Abstract
Synthesis of chiral cyanohydrins is performed in a monophasic micro-aqueous reaction system using whole recombinant Escherichia coli cells expressing the Arabidopsis thaliana hydroxynitrile lyase (AtHNL). Microscopy studies employing a fusion of AtHNL with a flavin-based fluorescent protein (FbFP) reveal that the cells remain intact in the reaction system.
INTRODUCTION
The synthesis of enantiopure cyanohydrins from aldehydes or ketones and hydrogen cyanide (HCN) catalyzed by hydroxynitrile lyases (HNLs) represents an industrially important biocatalytic route (5) (Fig. 1). Cyanohydrins serve as valuable building blocks in synthetic chemistry and the pharmaceutical industry (10). HNLs are usually applied in aqueous-organic two-phase systems at pH values equal to or below 4.5 in order to suppress the noncatalytic side reaction yielding racemic products (3).
Fig 1.
HNL-catalyzed hydrocyanation reaction in monophasic micro-aqueous methyl tert-butyl ether (MTBE).
However, such low pH values are not always tolerated by the enzymes. For example, the (R)-selective HNL from Arabidopsis thaliana (AtHNL) is unstable at low pH values (9), which severely limits its application in conventional aqueous-organic reaction systems. Application of the HNLs in organic solvent, with minimal water activity, represents an attractive alternative which has recently been explored using precipitated and immobilized enzyme preparations (14). The organic solvent system facilitates the solubility of aromatic substrates and products and suppresses the noncatalytic side reaction due to the low water content. However, the precipitated enzyme cannot be recycled and the preparation of recyclable immobilizates (i.e., enzyme formulations in which the enzyme is covalently linked or noncovalently bound to an inorganic material) requires additional steps and results in a partial loss of enzyme activity. In this respect, the use of whole cells in a monophasic micro-aqueous reaction system (6) would essentially eliminate all the aforementioned problems and thus represent a cost-efficient alternative to the use of purified and/or immobilized enzyme preparations. In order to test the applicability of such a system for the synthesis of chiral cyanohydrins, whole recombinant Escherichia coli cells expressing the AtHNL are used here as a whole-cell biocatalyst in a monophasic organic solvent. The cells and solvent can be recycled, and the product can easily be recovered from the solvent. Despite the significant potential of such an approach, only a few studies on the application of whole cells in monophasic organic solvents have so far been reported (6). Most of these studies focus on the stereoselective reduction of ketones by alcohol dehydrogenases using lyophilized yeast cells (6, 15). To the best of our knowledge, only two studies on the use of whole recombinant E. coli cells in water-free or micro-aqueous reaction systems have been published. In one study, lyophilized E. coli cells overexpressing an alcohol dehydrogenase from Rhodococcus ruber were used in a micro-aqueous system with 99% (vol/vol) isopropanol (6). A related strategy has recently been described, with lyophilized E. coli cells expressing the Candida parapsilosis carbonyl reductase, which were applied in a reaction system containing only neat substrates without the addition of an explicit solvent (11). While E. coli has not been used extensively in neat organic solvents, other bacteria such as solvent-tolerant Pseudomonas and Rhodococcus species have been applied for biotransformations under such conditions (13, 16). Classic and more recent examples include the production of cholest-4-ene-3-one by Nocardia species in neat carbon tetrachloride (2) and the synthesis of indigo by a Rhodococcus strain in pure bis(2-ethylhexyl) phthalate (19). While solvent-tolerant bacteria are promising candidates for nonaqueous whole-cell catalysis (12), they simply cannot rival E. coli in terms of easy handling and availability of molecular tools.
