiV), and Hendra virus. Paramyxoviruses possess accessory proteins that are not essential for minimal virus growth but are essential for efficient and pathogenic infection. Sendai virus (SeV), which belongs to the family Paramyxoviridae, is a respiratory tract pathogen of rodents and has accessory proteins C and V. The SeV genome, which is approximately 15.4 kb, comprises six genes, N, P, M, F, HN, and L, and the genes individually encode one polypeptide. The P gene exceptionally encodes the C and V proteins as well as the P protein (27, 32).
The C proteins, C′, C, Y1, and Y2, are synthesized from the colinear transcript of the P gene in an open reading frame (ORF) shifted from that of the P protein by alternative translational starts and a common stop codon. The C proteins have multiple functions, including interruption of the Jak/Stat pathway to inhibit interferon (IFN)-responsive gene activation (8, 10), inhibition of apoptosis (13, 26), viral RNA synthesis regulation (2, 5, 6, 14, 45), and assistance with virus particle formation (15, 38, 41). Inhibition of apoptosis may be linked to proper regulation of viral RNA synthesis by viral polymerase with the assistance of the C protein (13, 43).
The V protein is synthesized from an additional mRNA, which is generated from the P gene by inserting a pseudotemplated G residue at the specific editing site in the middle of the gene (46, 47). Consequently, the P and V proteins share the same 317 residues at the amino terminus (P/V common region), and the V protein has a unique 67-residue carboxyl terminus (Vu region). The Vu region contains 15 highly conserved amino acids in almost all of the members of the subfamily Paramyxovirinae. V-deficient SeV (SeV V−) was generated by introducing mutations at the RNA editing site of the P gene (17). SeV V− efficiently propagated in cultured cells, indicating that the V protein was a nonessential accessory protein. However, SeV V− was cleared from mouse lungs at an early stage of infection (17). SeV V− is remarkably attenuated in virulence in mice, indicating the importance of the V protein in SeV pathogenesis in vivo (reviewed in references 31, 32, and 37).
On the other hand, SeV V− propagated as efficiently as wild-type (WT) SeV in interferon regulatory factor 3 (IRF3)-knockout (KO) mice (23). The SeV V protein is thus thought to counteract an early anti-SeV innate immunity through signal transduction via IRF3. However, the innate immunity counteracted by the V protein was presumed to not involve IFNs, because large amounts of type I IFNs were produced even in IRF3-KO mice and because clearance of SeV V− was also observed in IFN α/β receptor-KO mice and Stat-1-KO mice (23).
Paramyxovirus V proteins, including the V protein of SeV, have been shown to interact with an intracellular viral RNA sensor, melanoma differentiation-associated gene 5 (MDA5), and inhibit the downstream IRF3 and beta IFN (IFN-β) activation in cultured cells (1, 3, 4, 50). We have recently analyzed the interaction of melanoma differentiation-associated gene 5 (MDA5) with V proteins derived from SeV mutants with different pathogenicities and have shown that SeV pathogenicity appears to be related to interaction of the V protein with MDA5 (36). It has also been reported that MDA5 is involved in activation of innate immunity in mice (9). On the other hand, infection experiments using gene-knockout mice revealed that infection of SeV is recognized by another intracellular RNA sensor, retinoic acid-inducible gene I (RIG-I), and not by MDA5 in cultured cells (19, 30) and in mice (20). Therefore, it is unknown whether the interaction between the SeV V protein and MDA5 has significance in V-mediated SeV pathogenesis in vivo.
In the present study, we first investigated the growth and pathogenicity of SeV V− in RIG-I- or MDA5-knockout mice and found that MDA5 was probably not a critical target of the V protein in vivo. We then focused on interaction of the V protein with IRF3 and found that the V protein inhibited translocation of activated IRF3 into the nucleus, and we further investigated the inhibition of IRF3 by the V proteins of other paramyxoviruses.
MATERIALS AND METHODS
Cells and viruses.
LLC-MK2 cells (rhesus monkey kidney-derived cells), 293T cells (human renal epithelial cells expressing the simian virus 40 large T antigen), L929 cells (mouse fibroblast-like cells), and HeLa cells (human cervical cancer-derived cells) were propagated in 10% fetal calf serum containing Dulbecco's minimum essential medium (DMEM). Mouse embryonic fibroblasts (MEFs) were prepared from mouse fetuses or newborn mice of gene-knockout mice as described previously (22).
SeV Z strain and V-deficient SeV (SeV V−) with mutations at the RNA editing site (17) were propagated in embryonated chicken eggs. Virus infectivity was measured by an immunofluorescent infectious focus assay using LLC-MK2 cells and expressed by the number of cell-infecting units (CIU)/ml (24).
Mice.
MDA5-KO mice in the C57BL/6 mouse background and RIG-I-KO mice in the ICR background were provided by S. Koike (Tokyo Metropolitan Institute for Neuroscience, Japan). MDA5/RIG-I-double-KO mice in the ICR background were obtained by mating MDA5−/− and RIG-I+/− mice in the ICR background and by selecting double-KO mice from the F1 progeny with PCR genotyping. MDA5−/− and RIG-I+/− ICR mice were provided by O. Takeuchi (Research Institute for Microbial Diseases, Osaka University, Japan). IRF3-KO mice were provided by Riken Bioresources Bank as described previously (23). C57BL/6 and ICR/Crj (CD-1) mice were purchased from Charles River Laboratories Japan, Ltd. (Atsugi, Japan).
Infection of mice with SeV.
Three-week-old mice were intranasally inoculated with 106 CIU of SeV per mouse under mild anesthesia with ether, and their body weights and clinical symptoms were checked daily. Two mice were sacrificed at certain time intervals, their lungs were homogenized in 1 ml of minimal essential medium (MEM) per mouse, and then virus infectivity was measured. Alternatively, four or five mice in a group were inoculated with 103, 104, 105, 106, 107, or 108 CIU of SeV per mouse and observed over a period of 2 weeks. On the basis of the lethality for mice, 50% mouse lethal doses (MLD50s) were calculated as described previously (11).
Infection experiments were performed under physical containment of level 3 at the animal facility in the Natural Science Center for Basic Research and Development, Hiroshima University, and were carried out in strict accordance with the Guidelines for Animal Experimentation of the Japanese Association for Laboratory Animal Science. The protocol was approved by the Committee on the Ethics of Animal Experiments of Hiroshima University. All efforts were made to minimize suffering.
Plasmids.
pCAG-V, an expression plasmid for the full-length SeV V protein, was generated by a standard PCR technique using pSeV(+)-4C(−), which was provided by A. Kato, as a template. The intrinsic ORF of the C protein in pCAG-V is disrupted, and no C proteins are generated from pCAG-V. For expression of SeV truncated V proteins, pCAG-P/V and pCAG-myc-Vu were constructed from pKS-P/V and pKS-myc-Vu (36), respectively. The full-length cDNA clone of NiV V protein was provided by L.-F. Wang (CSIRO Livestock Industries, Australia) through the courtesy of B. Gotoh (Shiga University of Medical Science, Japan). The cDNA of MeV V protein was constructed by two-step PCR from the genomic cDNA of the IC-B strain, which was provided by K. Takeuchi (Tsukuba University, Japan): the P/V common region and the V unique region were individually amplified in the reverse transcription (RT) and the 1st-step PCR, and the two fragments were connected in the 2nd-step PCR. The full-length cDNA fragment of the V protein of NDV was generated by RT and two-step PCR using genomic RNA purified from the NDV Herts strain as a template. The full-length cDNA fragment of the V protein of MuV was generated by RT-PCR using genomic RNA purified from the MuV RW strain as a template. For V cDNA of NiV, MeV, and NDV, a myc tag was attached at the N terminus by using PCR. These cDNAs were then subcloned into the pCAGGS.MCS vector under the chicken β-actin promoter (34). The resultant plasmids were designated pCAG-myc-NiV-V, pCAG-myc-MeV-V, pCAG-MuV-V, and pCAG-myc-NDV-V, respectively.
