Abstract
Mycophenolic acid (MPA) is a fungal secondary metabolite and the active component in several immunosuppressive pharmaceuticals. The gene cluster coding for the MPA biosynthetic pathway has recently been discovered in Penicillium brevicompactum, demonstrating that the first step is catalyzed by MpaC, a polyketide synthase producing 5-methylorsellinic acid (5-MOA). However, the biochemical role of the enzymes encoded by the remaining genes in the MPA gene cluster is still unknown. Based on bioinformatic analysis of the MPA gene cluster, we hypothesized that the step following 5-MOA production in the pathway is carried out by a natural fusion enzyme MpaDE, consisting of a cytochrome P450 (MpaD) in the N-terminal region and a hydrolase (MpaE) in the C-terminal region. We verified that the fusion gene is indeed expressed in P. brevicompactum by obtaining full-length sequence of the mpaDE cDNA prepared from the extracted RNA. Heterologous coexpression of mpaC and the fusion gene mpaDE in the MPA-nonproducer Aspergillus nidulans resulted in the production of 5,7-dihydroxy-4-methylphthalide (DHMP), the second intermediate in MPA biosynthesis. Analysis of the strain coexpressing mpaC and the mpaD part of mpaDE shows that the P450 catalyzes hydroxylation of 5-MOA to 4,6-dihydroxy-2-(hydroxymethyl)-3-methylbenzoic acid (DHMB). DHMB is then converted to DHMP, and our results suggest that the hydrolase domain aids this second step by acting as a lactone synthase that catalyzes the ring closure. Overall, the chimeric enzyme MpaDE provides insight into the genetic organization of the MPA biosynthesis pathway.
INTRODUCTION
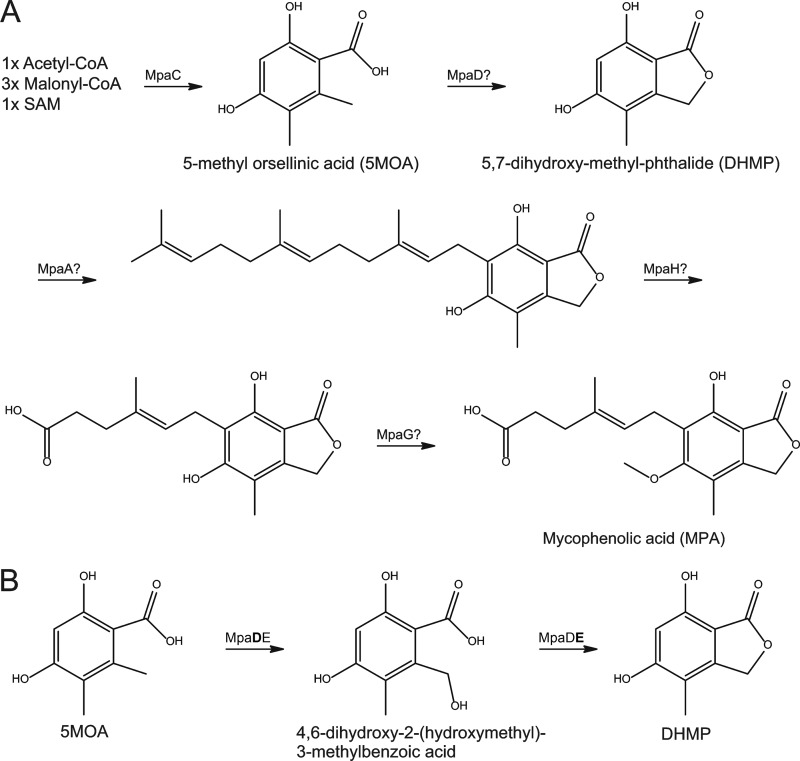
Fungi are among the most elaborate chemical producers in nature, producing a range of secondary metabolites, including some that are mycotoxins, food additives, or pharmaceutical drugs. Among the latter is mycophenolic acid (MPA), which is the active component in several immunosuppressants. MPA has also been associated with antiviral, antifungal, antibacterial, and antitumor activities (11). Consequently, MPA biosynthesis has received considerable research interest, and a biosynthetic route has been established through chemical labeling and culture feeding studies (3). MPA is a meroterpenoid proposed to be derived from a nonreduced tetraketide moiety through a five-step process involving oxidation, lactonization, and condensation with a farnesyl residue, followed by oxidative cleavage of the terpene part and a final methylation step (Fig. 1A). However, the biosynthesis remained unelucidated at the genetic level until the recent discovery of a putative MPA biosynthetic cluster in Penicillium brevicompactum (23). The defined cluster contained eight putative open reading frames (ORFs), including mpaC, which encodes a polyketide synthase catalyzing the production of 5-methylorsellinic acid (5-MOA), the first step in MPA biosynthesis (9, 23). Furthermore, it was recently demonstrated that mpaF encodes an MPA-insensitive inosine-5′-monophosphate dehydrogenase (IMPDH) conferring self-resistance toward MPA (8, 10). These studies utilized Aspergillus nidulans as a fungus of choice for the heterologous expression of mpaC and mpaF. A. nidulans provides a good model system to study the biosynthesis of MPA, since it can produce polyketides and does not produce any of the intermediates in MPA biosynthesis. In addition, the genome has been sequenced, and in the recent years the molecular biology toolbox has been greatly expanded (9, 16, 17, 19).
Fig 1.
(A) MPA biosynthetic pathway, including the enzymes proposed by Regueira et al. (23), to catalyze the individual reactions. Generation of 5-methylorsellinic acid by MpaC is the only enzymatic reaction that has been verified experimentally. Enzymes not accounted for in the prior study include MpaB and MpaE. (B) Hypothesized two biosynthesis steps from 5-methylorsellinic acid to 5,7-dihydroxy-4-methylphthalide involving MpaD and MpaE.
In the present study, we set out to identify and characterize the enzyme(s) responsible for the conversion of 5-MOA to 5,7-dihydroxy-4-methylphthalide (DHMP), which are the first and second known intermediates in MPA biosynthesis. A bioinformatics study of the MPA biosynthetic cluster, followed by heterologous expression in A. nidulans, showed that the conversion of 5-MOA to DHMP is catalyzed by a natural fusion of MpaD, a cytochrome P450, and MpaE, a putative lactone synthase.
MATERIALS AND METHODS
Strains and media.
The following strains were used in the present study: P. brevicompactum strain IBT23078 and A. nidulans strains NID210 (argB2 pyrG89 veA1 IS1::PgpdA-TtrpC::argB), NID211 (argB2 pyrG89 veA1 IS1::PgpdA-mpaC-TtrpC::argB), NID410 (argB2 pyrG89 veA1 nkuAΔ IS2::PgpdA-mpaDE-TtrpC::AFpyrG), NID416 (argB2 pyrG89 veA1 IS1::PgpdA-mpaC-TtrpC::argB IS2::PgdpA-mpaDE-TtrpC::AFpyrG), and NID944 (argB2 pyrG89 veA1 IS1::PgpdA-mpaC-TtrpC::argB IS2::PgdpA-mpaD-TtrpC::AFpyrG). Strains NID210 and NID211 were constructed in a previous study (9).
P. brevicompactum was grown on Czapek yeast extract agar (CYA) at 25°C. CYA is composed of 5 g of yeast extract (Biokar Diagnostics, Beauvais, France)/liter, 15 g of agar/liter, 35 g of Czapek dox broth/liter, 10 mg of ZnSO4·7H2O/liter, and 5 mg of CuSO4·5H2O/liter. The pH levels were adjusted to 6.5 with NaOH/HCl. A. nidulans strains were grown on minimal medium (MM) containing 1% glucose, 10 mM NaNO3, 1× salt solution (5), and 2% agar for solid media or YES medium containing 20 g of yeast extract (Biokar)/liter, 150 g of sucrose/liter, 0.5 g of MgSO4·H2O and ZnSO4·7H2O/liter, 5 mg of CuSO4·5H2O/liter, and 2% agar (pH 6). The MM was supplemented with 10 mM uridine (Uri), 10 mM uracil (Ura), and 4 mM l-arginine (Arg) when necessary.