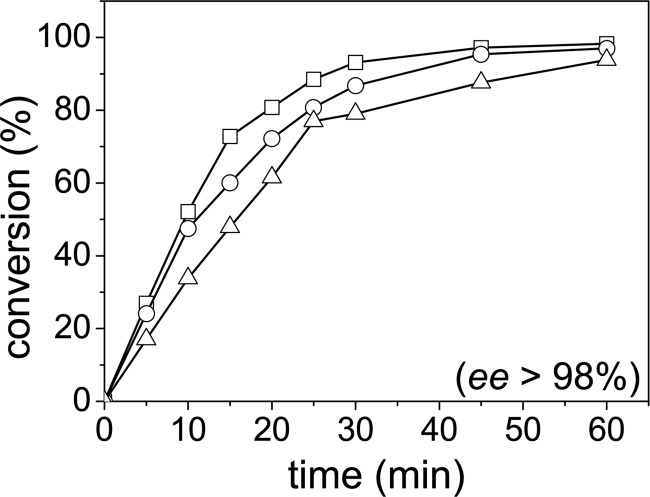
The feasibility of a monophasic micro-aqueous whole-cell reaction system for the production of chiral cyanohydrins was initially tested by the synthesis of (R)-mandelonitrile from benzaldehyde and hydrogen cyanide (HCN). The reaction was carried out at 20°C in a reaction system consisting of 1 ml of 2 M HCN solution in buffer-saturated methyl tert-butyl ether (MTBE) containing 350 mg of wild-type AtHNL-expressing E. coli BL21(DE3) cells. The reaction was started by the addition of 0.5 mmol benzaldehyde to the reaction system and monitored over 60 min by chiral gas chromatography (GC) (Fig. 2). MTBE is widely used in biocatalysis and has previously been shown to be advantageous for use with free and immobilized AtHNL preparations (14). To recycle the cells, fresh or frozen E. coli BL21(DE3) cells expressing wild-type AtHNL were placed in nylon mesh. Thus, the cells can easily be removed from the reaction vessel facilitating easy cell recycling. After each conversion, the cells were washed with MTBE, placed in a new vessel, and MTBE and fresh substrates were added. Both freshly prepared and frozen cells displayed very similar conversion rates and enantioselectivities over as many as three reaction cycles (Fig. 2; see also Fig. S1 in the supplemental material). Although initial reaction rates decreased afterwards, subsequent biotransformations still resulted in nearly complete conversion of benzaldehyde after a reaction time of 60 min. After 5 reaction cycles, AtHNL-expressing cells still converted 85% benzaldehyde with an enantiomeric excess (ee) of 98% within 60 min (data not shown).
Fig 2.
Conversion of benzaldehyde and HCN to (R)-mandelonitrile by whole E. coli BL21(DE3) cells expressing wild-type AtHNL. All reactions were performed at 20°C using 350 mg of wet cells placed in nylon mesh (pore size, 0.4 μm). The reaction system consisted of 1 ml of 2 M HCN in buffer-saturated MTBE mixed with 0.5 mmol of benzaldehyde as the substrates. After each conversion round, the cells in the nylon mesh were washed with MTBE and placed in a fresh reaction vessel, and the subsequent conversion was started by the addition of new substrates. Symbols: □, first reaction cycle; ○, second reaction cycle; △, third reaction cycle. During all reaction cycles the ee exceeded 98%.
Thus, recycling of whole cells resulted in slightly lower initial reaction rates and final yields in subsequent conversion rounds, however, without loss of enantioselectivity.
Cyanohydrin synthesis was further studied with different aldehyde substrates in order to evaluate the applicability of our reaction setup in more detail (Table 1). As before, 1 ml of 2 M HCN in MTBE mixed with 0.5 mmol of the respective aldehyde was used as the substrates. All reactions were carried out at 20°C. Fresh (wet) cells showed good conversion rates but gave only moderate ee values for 2-chloromandelonitrile and 2-fluoromandelonitrile (Table 1, reactions 2 and 3). In contrast, 2-furaldehyde (Table 1, reaction 4) was only poorly converted (50%) with low enantioselectivity (ee, 30%).
Table 1.