Protein expression plasmids pCAG-FL-RIG-I and pCAG-FL-MDA5 were described previously (36). An expression plasmid for a C-terminally truncated FLAG-tagged RIG-I (residues 1 to 129) was constructed to generate pCAG-FL-RIG-I-CA (the constitutive active [CA] form) according to a previous report (51). An expression plasmid for a constitutive active form of FLAG-tagged Toll–interleukin-1 receptor domain-containing adaptor-inducing β-IFN (TRIF) possessing an N-terminal 541-amino-acid region (pCAG-FL-TRIF-CA) was constructed by a standard PCR technique using pEFneo-hTRIF (25) as a template according to a report by Yamamoto et al. (48). The full-length cDNA clone of IRF3 was amplified from mRNA of 293T cells by using an RT-PCR technique with specific primers and simultaneously adding a hemagglutinin (HA) tag at the N terminus and subcloned into the pCAGGS vector. The cDNA was processed for three rounds of in vitro mutagenesis to generate IRF3-5D (S396D, S398D, S402D, T404D, and S405D) by using an AMAP multisite-directed mutagenesis kit (Amalgaam, Tokyo, Japan) following the manufacturer's instructions. A reporter plasmid, p-55C1B-EGFP, that has 8 tandem IRF3-binding motifs upstream of the enhanced green fluorescent protein (EGFP) gene was described previously (36).
Antibodies.
Mouse monoclonal antibodies against the HA tag (HA.C5; Applied Biological Materials), FLAG tag (M2; Sigma-Aldrich), myc tag (9E10; Santa Cruz Biotechnology), green fluorescent protein (GFP; sc8334; Santa Cruz Biotechnology), and mouse actin (MA1501; Chemicon International) and rabbit polyclonal antibodies against the myc tag (sc788; Santa Cruz Biotechnology) and human IRF3 (sc9082, Santa Cruz Biotechnology) were employed following each of the manufacturer's protocols. Rabbit antiserum against purified SeV P protein was provided by A. Kato (National Institute of Infectious Diseases, Japan), and that against MuV P protein was provided by K. Takeuchi (Tsukuba University, Japan). Mouse monoclonal antibody against SeV N protein was provided by E. Suzuki (National Institute of Infectious Diseases, Japan). Rabbit serum against SeV M protein was described previously (12), and that against purified SeV particles was also described previously (24). Alexa Fluor 488-conjugated anti-mouse IgG and Alexa Fluor 546-conjugated anti-rabbit IgG goat polyclonal antibodies (Invitrogen) and horseradish peroxidase (HRP)-conjugated anti-mouse IgG and HRP-conjugated anti-rabbit IgG goat polyclonal antibodies (Santa Cruz Biotechnology) were used according to the protocols of the suppliers. An enzyme-linked immunosorbent assay (ELISA) kit was used to measure mouse IFN-β (VeriKine mouse IFN-β ELISA kit; PBL Biomedical Laboratories, Piscataway, NJ).
IP-Western blotting.
Immunoprecipitation (IP)-Western blotting was performed basically as described previously (16). Briefly, 293T cells cultured in 6-well plates were cotransfected with the indicated plasmids. After 24 h, cells were solubilized in cell lysis buffer (0.5% NP-40, 20 mM Tris-HCl [pH 7.4], 150 mM NaCl). Cell lysates were then immunoprecipitated with either anti-P or anti-myc antibody to precipitate SeV V protein, its truncated mutants, and other paramyxovirus V proteins. The immunoprecipitates were separated by SDS-PAGE, followed by Western blotting using anti-HA or anti-FLAG antibody to detect coprecipitated host proteins. Cell lysates were also subjected directly to Western blotting with individual antibodies to confirm expression of proteins. An experiment using a reverse combination of antibodies was also performed.
Immunofluorescent imaging.
Immunofluorescent staining was performed basically as described previously (15). HeLa cells cultured in 6-well plates containing glass coverslips were transfected with the indicated plasmids. After 24 h, cells were fixed, permeabilized, and then stained using the indicated antibodies as primary antibodies and Alexa Fluor 546-conjugated anti-rabbit IgG and/or Alexa Fluor 488-conjugated anti-mouse IgG antibodies as secondary antibodies. The coverslips were mounted on glass slides and observed using a Zeiss LSM 5 confocal microscope (Carl Zeiss).
IRF3 reporter assay.
The IRF3 reporter assay was performed as described previously (36). Briefly, subconfluent 293T cells in 6-well plates were transfected with p-55C1B-EGFP, an IRF3 signal-inducing plasmid such as pCAG-FL-MDA5, pCAG-FL-RIG-I-CA, pCAG-FL-TRIF-CA, or pCAG-HA-IRF3-5D, and an expression plasmid for the V protein. After 24 h, cell lysates were prepared and processed for Western blotting by using anti-GFP antibody. Cell lysates were also subjected to Western blotting with antibodies against the individual proteins to confirm protein expression.
Poly(I·C) treatment and immunofluorescence microscopy.
HeLa cells cultured on glass coverslips were infected with SeV mutants at an input multiplicity of infection (MOI) of 5. After 1 h, inocula were removed and cells were washed with phosphate-buffered saline (PBS) three times and incubated with serum-free DMEM. At 12 h postinfection, the culture medium was replaced with fresh serum-free DMEM, and 5 μg of poly(I·C) (GE Healthcare) was transfected using Lipofectamine 2000 (Invitrogen). After an additional 6 h, cells were fixed with the 0.5% formaldehyde solution and treated with 0.1% Triton X-100 in PBS. Cells were then stained using a monoclonal antibody against SeV N and a polyclonal antibody against human IRF3 as primary antibodies and Alexa Fluor 546-conjugated anti-mouse IgG and Alexa Fluor 488-conjugated anti-rabbit IgG goat polyclonal antibodies as secondary antibodies. Coverslips were mounted on glass slides and observed using a confocal microscope.
RESULTS
Propagation and pathogenesis of SeV V− in MDA5-, RIG-I-, and RIG-I/MDA5-KO mice.
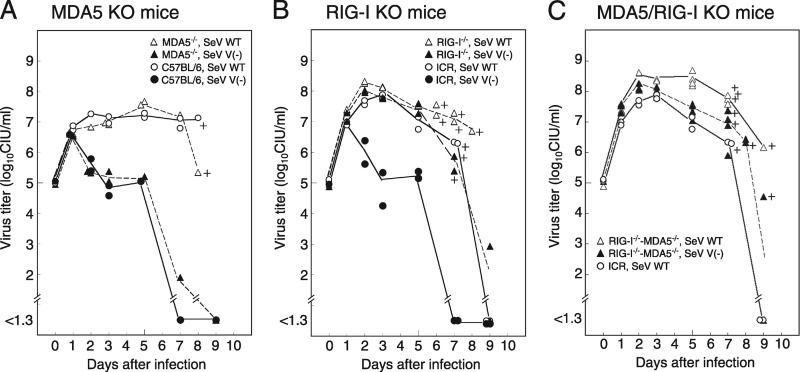
Mice were infected with SeV WT or SeV V− and sacrificed at 0 to 9 days postinfection to investigate virus growth in the lungs (Fig. 1A to C). In MDA5-KO mice, SeV WT replicated efficiently in the lungs, while SeV V− had not replicated at 2 days postinfection or later. These patterns were similar to those in parent C57BL/6 mice (Fig. 1A). In RIG-I-KO mice, however, SeV V− could replicate as efficiently as SeV WT (Fig. 1B). In RIG-I- and MDA5-double-KO mice, the growth of SeV V− was similar to that in RIG-I-KO mice (Fig. 1C).
Fig 1.
SeV V− propagation in lungs from gene-knockout mice. (A to C) Three-week-old gene-knockout mice and their parent strains, MDA5-KO C57BL/6 mice (A), RIG-I-KO ICR mice (B), and MDA5/RIG-I-KO ICR mice (C), were intranasally inoculated with 106 CIU of SeV WT or SeV V− per mouse and were sacrificed at certain time intervals. Their lungs were homogenized in 1 ml of MEM, followed by measurement of virus infectivity. Average infectivity is plotted in the graph, and a plus sign indicates death of a mouse.