RNA purification and cDNA synthesis.
Spores from P. brevicompactum IBT23078 were harvested and used to inoculate 200 ml of YES medium in 300 ml of shake flasks without baffles. P. brevicompactum was grown at 25°C and 150 rpm with shaking. After 48 h, the mycelium was harvested, and the RNA was purified using the fungal RNA purification miniprep kit (EZNA) according to the manufacturer's instructions. cDNA was synthesized from the RNA using a Finnzymes Phusion RT-PCR kit according to the manufacturer's instructions. The mpaDE transcript was amplified with the primer pair 657 and 660 (657, ATGAAGTCTTTGTCGCTAAC; 660, TTACTTCTGTCCTTCTATGG) and cloned into pJET1.2/blunt using a CloneJET PCR cloning kit (Fermentas) according to the manufacturer's instructions, resulting in pJet_mpaD_mpaE.
Plasmid construction.
Amplification of DNA by PCR to produce DNA fragments suitable for USER cloning was performed in 30 PCR cycles using PfuX7 (21) in 50 μl. USER cloning was performed as previously described (9, 22), with minor modifications. The USER vectors were digested for 6 h with AsiSI for the AsiSI/Nb.BsmI and AsiSI/Nb.BtsI USER cassettes or with PacI for the PacI/Nt.BbvCI USER cassettes A and B, followed by digestion with the appropriate nicking endonuclease for 1 h. Then, 0.1 pmol of purified digested vector was mixed with 1 pmol of purified PCR products amplified with primers that were extended by the appropriate tails for USER cloning into a designated USER cassette.
The PgpdA::USER cassette (AsiSI/Nb.BtsI)::TtrpC fragment was amplified from pU1111-1 (9) with the primer pair 556/559 (556, CGTGCGAUCTCTACACACAGGCTCAAAT; 559, CACGCGAUGCATTCTGGGTAAACGACTC) and USER cloned into the AsiSI/Nb.BsmI USER cassette in BGHA P147, generating BGHA P123. mpaDE was amplified from pJet_mpaDE with the primer pair 570/573 (570, AGAGCGAUATGAAGTCTTTGTCGCTAAC; 573, TCTGCGAUTTACTTCTGTCCTTCTATGG), and mpaD was amplified with the primer pair 570/709 (709, TCTGCGAUTACTCAATGGTATCATCTCG) and USER cloned into the AsiSI/Nb.BtsI USER cassette in BGHA P123, generating BGHA P127 and BGHA151.
Genetic transformation and cross.
Protoplasting and gene-targeting procedures were performed as described previously (19). A total of 5 μg of BGHA P146, BGHA P151, and BGHA P127 was digested with SwaI to liberate the gene-targeting substrates for bipartite transformation (19). Fragments from BGHA P146 and BGHA P127 or from BGHA P151 and BGHA P146 were used as bipartite transformation substrates for the transformation of A. nidulans NID1 (BGHA P146 and BGHA P127), resulting in NID410 or NID211 (BGHA P151 and BGHA P146) resulting in NID944 (see Fig. S1 in the supplemental material). Correct integration was confirmed by using the primer pairs 157/183 (157, TACTCCCCACCAGCGACTAC; 183, CATTCGGAGATCCTCAGAGC) and 61/156 (61, GGCTACGCTAGCGGATCCACTTAACGTTACTGAA; 156, GTCTCTGACTCTCCGCATCC). Strains NID211 and NID410 were crossed according to the protocol previously published (28), resulting in strain NID416.
UHPLC-HRMS analysis.