Conversion of different aldehydes to the respective cyanohydrines by whole E. coli cells expressing wild-type AtHNLa
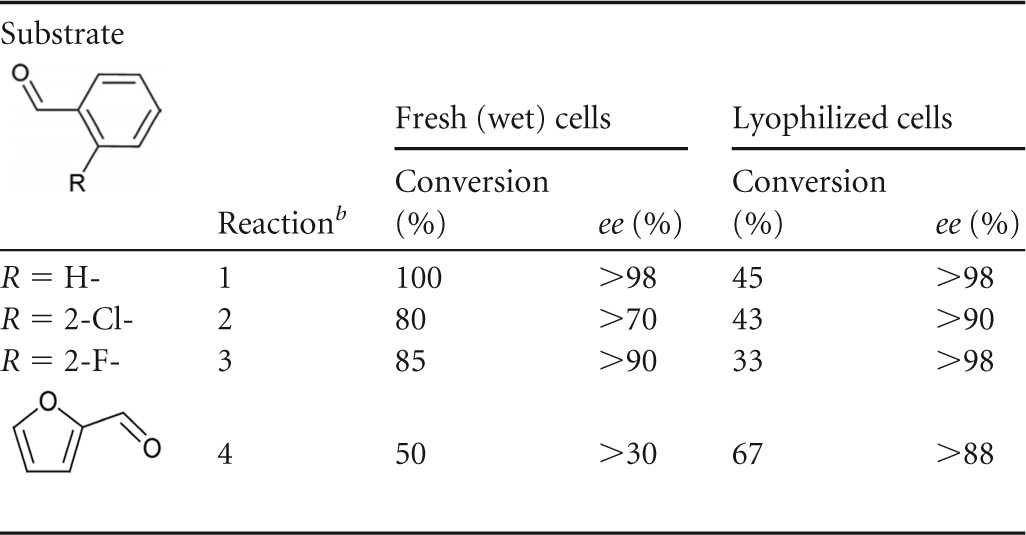
All conversions were carried out at 20°C using 350 mg of fresh (wet) cells or the corresponding amount of lyophilized cells. The reaction system consisted of 1 ml of 2 M HCN in buffer-saturated MTBE mixed with 0.5 mmol of the respective aldehyde. Conversion rates and ee values were determined by chiral GC from the relative peak areas of the aldehydes and the cyanohydrin derivatives.
1, benzaldehyde; 2, 2-chlorobenzaldehyde; 3, 2-fluorobenzaldehyde; 4, 2-furaldehyde. Reactions were observed for 60 min.
The difference between enzymatic and nonenzymatic reaction rates for the respective aldehydes may provide a reasonable explanation for the reduced enantioselectivities observed in comparison to those during benzaldehyde conversion. It has previously been demonstrated that, e.g., for 2-chlorobenzaldehyde and 2-fluorobenzaldehyde, the nonenzymatic side reaction in an aqueous reaction system is much faster than for benzaldehyde (1). Consequently, when dry AtHNL immobilizates were used for the conversion of 2-chlorobenzaldehyde and 2-furaldehyde, excellent ee values [>98% (R) enantiomer] could be obtained (14). Therefore, it appears that the water content in the MTBE-washed cell pellet (or in the E. coli cells) compromises the enantioselectivity in our whole-cell reaction system. To elucidate this possibility, the respective E. coli cells were lyophilized and used for the conversion of the same aldehyde substrates. Not surprisingly, conversion was lower than when the same amount of fresh cells was used; however, ee values for all products increased significantly. In particular, 2-furaldehyde could be converted with an ee value of 88% (Table 1, reaction 4). Likewise, 2-chlorobenzaldehyde that was previously converted with a moderate ee of 70% by fresh (wet) cells was now converted with an ee of 90% by lyophilized cells (Table 1, reaction 2).
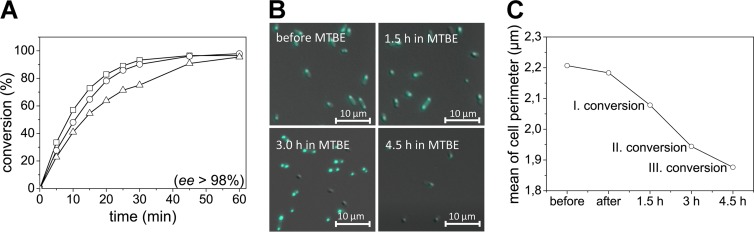
Bacterial cells are evolved to be stable in an aqueous environment. Hence, the impact of prolonged incubation in MTBE remains unclear. In order to evaluate the influence of the reaction system on cell integrity, a translational fusion of a flavin-based fluorescent protein (FbFP) with the AtHNL was generated. In the fusion construct, nFbFP-AtHNL, the FbFP module is attached N-terminally to the AtHNL by molecular biological methods. FbFPs were initially derived from the sensor domain of plant and bacterial light-oxygen-voltage (LOV) photoreceptors (7). They represent a promising new class of fluorescent reporter proteins with high potential for biotechnological and cell biological application (4, 7, 8, 17, 18). The resulting fluorescent cells expressing the fusion enzyme can be used similarly to the AtHNL-expressing E. coli cells for cyanohydrin synthesis. Further, the use of a fluorescent whole-cell biocatalyst allows the application of fluorescence microscopy and spectroscopy to track the fate of the cells in the reaction system. Wild-type AtHNL and the fusion with the fluorescent protein (nFbFP-AtHNL) are expressed at comparable levels in E. coli BL21(DE3) (data not shown). With respect to (R)-mandelonitrile synthesis and cell recycling, nFbFP-AtHNL-expressing E. coli cells behave identically to the wild-type AtHNL-expressing cells (Fig. 3A). Fluorescence microscopy revealed cell shrinkage by about 20% during three consecutive reaction cycles in MTBE (Fig. 3B and C). Interestingly, this small change in cell morphology coincides with an overall slightly decreased biocatalytic performance in the next cycle (Fig. 3A).