The MLD50 of SeV V− in C57BL/6 mice was almost 30 times higher than that of SeV WT in C57BL/6 mice, indicating attenuation of the virulence of SeV V− compared with that of SeV WT (Table 1, upper half). Comparison of the MLD50 values in C57BL/6 and MDA5-KO mice showed that the MLD50 values of SeV V− were almost the same (ratio of 1:1.5, as shown in parentheses in Table 1). This corresponds to similar virus replication in mouse lungs (Fig. 1A).
Table 1.
MLD50s of SeV V− in gene-knockout micea
| Mouse strain | MLD50 (no. of CIU/mouse) |
|
|---|---|---|
| SeV WT | SeV V− | |
| C57BL/6 | 3.2 × 105 (1) | 1.0 × 107 (1) |
| C57BL/6 MDA5−/− | 3.2 × 105 (1) | 1.5 × 107 (1.5) |
| ICR | 7.9 × 105 (1) | 1.3 × 108 (1) |
| ICR RIG-I−/− | 5.0 × 103 (1/158) | 3.2 × 104 (1/4,063) |
| ICR RIG-I−/− MDA5−/− | 3.2 × 103 (1/247) | 3.2 × 104 (1/4,063) |
Four or five 3-week-old mice in a group were intranasally inoculated with 103, 104, 105, 106, 107, or 108 CIU of SeV per mouse and observed for 2 weeks. The MLD50 values were calculated on the basis of the lethality for the mice. The numbers in parentheses are ratios of the MLD50 in gene-knockout mice to that in wild-type mice (C57BL/6 or ICR) in the same column.
In ICR mice, the MLD50 of SeV V− was higher than that of SeV WT, also indicating attenuation of the virulence of SeV V− (Table 1, lower half). In RIG-I-KO mice, the MLD50 of SeV WT was ca. 150 times smaller than that in ICR mice. These results indicate a negative effect of RIG-I on SeV V− replication. The MLD50 of SeV V− in RIG-I-KO mice was ca. 4,100 times smaller than the MLD50 of SeV V− in ICR mice (Table 1). These results suggest that the immunity that the V protein restricts is largely induced by RIG-I and not MDA5 in vivo.
On the other hand, when SeV WT replication was compared in RIG-I-KO and MDA5/RIG-I-KO mice (Fig. 1B and C), the peak titer of SeV WT was higher at 4 and 5 days postinfection in the lungs of MDA5/RIG-I-KO mice, suggesting a role of MDA5 in the suppression of SeV WT growth. The MLD50 value of SeV WT in MDA5/RIG-I-KO mice was slightly smaller than that in RIG-I-KO mice (Table 1).
IFN-β secretion from MEFs derived from RIG-I-KO and MDA5-KO mice infected with SeV.
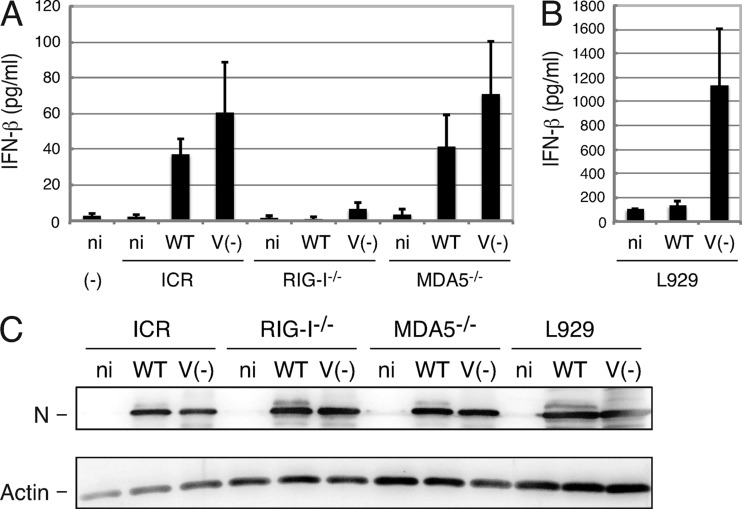
MEFs were prepared from gene-knockout mice for RIG-I and MDA5 and infected with SeV V− or SeV WT at an input MOI of 10. Mouse IFN-β in the culture medium was measured by using ELISA at 24 h postinfection (Fig. 2A). IFN-β secretion was restricted in RIG-KO mouse-derived cells. There was a tendency for IFN-β secretion to be greater in SeV V− infection than in SeV WT infection in MDA5-KO and parental ICR-derived cells. In mouse fibroblast-like cells, L929 cells, IFN-β secretion in SeV V− infection was much greater than that in SeV WT infection (Fig. 2B). Western blotting showed similar replication of both viruses in these cells (Fig. 2C). Secretion of IFN-β was not suppressed in SeV V−-infected cultured cells. The V protein may suppress a point in the pathway starting from RIG-I to IRF3 activation and IFN-β secretion.
Fig 2.
IFN-β secretion from MEFs derived from RIG-I-KO and MDA5-KO mice infected with SeV. MEFs derived from ICR, RIG-I-KO, and MDA5-KO mice or L929 cells were infected with SeV WT or SeV V− at an input MOI of 10, and after 24 h, the culture medium and cell lysates were harvested. (A and B) IFN-β in the medium was measured by using ELISA. Average values of three dishes were plotted in the graph. Error bars represent standard deviation. ni, mock infected; (−), no cells. (C) A set of cell lysates was processed for immunoblotting using anti-SeV antiserum, and the N protein bands are shown. Anti-actin antibody was also used for immunoblotting.
Interactions of SeV V protein with IRF3, RIG-I, and MDA5.
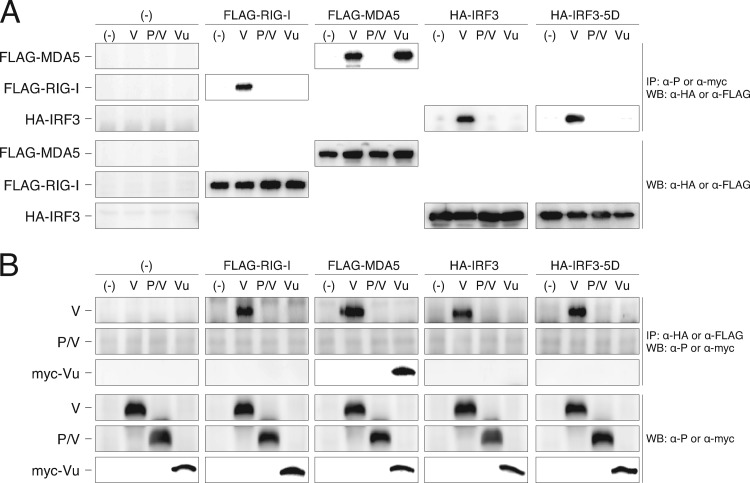
We previously showed interactions of multiple molecules, including RIG-I, MDA5, and IRF3, with the V protein by coimmunoprecipitation assays (36). Here we reproduced those results showing that the SeV V protein physically interacted with RIG-I and IRF3 as well as MDA5 by IP-Western blotting (Fig. 3). A constitutive active form of IRF3, IRF3-5D, also interacted with the V proteins in this overexpression experiment (Fig. 3). When the truncated forms, the N-terminal P/V common region (P/V) and C-terminal V unique region (Vu), were investigated, MDA5 also interacted with the Vu region of the V protein, indicating that interaction of the V protein with MDA5 is Vu dependent. The other proteins, however, appeared to require the entire V protein (P/V plus Vu) for the interaction.
Fig 3.
