Abstract
Human milk oligosaccharides (HMO), which constitute a major component of human milk, promote the growth of particular bacterial species in the infant's gastrointestinal tract. We hypothesized that HMO also interact with the bacterial communities present in human milk. To test this hypothesis, two experiments were conducted. First, milk samples were collected from healthy women (n = 16); culture-independent analysis of the bacterial communities was performed, HMO content was analyzed, and the relation between these factors was investigated. A positive correlation was observed between the relative abundance of Staphylococcus and total HMO content (r = 0.66). In a follow-up study, we conducted a series of in vitro growth curve experiments utilizing Staphylococcus aureus or Staphylococcus epidermidis and HMO isolated from human milk. HMO exhibited stimulatory effects on bacterial growth under various nutritional conditions. Analysis of culture supernatants from these experiments revealed that HMO did not measurably disappear from the culture medium, indicating that the growth-enhancing effects were not a result of bacterial metabolism of the HMO. Instead, stimulation of growth caused greater utilization of amino acids in minimal medium. Collectively, the data provide evidence that HMO may promote the growth of Staphylococcus species in the lactating mammary gland.
INTRODUCTION
Human milk oligosaccharides (HMO) are complex glycans highly abundant (∼5 to 15 g/liter) in human milk (5, 6, 24). Representing a diverse collection of structures, almost 150 distinct forms of HMO have been identified (42, 43). Based upon their structures, HMO can be classified as simple (sialylated or fucosylated lactose compounds made of three sugar units) or complex (composed of differentially sialylated or fucosylated repeats of lacto-N-biose or N-acetyllactosamine linked to a lactose motif). Interestingly, HMO patterns are highly unique to individual women, and their concentrations vary over the course of lactation, with levels peaking in colostrum and lower concentrations in mature milk (8, 12, 15, 39).
From an evolutionary standpoint, it is logical that because the mammary gland exerts a substantial amount of energy to produce these compounds, HMO likely promote the fitness of the offspring even though the infant is largely unable to metabolize them (1, 14, 16). Research from several groups suggests the possibility that HMO are capable of interacting with an infant's gastrointestinal microbiota to promote his or her health (10, 27, 30, 35, 38). Indeed, some of the first studies in this area noted the capacity of HMO to promote growth of Bifidobacterium bifidum, a species overrepresented in the gastrointestinal tract of the breast-fed, but not formula-fed, infant (17, 29, 40). Over 50 years later, research has confirmed that the genetic capacity to metabolize these compounds is conserved among bifidobacteria species common in the infant's gastrointestinal tract (38). These findings suggest that HMO may serve a prebiotic function in selectively promoting the presence of certain types of gastrointestinal bacteria.
Several recent reports have confirmed that human milk contains bacterial communities (19, 28, 32). For instance, we have described a relatively complex milk microbiome with several bacterial genera collectively representing ≥5% of the overall microbial community (19). We also reported that the composition of human milk bacterial communities is somewhat stable within a woman over time. Considering the aforementioned effects of HMO on the bacterial communities of the infant's gastrointestinal microbiota, we hypothesized that HMO may influence the milk microbiome. Fueling this hypothesis was the observation that both HMO profiles and milk bacterial communities are personalized to the woman who produces the milk, suggesting a potential connection between HMO and bacterial community patterns in human milk. To test our hypothesis, we analyzed and related the bacterial communities and HMO content of milk produced by 16 healthy women. We then conducted a series of in vitro experiments to gain a more detailed understanding of our in vivo observations.
MATERIALS AND METHODS
Subjects and experimental design of in vivo study.
Human milk was obtained from 16 healthy, lactating women and analyzed for its bacterial content as described previously (19). Briefly, DNA was extracted from each sample, and PCRs with bar-coded primers were carried out to amplify the V1-V2 segment of the 16S rRNA gene. Pyrosequencing of the amplicons was performed with the Roche 454 FLX platform, and quality control measures were employed to remove sequences of questionable quality. Sequences were then assigned phylogeny at the genus level using the Ribosomal Database Project Bayesian classifier (41). One sample from each woman was selected for analysis for HMO content as described below. All procedures for this portion of the study were approved by the University of Idaho and the Washington State University Institutional Review Boards.
HMO purification for in vitro studies.
To obtain sufficient amounts of HMO for the in vitro studies, additional milk was obtained from 16 healthy volunteers, different from the subjects described above, who were recruited at the University of California San Diego Medical Center, San Diego, CA, after approval by the University's Institutional Review Board. The milk samples from these volunteers were pooled, and oligosaccharides were isolated and purified as previously described (20, 21). The lipid layer was removed after centrifugation, and proteins were precipitated from the aqueous phase by addition of ice-cold ethanol and subsequent centrifugation. Ethanol was removed from the HMO-containing supernatant by roto-evaporation. Lactose and salts were removed by gel filtration chromatography over a Bio-Rad P2 column (100 cm by 16 mm; Bio-Rad, Hercules, CA) using a semiautomated fast protein liquid chromatography (FPLC) system. Endotoxins were removed by affinity chromatography over polymyxin B gravity columns (Thermo Scientific, Rockford, IL).
HMO analysis of milk samples and culture supernatants.
