Abstract
The adaptation of Lactobacillus sakei to a meat environment is reflected in its metabolic potential. For instance, the ability to utilize arginine through the arginine deiminase (ADI) pathway, resulting in additional ATP, represents a competitive benefit. In L. sakei CTC 494, the arc operon (arcABCTDR) shows the same gene order and organization as that in L. sakei 23K, the genome sequence of which is known. However, differences in relative gene expression were found, and these seemed to be optimal in different growth phases, namely, the highest relative gene expression level was in the end exponential growth phase in the case of L. sakei CTC 494 and in the mid-exponential growth phase of L. sakei 23K. Also, the environmental pH influenced the relative expression level of the arc operon, as shown for L. sakei CTC 494, with the highest relative expression level occurring at the optimal pH for growth (pH 6.0). Deviations from this optimal pH (pH 5.0 and pH 7.0) resulted in an overall decline of the relative expression level of all genes of the arc operon. Furthermore, a differential relative expression of the individual genes of the arc operon was found, with the highest relative gene expression occurring for the first two genes of the arc operon (arcA and arcB). Finally, it was shown that some L. sakei strains were able to convert agmatine into putrescine, suggesting an operational agmatine deiminase pathway in these strains, a metabolic trait that is undesirable in meat fermentations. This study shows that this metabolic trait is most probably encoded by a previously erroneously annotated second putative arc operon.
INTRODUCTION
Lactobacillus sakei is a facultative heterofermentative lactic acid bacterium (LAB) that plays an important role in sausage fermentations, where it is often applied as a starter culture (17, 28). The fact that this species is frequently isolated as the dominant LAB species from spontaneously fermented sausages has been attributed to its adaptation to the meat matrix (2). Fresh meat contains a relatively restricted amount and diversity of carbohydrates, consisting mostly of glucose at levels of between 0.2% and 1.0% (16, 20). To effectively cope with this environment, a flexible use of all available energy sources and nutrients present in meat is of importance (20, 24). In this sense, an efficient catabolism of nucleosides by L. sakei provides this species with an additional energy source (31). Also, conversion of certain amino acids, such as arginine through the arginine deiminase (ADI) pathway, generates ATP, protects this species from acid stress, and hence, provides it with a competitive advantage (30). As a result, the ADI pathway improves survival of L. sakei in the stationary growth phase (5, 30).
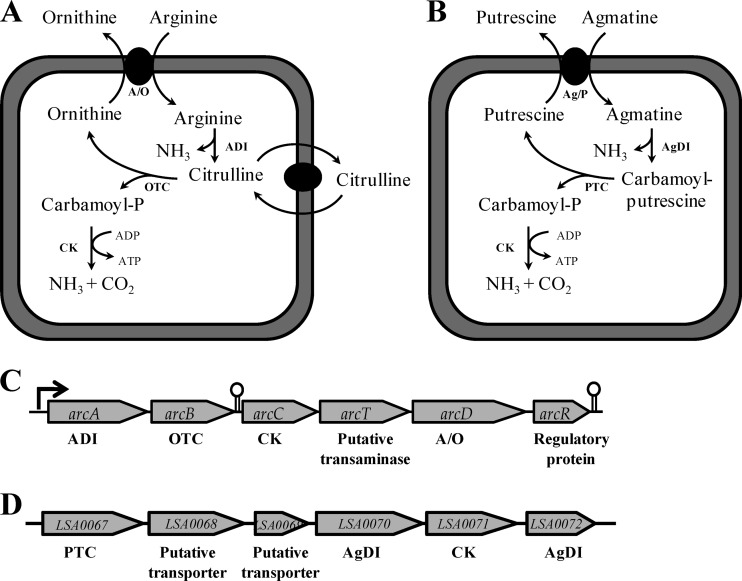
The ADI pathway comprises three cytoplasmic enzymes (Fig. 1A): (i) arginine deiminase, converting arginine into citrulline and ammonia, (ii) catabolic ornithine carbamoyltransferase (OTC), converting citrulline into carbamoyl phosphate and ornithine, and (iii) carbamate kinase (CK), converting carbamoyl phosphate into ammonia, carbon dioxide, and ATP (9). Thus, the ADI pathway results in the conversion of arginine into ornithine, ammonia, and carbon dioxide. In addition, it generates 1 mole of ATP per mole of arginine consumed. For this reason, a competitive advantage can be surmised, as ATP provides additional energy, whereas ammonia production offers an advantage under acid stress conditions (11, 41).
Fig 1.
(A and B) Catabolic pathways for arginine and agmatine in L. sakei through the ADI pathway (A) and the AgDI pathway (B), respectively; (C) organization of the arc operon in L. sakei CTC 494 (arcA, arginine deiminase; arcB, ornithine carbamoyltransferase [OTC]; arcC, carbamate kinase [CK]; arcT, putative transaminase; arcD, arginine/ornithine antiporter [A/O]; arcR, regulatory protein); (D) organization of a putative second arc operon or AgDI operon in L. sakei 23 K. The ADI pathway is present in all L. sakei strains, whereas an operational AgDI pathway was present in only 2 of the 29 L. sakei strains tested. PTC, putrescine carbamoyltransferase; Ag/P, agmatine/putrescine antiporter.
The genes of the ADI pathway are widely distributed in the microbial world (43). Most microorganisms studied so far possess ADI genes organized in a single operon, the so-called arc operon. However, a diversity in the gene organization of the arc operons occurs, with a particular complexity occurring among LAB species (43). The main cytoplasmic enzymes of the ADI pathway are encoded by the arcA, arcB, and arcC genes, coding for the enzymes ADI, OTC, and CK, respectively. In addition to these genes, other genes may be present in arc operons of LAB, such as those coding for an arginine/ornithine antiporter (arcD) or yet to be characterized transporters, putative aminotransferases or transaminases (arcT), and regulatory genes (arcR) (11, 42).
In the case of L. sakei 23K, the arc operon is comprised of six genes, arcABCTDR (41, 42). Moreover, genome analysis of L. sakei 23K has shown that this strain harbors a putative second arc operon (2, 24), suggesting that L. sakei heavily relies on the ADI pathway for competitiveness and survival in a meat environment. Initial annotation of this putative second arc operon predicted two putative peptidyl arginine deiminases (releasing peptide-bound arginine), a putative catabolic ornithine carbamoyltransferase, a putative carbamate kinase, and two putative transport proteins (2). The physiological role of this operon has not yet been verified.
The regulation of the genes of the ADI pathway differs among LAB, and these differences seem to be connected to their adaptation to different habitats (11). The signal responsible for ADI induction is probably linked to energy depletion of the cells (11). In addition, transcriptional studies have shown that the expression of the arc operon in L. sakei is induced by arginine and subjected to glucose repression (11, 42), suggesting carbon catabolite repression as a regulatory mechanism (11, 41). Moreover, the expression of the ADI pathway in L. sakei is stimulated by anaerobiosis (11). All of these factors seem to match the conditions prevailing in the natural meat environment, and it is likely that L. sakei has developed regulatory mechanisms particularly well adapted to this environment (5).
With respect to fermented meats, the environmental pH is an effective modulator of the metabolite kinetics of the ADI pathway of L. sakei (30). Large amounts of the intermediate citrulline are secreted into the environment under optimal pH conditions, whereas under acid stress conditions, arginine is almost exclusively converted into ornithine. When all arginine is depleted, citrulline is taken up and further converted into ornithine. The latter conversion is pH dependent in L. sakei CTC 494, operating maximally at about the optimal pH for growth (30). However, it was not yet clear what the impact of the environmental pH is on the expression of the individual genes of the ADI pathway.