Interactions of SeV V proteins with RIG-I, MDA5, IRF3, and IRF3-5D revealed by IP-Western blotting. (A) 293T cells were cotransfected with the plasmids indicated at the top of the panel. After 24 h, cells were suspended in cell lysis buffer (0.5% NP-40, 20 mM Tris-HCl [pH 7.4], 150 mM NaCl) and then immunoprecipitated with either anti-P or anti-myc antibody to precipitate SeV V protein and its truncated mutants. The immunoprecipitates were separated by SDS-PAGE, followed by Western blotting using anti-HA or anti-FLAG antibody to detect coprecipitated epitope-tagged host proteins, FLAG-RIG-I, FLAG-MDA5, HA-IRF3, and HA-IRF3-5D. Cell lysates were also subjected to Western blotting to confirm expression of proteins. (B) An experiment using a reverse combination of antibodies was also performed.
Intracellular localization of SeV V protein and IRF3, IRF3-5D, RIG-I, and MDA5.
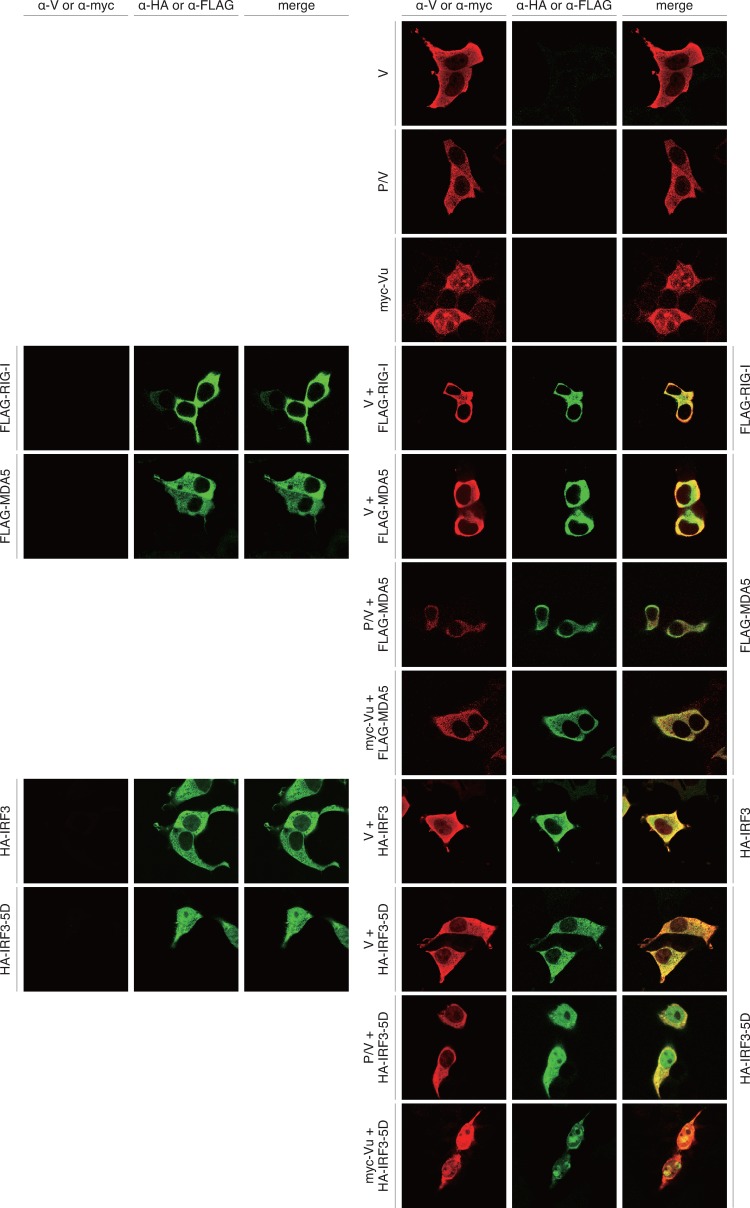
Protein expression and immunofluorescent microscopy demonstrated intracellular localization of the V protein and IRF3-related host factors (Fig. 4). When only the V protein was expressed, it appeared to be dispersed in the cytosol. Truncated V proteins, P/V and Vu, localized in the cytosol and in both the cytosol and the nucleus, respectively. IRF3, RIG-I, and MDA5 localized in the cytosol in the case of expression of an individual protein. IRF3-5D, an active form of IRF3, was found in the nucleus as well as the cytosol (Fig. 4).
Fig 4.
Intracellular localization of SeV V proteins and RIG-I, MDA5, IRF3, and IRF3-5D. HeLa cells on glass coverslips were transfected with the indicated plasmids. After 24 h, cells were fixed, permeabilized, and then stained using the indicated antibodies as primary antibodies and Alexa Fluor 546-conjugated anti-rabbit IgG and/or Alexa Fluor 488-conjugated anti-mouse IgG antibodies as secondary antibodies. The coverslips were mounted on glass slides and observed using a Zeiss LSM 5 confocal microscope.
In coexpression of the V protein and one of the host factors, IRF3, RIG-I, and MDA5, both proteins appeared to be colocalized in the cytosol. IRF3-5D, which was found in the nucleus in the case of single expression, was observed only in the cytosol in the case of coexpression with the V protein. The V protein appeared to retain IRF3-5D in the cytosol by a physical interaction between them. In coexpression of MDA5 and RIG-I with the V protein, a relatively intense yellow color, indicating colocalization, appeared.
In coexpression of truncated Vu with MDA5, the Vu protein was exclusively retained in the cytosol probably by interaction with MDA5. In coexpression of Vu and IRF3-5D and that of P/V and IRF3-5D, protein localization appeared to reflect localization of each protein when expressed individually.
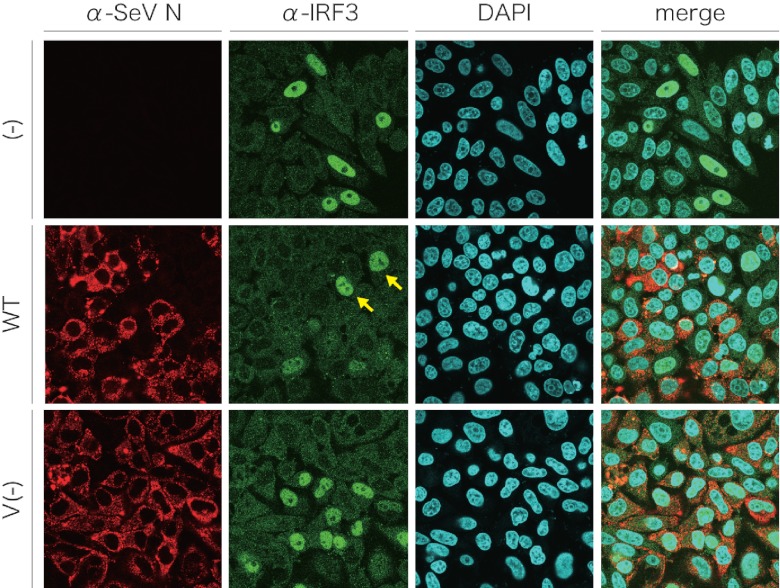
HeLa cells were infected with SeV and transfected with poly(I·C), and the distribution of native IRF3 was observed (Fig. 5). IRF3 accumulated in the nucleus of some uninfected cells [Fig. 5, (−)]. One reason that not all of the cells showed nuclear accumulation of IRF3 was probably a low transfection efficiency of poly(I·C). In SeV WT-infected cells, nuclear staining of IRF3 was not observed, and some cells with nuclear staining were uninfected cells (Fig. 5, WT, yellow arrows). In contrast, IRF3 nuclear staining was observed in SeV V−-infected cells (Fig. 5, V−). These results suggest that the V protein also retained IRF3 in the cytosol in SeV-infected cells.
Fig 5.