HMO were analyzed as previously described (20, 21). Briefly, raffinose was added to 40 μl of milk or culture supernatant to serve as an internal standard through sample processing and analysis. Lipids and proteins were removed from the samples by centrifugation and chloroform-methanol extraction. Lactose was removed by overnight incubation on lactase-immobilized beads (Invitrogen, Carlsbad, CA) at 37°C. Residual peptides and salt were removed over Sep-Pak C18 cartridges followed by porous graphitized carbon (PGC) cartridges. The reducing ends of the dried oligosaccharides were labeled with the fluorescent tag 2-aminobenzamide (2AB) for 2 h at 65°C. Free 2AB label was separated from the 2AB-labeled oligosaccharides using silica gel cartridges. 2AB-labeled oligosaccharides were analyzed by high-performance liquid chromatography (HPLC) on an amide-80 column (4.6-mm internal diameter by 25 cm; particle size, 5 μm; Tosoh Bioscience, Tokyo, Japan) with a 50 mM ammonium formate-acetonitrile buffer system. Separation was performed at 25°C and monitored with a fluorescence detector at 360-nm excitation and 425-nm emission. Peak annotation was based on standard retention times and mass spectrometric (MS) analysis on a Thermo LCQ Duo Ion trap mass spectrometer equipped with a nano-electrospray ionization (ESI) source. Total concentration of HMO was calculated as the sum of most common oligosaccharides, including 2′-fucosyllactose, 3′-fucosyllactose, 3′-sialyllactose, lacto-N-tetraose, lacto-N-neotetraose, lacto-N-fucopentaose I, lacto-N-fucopentaose II, and lacto-N-fucopentaose III, which collectively represent approximately 80% of all HMO in each sample.
To account for the effects of evaporation on the HMO concentrations of culture supernatants collected after incubation with bacteria, controls were run without the addition of bacteria to the medium. HMO values were then normalized for evaporation loss using these controls.
Isolation of Staphylococcus strains from human milk.
Milk samples were plated on sheep's blood agar and incubated for 24 h at 37°C. Individual colonies were selected for overnight growth in tryptic soy broth (TSB), and the subsequent cultures were streaked on mannitol salt agar (MSA) to determine if any of the cultures were Staphylococcus. Isolates that grew successfully on MSA were then selected for sequencing of the 16S gene to confirm their identities using previously described methods (45). One strain each of Staphylococcus epidermidis and Staphylococcus aureus was selected for use in the series of in vitro experiments described next.
Growth curve culture conditions.
To fully elucidate the effects of HMO on Staphylococcus growth, experiments were carried out in a variety of nutritional media, including TSB (EMD chemicals, Gibbstown, NJ), glucose-free Dulbecco's modified Eagle Medium (DMEM; Mediatech, Manassas, VA), a carbohydrate-free modification of a previously described defined medium for Staphylococcus growth (11), (see Table S1 in the supplemental material), and Similac 60/40 infant formula (Abbott Nutrition, Columbus, OH). Carbohydrate treatments (HMO, glucose, and lactose) were prepared by adding the component of interest (10 g/liter) to the culture medium before sterile filtration. All experiments were initiated by spiking cultures with ∼104 CFU/ml to approximate the concentration of bacteria found in human milk (2, 9). Cultures were aerobically incubated at 37°C with agitation at 250 rpm. At multiple time points, serial dilutions of each replicate were performed before plating and incubation of the appropriate dilutions to enumerate CFU/ml. Experiments were conducted twice in triplicate (n = 6).
Amino acid analysis.
Polar metabolites were extracted from culture medium samples using cold methanol extraction (44). Culture medium (100 μl) was mixed with 500 μl of cold 50% methanol containing 100 μM l-norvaline as an internal standard. Samples were vortexed, and 300 μl of chloroform was added. The samples were then vortexed for 15 s and placed on ice for 45 s; this vortex and cooling step was repeated five times. Samples were then centrifuged for 5 min at 14,000 × g at 4°C. A total of 400 μl of the top layer (methanol extract) was dried overnight, as was 0.05 to 25 nmol of a standard. After the addition of 50 μl of pure pyridine (Sigma-Aldrich) and 50 μl of N-tert-butyldimethylsilyl-N-methylfluoride-acetamide (PBDMS; Sigma-Aldrich), dried samples and standards were derivatized at 80°C for 60 min. Samples were vortexed several times during the heating process to dissolve the pellet. Finally, samples were centrifuged at 13,000 × g for 10 min, and the supernatant was transferred to glass vials for analysis by gas chromatography-mass spectrometry (GC-MS) as previously described (37). A Shimadzu QP2010 Plus GC-MS was utilized at an injection temperature of 250°C with an injection split ratio of 1/10 and an injection volume of 1 to 2 μl. GC oven temperature began at 130°C for 4 min, increasing to 230°C at 4°C/min and to 280°C at 20°C/min, with a final hold at this temperature for 2 min. GC flow rate with helium carrier gas was 50 cm/s over a 15-m by 0.25-mm by 0.25-μm SHRXI-5ms Shimadzu column. The mass spectrometer scanned an m/z range of 50 to 600, with ∼1 kV detector sensitivity (modified as necessary).
Statistical analysis.
R statistical software (33) was utilized for the following statistical tests. The Pearson correlation coefficient was calculated to test the associations between the relative abundance of the 15 most prevalent bacterial genera and HMO content in the milk samples collected in the in vivo portion of this study. An analysis of variance was performed on culture pH values measured at the termination of the growth curves. Tukey's honestly significant difference (HSD) test was then used to determine which treatments had elicited statistically different alterations in pH at the termination of the growth curves. One-sided t tests were performed to determine if the concentrations of individual HMO as well as individual amino acids after incubation with bacteria were significantly decreased from their starting values.
Results from the bacterial growth curves were fitted to separate mathematical models to estimate growth parameters using generalized nonlinear regression algorithms. Because growth dynamics varied greatly depending on the culture medium employed, data were analyzed independently for each growth medium. After testing several types of growth models, we determined that the Weibull model (34) was the most appropriate. Thus, the following growth model was used: Y = C + (D − C) × exp[B × (log(t) − log(I))]. In this equation Y is bacterial growth (log CFU/ml), t is time (in hours), C is the bacterial concentration at the initiation of the experiment (t = 0), D is the maximum growth value achieved, B is the rate of growth, and I is the time of the inflection point when the growth rate began to decrease.