Fig 3.
(A) Conversion of benzaldehyde and HCN to (R)-mandelonitrile by whole E. coli BL21(DE3) cells expressing nFbFP-AtHNL. All reactions were carried out at 20°C using 350 mg of wet E. coli cells. The reaction system consisted of 1 ml of 2 M HCN in buffer-saturated MTBE mixed with 0.5 mmol of benzaldehyde as the substrates. After each conversion round, the cells were washed with MTBE, and the subsequent conversion was started by the addition of new substrates. Symbols: □, first reaction cycle; ○, second reaction cycle; △, third reaction cycle. During all reaction cycles the ee exceeded 98%. (B) E. coli cells expressing nFbFP-AtHNL imaged with fluorescence microscopy. Cells were incubated in buffer-saturated MTBE for up to 4.5 h. Samples were taken at time points corresponding to the respective conversion cycle. (C) Change in relative E. coli cell perimeter during incubation in MTBE. Data were derived from statistical evaluation of microscopy pictures (see the supplemental material for details).
The fluorescence signal of the fused reporter protein, however, remained stable during several hours of incubation (see Fig. S2 in the supplemental material), indicating proper reporter protein integrity within the cell. Despite the observed cell shrinkage during incubation in MTBE, cellular integrity was essentially retained. Mechanistically, the reduced conversion rate during recycling may thus be due to limited leakage of the enzyme from the cells, the loss of cell material from the nylon mesh during washing and recycling steps, or could be caused by partial inactivation of the intracellular enzyme by the substrates.
Conclusions.
Standard E. coli BL21(DE3) cells, expressing the Arabidopsis thaliana HNL, were used as whole-cell biocatalyst for the synthesis of chiral cyanohydrins in monophasic micro-aqueous MTBE. The use of lyophilized cells makes this approach also suitable for the conversion of aldehydes showing fast noncatalyzed racemic product formation in aqueous systems. Due to its simplicity (use of whole cells, no protein purification, monophasic organic solvent), this novel process for the production of chiral cyanohydrins could also be an interesting alternative for industrial applications. In general, the application of whole-cell biotransformations in pure organic solvents represents a promising alternative to conventional biotransformations in aqueous media. The use of monophasic microaqueous organic solvents may be particularly interesting when poor substrate or product solubility limits their application or when substrates and/or products are unstable in water. Our approach should be directly transferable to any HNL that can be expressed in E. coli.
Moreover, our microscopic analyses revealed that standard E. coli cells do not lose their cellular integrity when incubated in MTBE. Hence, it can be expected that the transfer of the presented whole-cell biotransformation approach to other enzymatic systems could open up new biocatalytic routes even for the synthesis of novel products which are currently not accessible by biocatalysis.
Likewise, the use of fluorescent reporter techniques in such studies allows an in situ monitoring of the respective biocatalytic system. In this respect, we currently evaluate the potential of FbFPs and other fluorescent proteins as reporters for further biotechnological applications, including solvent screening and localization of enzymes in immobilization carrier materials and micellar systems.
Supplementary Material
ACKNOWLEDGMENT
This work was funded by the DFG research training group GK1166 “Biocatalysis using non-conventional media” (BioNoCo).