Previous studies on the expression of the arc operon in L. sakei 23K, using a Northern blot analysis, revealed a regulating effect by arginine and glucose on the expression of the arc operon (41, 42). Moreover, a differential expression pattern of the different arc operon genes was shown (41, 42). In addition, environmental pH was shown to display a modulating effect on the ADI pathway activity of L. sakei CTC 494 (30), an isolate from Spanish spontaneously fermented dry sausage (13), which is an interesting starter culture (28). The central aim of the present study was to obtain a better insight into the modulation of the ADI pathway in L. sakei, by comparing the relative expression levels of the different arc operon genes of L. sakei CTC 494 as a function of environmental pH. To this end, a quantitative real-time reverse transcription-PCR (RT-qPCR) methodology was applied to allow comparison with metabolite data and to detect differences between pH values. Also, a comparison with the L. sakei 23K meat strain was performed, to assess differences in the level of gene expression between strains. Furthermore, it was envisaged that the functionality of the putative second arc operon of certain L. sakei strains would be characterized.
MATERIALS AND METHODS
Strains and media.
Twenty-nine L. sakei strains, isolated from different fermented food products, were used during this study (Table 1). All L. sakei strains were stored at −80°C in de Man-Rogosa-Sharpe (MRS) medium (Oxoid Ltd., Basingstoke, Hampshire, United Kingdom) supplemented with 25% (vol vol−1) glycerol as a cryoprotectant. The authenticity of these strains was confirmed through 16S rRNA gene sequencing. All strains were used for the screening experiments, whereas L. sakei CTC 494 and L. sakei 23K were used for the fermentation and gene expression experiments as well.
Table 1.
Strains of Lactobacillus sakei used in the present study
| Strain | Origin | Reference or source |
|---|---|---|
| Lactobacillus sakei CTC 494 | Spanish artisan fermented sausage | 13 |
| Lactobacillus sakei 23K | French fermented sausage | 4 |
| Lactobacillus sakei F2P1 | French fermented sausage | IMDO laboratory collection |
| Lactobacillus sakei FCP1 | French fermented sausage | IMDO laboratory collection |
| Lactobacillus sakei 706 | German fermented sausage | 33 |
| Lactobacillus sakei PTD R9 | Spontaneously fermented sausage | 14 |
| Lactobacillus sakei PTD R14 | Spontaneously fermented sausage | IMDO laboratory collection |
| Lactobacillus sakei DBX R8 | Spontaneously fermented sausage | IMDO laboratory collection |
| Lactobacillus sakei DBX R11 | Spontaneously fermented sausage | IMDO laboratory collection |
| Lactobacillus sakei PTE R4 | Spontaneously fermented sausage | 14 |
| Lactobacillus sakei DFL R6 | Spontaneously fermented sausage | IMDO laboratory collection |
| Lactobacillus sakei VKE R7 | Spontaneously fermented sausage | 14 |
| Lactobacillus sakei DFL R13 | Spontaneously fermented sausage | IMDO laboratory collection |
| Lactobacillus sakei DBL R6 | Spontaneously fermented sausage | IMDO laboratory collection |
| Lactobacillus sakei PTE R19 | Spontaneously fermented sausage | 14 |
| Lactobacillus sakei VK5 R14 | Spontaneously fermented sausage | 14 |
| Lactobacillus sakei VK5 R23 | Spontaneously fermented sausage | 14 |
| Lactobacillus sakei VKD R7 | Spontaneously fermented sausage | 14 |
| Lactobacillus sakei SA201-1 | Industrial meat starter culture | Danisco, Copenhagen, Denmark |
| Lactobacillus sakei SA241-1 | Industrial meat starter culture | Danisco, Copenhagen, Denmark |
| Lactobacillus sakei Texel Lyoflore 2 M | Industrial meat starter culture | Danisco, Copenhagen, Denmark |
| Lactobacillus sakei BioAgro SA8-100 M | Industrial meat starter culture | Chr. Hansen, Hørsholm, Denmark |
| Lactobacillus sakei 2MA4 | Unspecified Cacciatore starter culture | 27 |
| Lactobacillus sakei CG1 | Spontaneous laboratory wheat sourdough fermentation | 29 |
| Lactobacillus sakei LMG 13558 | Sake starter culture | BCCM/LMG Bacteria Collection, Ghent, Belgium |
| Lactobacillus sakei 329 | Spontaneous leek fermentation | IMDO laboratory collection |
| Lactobacillus sakei 481 | Spontaneous leek fermentation | IMDO laboratory collection |
| Lactobacillus sakei 614 | Spontaneous leek fermentation | IMDO laboratory collection |
| Lactobacillus sakei R366 | Romanian mixed-vegetable fermentation | IMDO laboratory collection |
For DNA isolation, all L. sakei strains were grown in MRS medium (Oxoid). All other experiments were performed in modified MRS (mMRS) medium, i.e., MRS medium (10) without glucose and supplemented with the appropriate energy source, either glucose or arginine. For the fermentation experiments, mMRS medium was supplemented with 3 g liter−1 (17.2 mM) of arginine (mMRSarg). Furthermore, control fermentations with mMRS medium containing 5 g liter−1 (27.7 mM) of glucose (mMRSglc) as the sole added energy source were carried out. For the screening experiments, mMRS medium was supplemented with 1 g liter−1 (7.7 mM) of agmatine (further referred to as mMRSagm). Solid MRS medium was prepared by adding 1.5% (wt vol−1) agar (Oxoid) to MRS medium (Oxoid). All chemicals were purchased from VWR International (Darmstadt, Germany).
DNA isolation, PCR assays, and sequencing of the arc operon of Lactobacillus sakei CTC 494.
DNA from L. sakei CTC 494 was isolated from an overnight culture in MRS medium at 30°C. Cell pellets were obtained by centrifugation of 1 ml of this culture (21,036 × g for 20 min) and were washed with TES buffer (50 mM Tris-base, 1 mM EDTA, 6.7% [wt vol−1] sucrose, pH 8.0). Cell pellets were then mixed with a lysis solution (20 mM Tris, 2 mM EDTA, 1% [vol vol−1] Triton X-100, 20 μg μl−1 lysozyme [VWR International], and 0.5 U μl−1 mutanolysin [Sigma-Aldrich, Steinheim, Germany]), and the mixture was incubated at 37°C for 60 min. DNA was isolated using a Nucleospin 96 tissue kit (Macherey-Nagel GmbH, Düren, Germany). DNA concentrations were quantified using a Nanodrop 2000 spectrophotometer (Thermo Scientific, Wilmington, NC). Eleven primer pairs, based on gene sequences of the arc operon of L. sakei 23K (2), were designed using the SNPbox program (39) (see Table S1 in the supplemental material), resulting in overlapping PCR amplicons and covering the whole arc operon of L. sakei CTC 494 (RefSeq no. NC_007576; from positions 372772 to 380240). PCR amplifications were performed using a T3000 thermocycler (Biometra GmbH, Göttingen, Germany). The reaction mixtures contained 100 ng of genomic DNA, 200 μM each deoxynucleoside triphosphate (dNTP), 20 pmol of each primer, 5 μl of 10× PCR buffer, 1.25 U of Taq DNA polymerase (Roche, Basel, Switzerland), and sterile ultrapure water in a final volume of 50 μl. PCR conditions were set as follows: initial denaturation at 95°C for 300 s; 35 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 45 s, and extension at 72°C for 60 s; and a final extension at 72°C for 600 s. Following amplification, PCR product sizes (approximately 800 bp) were controlled on a 1.0% (wt vol−1) agarose gel, and the remaining reaction mixture was purified using a Wizard SV gel and PCR cleanup system (Promega, Madison, WI). Amplicons were sequenced in the Genetic Service Facility of the Flanders Institute for Biotechnology (VIB, Antwerp, Belgium), and the sequences obtained were assembled using Vector NTI Advance (version 10) software (Invitrogen, Carlsbad, CA). Sequence identities between the arc operon genes of L. sakei CTC 494 and L. sakei 23K were calculated using the BLASTX algorithm (National Center for Biotechnology Information [NCBI], http://www.ncbi.nlm.nih.gov/). Rho-independent terminators were predicted using the WebGeSTer program (http://pallab.serc.iisc.ernet.in/gester/rungester.html). Finally, regulatory protein recognition sites (in casu cre sites and ArcR binding sites) in the promoter region upstream of the arcA gene of L. sakei CTC 494 were indicated on the basis of consensus sequences (cre site, 5′-WGNAASCGNWWNCA-3′; consensus sequence for ArcR binding site, 5′-AAATGTGATCTAGATCACATTT-3′) previously reported for L. sakei 23K (41, 42), and a sequence comparison of these promoter regions between the two strains was made.