For growth of the S. epidermidis isolate in the carbohydrate-free defined medium, the model fit was adjusted to constrain the value of D (maximum bacterial growth achieved) to a log CFU value of 10. This approach was selected because when the experiment was terminated at 24 h, growth had not reached a maximum in three of the six treatments. Consequently, the model was not able to predict an appropriate estimate for D. The constraint was set at 10 because in the trials performed with this isolate, the maximum log CFU observed was consistently near this value. Furthermore, with this constraint in place, the model fit was adequate. The residuals for each model were assessed for normality and random distribution to ensure adequacy of fit.
The experiments carried out with TSB as the growth medium were also analyzed with the Weibull model. For the growth of the S. aureus isolate, the model fit was good, as determined by examining the residuals as described previously. Although the fit for the growth of the S. epidermidis isolate was not perfect, it was nonetheless deemed sufficient; the residuals were organized in an alternating pattern, relatively small in magnitude, and complied with the assumption of normality.
The segmented exponential regression model was the best fit to describe the growth of the isolates in infant formula, and the following growth model was employed: L = log(1 − C/M)/−B, where Y = C for t ≤ L; otherwise Y = M × [1 − exp(−B × t)]. In this equation Y is bacterial growth (log CFU/ml), L is lag phase before the initiation of exponential growth, t is time (in hours), C is the bacterial concentration at the initiation of the experiment (t = 0), M is the maximum growth value achieved, and B is the growth rate.
A full model dummy-variable regression procedure was employed for both isolates in each growth medium to perform preplanned contrasts to evaluate the effects of the carbohydrate treatments on the equality of estimated lines, growth rates, and maximum growth values. Significance was declared at a P value of <0.05. Analyses of growth curve data were carried out using the NLMIXED procedure of SAS (SAS Institute, Inc., Cary, NC).
RESULTS
A culture-independent analysis of the bacterial communities in the milk samples collected for the in vivo portion of this study revealed that the most abundant genera present were Streptococcus (25% ± 6%, mean ± standard error of the mean [SEM]), Staphylococcus (18% ± 6%), Serratia (8% ± 2%), and Corynebacterium (4% ± 2%). The total HMO content (7.7 ± 0.7 g/liter) was positively related (r = 0.66, P < 0.01) to the relative abundance of Staphylococcus. No relation was observed, however, between total HMO and any other highly abundant bacterial genera.
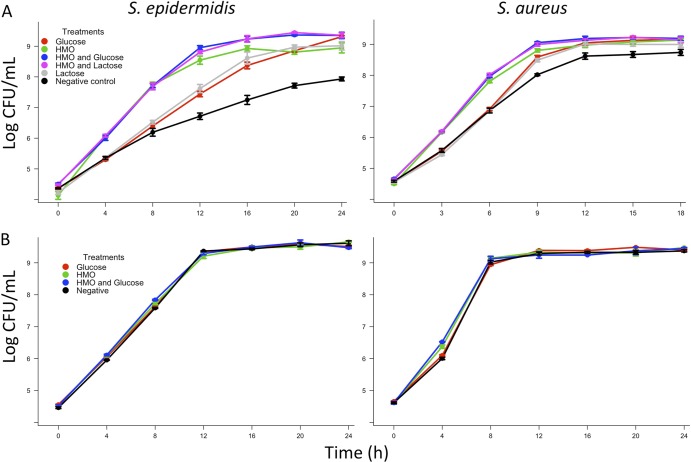
As described previously, to investigate the correlation between Staphylococcus and HMO, selective media were utilized to isolate bacterial strains from human milk, and one strain each of S. aureus and S. epidermidis obtained was selected for use in all experiments. Bacterial growth curves were then completed in a variety of media with or without HMO to elaborate the effects of these complex glycans on Staphylococcus growth. In each experiment carbohydrate treatments were formulated at a concentration of 10 g/liter to emulate the reported concentration of HMO in mature human milk. The first series of experiments utilized a carbohydrate-free formulation of a defined medium for Staphylococcus aureus growth (11) (see Table S1 in the supplemental material). Treatments included a negative control with no carbohydrate added, medium supplemented with HMO, medium supplemented with glucose or lactose, and medium supplemented with both HMO and glucose (HMO+G) or HMO and lactose (HMO+L). Comparison of the regression lines generated in this series of experiments demonstrated that the growth of both isolates was enhanced (P < 0.0001) by the HMO treatment compared to growth of the negative control (Table 1 and Fig. 1A). Additionally, in both isolates, the HMO+G and HMO+L treatments resulted in enhanced growth compared to treatments of only glucose or lactose.
Table 1.
Contrasts of regression lines among treatments
| Medium | Contrasted pair |
P value |
|
|---|---|---|---|
| S. epidermidis | S. aureus | ||
| Carbohydrate-free | HMO+glucose vs glucose | <0.0001 | <0.0001 |
| HMO+lactose vs lactose | <0.0001 | <0.0001 | |
| HMO vs negative control | <0.0001 | <0.0001 | |
| TSB | HMO+glucose vs glucose | 0.33 | <0.0001 |
| HMO vs negative control | 0.26 | <0.0001 | |
| Formula | HMO vs negative control | <0.0001 | <0.0001 |
Fig 1.
(A) One strain of S. epidermidis and one of S. aureus isolated from human milk were grown in a carbohydrate-free medium. The effects of various carbohydrate sources on growth were tested by adding them to the medium at a concentration of 10 g/liter. The growth of both isolates with HMO added was enhanced in comparison with the negative control (P < 0.0001, n = 6). (B) When the same strains were grown in TSB, HMO addition did not affect the growth of the S. epidermidis isolate (P = 0.26, n = 3) but altered the growth of the S. aureus isolate (P < 0.0001, n = 3) although the differences were subtle and occurred mainly in the first 4 h of growth.
In the second set of in vitro experiments, TSB (a nutritionally rich medium containing 2.5 g/liter glucose) was employed. The treatment effect of HMO addition reached statistical significance only in the S. aureus isolate (P < 0.0001), but the magnitude of the effect was less than that observed in the carbohydrate-free medium (Table 1 and Fig. 1B). In the S. epidermidis isolate no treatment effect was observed.