Footnotes
Published ahead of print 27 April 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Andexer J, et al. 2007. An R-selective hydroxynitrile lyase from Arabidopsis thaliana with an alpha/beta-hydrolase fold. Angew. Chem. Int. Ed. Engl. 46:8679–8681 [DOI] [PubMed] [Google Scholar]
- 2.Buckland BC, Dunnill P, Lilly MD. 1975. The enzymatic transformation of water-insoluble reactants in nonaqueous solvents. Conversion of cholesterol to cholest-4-ene-3-one by a Nocardia sp. Biotechnol. Bioeng. 17:815–826 [PubMed] [Google Scholar]
- 3.Bühler H, Effenberger F, Forster S, Roos J, Wajant H. 2003. Substrate specificity of mutants of the hydroxynitrile lyase from Manihot esculenta. Chembiochem 4:211–216 [DOI] [PubMed] [Google Scholar]
- 4.Chapman S, et al. 2008. The photoreversible fluorescent protein iLOV outperforms GFP as a reporter of plant virus infection. Proc. Natl. Acad. Sci. U. S. A. 105:20038–20043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dadashipour M, Asano Y. 2011. Hydroxynitrile lyases: insights into biochemistry, discovery, and engineering. ACS Catal. 1:1121–1149 [Google Scholar]
- 6.de Gonzalo G, Lavandera I, Faber K, Kroutil W. 2007. Enzymatic reduction of ketones in “micro-aqueous” media catalyzed by ADH-A from Rhodococcus ruber. Org. Lett. 9:2163–2166 [DOI] [PubMed] [Google Scholar]
- 7.Drepper T, et al. 2007. Reporter proteins for in vivo fluorescence without oxygen. Nat. Biotechnol. 25:443–445 [DOI] [PubMed] [Google Scholar]
- 8.Drepper T, et al. 2010. Flavin mononucleotide-based fluorescent reporter proteins outperform green fluorescent protein-like proteins as quantitative in vivo real-time reporters. Appl. Environ. Microbiol. 76:5990–5994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guterl JK, et al. 2009. Uneven twins: comparison of two enantiocomplementary hydroxynitrile lyases with alpha/beta-hydrolase fold. J. Biotechnol. 141:166–173 [DOI] [PubMed] [Google Scholar]
- 10.Hatti-Kaul R, Tornvall U, Gustafsson L, Borjesson P. 2007. Industrial biotechnology for the production of bio-based chemicals—a cradle-to-grave perspective. Trends Biotechnol. 25:119–124 [DOI] [PubMed] [Google Scholar]
- 11.Jakoblinnert A, et al. 2011. Asymmetric reduction of ketones with recombinant E. coli whole cells in neat substrates. Chem. Commun. (Camb.) 47:12230–12232 [DOI] [PubMed] [Google Scholar]
- 12.Klibanov AM. 2001. Improving enzymes by using them in organic solvents. Nature 409:241–246 [DOI] [PubMed] [Google Scholar]
- 13.Nikolova P, Ward OP. 1993. Whole-cell biocatalysis in nonconventional media. J. Ind. Microbiol. 12:76–86 [DOI] [PubMed] [Google Scholar]
- 14.Okrob D, et al. 2011. Hydroxynitrile lyase from Arabidopsis thaliana: identification of reaction parameters for enantiopure cyanohydrin synthesis by pure and immobilized catalyst. Adv. Synth. Catal. 353:2399–2408 [Google Scholar]
- 15.Rotthaus O, Krüger D, Demuth M, Schaffner K. 1997. Reductions of keto esters with baker's yeast in organic solvents—a comparison with the results in water. Tetrahedron 53:935–938 [Google Scholar]
- 16.Sardessai YN, Bhosle S. 2004. Industrial potential of organic solvent tolerant bacteria. Biotechnol. Prog. 20:655–660 [DOI] [PubMed] [Google Scholar]
- 17.Shu X, et al. 2011. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 9:e1001041 doi:10.1371/journal.pbio.1001041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tielker D, Eichhof I, Jaeger KE, Ernst JF. 2009. Flavin mononucleotide-based fluorescent protein as an oxygen-independent reporter in Candida albicans and Saccharomyces cerevisiae. Eukaryot. Cell 8:913–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamashita S, et al. 2007. Utilization of hydrophobic bacterium Rhodococcus opacus B-4 as whole-cell catalyst in anhydrous organic solvents. Appl. Microbiol. Biotechnol. 74:761–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.