Fermentation experiments with Lactobacillus sakei CTC 494 and 23K.
Batch fermentations were carried out in 15-liter Biostat C fermentors (Sartorius AG/B; Braun Biotech International, Melsungen, Germany) containing 10 liters of the appropriate mMRS medium. The pH of the medium was adjusted to the desired value prior to sterilization. The fermentor was sterilized in situ at 121°C and 2.1 bars for 20 min. The fermentation temperature was kept at 30°C. During fermentation, the pH of the medium was kept constant through automatic addition of a 10 M NaOH and a 5 M HCl solution to the fermentation medium. The stirring speed was fixed at 100 rpm to keep the medium homogeneous. No aeration was applied. Temperature, pH, and agitation were controlled online (Micro MFCS for Windows NT software; Sartorius AG/B, Braun Biotech International). The inoculum was prepared through three subcultures of the strain under study in the appropriate mMRS medium for 12 h each. The first two subcultures were carried out in 10 ml of the appropriate mMRS medium, followed by a third subculture in 100 ml of this medium. The transfer volumes were always 1% (vol vol−1).
Three fermentations were carried out in mMRSarg medium at different constant pH values (pH 5.0, pH 6.0, and pH 7.0) to investigate the influence of pH on growth, ADI pathway activity, and relative expression levels of the genes of the arc operon of L. sakei CTC 494. A fermentation in mMRSarg medium at a constant pH of 6.0 was performed with L. sakei 23K to compare the relative expression levels of the genes of its arc operon with those of the genes of the arc operon of L. sakei CTC 494 and to analyze the relative gene expression of the putative second arc operon. Also, control fermentations were carried out at pH 6.5 in mMRSglc medium with both L. sakei CTC 494 and L. sakei 23K, during which, due to carbon catabolite repression, no expression of the genes of the arc operons was assumed.
All fermentations were carried out in quadruplicate; the results and figures presented are representative for the four fermentations.
Analysis of growth and ADI pathway metabolites.
During the screening and fermentation experiments, samples were regularly withdrawn for the determination of bacterial growth and concentrations of ADI pathway metabolites.
(i) Determination of bacterial growth.
Growth was followed by the measurement of the numbers of CFU and the optical density at 600 nm (OD600). Cell counts (in numbers of CFU ml−1) were obtained by plating 10-fold serial dilutions of the samples in saline (0.85% [wt vol−1] NaCl solution) on MRS agar and incubating at 30°C for 24 h. Every measurement was performed on three independent samples. The errors of the measurements are represented as standard deviations.
(ii) Determination of arginine, citrulline, and ornithine concentrations.
Concentrations of arginine, citrulline, and ornithine were determined using a Waters 2695 liquid chromatograph coupled to a Quattro Micro mass spectrometer (Waters Corp., Milford, MA), as described previously (38). Due to matrix interference, all quantifications were carried out through standard addition; the original sample concentrations with corresponding errors were calculated as described previously (37).
Relative gene expression. (i) Primer design for quantitative real-time reverse transcription-PCR.
All primers for RT-qPCR were designed with Primer Express (version 2.0) software (Applied Biosystems, Carlsbad, CA), using the corresponding gene sequences of L. sakei CTC 494 for the target (arc operon) genes (arcA, arcB, arcC, arcT, arcD, and arcR) and reference genes (fusA, ileS, pcrA, pfk, recA, rpoB, and secY) (see Table S2 in the supplemental material).
The primers for L. sakei 23K were designed on the basis of its complete genome sequence (EMBL-EBI database accession number CR936503). Primers were designed for the target genes arcA, arcB, arcC, arcT, arcD, and arcR of its arc operon. Also, primers targeting the genes of a putative second arc operon were designed (LSA0067, putative ornithine carbamoyltransferase; LSA0068, putative amino acid/polyamine antiporter; LSA0069, putative amino acid/polyamine antiporter; LSA0070, putative peptidyl arginine deiminase; LSA0071, putative carbamate kinase; and LSA0072, putative peptidyl arginine deiminase) (see Table S3 in the supplemental material).
For all primers, melting temperatures were kept at between 58°C and 60°C. The total amplicon sizes were limited to values of between 50 bp and 150 bp, and the primers did not contain more than two G and/or C nucleotides in the last five bases. Hairpins, self-dimers, and cross-hybridizations were controlled with Primer Express (version 2.0) software (Applied Biosystems). The primers were synthesized by Integrated DNA Technologies (Leuven, Belgium).
(ii) RNA extraction and preparation of cDNA.
Total RNA from growing cultures of L. sakei CTC 494 was extracted at different time points, representing a continuum from the late exponential growth phase (OD600, 0.3) to the end exponential (OD600, 0.5), early stationary (OD600, 0.9), and late stationary (OD600, 1.0 after 24 h of fermentation) growth phases. For L. sakei 23K, RNA samples were taken at time points corresponding to the early exponential (OD600, 0.3), mid-exponential (OD600, 0.5), end stationary (OD600, 2.5), and stationary (OD600, 2.7 after 24 h of fermentation) growth phases. For the control fermentations, total RNA was extracted at different times points representing a continuum from mid-exponential growth phase (OD600, 1.5) to late exponential (OD600, 3.0), end exponential (OD600, 5.0), and late stationary (OD600, 3.5 after 24 h of fermentation) growth phases. After sampling, cell suspensions (2 ml) were immediately mixed with RNAprotect Bacteria reagent (4 ml; Qiagen, Hilden, Germany) and stored at −80°C. Sample handling and total RNA extraction were performed according to the standard protocol described in the RNAprotect Bacteria reagent handbook (Qiagen) with some modifications. The cell pellet was suspended in 200 μl TE buffer (10 mM Tris base, 1 mM EDTA, pH 8.0) to which 20 μg μl−1 lysozyme (VWR International) and 0.5 U μl−1 mutanolysin (Sigma-Aldrich) were added, followed by incubation at 37°C for 60 min. An RNase-free DNase treatment was carried out directly on an RNeasy column (Qiagen). RNA concentrations were quantified using a Nanodrop 2000 spectrophotometer (Thermo Scientific). To eliminate residual DNA contamination present in the RNA samples, a second DNase treatment (DNase I; Invitrogen) was introduced, prior to cDNA production, by adding 1 μl of DNase I reaction buffer, 1 μl of DNase I (1 U μl−1), and diethylpyrocarbonate (DEPC)-treated water (Invitrogen) to 1 μg of RNA in a final volume of 10 μl. After an incubation period of 15 min at room temperature, DNase I was inactivated by adding 2 μl of EDTA (25 mM; Invitrogen) to the reaction mixture and heating at 65°C for 10 min. The absence of DNA contamination in the RNA samples was verified by RT-qPCR amplification using the secY primers prior to cDNA synthesis. Total RNA (1 μg) was reverse transcribed into cDNA in a final volume of 50 μl with a High Capacity cDNA Archive kit (Applied Biosystems), as recommended by the manufacturer, using a T3000 thermocycler (Biometra GmbH). The reverse transcriptase reaction was applied using the following two-step temperature profile: incubation at 25°C for 10 min, followed by incubation at 37°C for 120 min. Finally, cDNA aliquots were stored at −80°C.
(iii) Quantitative real-time reverse transcription-PCR.