To create culture conditions that more closely emulated the nutritional conditions in the lactating mammary gland, a third series of growth curves was carried out using HMO-free infant formula (Similac 60/40; Abbott Nutrition) as a growth medium. This particular infant formula was selected because, like human milk, it contains almost no glucose. Rather, this formula contains lactose as a carbohydrate source. Addition of HMO elicited an alteration of growth in both isolates, as evidenced by the lack of coincidence of lines in the regression model (Table 1 and Fig. 2). Specifically, HMO enhanced growth by increasing the rate and maximum growth achieved by the cultures (see Table S4 in the supplemental material).
Fig 2.
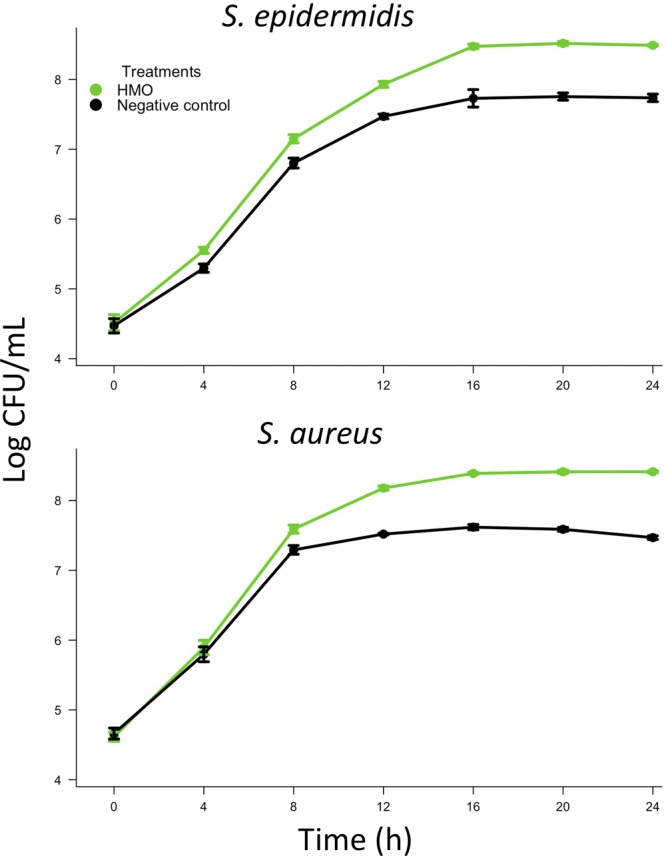
Staphylococcus isolates were grown in HMO-free infant formula with or without HMO (10 g/liter). The growth of both isolates was enhanced by HMO (P < 0.0001, n = 6).
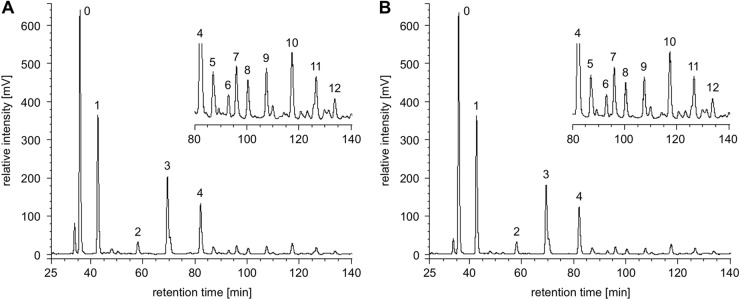
To determine if the effects of HMO on bacterial growth were associated with catabolism of HMO, culture supernatants were collected before the experiments and when growth had reached stationary phase, late in the growth curves. No measurable decrease in HMO concentration from either isolate's supernatants was observed between time zero and stationary phase (Fig. 3 and Table 2). Consequently, because no disappearance of substrate was observed, the HMO-associated enhancement of bacterial growth was not due to catabolism of HMO.
Fig 3.
Representative HPLC-fluorescence chromatogram of HMO in culture supernatant of the S. aureus isolate at t = 0 (A) and t = 30 (B). No alteration in HMO content was observed in supernatants from either the S. epidermidis or the S. aureus isolate. The numbered peaks represent the following oligosaccharides: 0, raffinose (internal standard); 1, 2′-fucosyllactose (2′FL); 2, 3′-sialyllactose (3′SL); 3, lacto-N-tetraose (LNT); 4, lacto-N-fucopentaose 1 (LNFP1); 5, lacto-N-fucopentaose 2 (LNFP2); 6, sialyl-lacto-N-tetraose b; 7, sialyl-lacto-N-tetraose c; 8, lacto-N-difuco-hexaose; 9, disialyl-lacto-N-tetraose; 10, fucosyl-lacto-N-hexaose; 11, difucosyl-lacto-N-hexaose; 12, fucosyl-disialyl-lacto-N-hexaose.
Table 2.
HMO concentrations in culture supernatant of the S. epidermidis and S. aureus isolates at t = 0 h and t = 30 h
| HMOa | HMO concn(μg/ml) atb: |
||
|---|---|---|---|
| t = 0 h |
t = 30 h |
||
| S. epidermidis | S. aureus | ||
| Total | 10.9 × 103 | (10.7 ± 0.3) × 103 | (11.4 ± 0.1) × 103 |
| 2′FL | 2,550 | 2,460 ± 73 | 2,526 ± 60 |
| 3′SL | 313 | 325 ± 9 | 331 ± 8 |
| LNT | 2,208 | 2,246 ± 70 | 2,309 ± 40 |
| LNnT | 318 | 313 ± 13 | 316 ± 9 |
| LNFP I | 2,721 | 2,740 ± 78 | 2905 ± 19 |
| LNFP II | 635 | 661 ± 15 | 696 ± 8 |
| LSTb | 172 | 187 ± 7 | 191 ± 6 |
| LSTc | 370 | 359 ± 10 | 383 ± 2 |
Abbreviations: 2′FL, 2′-fucosyllactose; 3′FL, 3′-sialyllactose; LNT, lacto-N-tetraose; LNFP I, lacto-N-fucopentaose 1; LNFP II, lacto-N-fucopentaose 2; LSTb, sialyl-lacto-N-tetraose b; LSTc, sialyl-lacto-N-tetraose c.