Three plugs were taken from each strain grown for 8 days at 37°C in three point inoculations on YES media (with supplements, if necessary) and transferred to a 2-ml vial. After the addition of 1 ml of methanol-dichloromethane-ethyl acetate (1:2:3 [vol/vol/vol]), the vials were capped and subjected to ultrasonication for 60 min. The supernatants were transferred to clean vials, and the organic phase was evaporated under an N2 flow at 30°C. The residues were redissolved in 200 μl of acetonitrile-water (1:1 [vol/vol]) (with subjection to ultrasonication) and filtered through a 0.22-μm-pore-size PFTE syringe filter.
UHPLC-HRMS was performed on a maXis G3 quadrupole time-of-flight mass spectrometer (Bruker Daltonics, Bremen, Germany) equipped with an electrospray ionization (ESI) ion source. The mass spectrometer was connected to an Ultimate 3000 UHPLC system (Dionex, Sunnyvale, CA) equipped with a diode-array detector scanning 200 to 700 nm. Separation of 1- to 3-μl samples were performed at 40°C on a Kinetex C18 column (100 by 2.1 mm [inner diameter], 2.6 μm; Phenomenex, Torrance, CA) using a linear water-acetonitrile gradient (both buffered with 20 mM formic acid) at a flow of 0.4 ml min−1 starting from 10% acetonitrile and increasing to 100% in 10 min, remaining at 100% acetonitrile for 3 min. Mass spectrometry (MS) analyses were performed in both ESI+ and ESI− (separate runs) with a data acquisition range of m/z 100 to 1,000, and the MS was calibrated using sodium formate automatically infused prior to each analytical run, providing a mass accuracy of <1.5 ppm.
Reference standards of nuclear magnetic resonance (NMR)-validated DHMP and 5-MOA (see below) and 3-methylorsellinic acid (Ambinter, Paris, France), as well as orsellenic acid (Apin Chemicals, Oxon, United Kingdom), were coanalyzed. 4,6-Dihydroxy-2-(hydroxymethyl)-3-methylbenzoic acid (DHMB) was tentatively identified in ESI− with a matching accurate mass (<1 ppm accuracy, no other candidate compositions) and isotope pattern (SigmaFit better than 10), loss of CO2 (diagnostic of a carboxylic acid), and UV absorptions at 210 nm (100%), 261 nm (30%), and 304 nm (25%). The retention time (1.61 min) compared to DHMP (2.78 min) fit well with the calculated LogD values of 0.60 and 2.39, respectively (at a pH of 3.2 as in the solvent) (18). Extracted ion chromatograms of the [M+H]+ ions (± m/z 0.001) for all target peaks were constructed for all extracts to exclude that the compounds were produced in various quantities or present in trace amounts in the medium or the wild-type strains. DHMB was detected as the [M-H]− ions. The target compounds were identified by the whole UV/VIS spectrum, the retention time (±0.02 min), the accurate masses (±1.5 ppm), and the relative intensities of the [M+H]+, [M+Na]+, [M+H-H2O]+, [M-H]−, and [M-H-CO2]− ions, as well as by their respective isotope patterns.
Structure verification of 5-MOA and DHMP.
5-MOA was isolated from a large-scale ethyl acetate extract prepared from 100 petri dishes with MM+Ura+Uri agar after cultivation of strain NID211 for 4 days at 37°C. Similarly, DHMP was isolated from a large-scale ethyl acetate extract prepared from 100 petri dishes with MM+Ura+Uri agar after cultivation of strain NID416 for 4 days at 37°C. The two compounds were isolated to NMR purity using a 10-by-250-mm, 5-μm Phenomenex pentafluorophenyl column using a gradient from 15% CH3CN to 100% CH3CN (both containing 50 ppm trifluoroacetic acid) in 20 min using a flow of 5 ml/min to give 15 mg of 5-methylorsellinic acid and 11 mg of DHMP. The NMR spectra were acquired on a Varian Unity Inova 500 MHz spectrometer equipped with a 5-mm probe and by using standard pulse sequences. The signals of the residual solvent protons and solvent carbons were used as internal references (δH 2.50 ppm and δC 39.5 ppm for dimethyl sulfoxide). The 1H NMR data (see Fig. S2 and S3 and Table S2 in the supplemental material) and the HSQC and HMBC data (see Table S2 in the supplemental material) of the two compounds were in agreement with the literature (6).