RT-qPCR was performed using a 7300 real-time PCR system (Applied Biosystems). Each PCR assay mixture contained 12.5 μl of SYBR green master mix with carboxy-X-rhodamine (ROX) as a passive reference dye (Applied Biosystems), 2 μl of a 100-times-diluted cDNA solution, the forward and reverse primers at the optimal concentrations (see Tables S4 and S5 in the supplemental material), and sterile nuclease-free ultrapure water in a final volume of 25 μl. The first step of the RT-qPCR assay consisted of an initial hold at 95°C for 600 s, followed by 40 two-step cycles at 95°C for 15 s and 60°C for 60 s. All sample and primer set combinations were assessed in triplicate. In each run, negative controls (without cDNA) for each primer set were included. After each amplification run, a melting curve was obtained by increasing the temperature by 1°C from 65°C to 94°C every 20 s, to confirm primer specificity. The specificity of each primer set was checked by analyzing the dissociation curve and controlling the absence of unspecific and secondary products. The cycle threshold (CT) value, proportional to the concentration of gene transcripts, was determined using the auto-CT tool of the 7300 system SDS software (Applied Biosystems).
(iv) Relative quantification of gene expression levels.
Relative gene expression levels were calculated by taking into account the amplification efficiency of each primer set and a normalization factor based on the expression levels of multiple reference genes, using the efficiency-corrected delta delta CT method (27). The amplification efficiency of each primer set (target genes and reference genes) was calculated for the whole data set (for all fermentations in quadruplicate) using data analysis for real-time PCR (see Tables S4 and S5 in the supplemental material) (26). The GeNorm software was used to identify which reference genes out of six candidate genes showed the most stable expression ratios for this experimental setup (36). GeNorm determines a ranking of candidate reference genes using stability (Mj) values and selects the amount of reference genes necessary for proper normalization. Using this methodology, reference gene expression stabilities were determined for seven reference genes (fusA, ileS, pcrA, pfk, recA, rpoB, and secY) and were evaluated for different growth phase time points in the case of L. sakei CTC 494 during fermentations at different pH values (pH 5.0, pH 6.0, and pH 7.0) and for different growth phase time points in the case of L. sakei 23K during fermentation at pH 6.0. The resulting relative gene expression levels were calculated for four independent fermentation experiments. The errors of these measurements are represented as standard deviations. The relative expression levels of the arc operon in the different growth phases in mMRSarg at different pH values versus the control condition (glucose) were tested for significance by randomization and bootstrapping techniques carried out by applying the relative expression software tool (REST2005), using 50,000 random reallocations.
Genomic screening for the presence of a putative second arc operon.
Twenty-nine L. sakei strains were screened for the presence of a putative second arc operon (Table 1). Therefore, DNA from all L. sakei strains was isolated from overnight cultures in MRS medium at 30°C, as described above. Primer pairs resulting in overlapping PCR amplicons and covering the whole putative second arc operon of these strains were designed on the basis of the sequence of the putative second arc operon of L. sakei 23K (2) using SNPbox software (39) (see Table S6 in the supplemental material). PCR amplifications were performed using a DNA T3000 thermocycler (Biometra GmbH), containing 100 ng of genomic DNA, 200 μM each dNTP, 20 pmol of each primer, 5 μl of 10× PCR buffer, 1.25 U of Taq DNA polymerase (Roche), and sterile ultrapure water in a final volume of 50 μl. PCR conditions were set as follows: initial denaturation at 95°C for 300 s; 35 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 45 s, and extension at 72°C for 60 s; and a final extension at 72°C for 600 s. Following amplification, PCR product sizes (approximately 800 bp) were controlled on a 1.0% (wt vol−1) agarose gel (Invitrogen).
For the positive L. sakei strains, the genomic region from position 64776 to position 66663 in the genome of L. sakei 23K, covering the genes LSA0068 and LSA0069, was sequenced (VIB) and assembled using Vector NTI Advance (version 10) software (Invitrogen). Open reading frames were predicted using the Artemis program (32); contig sequences as well as predicted amino acid sequences were aligned using the ClustalW program (15).
To assess the relatedness of the genes of the putative second arc operon with genes of the arc operon and/or agmatine deiminase (AgDI) operon, clustering trees were constructed using the MEGA (version 4.0) program (35) and the neighbor-joining clustering method and were based on amino acid sequences of the following species: Pseudomonas aeruginosa PAO1, LASB58, UCBPP-PA14, and PA7; Enterococcus faecalis 62, V583, and ATCC 4200; L. sakei 23K; Lactobacillus hilgardii ATCC 8290; Lactobacillus brevis ATCC 367; and Pediococcus pentosaceus ATCC 25745.
Physiological screening for the ability to convert agmatine into putrescine.
All L. sakei strains were grown in test tubes containing 10 ml of mMRSagm medium (pH 6.5) at 30°C for 24 h. Concentrations of agmatine and putrescine were determined after 24 h of fermentation, using a Waters Acquity ultrapure liquid chromatography-tandem mass spectrometry apparatus (Waters Corp.). A hydrophilic interaction chromatography (HILIC) column (Waters Corp.) was used for separation and was kept at 30°C. The mobile phase, used at a flow rate of 1.0 ml min−1, was composed of 10 mM ammonium acetate with 0.2% (vol vol−1) formic acid and 5% (vol vol−1) acetonitrile in ultrapure water at pH 4.0 (eluent A) and acetonitrile with 0.2% (vol vol−1) formic acid (eluent B). The following gradient was used (composition [vol vol−1] at different time points with a linear profile imposed in between): 0.0 min, 5% eluent A and 95% eluent B; 0.5 min, 5% A and 95% B; 3.0 min, 50% A and 50% B; 3.1 min, 5% A and 95% B; and 6.0 min, 5% A and 95% B. Settings of the mass spectrometer were as follows: capillary voltage, 3.50 kV; cone voltage, 20 V; source temperature, 120°C; desolvation temperature, 350°C; cone gas flow, 50 liters h−1; desolvation gas flow, 550 liters h−1; and collision energy, 15 eV. The following mother and daughter ions were followed: agmatine at m/z 131.00 to 72.02 and putrescine at m/z 89.00 to 71.99. Due to matrix interference, all quantifications were carried out through standard addition. Four standard solutions with the following composition were prepared: solution A consisted of ultrapure water, solution B consisted of agmatine at 0.15 g liter−1 and putrescine at 0.06 g liter−1, solution C consisted of agmatine at 0.30 g liter−1 and putrescine at 0.12 g liter−1), and solution D consisted of agmatine at 0.45 g liter−1 and putrescine at 0.17 g liter−1. For each time point, 100 μl of cell-free culture supernatant was mixed with 300 μl of ultrapure water and 100 μl of solution A, B, C, or D, filtered (0.2-μm-pore-size filters; Minisart high flow; Sartorius AG, Göttingen, Germany) into a vial, and injected (3 μl) into the column. The concentrations of the original samples were calculated as described before (37).
Nucleotide sequence accession numbers.
The sequence of the arc operon was submitted to the EMBL database (EMBL-EBI, Hinxton, United Kingdom) with accession number FR870226. The sequences of the genomic regions covering the putative agmatine transporter gene for all positive strains except L. sakei 23K were submitted to the EMBL database with accession numbers HE681913 (L. sakei PTD R9), HE681914 (L. sakei PTD R14), HE681915 (L. sakei PTE R4), HE681916 (L. sakei DFL R6), HE681917 (L. sakei DBX R8), HE681918 (L. sakei DBX R11), and HE681919 (L. sakei F2P1).
RESULTS
Identification and characterization of the arc operon of Lactobacillus sakei CTC 494.
