Abstract
Two independent twin-arginine translocases (Tat) for protein secretion were previously identified in the Gram-positive bacterium Bacillus subtilis. These consist of the TatAd-TatCd and TatAy-TatCy subunits. The function of a third TatA subunit named TatAc was unknown. Here, we show that TatAc can form active protein translocases with TatCd and TatCy.
TEXT
Protein transport from the cytoplasm to different bacterial compartments or the external milieu is facilitated by dedicated molecular machines (6). Among these protein translocases, the twin-arginine translocases (Tat) stand out because they permit the passage of tightly folded proteins across the cytoplasmic membrane. The proteins translocated by Tat are synthesized with signal peptides that contain a well-conserved twin-arginine (RR) motif for specific targeting to a membrane-embedded Tat translocase (13, 17, 23). The Tat translocases of Gram-negative bacteria, such as Escherichia coli, are composed of three subunits named TatA, TatB, and TatC (4, 18). The formation of an active protein-conducting channel is believed to require the formation of a supercomplex composed of a TatABC heterotrimeric complex and homo-oligomeric TatA complexes (1, 8). In contrast, most Gram-positive bacteria possess minimized Tat translocases that contain only TatA and TatC subunits. Nevertheless, various studies indicate that these TatAC translocases employ a mechanism similar to that of the TatABC translocases of Gram-negative bacteria (10, 17).
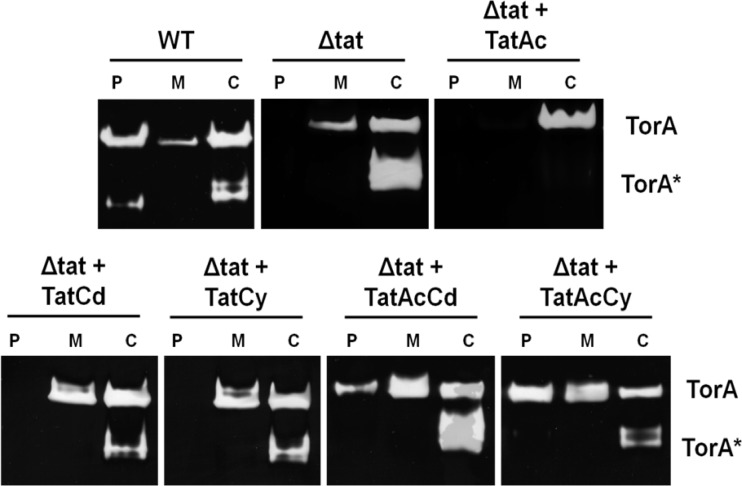
The Gram-positive bacterium Bacillus subtilis is a well-known “cell factory” for secretory protein production (20, 21). In this organism, two Tat translocases are known to operate in parallel. The TatAdCd translocase consists of the TatAd and TatCd subunits, and the TatAyCy translocase consists of the TatAy and TatCy subunits (11, 12, 15). While the TatAdCd translocase is produced mainly under conditions of phosphate starvation (12, 14, 15), the TatAyCy translocase is expressed under all conditions tested (12, 14). Interestingly, B. subtilis produces a third TatA subunit named TatAc (12). The function of TatAc has remained enigmatic due to the fact that no phenotype was so far detectable for tatAc mutant B. subtilis cells (11, 12, 20). Therefore, the present studies were aimed at determining whether TatAc can actually form active translocases in combination with TatCd or TatCy. This possibility was tested by expressing the respective tat genes in E. coli, because the activity and assembly of Bacillus Tat translocases can be assayed more readily in this organism than in B. subtilis (2). For this purpose, the tatAc gene was amplified from the B. subtilis genome (GenBank/EMBL/DDBJ accession number AL009126) and cloned into plasmid pBAD24, resulting in pBAD-Ac. Next, the tatCd and tatCy genes were PCR amplified such that the respective proteins contain a C-terminal StrepII tag. The amplified tatCd-StrepII and tatCy-StrepII genes were cloned into pBAD-Ac, resulting in pBAD-AcCd-Strep and pBAD-AcCy-Strep, respectively. These vectors were subsequently used to transform E. coli ΔtatABCDE cells, which lack all E. coli tat genes. Next, the resulting strains were tested for their ability to transport the previously identified E. coli Tat substrates TorA, AmiA, and AmiC (3, 5, 9). To monitor TorA export to the periplasm, E. coli cells were grown anaerobically until mid-exponential growth phase, and these cells were then subjected to subcellular fractionation as previously described (16). The periplasmic, cytoplasmic, and membrane fractions thus obtained were separated on a 10% native polyacrylamide gel that was subsequently assayed for trimethylamine N-oxide (TMAO) reductase activity as described previously (5, 19). The results in Fig. 1 show that the ΔtatABCDE cells producing TatAc plus TatCd or TatAc plus TatCy were capable of transporting active TorA to the periplasm. In contrast, ΔtatABCDE cells expressing only tatAc, tatCy, or tatCd were not able to export active TorA to this subcellular location. This showed for the first time that TatAc was able to form active translocases in combination with TatCd or TatCy. To further investigate the activity of these translocases, we tested the export of AmiA and AmiC, which are both required for cell wall biosynthesis in E. coli (3, 9). Cells that do not export these molecules to the periplasm grow in long chains, as is observed for the E. coli ΔtatABCDE strain (Fig. 2A and B) (9). As shown by phase-contrast microscopy, the bacteria producing TatAc plus TatCd or TatAc plus TatCy showed the wild-type phenotype, although some slightly longer chains were still detectable (Fig. 2C and D). This is indicative of active export of AmiA and/or AmiC to the periplasm, providing further support for the idea that active TatAcCd and TatAcCy complexes can be formed in E. coli.