RESULTS AND DISCUSSION
Remote homology modeling: mpaD encodes a putative hydroxylase.
Initial annotation of the eight proposed ORFs that constitutes the putative MPA biosynthetic cluster in P. brevicompactum suggested the following candidates for the established biosynthetic reactions: (i) polyketide synthase (PKS), mpaC; (ii) cytochrome P450, mpaD; (iii) prenyl transferase, mpaA; (iv) oxidative cleaving, mpaH; (v) and finally O-methyltransferase, mpaG (23). However, the lactonization occurring at the second step in MPA biosynthesis is not a standard cytochrome P450 reaction, which prompted us to investigate this step in detail. In order to pinpoint what type of reaction MpaD catalyzes, we conducted a BLASTP search to identify homologs with known functions. The search identified putative homologs with >50% identity, indicating that orthologs are present in other filamentous fungi (data not shown). However, none of these orthologs had a known function. We therefore made a search using HHpred (26) in an attempt to identify remote homologs with known function. Subjection of MpaD as a query for HHpred analysis resulted in more than 10 hits crossing the recommended 95% score threshold (data not shown). Among the topmost hits were three cytochrome P450 enzymes, all known to catalyze hydroxylations. These hits included the Rattus norvegicus CYP24A1 catalyzing hydroxylation of 25-hydroxyvitamin D3 (1), the human CYP2D6 catalyzing hydroxylation of debrisoquine (24), and P450 BM3 from Bacillus megaterium that catalyzes hydroxylation of several long-chain fatty acids (7). Multiple sequence alignments of MpaD with the three aforementioned CYP strains, as well as Talaromyces stipitatus TSTA_060710 and Phaeosphaeria nodorum SNOG_06679, revealed that the P450 heme signature motif and the EXXR motif of the P450 heme thiolate enzymes are conserved in MpaD (Fig. 2). Based on these predictions, MpaD was assigned as CYP631B5, TSTA_060710 was assigned as CYP631B4, and SNOG_06679 was assigned as CYP631C2 by D. R. Nelson of the P450 Nomenclature Committee (Department of Molecular Sciences, University of Tennessee). The output from HHpred indicated that MpaD is very likely to catalyze a hydroxylation reaction, and we hypothesized that the target is the methyl group in ortho position to the carboxylic acid group on 5-MOA (Fig. 1B). Lactonization then yields DHMP. We considered it unlikely that this reaction is also catalyzed by MpaD and turned our attention toward the remaining unassigned two putative ORFs in the gene cluster, mpaB and mpaE, annotated as encoding a protein of unknown function and a zinc-dependent hydrolase, respectively. Since the lactone formation from hydroxylated 5-MOA to DHMP requires what resemble a reverse hydrolysis, we turned our attention to mpaE.
Fig 2.
Multiple sequence alignment of conserved domains in MpaDE and selected orthologs. The P450 heme signature motif, the EXXR motif of P450 heme thiolate enzymes, and the Zn-hydrolase motif HXHXDH are boxed. Conserved residues are written in red in the alignment. MpaD orthologs: PDB code 2ij2 (Bacillus megaterium P450 BM3), PDB code 3k9v (Rattus norvegicus CYP24A1), PDB code 2f9q (Homo sapiens CYP2D6), TSTA_060710 (Talaromyces stipitatus TSTA_060710), SNOG_06679 (Phaeosphaeria nodorum SNOG_06679). MpaE orthologs: PBD code 3dha (Bacillus thuringiensis lactone hydrolase), PBD code 2r2d (Agrobacterium tumefaciens lactone hydrolase), PBD code 1sml (Stenotrophomonas maltophilia lactone hydrolase), TSTA_060680 (Talaromyces stipitatus TSTA_060680), SNOG_06681 (Phaeosphaeria nodorum SNOG_06681).