Subcellular distribution of cellular IRF3 in SeV-infected cells after induction by poly(I·C). HeLa cells were mock infected [(−)] or infected with SeV WT or SeV V−. At 12 h postinfection, cells were transfected with 5 μg of poly(I·C) and incubated for another 6 h. Cells were fixed, permeabilized, and then stained with anti-IRF3 polyclonal antibody and anti-SeV N monoclonal antibody containing 4′,6-diamino-2-phenylindole (DAPI), together with Alexa Fluor 546-conjugated anti-mouse IgG and Alexa Fluor 488-conjugated anti-rabbit IgG goat polyclonal antibodies. Cells were observed under a Zeiss LSM 5 confocal microscope. Yellow arrows, nuclei of poly(I·C)-treated and uninfected cells.
Effect of SeV V protein on IRF3 activation.
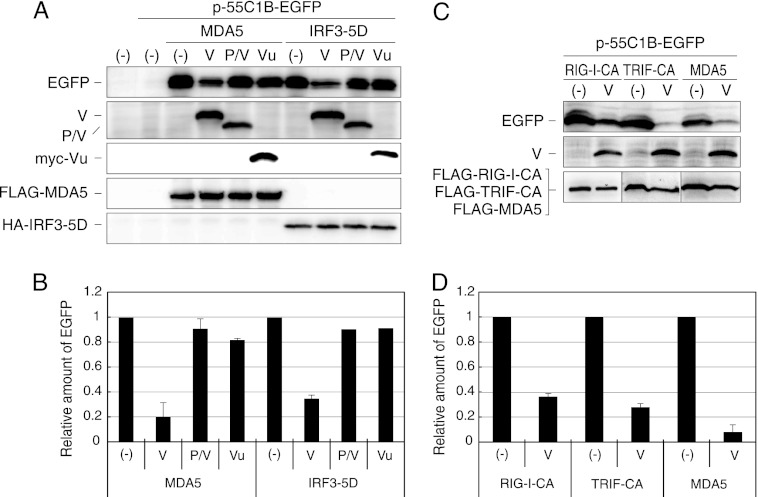
IRF3 activation in the presence of the V protein was investigated by using a reporter plasmid, p-55C1B-EGFP (Fig. 6). Here we focused on the interaction of IRF3 with the V protein, since IRF3 is in the most downstream location in this signal transduction pathway and since translocation of IRF3-5D to the nucleus was likely to be inhibited by the V protein (Fig. 4).
Fig 6.
IRF3 reporter assay in the presence of V proteins. (A) 293T cells were transfected with p-55C1B-EGFP together with pCAG-FL-MDA5 or pCAG-IRF3-5D and a plasmid expressing the V, P/V, or myc-Vu protein, as indicated. After 24 h, cells were solubilized and processed for Western blotting using anti-GFP antibody (EGFP), anti-P antibody (V, P/V), anti-myc antibody (myc-Vu), anti-FLAG antibody (FLAG-MDA5), or anti-HA antibody (HA-IRF3-5D). (B) The density of EGFP bands was quantitated and plotted in a graph. The value is the ratio against that without V protein [(−)]. Experiments were repeated at least three times, and the average value is shown. Error bars represent standard deviation. (C and D) 293T cells were transfected with p-55C1B-EGFP together with pCAG-FL-RIG-I-CA, pCAG-FL-TRIF-CA, or pCAG-FL-MDA5 and a plasmid expressing the V protein. After 24 h, cells were solubilized and processed for Western blotting, and the density of EGFP bands was quantitated and plotted in a graph as was done for panel B.
Overexpression of MDA5 induced IRF3 activation (Fig. 6A and B). The activation was reduced in the presence of the V protein, was slightly reduced in the presence of the Vu protein, and was not reduced in the presence of the P/V region (Fig. 6A and B) (36). The slight suppression of IRF3 activation by Vu may be due to the interaction of Vu with MDA5.
When the signal was induced by IRF3-5D, the IRF3 activation was restricted in the presence of the V protein but not in the presence of the P/V and Vu proteins. Inhibition of the IRF3-5D-induced IRF3 transcriptional activation by the V protein appeared to correspond to the restriction of nuclear localization of IRF3-5D by coexpression with the V protein (Fig. 4).
When the signal was induced by a constitutive active form of RIG-I, RIG-I-CA, or a constitutive active form of TRIF, TRIF-CA, the V protein also inhibited IRF3 activation, though the extent of inhibition appeared to be less than that from overexpression of MDA5 (Fig. 6C and D). This result suggests that the V protein suppresses IRF3 activation even if the signal is induced by a distinct RNA sensor, RIG-I, Toll-like receptor 3 (TLR3), or MDA5.
Interactions of paramyxovirus V proteins with IRF3, RIG-I, and MDA5.
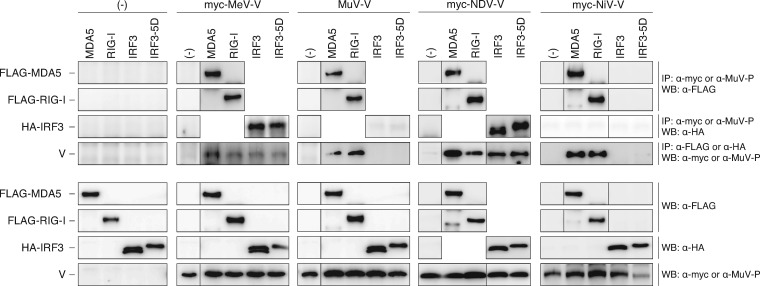
We next investigated interactions of the V proteins of other paramyxoviruses, MeV, MuV, NiV, and NDV, with host factors (Fig. 7). IP-Western blotting demonstrated that all of the V proteins investigated interacted with both MDA5 and RIG-I. However, results of binding with IRF3 and IRF3-5D were different. The V proteins of MuV and NiV did not interact with either IRF3 or IRF3-5D, while the V proteins of MeV and NDV interacted with both IRF3 and IRF3-5D.
Fig 7.
Interactions of paramyxovirus V proteins with RIG-I, MDA5, IRF3, and IRF3-5D as revealed by IP-Western blotting. 293T cells were cotransfected with the indicated plasmids. After 24 h, cells were suspended in cell lysis buffer and then immunoprecipitated with anti-myc or anti-MuV P antibody to precipitate paramyxovirus V proteins. The immunoprecipitates were separated by SDS-PAGE, followed by Western blotting using anti-FLAG or anti-HA antibody to detect coprecipitated epitope-tagged host proteins, FLAG-MDA5, FLAG-RIG-I, HA-IRF3, and HA-IRF3-5D. Cell lysates were also subjected to Western blotting with individual antibodies to confirm expression of proteins. An experiment using a reverse combination of antibodies was also performed.
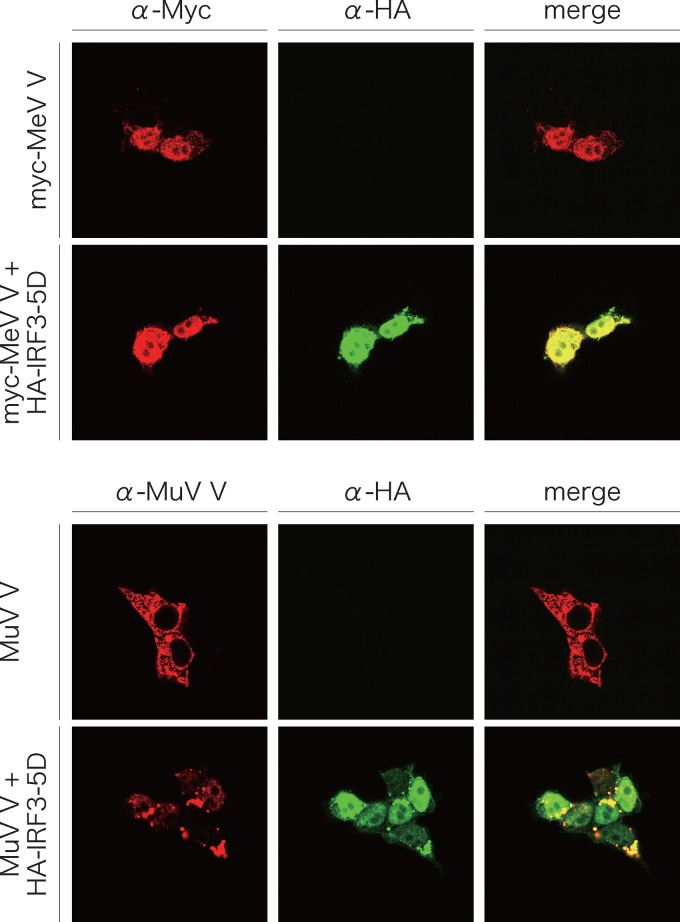
Immunofluorescent staining demonstrated that the MuV V protein was present in the cytosol and that IRF3-5D was present in the nucleus (Fig. 4 and 8). Even with coexpression of the two proteins, the subcellular localization in the case of expression of an individual protein did not change. The MeV V protein was unexpectedly found in the nucleus, as was IRF3-5D when expressed alone. In coexpression with IRF3-5D, the two proteins appeared to colocalize in the nucleus (Fig. 8).