Fig 1.
B. subtilis TatAcCd and TatAcCy facilitate TorA export in E. coli. Cells of E. coli ΔtatABCDE were subjected to subcellular fractionation. Proteins in the periplasmic (P), membrane (M), and cytoplasmic (C) fractions obtained were separated by native PAGE, and the gels were subsequently analyzed for TMAO reductase (TorA) activity. Strains used in this analysis were E. coli MC4100 (WT), E. coli ΔtatABCDE (Δtat), or E. coli ΔtatABCDE expressing B. subtilis TatAc from plasmid pBAD-Ac-Strep (Δtat + TatAc), B. subtilis TatCd from plasmid pBAD-Cd-Strep (Δtat + TatCd), B. subtilis TatCy from plasmid pBAD-Cy-Strep (Δtat + TatCy), B. subtilis TatAcCd from plasmid pBAD-AcCd-Strep (Δtat + TatAcCd), or B. subtilis TatAcCy from plasmid pBAD-AcCy-Strep (Δtat + TatAcCy). The position of active full-length TorA is indicated. TorA* indicates a faster-migrating form of TorA (22).
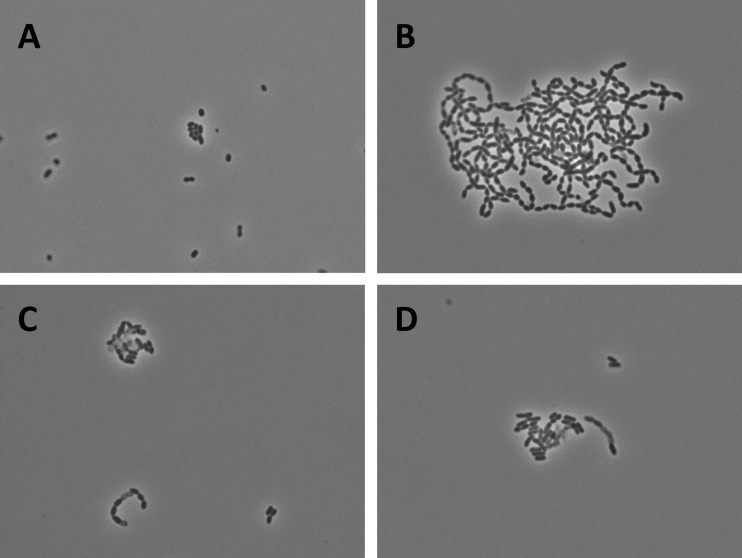
Fig 2.
B. subtilis TatAcCd and TatAcCy facilitate AmiA and AmiC export in E. coli. The export of AmiA and AmiC in E. coli was assayed indirectly by assessing the chain length of exponentially growing cells. (A) E. coli MC4100 (WT); (B) E. coli ΔtatABCDE forms long chains due to the mislocalization of AmiA and AmiC (9); (C) E. coli ΔtatABCDE producing TatAcCd from plasmid pBAD-AcCd-Strep; (D) E. coli ΔtatABCDE producing TatAcCy from plasmid pBAD-AcCy-Strep. As evidenced by the significantly reduced chain length, the export of AmiA and AmiC in E. coli ΔtatABCDE is at least partially restored by the production of TatAcCd or TatAcCy.