None of the HMO fractions measured decreased after incubation with bacteria. The values for t = 0 represent both isolates.
To gain more insight into the effects of HMO on bacterial metabolism, pH was assessed at each time point during the growth curves performed in the carbohydrate-free medium (Fig. 4). In cultures of both isolates, the HMO+G treatment resulted in a drastic drop in pH, whereas the glucose-only treatment was associated with a less pronounced reduction. Likewise, in cultures of the S. epidermidis isolate, the HMO+L treatment caused a substantial decrease in pH, and the lactose-only treatment caused a less drastic reduction. However, in cultures of the S. aureus isolate, the HMO+L and lactose-only treatments elicited only mild decreases in pH. Most importantly, the HMO treatment in both isolates resulted in very little alteration in the culture pH.
Fig 4.
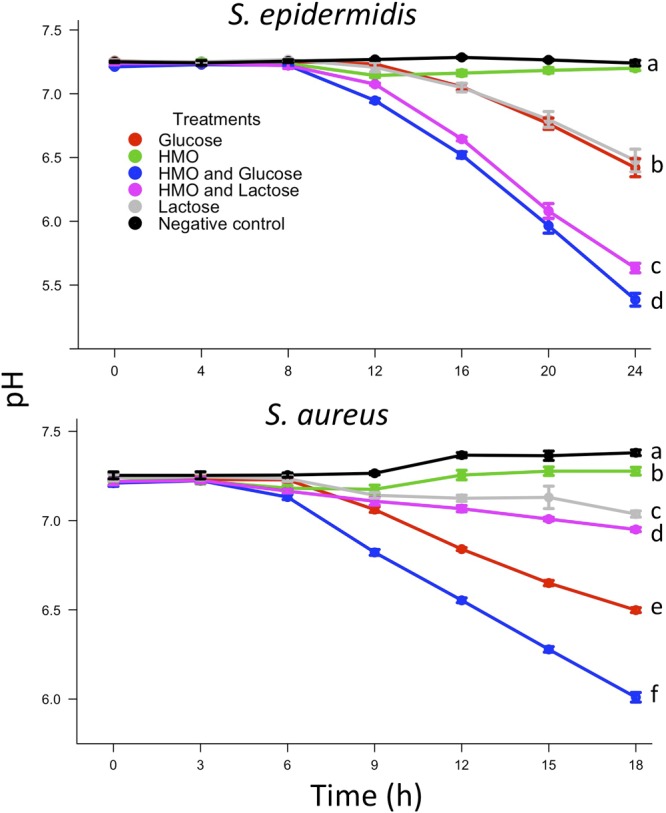
The pH of the culture supernatants from Staphylococcus isolates grown in the carbohydrate-free medium with various carbohydrate treatments (10 g/liter) was measured over time. Letters indicate significant treatment differences (P < 0.05) at the termination of the growth curves.
To investigate the effects of HMO on bacterial catabolism of amino acids, cultures were grown in glucose-free DMEM (Mediatech) composed of amino acids and salts to facilitate HPLC analysis of culture supernatants. Treatment effects of HMO on growth in this medium were similar to those observed in the carbohydrate-free medium (data not shown). Supernatants were collected at the start of incubation and again at stationary phase (18 h for S. aureus and 20 h for S. epidermidis) to compare the concentrations of amino acids in the cultures before and after growth. In both isolates, the HMO treatment resulted in the depletion of several amino acids (Fig. 5), suggesting that the enhanced bacterial growth observed was accompanied by increased amino acid metabolism and not catabolism of HMO.
Fig 5.
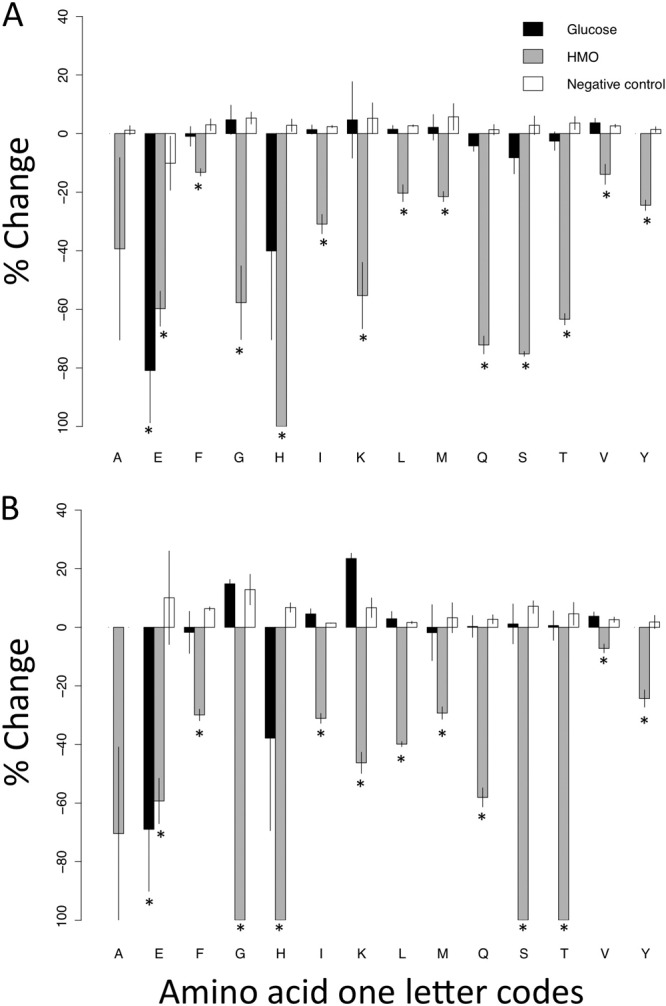
Relative change in amino acid concentration in culture supernatants of the S. epidermidis (A) and S. aureus (B) strains at t = 18 h and t = 20 h, respectively, compared to t = 0. Bars represent means and SEM of three independent experiments for cultures without carbohydrate source, with glucose as the sole carbohydrate source, or with HMO as the sole carbohydrate source. ⁎, significant decrease (P < 0.05).