The 8,379-bp arc operon of L. sakei CTC 494 had the same order of genes (arcABCTDR; Fig. 1C) as that of L. sakei 23K, with arcA (1,236 bp) coding for an arginine deiminase (99% nucleotide sequence identity with arcA of L. sakei 23K; one nonsynonymous mutation at nucleotide position 1,133 resulted in an altered amino acid sequence, i.e., a cysteine for arginine deiminase of L. sakei CTC 494 versus a tyrosine for arginine deiminase of L. sakei 23K), arcB (1,017 bp) coding for an ornithine carbamoyltransferase (100% identity with arcB of L. sakei 23K), arcC (995 bp) coding for a carbamate kinase (100% identity with arcC of L. sakei 23K), arcT (1,116 bp) coding for a putative aminotransferase (100% identity with arcT of L. sakei 23K), arcD (1,428 bp) coding for an arginine/ornithine antiporter (100% identity with arcD of L. sakei 23K), and arcR (702 bp) coding for a regulatory protein (100% identity with arcR of L. sakei 23K). As for L. sakei 23K (41), one putative rho-independent terminator was found in the arc operon, namely, between the arcB and arcC genes. Moreover, nucleotide sequence alignment of the promoter regions upstream of the arcA genes for both L. sakei CTC 494 and L. sakei 23K showed the presence of two putative 14-bp cre sites (58 bp and 138 bp upstream of the ATG start codon) and three putative ArcR binding sites (54 bp, 74 bp, and 95 bp upstream of the ATG start codon) located at the same positions for both strains. Sequence alignment of both promoter regions indicated two sequence variations between L. sakei 23K and L. sakei CTC 494: one located in the first predicted cre box (position −129, C in L. sakei 23K versus T in L. sakei CTC 494) and one located in a predicted ArcR binding site (position −45, G in L. sakei 23K versus A in L. sakei CTC 494).
Influence of pH on growth, ADI pathway activity, and relative gene expression level of arc operon in Lactobacillus sakei CTC 494.
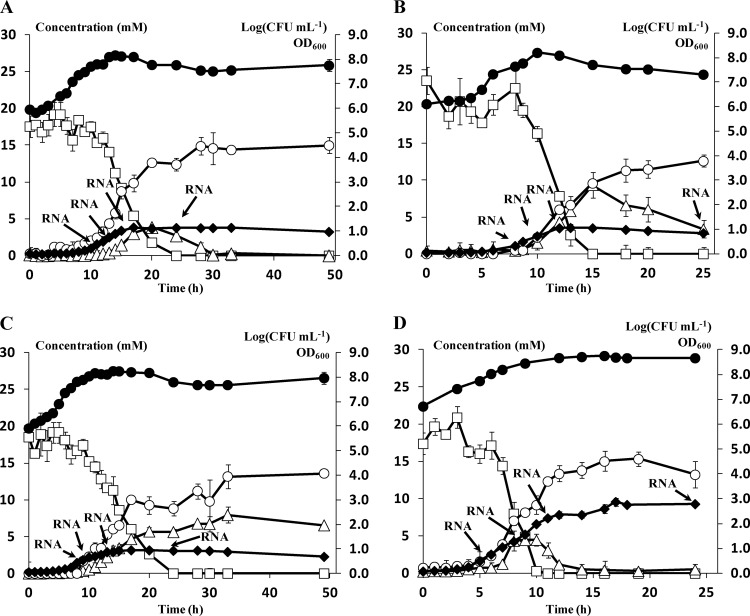
The pH had a pronounced effect on the growth and the kinetics of arginine conversion in L. sakei CTC 494 (Fig. 2A to C). Arginine conversion started after approximately 14 h (pH 5.0), 8 h (pH 6.0), and 8 h (pH 7.0) of fermentation and for all pH values resulted in the production of both citrulline and ornithine. The maximal extracellular concentration of the intermediate citrulline was found at pH 6.0. After all arginine was depleted, a further conversion of citrulline into ornithine took place at pH 5.0 and pH 6.0. For the fermentations at pH 7.0, no citrulline-into-ornithine conversion took place when all arginine was depleted. As a result, a constant and higher ratio of citrulline to ornithine was found in the fermentation medium in comparison with the fermentations at lower pH.
Fig 2.
(A to C) Growth (●, log CFU ml−1; ⧫, OD600) of Lactobacillus sakei CTC 494 in modified MRS medium supplemented with 3 g liter−1 (17.2 mM) of arginine at different constant pH values: pH 5.0 (A), pH 6.0 (B), and pH 7.0 (C). (D) Growth (●, log CFU ml−1; ⧫, OD600) of L. sakei 23K in modified MRS medium supplemented with 3 g liter−1 (17.2 mM) of arginine at a constant pH of 6.0. Extracellular concentrations (mM) of arginine (□), citrulline (Δ), and ornithine (○) are also shown. Data shown are representative for four independent fermentation experiments.
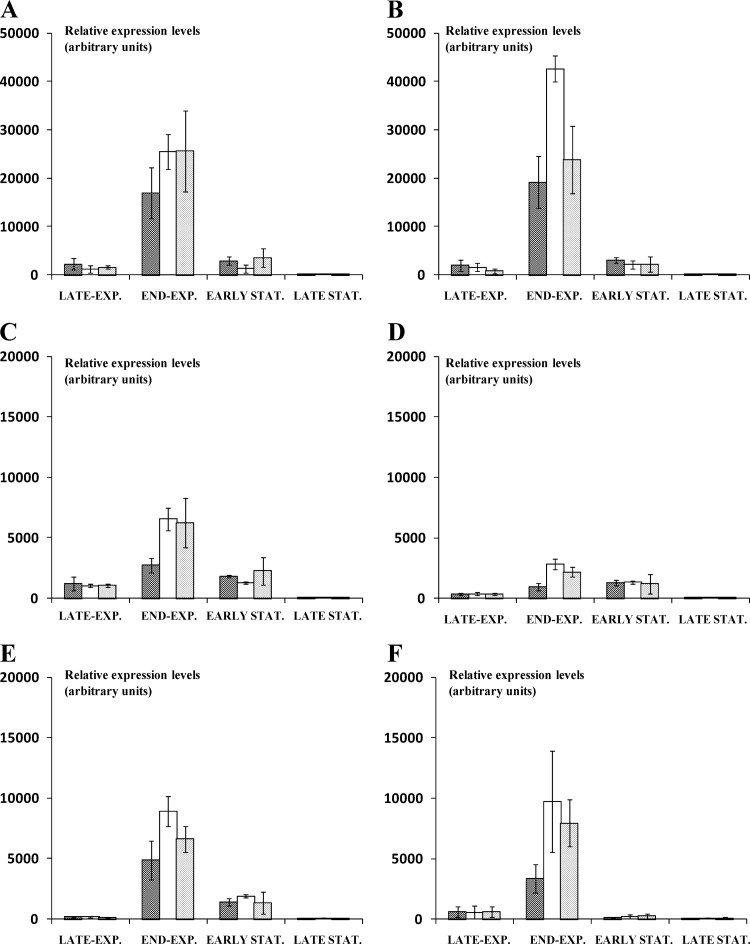
The most stable reference gene was pcrA, with an Mj value of 1.294, whereas the least stable reference gene was secY, with an Mj value of 1.812. On the basis of these stability values, the three most stable reference genes for L. sakei CTC 494 (i.e., fusA, ileS, and pcrA) were selected for proper normalization. For all genes of the arc operon of L. sakei CTC 494, a clear induction of gene expression, compared to the control fermentation in mMRSglc medium, was found at all pH values and for all growth phases investigated during fermentation under various pH conditions. Relative gene expression levels ranged from 10.8 ± 8.8 to 42,597.6 ± 2,658.9 (Fig. 3). The highest relative expression level was found in the case of the first two genes of the arc operon (arcA and arcB), whereas a progressively declining relative expression level was found for the genes further downstream in the arc operon. For all genes, the relative expression level was highest when the culture reached the end of the exponential growth phase (Fig. 3). Upon further fermentation, a decline in overall relative expression level was found for all genes from the start of the stationary growth phase on.
Fig 3.
Relative expression of the genes involved in the arginine deiminase pathway of Lactobacillus sakei CTC 494 when grown in modified MRS medium supplemented with 3 g liter−1 (17.2 mM) of arginine as a function of the growth phase (late exponential [EXP.; OD600, 0.3], end exponential [OD600, 0.5], early stationary [STAT.; OD600, 0.9], and late stationary [OD600, 1.0 after 24 h of fermentation] growth phases) and environmental pH (pH 5.0, pH 6.0, and pH 7.0, indicated from left to right, respectively, for each set of bars): arcA (A), arcB (B), arcC (C), arcT (D), arcD (E), and arcR (F). The data shown are mean values ± standard deviations from four independent fermentation experiments, whereby the probability of expression levels different from those in the control fermentation was calculated by the pairwise fixed reallocation randomization test (50,000 permutations) between samples and control groups (P < 0.001).