Remote homology modeling: mpaE encodes a putative lactone synthase.
MpaE was subjected to bioinformatic analysis as described for MpaD with similar results. The highest-scoring hits from HHpred for MpaE were acyl homoserine lactone (AHL) lactonases from Stenotrophomonas maltophilia (29), Agrobacterium tumefaciens (15), and Bacillus thuringiensis (14). This homology indicates that MpaE has the same structure as enzymes cleaving a lactone bond. Furthermore, multiple sequence alignments confirmed that the signature sequence HXHXDH, which is completely conserved and essential for activity in all Zn-dependent hydrolases, is present in MpaE (Fig. 2). We hypothesized that MpaE catalyzes the reverse reaction, i.e., the formation of a lactone through dehydration and is thereby a lactone synthase. The predicted activity of MpaD and MpaE would in combination result in the conversion of 5-MOA to the second intermediate DHMP in MPA biosynthesis in fungi (3).
mpaD and mpaE is a single gene that encodes a fusion protein.
We decided to undertake a heterologous expression approach to investigate our hypothesis. We have previously used such an approach successfully for expressing the MPA PKS in the MPA-nonproducer fungus A. nidulans (9). The putative ORFs of mpaD and mpaE are located in tandem within the defined MPA cluster and were annotated as separate ORFs (23) using GenScan software (4). GenScan, however, is based on invertebrate sequences and, to be more certain that we would clone the correct and full sequences of the genes, we decided to confirm the proposed annotations with two additional algorithms, FgeneSH (25) and Augustus (27). Both of these programs have been trained on fungal sequences. Surprisingly, predictions from both of these two programs suggested that mpaD and mpaE are a single gene that encodes a single polypeptide (data not shown), hereafter named mpaDE. Curiously, the MpaD homologs P450 BM3 and CYP505A1 (12) are natural fusion proteins as well, unlike almost every other known cytochrome P450s. In P450 BM3 and CYP505A1, the CYP domain is fused with an electron-donating domain. To establish whether mpaD and mpaE are transcribed as two separate or one fused ORF, RNA was extracted from P. brevicompactum under MPA producing conditions. PCR performed on the corresponding cDNA yielded a 2,562-bp product corresponding to the mpaDE fusion. mpaDE is therefore indeed a natural fusion gene containing six introns (GenBank number BK008023). An alignment of the full-length mpaDE with the previously identified homologs can be found in Fig. S4 in the supplemental material.
MpaDE catalyzes the conversion of 5-MOA to DHMP.
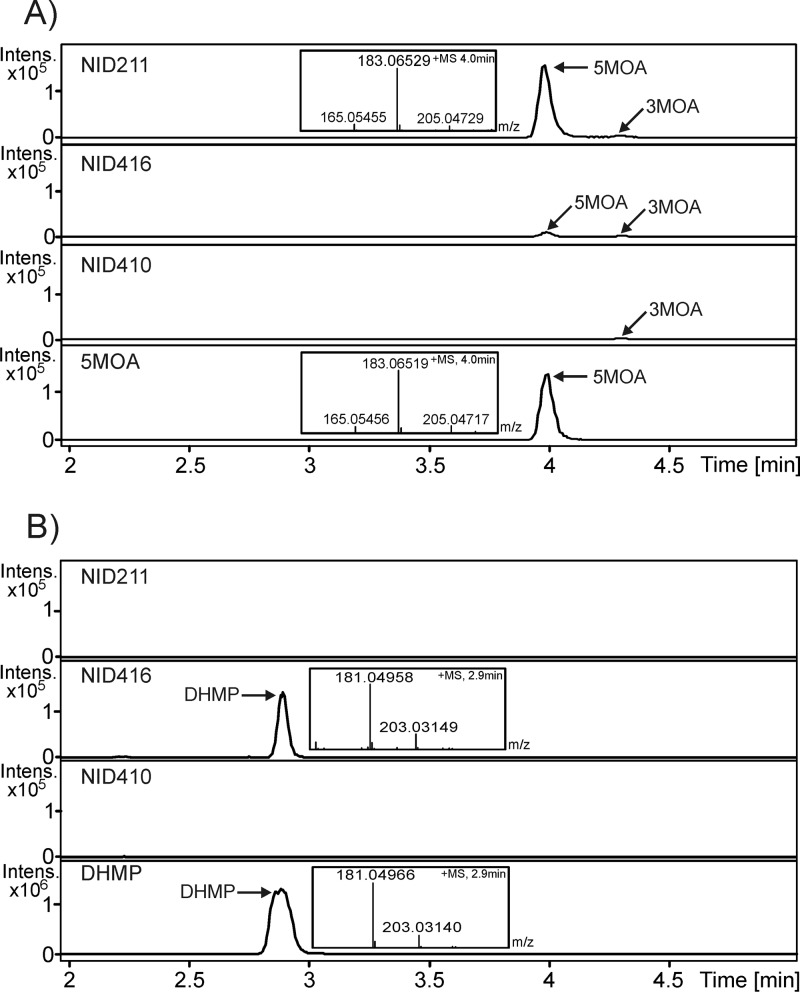