DISCUSSION
Numerous reports have detailed the effects of HMO on the growth of certain types of bacteria associated with the infant's gastrointestinal tract and, in particular, members of the genus Bifidobacterium (10, 27, 30, 35, 38). This is the first report, however, that investigates the effects of HMO on the bacteria in the lactating mammary gland and, consequently, provides novel evidence of a connection between HMO and the bacterial profile of a woman's milk. The positive correlation observed between Staphylococcus relative abundance and HMO content in human milk was relatively strong and highly significant. Furthermore, there was no evidence of a similar correlation between HMO and any of the other prevalent bacterial genera. No correlation was observed between HMO and Bifidobacterium because very few members of this genus were observed in the milk samples (19). Other investigators have identified bifidobacteria in milk samples (9), but in our samples it was not present. The subsequent in vitro analysis supported the in vivo observations as HMO stimulated the growth “profile” of Staphylococcus in a variety of media, which is similar to HMO stimulation of the growth of bifidobacteria in previous studies (3, 26).
It is of particular interest to note that the effects of HMO on bacterial growth varied depending on the nutritional composition of the growth medium. Addition of HMO to cultures grown in the nutritionally rich TSB elicited less pronounced (S. aureus) or nonexistent (S. epidermidis) effects in comparison with those observed in the carbohydrate-free medium. When TSB was used, the treatment effects may have been somewhat ameliorated because cultures were already growing to their maximum potential due to the nutritionally rich nature of the growth medium. This reduced effect is noteworthy because it illustrates the importance of choosing culture conditions that are relevant to physiological situations.
Therefore, to emulate nutritional conditions in the lactating mammary gland more closely, experiments were performed with HMO-free infant formula. Ideally, these experiments would be conducted using human milk as the growth medium as this approach would account for the bioactive compounds (e.g., secretory IgA, lactoferrin, and lysozyme) contained in milk that potentially influence bacterial growth in the mammary gland. However, removal of HMO from human milk to provide an appropriate negative control is technically difficult. Hence, infant formula represents the most suitable alternative. Importantly, the addition of HMO to cultures grown under these nutritionally similar conditions resulted in enhanced bacterial growth in terms of rate and maximum growth achieved. This provides more evidence that HMO stimulate the growth of Staphylococcus in the lactating mammary gland.
The HMO-based augmentation of the growth of both Staphylococcus isolates occurred without any change in the HMO content of culture supernatants. This suggests that the bacteria did not metabolize the HMO. In contrast, previously published studies demonstrated that HMO-based promotion of bifidobacteria growth was accompanied by bacterial catabolism of HMO (3, 26). In the previous work, the authors investigated the genomes of the HMO-utilizing strains and determined that they contained several genes coding for glycoside hydrolases that might have been important to HMO degradation, such as α-galactosidase, β-N-acetylgalactosaminidase, β-hexosaminidase, α-l-fucosidase, sialidase, β-galactosidase, and α1,2-l-fucosidase (26). The experiments in the present study were performed using Staphylococcus strains isolated directly from human milk samples. This approach was taken due to the possibility that the genetic components associated with any response to HMO may be unique to isolates that have adapted to the mammary environment. One limitation of this approach is that the genetic makeup of the isolates employed is unknown, so at present we cannot draw any conclusions about the genetic basis of HMO-associated enhancement of Staphylococcus growth. However, examination of the enzyme-coding genes in the Staphylococcus genomes available on the Carbohydrate-Active enZYmes (CAZy) database (7) revealed that none of the 27 S. aureus or 2 S. epidermidis genomes contained any of the aforementioned glycosidic enzymes. Interestingly, the genomes of other Staphylococcus species, including Staphylococcus carnosus, Staphylococcus haemolyticus, Staphylococcus lugdunensis, Staphylococcus pseudintermedius, and Staphylococcus saprophyticus, contained at least one copy of the potentially HMO-degrading enzymes. Nonetheless, S. aureus and S. epidermidis were the species tested in the present study, so the observed lack of genetic basis for the HMO-specific glycoside hydrolases in isolates of these species may provide more evidence that the effects of HMO are due to a mechanism independent of bacterial metabolism of these glycans.
Furthermore, cultures grown in HMO-supplemented medium demonstrated very little pH alteration after several hours of growth. A drop in culture pH is generally indicative of carbohydrate catabolism and the buildup of the acidic intermediates associated with glycolysis. The intensified decrease in pH associated with the HMO+G treatment in both isolates and the HMO+L treatment in the S. epidermidis isolate is logical because the addition of HMO enhanced the bacterial growth rate, likely leading to greater utilization of glucose or lactose. Nonetheless, in the treatment with only HMO added, very little (if any) pH alteration was observed. These observations provide further evidence that the HMO-associated enhancement of bacterial growth was not a result of HMO catabolism.
Recent work employing a biosensor-based assay demonstrated that a strain of Staphylococcus may interact with 2′-fucosyllactose (25), which is one of the most abundant oligosaccharides in milk. These findings presented novel information that at least some strains of Staphylococcus may be able to bind to specific HMO. Interestingly, in the present study a positive correlation was observed between Staphylococcus and 2′-fucosyllactose in vivo (data not shown). Our in vitro data suggest that the stimulatory effects of HMO on Staphylococcus growth were not associated with metabolism of the HMO, so it is possible that the observed effects were connected to this type of specific binding event. Furthermore, if HMO binding were occurring, it might be connected to the activation of a growth-promoting signaling cascade.