Fig 8.
Intracellular localization of MeV V or MuV V proteins and IRF3-5D. HeLa cells on glass coverslips were transfected with the indicated plasmids. After 24 h, cells were fixed and permeabilized and then stained as described in the legend to Fig. 4.
Effects of paramyxovirus V proteins on IRF3 activation.
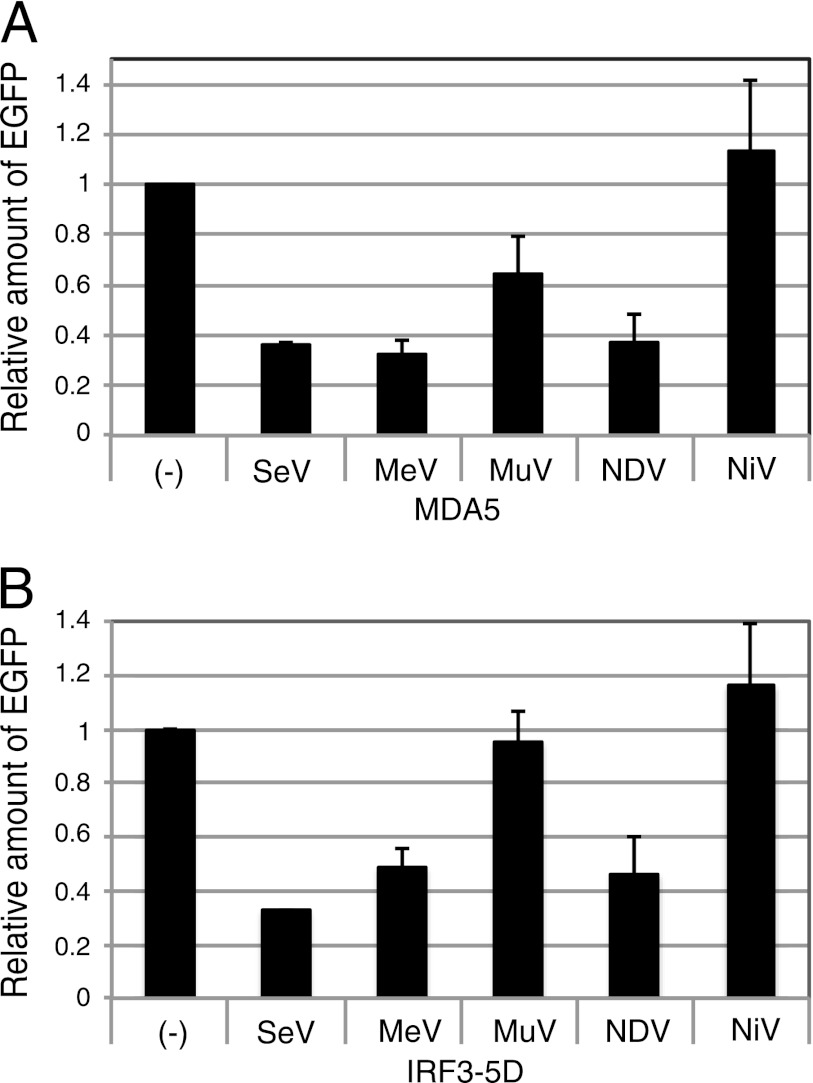
In reporter assays, the V proteins of SeV, MeV, MuV, and NDV restricted the IRF3 activation when the signal was induced by overexpression of MDA5. However, the NiV V protein did not restrict the IRF3 activation (Fig. 9A), though the NiV V protein interacted with MDA5 (Fig. 5). When the signal was induced by IRF3-5D, the V proteins of SeV, MeV, and NDV inhibited IRF3 activation, while those of MuV and NiV did not (Fig. 9B). This inhibition of the IRF3-5D-induced IRF3 activation was correlated with interaction of the V proteins with IRF3-5D.
Fig 9.
IRF3 reporter assay in the presence of paramyxovirus V proteins. 293T cells were transfected with p-55C1B-EGFP together with pCAG-FL-MDA5 (A) or pCAG-HA-IRF3-5D (B) and a plasmid expressing the paramyxovirus V protein, as indicated. After 24 h, cells were solubilized and processed for Western blotting using an anti-GFP antibody. The density of EGFP bands was quantitated and plotted in a graph. The value is the ratio against that without V protein [(−)]. Experiments were repeated at least three times, and the average value is shown. Error bars indicate standard deviation.
DISCUSSION
The SeV V protein is essential for efficient virus growth in mouse lungs and for viral pathogenicity. The V protein counteracts innate immunity, and the immunity is exerted through activation of IRF3 (23). In the present study, we infected MDA5-KO mice, RIG-I-KO mice, and double-KO mice with SeV V− in order to determine whether the immunity related to the SeV V protein was induced by RIG-I or by MDA5. The growth of SeV V− was restricted in MDA5-KO mice as well as in the parental wild-type mice. In contrast, SeV V− efficiently propagated in RIG-I-KO and double-KO mice. Virus pathogenicity revealed by measurement of the MLD50 corresponded to virus propagation in mouse lungs. IRF3-KO mice and RIG-I-KO mice allow growth of SeV V− in the lungs and are highly vulnerable to SeV V−. It is thus thought that the innate immunity that the V protein counteracts is induced by detection of virus infection with RIG-I and subsequent signal transduction through IRF3.
Involvement of MDA5 in induction of innate immunity in SeV infection has been suggested by a study of infection of MDA5-KO mice with SeV (9). It has been demonstrated that the V protein, especially the C-terminal Vu domain, is essential for counteracting antiviral innate immunity and that it facilitates virus growth in mouse lungs (7, 11, 18). We have investigated the interaction between MDA5 and the SeV V proteins that have mutations at the Vu region, and we found that the interaction correlated well with virus pathogenesis (36). When the time courses of SeV WT replication were compared in RIG-I-KO and MDA5/RIG-I-KO mice, the SeV WT replicated more efficiently at 4 and 5 days postinfection in the lungs of MDA5/RIG-I-KO mice (Fig. 1B and C). Gitlin et al. (9) also observed efficient replication of SeV WT in the lungs of MDA5-KO mice, despite a difference in the virus strain used (Z strain versus Fushimi strain). It seems that MDA5 suppresses SeV WT replication. MDA5 may play a more significant role in triggering innate immunity to SeV in a situation such as the appearance of defective interfering (DI) genomes in SeV infection, since SeV containing DI genomes is recognized by MDA5 in dendritic cells (52).
However, the present work showed that RIG-I is more important for virus pathogenesis in relation to the SeV V protein. A profound effect of RIG-I in infection of mice with SeV was shown in a previous study (20). Then, the next question is how the V protein inhibits IRF3 activation. A search for V-interacting molecules revealed IRF3-activating molecules, including RIG-I, IκB kinase ε (IKKε), and IRF3, other than MDA5, as interacting partners in a coimmunoprecipitation assay (36) (Fig. 2), although this apparent stickiness of the V protein may be due to overexpression of proteins. Among the molecules identified, IRF3 was further investigated in the present study, since IRF3 is located at the most downstream site in the signal pathway. Even if molecules upstream of IRF3 are functionally disturbed by the V protein, IRF3 disturbance should be dominant. Although interaction of the V protein with RIG-I was interesting, we have not performed further investigation of this interaction. The SeV V protein was found to interact with IRF3 as well as its active form, IRF3-5D. Translocation of IRF3-5D into the nucleus appeared to be inhibited by trapping of IRF3-5D in the cytosol through its interaction with the SeV V protein. Therefore, the V protein inhibits IRF3 activation probably through interaction with IRF3. However, there is a possibility that the V protein interacts with a host protein that is downstream of IRF3 in the IRF3/IFN expression pathway.