The pH of the fermentation medium influenced the overall relative expression level of the arc operon in L. sakei CTC 494, with the highest relative gene expression levels occurring at the optimal pH for growth (pH 6.0). Decreasing or increasing the pH resulted in an overall decrease of the relative expression level of all genes. In general, a 2- to 3-fold decrease in relative expression level of all genes of the arc operon was found at pH 5.0 compared to that at pH 6.0. At pH 7.0, overall, a less pronounced decrease of relative gene expression levels was found compared to those at pH 6.0. The arcT gene, encoding a putative aminotransferase, showed the same expression pattern found for all other genes of the arc operon, namely, the highest relative gene expression level at the end of the exponential growth phase in the case of arginine supplementation during fermentation at pH 6.0.
Influence of pH on growth, ADI pathway activity, and relative gene expression level of arc operon in Lactobacillus sakei 23K.
The conversion of arginine by L. sakei 23K showed a sequential pattern similar to that for L. sakei CTC 494 but with a different timing (Fig. 2D). Whereas for L. sakei CTC 494 arginine conversion started at the end of the exponential growth phase (Fig. 2A to C), arginine conversion by L. sakei 23K had already started in the mid-exponential growth phase (Fig. 2D). After all arginine was depleted, a further conversion of citrulline into ornithine took place, and ornithine in turn was excreted into the medium as the main end product at the end of the fermentation.
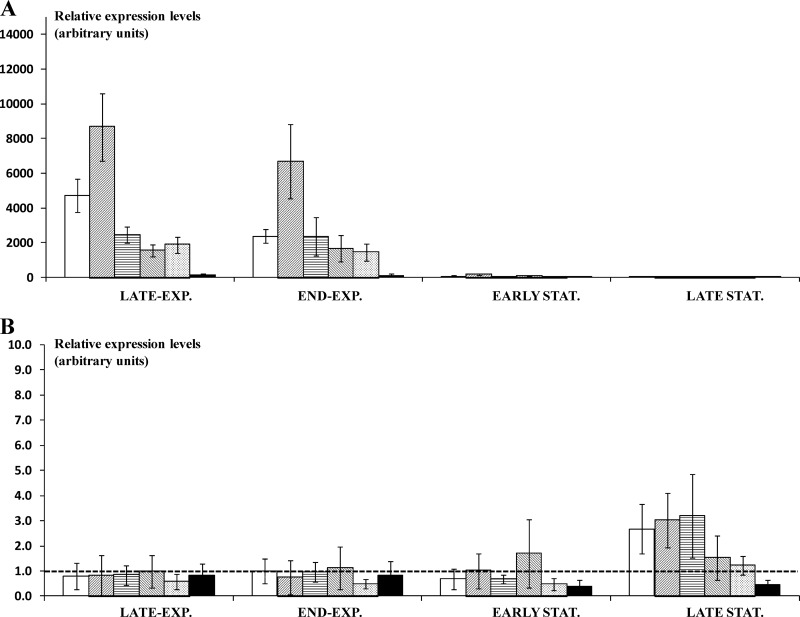
For L. sakei 23K, two genes (fusA and ileS) were sufficient for a proper normalization of the RT-qPCR data. The Mj values for the reference genes ranged from 0.096 (recA) to 0.045 (ileS). The relative expression of all arc genes compared to that during the control fermentation with glucose was again induced in all growth phases analyzed (Fig. 4A). Relative gene expression levels ranging from 23.6 ± 7.0 to 8,660.4 ± 1,928.4 were found. Once the cells entered the stationary growth phase, induction ceased, with relative gene expression levels being between 0.5 ± 0.3 and 6.0 ± 4.9. Relative expression levels of the arc genes at pH 6.0 were found to be much lower in L. sakei 23K than in L. sakei CTC 494. The relative gene expression level of the arc genes was highest in the late exponential growth phase and declined upon further fermentation, with the highest relative expression level found for both the arcA and arcB genes.
Fig 4.
RT-qPCR analysis of the arc genes (arcA, arcB, arcC, arcT, arcD, and arcR [from left to right, respectively, for each set of bars]) (A) and a putative second arc operon (LSA0067, LSA0068, LSA0069, LSA0070, LSA0071, and LSA0072 [from left to right, respectively, for each set of bars]) (B) in Lactobacillus sakei 23K in modified MRS medium supplemented with 3 g liter−1 (17.2 mM) of arginine during different growth phases (late exponential [OD600, 0.3], end exponential [OD600, 0.5], early stationary [OD600, 0.9], and late stationary [OD600, 1.0 after 24 h of fermentation] growth phases) at pH 6.0. Data shown are mean values ± standard deviations from four independent fermentation experiments. No significant differences were found for the relative expression levels of the genes of the second putative arc operon compared to the control fermentations, except for LSA0072, which showed a significant (P < 0.001) decrease of expression in the stationary growth phase.
Also, the relative gene expression of a putative second arc operon in L. sakei 23K was followed during growth in mMRSarg medium at a constant pH of 6.0 and in different growth phases. An increase of the relative expression levels was not found for any of the genes of the putative second arc operon (Fig. 4B), with fold changes being close to 1 in all genes and for all growth phases examined.
Presence and function of a putative second arc operon in Lactobacillus sakei.
Eight L. sakei strains (L. sakei 23K, L. sakei PTD R9, L. sakei PTD R14, L. sakei DBX R8, L. sakei DBX R11, L. sakei PTE R4, L. sakei DFL R6, and L. sakei F2P1) out of the 29 ones tested contained a putative second arc operon, as amplification using all primer sets designed was successful.
The second putative arc operon of L. sakei 23K comprised five genes (Fig. 1). Sequence comparison between the proteins encoded by LSA0067 and arcB showed an amino acid sequence identity of only 33%. Similarly, sequence comparison between the proteins encoded by LSA0070 and arcA showed an amino acid sequence identity of 57%. Therefore, a clustering tree based on a multiple-sequence alignment at the protein level of the gene product of LSA0067 (putative ornithine carbamoyltransferase) with other well-characterized ornithine carbamoyltransferases and putrescine carbamoyltransferases was constructed, as was a clustering tree based on multiple-sequence alignments at the protein level of the gene product of LSA0070 (putative agmatine deiminase) with other well-characterized arginine deiminases and agmatine deiminases (see Fig. S1 and S2 in the supplemental material). The gene product of LSA0067 clearly clustered together with other well-characterized putrescine carbamoyltransferases rather than with other ornithine carbamoyltransferases. Similarly, the gene product of LSA0070 clustered together with other well-characterized agmatine deiminases rather than with other arginine deiminases.
As the expression of the putative second arc operon was not induced by arginine in L. sakei 23K (see above), an additional screening experiment was set up to further examine if the putative second arc operon was perhaps correlated with AgDI pathway activity rather than with ADI pathway activity. Therefore, the eight L. sakei strains containing a putative second arc operon were grown in mMRSagm medium to assess their capacity to convert agmatine into the biogenic amine putrescine. Only two of the eight strains tested (L. sakei PTD R14 and L. sakei DBX R11) were able to convert agmatine into putrescine after 24 h of fermentation, resulting in the production of 0.48 ± 0.09 mM and 2.52 ± 0.04 mM putrescine, respectively. Finally, sequence analysis of the genomic region covering the putative agmatine transporter gene for these eight strains of L. sakei revealed the presence of a frameshift mutation in this transporter gene for four strains (L. sakei 23K, F2P1, PTE R4, and DFL R6), which were the strains that were not able to convert agmatine into putrescine phenotypically. Of the four other strains (L. sakei PTD R9, PTD R14, DBX R11, and DBX R8) that did not contain such a frameshift mutation, two strains (L. sakei PTD R14 and DBX R11) displayed agmatine-into-putrescine conversion after 24 h of fermentation. These findings suggest that the putative second arc operon in L. sakei 23K might encode an AgDI operon rather than a second arc operon (Fig. 1B).