Staphylococcus is a genus that profoundly influences lactation-associated health outcomes as it encompasses several pathogenic species implicated in lactational mastitis (13, 31). Affecting up to 30% of lactating women (4), mastitis is defined as inflammation of the breast that is associated with redness, swelling, and painful lactation that may be accompanied by fever or flu-like symptoms. Considering the pain that is associated with these symptoms, it is not surprising that women who have mastitis are more likely to abandon their efforts to breast-feed, thereby depriving the mother-infant dyad of the numerous health benefits associated with lactation and breast-feeding (36). It is therefore noteworthy that, under nutritional conditions similar to those in the lactating mammary gland, HMO stimulated growth of both Staphylococcus isolates. Although the mechanism has yet to be elucidated, this interaction may be an important factor in the development of mastitis and, subsequently, the likelihood of a woman successfully breast-feeding her infant.
Indeed, S. aureus and S. epidermidis are both common mastitis pathogens, yet in several studies examining the bacterial communities of milk produced by women free from mastitis, the presence of both of these species has been reported (18, 22, 28). It is therefore of great interest to understand why these species are seemingly harmless commensal organisms at some times and pathogens at others. Considering the striking interpersonal variation in HMO content and the positive effects of HMO on Staphylococcus growth reported here, we suggest the possibility that a woman's HMO profile may be one factor that influences the growth of Staphylococcus species involved in lactational mastitis.
A cocktail of heterogeneous compounds, HMO vary drastically in size, charge, and conformation (5, 6, 23, 24). After observing the in vitro correlation between HMO and Staphylococcus in milk, we chose to perform our experiments with HMO purified from pooled donor milk collected from several individuals. We took this approach so our experiments would include a large variety of structures because it was unknown which type of compound would influence Staphylococcus growth in vitro. However, considering the diversity of structures that are contained in HMO, it is possible that not all HMO were responsible for the observed effects. Rather, it is more likely that bioactivity is limited to a specific structure, or set of structures, with a particular charge, size, or conformational characteristic. This concept was recently demonstrated in work with a neonatal rat model of necrotizing enterocolitis (NEC) (20). The authors initially determined that administration of whole HMO to formula-fed pups decreased the incidence of NEC-like symptoms to a level similar to that observed in pups fed with mother's milk. Further investigation found that, of the numerous structures that compose HMO, the protective effects were exclusive to one specific glycan: disialyllacto-N-tetraose. It is possible that a similar scenario is responsible for the effects of HMO on Staphylococcus growth reported here. Consequently, further work is needed to determine the exact structures required to elicit the effects described in this study.
In conclusion, this study provides novel evidence that HMO act to stimulate growth of S. aureus and S. epidermidis in vitro, confirming the positive relation detected in human milk. Growth of S. aureus and S. epidermidis occurred without metabolism of HMO, suggesting that HMO act as growth stimulants. Therefore, the effects of HMO on human health during lactation may not be limited to the infant, but, rather, their influence may also extend to the mammary health of the lactating mother. These data suggest that HMO may interact with the bacterial communities of a mother's mammary gland before ever being introduced to the nursing infant and provoke the question as to how this interaction influences the success of lactation.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the United Dairymen of Idaho, National Institutes of Health grants P20 RR15587 (M.A.M.), P20 RR016454 (M.A.M.), and R00DK078668 (L.B.), and the Initiative for Bioinformatics and Evolutionary Studies at the University of Idaho.
Footnotes
Published ahead of print 4 May 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Albrecht S, et al. 2011. Oligosaccharides in feces of breast- and formula-fed babies. Carbohydr. Res. 346:2173–2181 [DOI] [PubMed] [Google Scholar]
- 2.Arroyo R, et al. 2010. Treatment of infectious mastitis during lactation: antibiotics versus oral administration of lactobacilli isolated from breast milk. Clin. Infect. Dis. 50:1551–1558 [DOI] [PubMed] [Google Scholar]
- 3.Asakuma S, et al. 2011. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J. Biol. Chem. 286:34583–34592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbosa-Cesnik C, Schwartz K, Foxman B. 2003. Lactation mastitis. JAMA 289:1609–1612 [DOI] [PubMed] [Google Scholar]
- 5.Bode L, Beermann C, Mank M, Kohn G, Boehm G. 2004. Human and bovine milk gangliosides differ in their fatty acid composition. J. Nutr. 134:3016–3020 [DOI] [PubMed] [Google Scholar]
- 6.Bode L. 2009. Human milk oligosaccharides: prebiotics and beyond. Nutr. Rev. 67(Suppl 2):S183–S191 [DOI] [PubMed] [Google Scholar]
- 7.Cantarel BL, et al. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaturvedi P, et al. 2001. Fucosylated human milk oligosaccharides vary between individuals and over the course of lactation. Glycobiology 11:365–372 [DOI] [PubMed] [Google Scholar]
- 9.Collado MC, Delgado S, Maldonado A, Rodríguez JM. 2009. Assessment of the bacterial diversity of breast milk of healthy women by quantitative real-time PCR. Lett. Appl. Microbiol. 48:523–528 [DOI] [PubMed] [Google Scholar]
- 10.Coppa GV, et al. 2011. Oligosaccharides in 4 different milk groups, Bifidobacteria, and Ruminococcus obeum. J. Pediatr. Gastroenterol. Nutr. 53:80–87 [DOI] [PubMed] [Google Scholar]
- 11.Crossley KB, Archer G. 1997. Biology, p 1–38 In Crossley KB, Archer GL. (ed), Staphylococci in human disease. Churchill and Livingstone Inc., New York, NY [Google Scholar]