In acute or chronic infection with SeV in cultured cells, it was shown that IRF3 was degraded and IFN-β production was restricted and that these phenomena were due to synthesis of the V protein (49). Translocation of IRF3 into the nucleus was inhibited in SeV WT-infected HeLa cells but not in SeV V−-infected cells at 18 h postinfection, and degradation of IRF3 was not prominent under this condition. However, since we did the experiment at only one time point, we cannot exclude the possibility of degradation of IRF3 by the V protein.
The V proteins of other paramyxoviruses, MeV, MuV, NDV, and NiV, all interacted with MDA5. Furthermore, almost all of the V proteins inhibited IRF3 activation induced by overexpression of MDA5. One exception was the NiV V protein, which did not inhibit IRF3 activation. The reason for this is unknown. Shaw et al. (40) reported that IRF3-responsive gene expression is inhibited by the NiV V protein under the condition of stimulation by SeV infection but not under the condition of stimulation by TLR3. The NiV V protein may inhibit a specific point in the IRF3 activation pathway. The V protein of wild-type MeV has been reported to bind to MDA5 and inhibit IFN-β induction (33). In contrast, the V protein of an MeV vaccine strain does not interact with MDA5 due to a single amino acid mutation of the V protein (42). The MeV V protein used in this study that interacted with MDA5 was derived from a wild-type MeV IC-B strain (44).
The V proteins of SeV, MeV, and NDV interacted with IRF3 and IRF3-5D and inhibited IRF3 activation induced by IRF3-5D. The SeV V protein was localized in the cytosol, and coexpressed IRF3-5D appeared to be trapped in the cytosol. The MeV V protein was localized in the nucleus, colocalizing with IRF3-5D. The interaction of the two proteins in the nucleus seems to suppress IRF3 transcription. On the other hand, the V proteins of MuV and NiV did not interact with IRF3 or IRF3-5D and did not inhibit IRF3 activation induced by IRF3-5D. The MuV V protein binds to IKKε and functions as a decoy of phosphorylation by TANK binding kinase-1 (TBK1)/IKKε, inhibiting IRF3 activation (29). The V protein of MuV restricts IRF3 activation at a point upstream of IRF3. Suppression of IRF3 by paramyxovirus V proteins depends on the virus species.
It has also been reported that IRF7, which is structurally and functionally similar to IRF3, is inhibited by paramyxovirus V proteins. In plasmatoid dendritic cells (pDCs), the MeV V protein binds to IKKα and is phosphorylated, resulting in inhibition of IRF7 phosphorylation (35). The MeV V protein also binds to IRF7 and inhibits its translocation to the nucleus (35). It has also been reported that paramyxovirus V proteins, including human parainfluenza virus type 2 (hPIV2), SeV, bovine parainfluenza virus type 3, MeV, and NiV, interact with IRF7, inhibiting the IFN-α promoter in a reconstituted pDC model (21). Our preliminary experiments also suggested that the SeV V protein inhibited IRF7-dependent gene expression (data not shown).
The MeV V protein binds to p65 (RelA) to suppress NF-κB activation (39). Furthermore, the PIV5 V protein inhibits IL-6 expression (28). The restriction of IRF7, NF-κB, and IL-6 is dependent on the Vu region, which is highly conserved among members of the subfamily Paramyxovirinae in the family Paramyxoviridae. Suppression of innate immunity by the V protein seems to be common in paramyxoviruses. The inhibition of IRF3 by paramyxovirus V proteins revealed in this study is a novel mechanism for the suppression of innate immunity.
ACKNOWLEDGMENTS
We thank S. Koike (National Center of Neurology and Psychiatry, Japan) for providing MDA5-KO mice and RIG-I-KO mice and O. Takeuchi (Research Institute for Microbial Diseases, Osaka University, Japan) for providing MDA5−/− RIG-I+/− ICR mice. We also thank A. Kato (National Institute of Infectious Diseases, Japan) for providing anti-SeV P antiserum and pSeV(+)-4C(−), K. Takeuchi (Tsukuba University, Japan) for providing anti-MuV P antiserum and genomic cDNA of the measles virus IC-B strain, E. Suzuki (National Institute of Infectious Diseases, Japan) for providing anti-SeV N monoclonal antibody, and L.-F. Wang (CSIRO Livestock Industries, Australia) and B. Gotoh (Shiga University of Medical Science, Japan) for providing the full-length cDNA clone of NiV V protein. We also thank Y. Ito (Chubu University, Japan) for providing the mumps virus RW strain and T. Komatsu (Aichi Medical University School of Medicine, Japan) for providing pEFneo-hTRIF. We also thank the staff of the animal facility in Natural Science Center for Basic Research and Development, Hiroshima University, and in the Research Center for Molecular Medicine and the Analysis Center of Life Science, Hiroshima University, for the use of their facilities.
This work was supported by grants-in-aid for scientific research from the Japan Society for the Promotion of Science.
Footnotes
Published ahead of print 24 April 2012
REFERENCES
- 1. Andrejeva J, et al. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. U. S. A. 101:17264–17269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cadd T, et al. 1996. The Sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter-specific fashion. J. Virol. 70:5067–5074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Childs K, et al. 2007. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology 359:190–200 [DOI] [PubMed] [Google Scholar]
- 4. Childs KS, Andrejeva J, Randall RE, Goodbourn S. 2009. Mechanism of mda-5 inhibition by paramyxovirus V proteins. J. Virol. 83:1465–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Curran J, Boeck R, Kolakofsky D. 1991. The Sendai virus P gene expresses both an essential protein and an inhibitor of RNA synthesis by shuffling modules via mRNA editing. EMBO J. 10:3079–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Curran J, Marq JB, Kolakofsky D. 1992. The Sendai virus nonstructural C proteins specifically inhibit viral mRNA synthesis. Virology 189:647–656 [DOI] [PubMed] [Google Scholar]
- 7. Fukuhara N, Huang C, Kiyotani K, Yoshida T, Sakaguchi T. 2002. Mutational analysis of the Sendai virus V protein: importance of the conserved residues for Zn binding, virus pathogenesis, and efficient RNA editing. Virology 299:172–181 [DOI] [PubMed] [Google Scholar]
- 8. Garcin D, Latorre P, Kolakofsky D. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 73:6559–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gitlin L, et al. 2010. Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo. PLoS Pathog. 6:e1000734 doi:10.1371/journal.ppat.1000734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gotoh B, et al. 1999. Knockout of the Sendai virus C gene eliminates the viral ability to prevent the interferon-alpha/beta-mediated responses. FEBS Lett. 459:205–210 [DOI] [PubMed] [Google Scholar]
- 11. Huang C, et al. 2000. Involvement of the zinc-binding capacity of Sendai virus V protein in viral pathogenesis. J. Virol. 74:7834–7841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Irie T, Inoue M, Sakaguchi T. 2010. Significance of the YLDL motif in the M protein and Alix/AIP1 for Sendai virus budding in the context of virus infection. Virology 405:334–341 [DOI] [PubMed] [Google Scholar]
- 13. Irie T, Nagata N, Igarashi T, Okamoto I, Sakaguchi T. 2010. Conserved charged amino acids within Sendai virus C protein play multiple roles in the evasion of innate immune responses. PLoS One 5:e10719 doi:10.1371/journal.pone.0010719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Irie T, Nagata N, Yoshida T, Sakaguchi T. 2008. Paramyxovirus Sendai virus C proteins are essential for maintenance of negative-sense RNA genome in virus particles. Virology 374:495–505 [DOI] [PubMed] [Google Scholar]