DISCUSSION
Lactobacillus sakei has evolved to include metabolic pathways and stress response mechanisms specific for fresh and, particularly, fermented meats (2, 4, 19). For example, the conversion of arginine, an alternative energy source abundantly present in meat due to endogenous protease activity, through the ADI pathway renders this species with an improved survival in the stationary growth phase (4, 5, 31, 43). Arginine conversion is a common metabolic trait for the species L. sakei and is encoded by the arc operon (24, 41, 42). In L. sakei CTC 494, the arc operon (arcABCTDR) shows the same gene order and organization as that of L. sakei 23K (42). Genetic studies have shown that the organization of arc operons in LAB is, in general, very complex, due to the presence of additional and duplicated genes next to arcA, arcB, arcC, and arcD, and that the order of these genes may vary in different species (11, 43).
Environmental pH is an important regulator of the ADI pathway in LAB (25, 30, 38). The present results underline this regulating effect of environmental pH on the expression profiles of the arc genes in L. sakei CTC 494. Overall, the expression of the ADI pathway genes was highest at the optimal pH for growth (pH 6.0) but decreased when it shifted away from this value, in particular, toward low pH (pH 5.0). These effects on the levels of gene expression corresponded with the conversion kinetics of the different metabolites of the ADI pathway as a function of pH, with the highest (and similar) catabolic activities being at pH 6.0 and pH 7.0 and the slowest conversion being at pH 5.0 (30). In contrast, low pH values induced gene expression of the arc operon of Lactococcus lactis subsp. lactis ML3, Lactobacillus hilgardii X1B, Lactobacillus brevis IOEB 9809, and Streptococcus gordonii DL1 (1, 7, 18), suggesting a protective mechanism against acid stress due to ammonia production through the ADI pathway. For L. sakei, improved survival in the stationary growth phase was probably not due to a protective effect against acid conditions (5) but rather was due to the generation of extra ATP (9, 11, 30). The mechanism responsible for pH-mediated changes in gene expression is difficult to unravel, as a diverse and interdependent set of signals is generated when the external or internal pH changes (25). Moreover, genes that show pH dependence are often coinduced by other factors, such as the growth phase, carbon source, and anaerobiosis (40).
The expression of the arc operon of L. sakei is subjected to carbon catabolite repression, whereas anaerobic conditions and the presence of arginine enhance the expression of the genes of the arc operon (5, 11, 21, 41, 42). Moreover, this study revealed the presence of strain-dependent sequence variations located in a putative cre site and a putative ArcR binding site in the promoter region of the arc operon of L. sakei CTC 494 and 23K. Such differences in promoter regions of the arc operon might therefore be responsible for the different onset of arginine conversion and arc operon expression between the two strains. Nevertheless, the actual DNA binding activity by ArcR and its recognition site have not been determined and thus remain to be investigated. In this study, it was shown that the highest expression of the arc genes was found when L. sakei CTC 494 reached the end of the exponential growth phase, whereas arginine conversion by L. sakei 23K started earlier, with the highest expression level of the arc genes found in the mid-exponential growth phase. Therefore, arginine conversion can contribute to growth and survival of L. sakei, depending on the strain, yet growth differences on complex substrates may determine this different onset of arginine conversion as well. The exact mechanism for these differences in ADI pathway activity between strains needs further investigation. As meat is a dynamic environment representing a broad range of complex niches, intraspecies variations within L. sakei may lead to different genotypes successively dominating the fermented meat matrix (3). However, information on the relationship between differences on a genomic level and the general competitiveness of specific L. sakei strains is still lacking.
Differential expression of the individual genes within the arc operon was found, with the highest (and similar) expression found for the first two genes of the arc operon (i.e., arcA and arcB) and a progressively declining expression found for the other genes downstream of these genes. This differential expression might be explained by the presence of a putative rho-independent terminator directly downstream of the arcB gene. Indeed, in the case of L. sakei 23K, a complex pattern of RNA transcripts of different lengths has been found through Northern blot analysis (42). Similar complex transcriptional patterns of the arc operon have also been found for other bacteria, suggesting that processing and partial termination may play a role in the regulation of the expression of individual genes of the arc operon (6, 11, 12, 41, 42).
Genomic analysis revealed the presence of a putative second arc operon in L. sakei 23K, apparently emphasizing the importance of arginine as an alternative energy source (2). However, homology and phylogenetic analysis of the putative arginine deiminase and putative ornithine carbamoyltransferase enzymes with other well-characterized arc operons and AgDI operons revealed that this second putative arc operon resembles more an agmatine deiminase operon. Similarly, putrescine carbamoyltransferases have previously often been misannotated solely on the basis of sequence data (22, 23). Moreover, expression of the genes of the AgDI operon was not induced by arginine, further indicating a different physiological role. Among the 29 L. sakei strains tested in the current study, the whole operon was present in only 28% of these strains, all of which were isolates from fermented sausages. It has already been shown that pathways responsible for biogenic amine production are strain specific, with horizontal gene transfer being the mechanism responsible for this strain variation (8). Although extensive intraspecies genomic variability within L. sakei has been shown before (3, 24), its occurrence solely in meat isolates may suggest a putative role of the AgDI pathway in meat adaptation. The two L. sakei strains with an operational AgDI pathway were able to produce the biogenic amine putrescine from agmatine through the AgDI pathway, thereby producing ammonia toward acid stress and extra ATP to improve competitiveness. Also, this finding underlines the importance of the choice of starter cultures for fermented sausage production, as putrescine is one of the main biogenic amines found in these food products (34). However, the presence of the genes involved in the agmatine-into-putrescine conversion did not always imply agmatine deiminase pathway activity, as agmatine-into-putrescine conversion was detected for only two strains. The other L. sakei strains that contained all genes of the AgDI pathway but that were not able to convert agmatine were probably impaired in the transport of agmatine. Indeed, in the case of L. sakei 23K, a frameshift mutation occurred in the putative transporter genes of this operon (2). In the present study, similar frameshift mutations were also present in three other L. sakei strains that were also not able to convert agmatine into putrescine. However, for two other strains, no frameshift mutation was found in the transport protein-encoding gene, despite their inability to convert agmatine. This suggests that different regulation mechanisms or mutations present in other genes of the AgDI pathway may account as well for this finding.
In conclusion, the results obtained in this work underline the fact that the contribution of the ADI pathway to the general competitiveness in and adaptation of L. sakei to the meat environment is strain and pH dependent. Also, the fact that some strains possess an operational AgDI pathway underlines the importance of selection of appropriate starter cultures for meat fermentations. To this end, the relationship between the occurrence of AgDI operons with their expression and activity in L. sakei strains should be taken into account.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the financial support of the Research Council of the Vrije Universiteit Brussels and the Research Foundation—Flanders (FWO-Vlaanderen). T.R. and A.R. are supported by a predoctoral fellowship of the FWO-Vlaanderen.