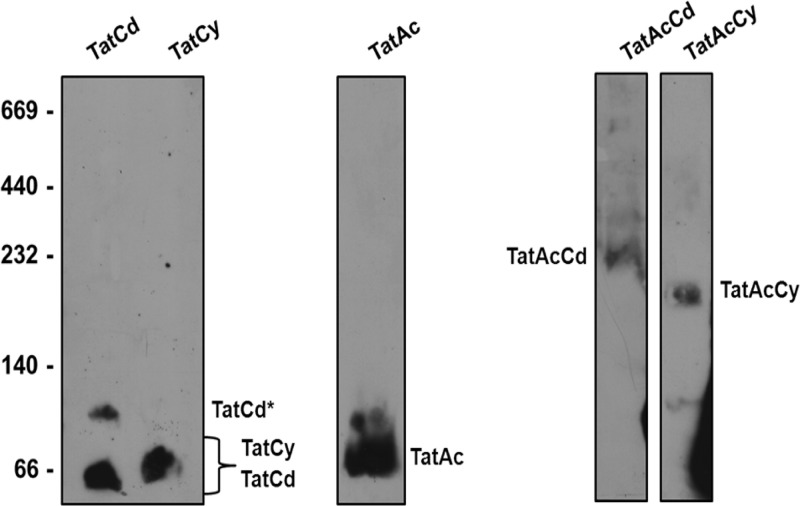
To demonstrate the formation of TatAcCd and TatAcCy complexes, a blue native (BN) PAGE analysis was performed. For this purpose, membranes were isolated from cells expressing TatCd-StrepII, TatCy-StrepII, TatAc-TatCd-StrepII, or TatAc-TatCy-StrepII. In addition, cells producing TatAc-StrepII from plasmid pBAD24 were included in the analyses. Upon solubilization in 2% digitonin, membrane proteins were separated by BN PAGE, followed by immunoblotting with antibodies against the StrepII tag. As shown in Fig. 3, TatCd-StrepII and TatCy-StrepII alone formed bands of ∼66 kDa. In addition, TatCd-StrepII formed a minor band of ∼100 kDa. TatAc-StrepII expressed by itself formed a small homogeneous complex of ∼100 kDa. Importantly, when TatAc (nontagged) was coexpressed with either TatCd-StrepII or TatCy-StrepII, bands of ∼230 kDa or ∼200 kDa, respectively, were observed. This showed that TatAc does indeed form membrane-embedded complexes with TatCd and TatCy.
Fig 3.
TatAcCd and TatAcCy form discrete complexes. To investigate complex formation, TatCd-StrepII, TatCy-StrepII, TatAc-StrepII, TatAc-TatCd-StrepII, or TatAc-TatCy-StrepII were expressed in E. coli ΔtatABCDE. Next, membranes from the respective cells were isolated. Membrane proteins were then solubilized in 2% digitonin and separated by blue native PAGE. The gels were immunoblotted using StrepII-specific antibodies and a secondary anti-mouse IgG-horseradish peroxidase conjugate. The EZ-ECL detection kit was used to visualize bound antibodies. The mobility of molecular mass markers (left panel) and StrepII-tagged proteins and complexes is indicated (kDa). TatCd* indicates a TatCd complex with higher molecular weight.
In conclusion, our present studies document for the first time that the hitherto enigmatic third TatA subunit of B. subtilis known as TatAc can engage in the formation of active TatAC-type translocases. The results also show that TatAc has no particular preference for partnering with TatCd or TatCy. It is noteworthy that the identified TatAcCd and TatAcCy complexes appear to be homogeneous and relatively small (∼230 to 200 kDa). Interestingly, previous studies in B. subtilis have shown that the coexpression of TatAc and TatCd or TatAc and TatCy does not facilitate the export of the known Tat substrates PhoD and YwbN (7). Furthermore, a recent tiling array analysis across 104 conditions has shown that tatAc is expressed under most conditions (14). These previous observations together with our present findings suggest that TatAcCd and TatAcCy translocases could be involved in the specific export of as-yet-unidentified Tat substrates in B. subtilis. However, it has to be noted here that our present observations on TatAc function were made upon heterologous expression in E. coli and that it remains to be assessed whether TatAc fulfills the same functions in B. subtilis.
ACKNOWLEDGMENTS
We acknowledge Ewoud Reilman for his help with the microscopy.
C.G.M., J.B., C.R., and J.M.V.D. were supported through the CEU projects PITN-GA-2008-215524 and -244093 and the transnational SysMO projects BACELL SysMO 1 and 2 through the Research Council for Earth and Life Sciences of the Netherlands Organization for Scientific Research.