- 12.Davidson B, Meinzen-Derr JK, Wagner CL, Newburg DS, Morrow AL. 2004. Fucosylated oligosaccharides in human milk in relation to gestational age and stage of lactation. Adv. Exp. Med. Biol. 554:427–430 [DOI] [PubMed] [Google Scholar]
- 13.Delgado S, et al. 2009. Staphylococcus epidermidis strains isolated from breast milk of women suffering infectious mastitis: potential virulence traits and resistance to antibiotics. BMC Microbiol. 9:82 doi:10.1186/1471-2180-9-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engfer MB, Stahl B, Finke B, Sawatzki G, Daniel H. 2000. Human milk oligosaccharides are resistant to enzymatic hydrolysis in the upper gastrointestinal tract. Am. J. Clin. Nutr. 71:1589–1596 [DOI] [PubMed] [Google Scholar]
- 15.Gabrielli O, et al. 2011. Preterm milk oligosaccharides during the first month of lactation. Pediatrics 128:e1520–1531 doi:10.1542/peds.2011-1206 [DOI] [PubMed] [Google Scholar]
- 16.Gnoth MJ, Kunz C, Kinne-Saffran E, Rudloff S. 2000. Human milk oligosaccharides are minimally digested in vitro. J. Nutr. 130:3014–3020 [DOI] [PubMed] [Google Scholar]
- 17.György P, Kuhn R, Rose CS, Zilliken F. 1954. Bifidus factor I. A variant of Lactobacillus bifidus requiring a special growth factor. Arch. Biochem. Biophys. 48:193–201 [DOI] [PubMed] [Google Scholar]
- 18.Heikkilä MP, Saris PEJ. 2003. Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J. Appl. Microbiol. 95:471–478 [DOI] [PubMed] [Google Scholar]
- 19.Hunt KM, et al. 2011. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS One 6:e21313 doi:10.1371/journal.pone.0021313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jantscher-Krenn E, et al. 2011. The human milk oligosaccharide disialyllacto-N-tetraose prevents necrotising enterocolitis in neonatal rats. Gut. [Epub ahead of print.] doi:10.1136/gutjnl-2011-301404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jantscher-Krenn E, et al. 2012. Human milk oligosaccharides reduce Entamoeba histolytica attachment and cytotoxicity in vitro. Br. J. Nutr. [Epub ahead of print.] doi:10.1017/S0007114511007392 [DOI] [PubMed] [Google Scholar]
- 22.Jiménez E, et al. 2008. Staphylococcus epidermidis: a differential trait of the fecal microbiota of breast-fed infants. BMC Microbiol. 8:143 doi:10.1186/1471-2180-8-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobata A. 2010. Structures and application of oligosaccharides in human milk. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 86:731–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunz C, Rudloff S, Baier W, Klein N, Strobel S. 2000. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu. Rev. Nutr. 20:699–722 [DOI] [PubMed] [Google Scholar]
- 25.Lane JA, Mehra RK, Carrington SD, Hickey RM. 2011. Development of biosensor-based assays to identify anti-infective oligosaccharides. Anal. Biochem. 410:200–205 [DOI] [PubMed] [Google Scholar]
- 26.Marcobal A, et al. 2010. Consumption of human milk oligosaccharides by gut-related microbes. J. Agric. Food. Chem. 58:5334–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcobal A, et al. 2011. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe 10:507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martín R, et al. 2007. Cultivation-independent assessment of the bacterial diversity of breast milk among healthy women. Res. Microbiol. 158:31–37 [DOI] [PubMed] [Google Scholar]
- 29.Moro E. 1900. Morphologie und bakteriologische Untersuchungen über die Darmbakterien des Säuglings: Die Bakterien-flora des normalen Frauenmilchstuhls. Jahrbuch Kinderh. 61:686–734 [Google Scholar]
- 30.Morrow AL, et al. 2004. Human milk oligosaccharides are associated with protection against diarrhea in breast-fed infants. J. Pediatr. 145:297–303 [DOI] [PubMed] [Google Scholar]
- 31.Osterman KL, Rahm V. 2000. Lactation mastitis: bacterial cultivation of breast milk, symptoms, treatment, and outcome. J. Hum. Lact. 16:297–302 [DOI] [PubMed] [Google Scholar]
- 32.Perez PF, et al. 2007. Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics 119:e724–732 doi:10.1542/peds.2006-1649 [DOI] [PubMed] [Google Scholar]
- 33. R Development Core Team 2011. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 34.Ritz C. 2010. Toward a unified approach to dose-response modeling in ecotoxicology. Env. Tox. Chem. 29:220–229 [DOI] [PubMed] [Google Scholar]
- 35.Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, Newburg DS. 2003. Campylobacter jejuni binds intestinal H(O) antigen (Fucα1, 2Galβ1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J. Biol. Chem. 278:14112–14120 [DOI] [PubMed] [Google Scholar]
- 36.Schwartz K, et al. 2002. Factors associated with weaning in the first 3 months postpartum. J. Fam. Pract. 51:439–444 [PubMed] [Google Scholar]
- 37.Scott DA, et al. 2011. Comparative metabolic flux profiling of melanoma cell lines: beyond the Warburg effect. J. Biol. Chem. 286:42626–42634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sela DA, Mills DA. 2010. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 18:298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stahl B, et al. 2001. Detection of four human milk groups with respect to Lewis-blood-group-dependent oligosaccharides by serologic and chromatographic analysis. Adv. Exp. Med. Biol. 501:299–306 [DOI] [PubMed] [Google Scholar]
- 40.Tissier H. 1900. Recherches sur la flora intestinale de nourissons (état normal et pathologique). Paris, France [Google Scholar]
- 41.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu S, Tao N, German JB, Grimm R, Lebrilla CB. 2010. Development of an annotated library of neutral human milk oligosaccharides. J. Proteome Res. 9:4138–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu S, Grimm R, German JB, Lebrilla CB. 2011. Annotation and structural analysis of sialylated human milk oligosaccharides. J. Proteome Res. 10:856–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang C, Richardson AD, Osterman A, Smith JW. 2008. Profiling of central metabolism in human cancer cells by two-dimensional NMR, GC-MS analysis, and isotopomer modeling. Metabolomics 4:13–29 [Google Scholar]
- 45.Zhou X, et al. 2004. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology 150:2565–2573 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.