Footnotes
Published ahead of print 27 April 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Arena MP, Russo P, Capozzi V, Beneduce L, Spano G. 2011. Effect of abiotic stress conditions on expression of the Lactobacillus brevis IOEB 9809 tyrosine decarboxylase and agmatine deiminase genes. Ann. Microbiol. 61:179–183 [DOI] [PubMed] [Google Scholar]
- 2.Chaillou S, et al. 2005. The complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23K. Nat. Biotechnol. 23:1527–1533 [DOI] [PubMed] [Google Scholar]
- 3.Chaillou S, et al. 2009. Intraspecies genomic diversity and natural population structure of the meat-borne lactic acid bacterium Lactobacillus sakei. Appl. Environ. Microbiol. 75:970–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Champomier-Vergès MC, Chaillou S, Cornet M, Zagorec M. 2001. Lactobacillus sakei: recent developments and future prospects. Res. Microbiol. 152:839–848 [DOI] [PubMed] [Google Scholar]
- 5.Champomier-Vergès MC, et al. 1999. Relationships between arginine degradation, pH and survival in Lactobacillus sakei. FEMS Microbiol. Lett. 180:297–304 [DOI] [PubMed] [Google Scholar]
- 6.Chopin A. 1993. Organisation and regulation of genes for amino acid biosythesis in lactic acid bacteria. FEMS Microbiol. Rev. 12:21–38 [DOI] [PubMed] [Google Scholar]
- 7.Chou LS, Weimer BC, Cutler R. 2001. Relationship of arginine and lactose utilization by Lactococcus lactis ssp. lactis ML3. Int. Dairy J. 11:253–258 [Google Scholar]
- 8.Coton E, Coton M. 2009. Evidence of horizontal transfer as origin of strain to strain variation of the tyramine production trait in Lactobacillus brevis. Food Microbiol. 26:52–57 [DOI] [PubMed] [Google Scholar]
- 9.Cunin R, Glansdorff N, Piérard A, Stalon V. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50:314–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Man JD, Rogosa M, Sharpe ME. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130–135 [Google Scholar]
- 11.Fernández M, Zúñiga M. 2006. Amino acid catabolic pathways of lactic acid bacteria. Crit. Rev. Microbiol. 32:155–183 [DOI] [PubMed] [Google Scholar]
- 12.Gamper M, Ganter B, Polito MR, Haas D. 1992. RNA processing modulates the expression of the arcDABC operon in Pseudomonas aeruginosa. J. Mol. Biol. 226:943–957 [DOI] [PubMed] [Google Scholar]
- 13.Hugas M, Garriga M, Aymerich T, Monfort JM. 1993. Biochemical characterization of lactobacilli from dry fermented sausages. Int. J. Food Microbiol. 18:107–113 [DOI] [PubMed] [Google Scholar]
- 14.Janssens M, Myter N, De Vuyst L, Leroy F. 2012. Species diversity and metabolic impact of the microbiota are low in spontaneously acidified Belgian sausages with an added starter culture of Staphylococcus carnosus. Food Microbiol. 29:167–177 [DOI] [PubMed] [Google Scholar]
- 15.Larkin MA, et al. 2007. ClustalW and ClustalX version 2. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 16.Lauret R, et al. 1996. Carbohydrate utilization in Lactobacillus sake. Appl. Environ. Microbiol. 62:1922–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leroy F, Verluyten J, De Vuyst L. 2006. Functional meat starter cultures for improved sausage fermentation. Int. J. Food Microbiol. 106:270–285 [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Dong Y, Chen YYM, Burne RA. 2008. Environmental and growth phase regulation of the Streptococcus gordonii arginine deiminase genes. Appl. Environ. Microbiol. 74:5023–5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marceau A, Zagorec M, Chaillou S, Mera T, Champomier-Vergès MC. 2004. Evidence for involvement of at least six proteins in adaptation of Lactobacillus sakei to cold temperatures and addition of NaCl. Appl. Environ. Microbiol. 70:7260–7268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLeod A, Zagorec M, Champomier-Vergès MC, Naterstad K, Axelsson L. 2010. Primary metabolism in Lactobacillus sakei food isolates by proteomic analysis. BMC Microbiol. 10:120 doi:10.1186/1471-2180-10-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montel MC, Champomier MC. 1987. Arginine catabolism in Lactobacillus sake isolated from meat. Appl. Environ. Microbiol. 53:2683–2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naumoff DG, Xu Y, Glansdorff N, Labedan B. 2004. Retrieving sequences of enzymes experimentally characterized but erroneously annotated: the case of the putrescine carbamoyltransferase. BMC Genomics 5:52 doi:10.1186/1471-2164-5-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naumoff DG, Xu Y, Stalon V, Glansdorff N, Labedan B. 2004. The difficulty of annotating genes: the case of putrescine carbamoyltransferase. Microbiology 150:3908–3911 [DOI] [PubMed] [Google Scholar]
- 24.Nyquist OL, et al. 2011. Comparative genomics of Lactobacillus sakei with emphasis on strains from meat. Mol. Genet. Genomics 285:297–311 [DOI] [PubMed] [Google Scholar]
- 25.Olson ER. 1993. Influence of pH on bacterial gene expression. Mol. Microbiol. 8:5–14 [DOI] [PubMed] [Google Scholar]
- 26.Peirson SN, Butler JN, Foster RG. 2003. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 31:e73 doi:10.1093/nar/gng073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfaffl MW. 2004. Quantification strategies in real-time PCR, p 87–120. In Bustin SA. (ed), A-Z of quantitative PCR. IUL Biotechnology Series, International University Line, La Jolla, CA [Google Scholar]
- 28.Ravyts F, et al. 2008. Competitiveness and antibacterial potential of bacteriocin-producing starter cultures in different types of fermented sausages. J. Food Prot. 71:1817–1827 [DOI] [PubMed] [Google Scholar]
- 29.Ravyts F, De Vuyst L. 2011. Prevalence and impact of single-strain starter cultures of lactic acid bacteria on metabolite formation in sourdough. Food Microbiol. 28:1129–1139 [DOI] [PubMed] [Google Scholar]
- 30.Rimaux T, et al. 2011. The kinetics of the arginine deiminase pathway in the meat starter culture Lactobacillus sakei CTC 494 are pH-dependent. Food Microbiol. 28:597–604 [DOI] [PubMed] [Google Scholar]
- 31.Rimaux T, Vrancken G, Vuylsteke B, De Vuyst L, Leroy F. 2011. The pentose moiety of adenosine and inosine is an important energy source for the fermented-meat starter culture Lactobacillus sakei CTC 494. Appl. Environ. Microbiol. 77:6539–6550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rutherford K, et al. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945 [DOI] [PubMed] [Google Scholar]
- 33.Schillinger U, Lücke FK. 1989. Antibacterial activity of Lactobacillus sake isolated from meat. Appl. Environ. Microbiol. 55:1901–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzzi G, Gardini F. 2003. Biogenic amines in dry fermented sausages: a review. Int. J. Food Microbiol. 88:41–54 [DOI] [PubMed] [Google Scholar]
- 35.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 36.Vandesompele J, et al. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034 doi:10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vrancken G, Rimaux T, De Vuyst L, Leroy F. 2008. Kinetic analysis of growth and sugar consumption by Lactobacillus fermentum IMDO 130101 reveals adaptation to the acidic sourdough ecosystem. Int. J. Food Microbiol. 128:58–66 [DOI] [PubMed] [Google Scholar]
- 38.Vrancken G, Rimaux T, Weckx S, De Vuyst L, Leroy F. 2009. Environmental pH determines citrulline and ornithine release through the arginine deiminase pathway in Lactobacillus fermentum IMDO 130101. Int. J. Food Microbiol. 135:216–222 [DOI] [PubMed] [Google Scholar]
- 39.Weckx S, De Rijk P, Van Broeckhoven C, Del-Favero J. 2004. SNPbox: Web-based high-throughput primer design from gene to genome. Nucleic Acids Res. 32:W170–W172 doi:10.1093/nar/gkh369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yohannes E, Barnhart DM, Slonczewski JL. 2004. pH-dependent catabolic protein expression during anaerobic growth of Escherichia coli K-12. J. Bacteriol. 186:192–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zúñiga M, Champomier-Verges M, Zagorec M, Perez-Martinez G. 1998. Structural and functional analysis of the gene cluster encoding the enzymes of the arginine deiminase pathway of Lactobacillus sake. J. Bacteriol. 180:4154–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zúñiga M, Miralles MC, Pérez-Martínez G. 2002. The product of arcR, the sixth gene of the arc operon of Lactobacillus sakei, is essential for expression of the arginine deiminase pathway. Appl. Environ. Microbiol. 68:6051–6058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zúñiga M, Pérez G, González-Candelas F. 2002. Evolution of arginine deiminase (ADI) pathway genes. Mol. Phylogenet. Evol. 25:429–444 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.