Footnotes
Published ahead of print 27 April 2012
REFERENCES
- 1.Baglieri J, Beck D, Vasisht N, Smith CJ, Robinson C. 2012. Structure of the TatA paralog, TatE, suggests a structurally homogeneous form of Tat protein translocase that transports folded proteins of differing diameter. J. Biol. Chem. 287:7335–7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett JP, et al. 2009. The twin-arginine translocation (Tat) systems from Bacillus subtilis display a conserved mode of complex organization and similar substrate recognition requirements. FEBS J. 276:232–243 [DOI] [PubMed] [Google Scholar]
- 3.Bernhardt TG, de Boer PA. 2003. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 48:1171–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogsch EG, et al. 1998. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J. Biol. Chem. 273:18003–18006 [DOI] [PubMed] [Google Scholar]
- 5.Bolhuis A, Mathers JE, Thomas JD, Barrett CML, Robinson C. 2001. TatB and TatC form a functional and structural unit of the twin-arginine translocase from Escherichia coli. J. Biol. Chem. 276:20213–20219 [DOI] [PubMed] [Google Scholar]
- 6.Desvaux M, Hébraud M, Talon R, Henderson IR. 2009. Secretion and subcellular localizations of bacterial proteins: a semantic awareness issue. Trends Microbiol. 17:139–145 [DOI] [PubMed] [Google Scholar]
- 7.Eijlander RT, Jongbloed JD, Kuipers OP. 2009. Relaxed specificity of the Bacillus subtilis TatAdCd translocase in Tat-dependent protein secretion. J. Bacteriol. 191:196–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gohlke U, et al. 2005. The TatA component of the twin-arginine protein transport system forms channel complexes of variable diameter. Proc. Natl. Acad. Sci. U. S. A. 102:10482–10486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ize B, Stanley NR, Buchanan G, Palmer T. 2003. Role of the Escherichia coli Tat pathway in outer membrane integrity. Mol. Microbiol. 48:1183–1193 [DOI] [PubMed] [Google Scholar]
- 10.Jongbloed JDH, van der Ploeg R, van Dijl JM. 2006. Bifunctional TatA subunits in minimal Tat protein translocases. Trends Microbiol. 14:2–4 [DOI] [PubMed] [Google Scholar]
- 11.Jongbloed JDH, et al. 2004. Two minimal Tat translocases in Bacillus. Mol. Microbiol. 54:1319–1325 [DOI] [PubMed] [Google Scholar]
- 12.Jongbloed JDH, et al. 2000. TatC is a specificity determinant for protein secretion via the twin-arginine translocation pathway. J. Biol. Chem. 275:41350–41357 [DOI] [PubMed] [Google Scholar]
- 13.Jongbloed JDH, et al. 2002. Selective contribution of the twin-arginine translocation pathway to protein secretion in Bacillus subtilis. J. Biol. Chem. 277:44068–44078 [DOI] [PubMed] [Google Scholar]
- 14.Nicolas P, et al. 2012. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335:1103–1106 [DOI] [PubMed] [Google Scholar]
- 15.Pop O, Martin U, Abel C, Müller JP. 2002. The twin-arginine signal peptide of PhoD and the TatAd/Cd proteins of Bacillus subtilis form an autonomous Tat translocation system. J. Biol. Chem. 277:3268–3273 [DOI] [PubMed] [Google Scholar]
- 16.Randall LL, Hardy SJ. 1986. Correlation of competence for export with lack of tertiary structure of the mature species: a study in vivo of maltose-binding protein in E. coli. Cell 46:921–928 [DOI] [PubMed] [Google Scholar]
- 17.Robinson C, et al. 2011. Transport and proofreading of proteins by the twin-arginine translocation (Tat) system in bacteria. Biochim. Biophys. Acta 1808:876–884 [DOI] [PubMed] [Google Scholar]
- 18.Sargent F, et al. 1998. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 17:3640–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silvestro A, Pommier J, Pascal MC, Giordano G. 1989. The inducible trimethylamine N-oxide reductase of Escherichia coli K12: its localization and inducers. Biochim. Biophys. Acta 999:208–216 [DOI] [PubMed] [Google Scholar]
- 20.Tjalsma H, Bolhuis A, Jongbloed JD, Bron S, van Dijl JM. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tjalsma H, et al. 2004. Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol. Mol. Biol. Rev. 68:207–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warren G, Oates J, Robinson C, Dixon AM. 2009. Contributions of the transmembrane domain and a key acidic motif to assembly and function of the TatA complex. J. Mol. Biol. 388:122–132 [DOI] [PubMed] [Google Scholar]
- 23.Yuan J, Zweers JC, van Dijl JM, Dalbey RE. 2010. Protein transport across and into cell membranes in bacteria and archaea. Cell. Mol. Life Sci. 67:179–199 [DOI] [PMC free article] [PubMed] [Google Scholar]