- 15. Irie T, Nagata N, Yoshida T, Sakaguchi T. 2008. Recruitment of Alix/AIP1 to the plasma membrane by Sendai virus C protein facilitates budding of virus-like particles. Virology 371:108–120 [DOI] [PubMed] [Google Scholar]
- 16. Irie T, Shimazu Y, Yoshida T, Sakaguchi T. 2007. The YLDL sequence within Sendai virus M protein is critical for budding of virus-like particles and interacts with Alix/AIP1 independently of C protein. J. Virol. 81:2263–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kato A, Kiyotani K, Sakai Y, Yoshida T, Nagai Y. 1997. The paramyxovirus, Sendai virus, V protein encodes a luxury function required for viral pathogenesis. EMBO J. 16:578–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kato A, et al. 1997. Importance of the cysteine-rich carboxyl-terminal half of V protein for Sendai virus pathogenesis. J. Virol. 71:7266–7272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kato H, et al. 2005. Cell type-specific involvement of RIG-I in antiviral response. Immunity 23:19–28 [DOI] [PubMed] [Google Scholar]
- 20. Kato H, et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105 [DOI] [PubMed] [Google Scholar]
- 21. Kitagawa Y, et al. 2011. A tryptophan-rich motif in the human parainfluenza virus type 2 V protein is critical for the blockade of Toll-like receptor 7 (TLR7)- and TLR9-dependent signaling. J. Virol. 85:4606–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kiyotani K, Sakaguchi T, Fujii Y, Yoshida T. 2001. Attenuation of a field Sendai virus isolate through egg-passages is associated with an impediment of viral genome replication in mouse respiratory cells. Arch. Virol. 146:893–908 [DOI] [PubMed] [Google Scholar]
- 23. Kiyotani K, Sakaguchi T, Kato A, Nagai Y, Yoshida T. 2007. Paramyxovirus Sendai virus V protein counteracts innate virus clearance through IRF-3 activation, but not via interferon, in mice. Virology 359:82–91 [DOI] [PubMed] [Google Scholar]
- 24. Kiyotani K, Takao S, Sakaguchi T, Yoshida T. 1990. Immediate protection of mice from lethal wild-type Sendai virus (HVJ) infections by a temperature-sensitive mutant, HVJpi, possessing homologous interfering capacity. Virology 177:65–74 [DOI] [PubMed] [Google Scholar]
- 25. Komatsu T, Takeuchi K, Gotoh B. 2007. Bovine parainfluenza virus type 3 accessory proteins that suppress beta interferon production. Microbes Infect. 9:954–962 [DOI] [PubMed] [Google Scholar]
- 26. Koyama AH, Irie H, Kato A, Nagai Y, Adachi A. 2003. Virus multiplication and induction of apoptosis by Sendai virus: role of the C proteins. Microbes Infect. 5:373–378 [DOI] [PubMed] [Google Scholar]
- 27. Lamb RA, Parks GD. 2006. Paramyxoviridae: the viruses and their replication, p 1449–1496 In Knipe DM, et al. (ed), Fields virology, 5th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 28. Lin Y, et al. 2007. Inhibition of interleukin-6 expression by the V protein of parainfluenza virus 5. Virology 368:262–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lu LL, Puri M, Horvath CM, Sen GC. 2008. Select paramyxoviral V proteins inhibit IRF3 activation by acting as alternative substrates for inhibitor of kappaB kinase epsilon (IKKe)/TBK1. J. Biol. Chem. 283:14269–14276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Melchjorsen J, et al. 2005. Activation of innate defense against a paramyxovirus is mediated by RIG-I and TLR7 and TLR8 in a cell-type-specific manner. J. Virol. 79:12944–12951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nagai Y. 1999. Paramyxovirus replication and pathogenesis. Reverse genetics transforms understanding. Rev. Med. Virol. 9:83–99 [DOI] [PubMed] [Google Scholar]
- 32. Nagai Y, Takakura A, Irie T, Yonemitsu Y, Gotoh B. 2011. Sendai virus: evolution from mouse pathogen to a state-of-the-art tool in virus research and biotechnology, p 115–173 In Samal SK. (ed), The biology of paramyxoviruses. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 33. Nakatsu Y, et al. 2008. Measles virus circumvents the host interferon response by different actions of the C and V proteins. J. Virol. 82:8296–8306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199 [DOI] [PubMed] [Google Scholar]
- 35. Pfaller CK, Conzelmann KK. 2008. Measles virus V protein is a decoy substrate for IkappaB kinase alpha and prevents Toll-like receptor 7/9-mediated interferon induction. J. Virol. 82:12365–12373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sakaguchi T, et al. 2011. Analysis of interaction of Sendai virus V protein and melanoma differentiation-associated gene 5. Microbiol. Immunol. 55:760–767 [DOI] [PubMed] [Google Scholar]
- 37. Sakaguchi T, Kato A, Kiyotani K, Yoshida T, Nagai Y. 2008. Studies on the paramyxovirus accessory genes by reverse genetics in the Sendai virus-mouse system. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 84:439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sakaguchi T, et al. 2005. AIP1/Alix is a binding partner of Sendai virus C protein and facilitates virus budding. J. Virol. 79:8933–8941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schuhmann KM, Pfaller CK, Conzelmann KK. 2011. The measles virus V protein binds to p65 (RelA) to suppress NF-kappaB activity. J. Virol. 85:3162–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shaw ML, Cardenas WB, Zamarin D, Palese P, Basler CF. 2005. Nuclear localization of the Nipah virus W protein allows for inhibition of both virus- and Toll-like receptor 3-triggered signaling pathways. J. Virol. 79:6078–6088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sugahara F, et al. 2004. Paramyxovirus Sendai virus-like particle formation by expression of multiple viral proteins and acceleration of its release by C protein. Virology 325:1–10 [DOI] [PubMed] [Google Scholar]
- 42. Takaki H, et al. 2011. Strain-to-strain difference of V protein of measles virus affects MDA5-mediated IFN-beta-inducing potential. Mol. Immunol. 48:497–504 [DOI] [PubMed] [Google Scholar]
- 43. Takeuchi K, Komatsu T, Kitagawa Y, Sada K, Gotoh B. 2008. Sendai virus C protein plays a role in restricting PKR activation by limiting the generation of intracellular double-stranded RNA. J. Virol. 82:10102–10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Takeuchi K, et al. 2002. Recombinant wild-type and Edmonston strain measles viruses bearing heterologous H proteins: role of H protein in cell fusion and host cell specificity. J. Virol. 76:4891–4900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tapparel C, et al. 1997. Inhibition of Sendai virus genome replication due to promoter-increased selectivity: a possible role for the accessory C proteins. J. Virol. 71:9588–9599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thomas SM, Lamb RA, Paterson RG. 1988. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell 54:891–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vidal S, Curran J, Kolakofsky D. 1990. Editing of the Sendai virus P/C mRNA by G insertion occurs during mRNA synthesis via a virus-encoded activity. J. Virol. 64:239–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yamamoto M, et al. 2002. Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J. Immunol. 169:6668–6672 [DOI] [PubMed] [Google Scholar]
- 49. Ye J, Maniatis T. 2011. Negative regulation of interferon-beta gene expression during acute and persistent virus infections. PLoS One 6:e20681 doi:10.1371/journal.pone.0020681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yoneyama M, et al. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851–2858 [DOI] [PubMed] [Google Scholar]
- 51. Yoneyama M, et al. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737 [DOI] [PubMed] [Google Scholar]
- 52. Yount JS, Gitlin L, Moran TM, Lopez CB. 2008. MDA5 participates in the detection of paramyxovirus infection and is essential for the early activation of dendritic cells in response to Sendai virus defective interfering particles. J. Immunol. 180:4910–4918 [DOI] [PubMed] [Google Scholar]