mpaDE was introduced into an A. nidulans strain (NID211) expressing MPA PKS (mpaC) and therefore capable of producing 5-MOA. In contrast to the reference strain NID211, this new strain (NID416) produced a compound eluting as a prominent peak at 2.9 min with the mass expected for DHMP (Fig. 3). The compound was purified and identified as DHMP by NMR (see Fig. S3 and Table S2 in the supplemental material) and by comparison with the published spectra. In addition, we note that, in contrast to NID211, the DHMP producing NID416 contained very little 5-MOA, which also points to that 5-MOA is converted by MpaDE (Fig. 3). We next addressed the question of whether the unique compound produced by NID416 is due to the mpaDE gene product converting 5-MOA to DHMP or whether it results from the conversion of an endogenous A. nidulans metabolite. We thus expressed mpaDE in an A. nidulans strain not producing 5-MOA. The resulting strain (NID410) did not produce detectable levels of DHMP. Finally, we constructed a strain NID944 where the mpaD part of mpaDE is expressed in a strain expressing mpaC. We found that strain NID944 contains a high amount of DHMB, whereas this metabolite was not detected in the other strains. This result conclusively shows that MpaD catalyzes the conversion of 5-MOA to DHMB. Interestingly, NID944 does produce DHMP, which shows that the conversion from DHMB can happen nonenzymatically or that A. nidulans has an endogenous enzyme that can catalyze the lactonization. AN0028 and AN2639 do both have high structural similarity to MpaE, and in HHpred the top hits are the same as for MpaE (data not shown). Therefore, they are good candidates for catalyzing the lactone formation when a high concentration of DHMB is available. Taken together, these results show that the natural fusion protein MpaDE catalyzes the formation of DHMP in MPA biosynthesis, and the buildup of DHMB in NID944 supports that MpaD catalyzes the hydroxylation (Fig. 1B and see Fig. S5 in the supplemental material). The results also suggest that the lactonization step is potentially aided by MpaE.
Fig 3.
MpaDE catalyzes the conversion of 5-MOA to DHMP. UHPLC-UV/VIS-HRMS analyses of the standards 5-MOA and DHMP and A. nidulans strains NID211, NID410, and NID416. (A) Extracted ion chromatogram, m/z 183.06519 ± 0.001 corresponding to the [M+H]+ ion of 5-methylorsellinic acid. The MS spectrum at 4.0 min is inserted in relevant chromatograms. (B) Extracted ion chromatogram (m/z 181.04954 ± 0.001) corresponding to the [M+H]+ ion of DHMP. The MS spectrum at 2.9 min is inserted in relevant chromatograms. 3-MOA, 3-methylorsellinic acid; 5-MOA, 5-methylorsellinic acid; DHMP, 5,7-dihydroxy-4-methylphthalide.
MpaD and MpaE orthologs involved in lactonization in other fungi?
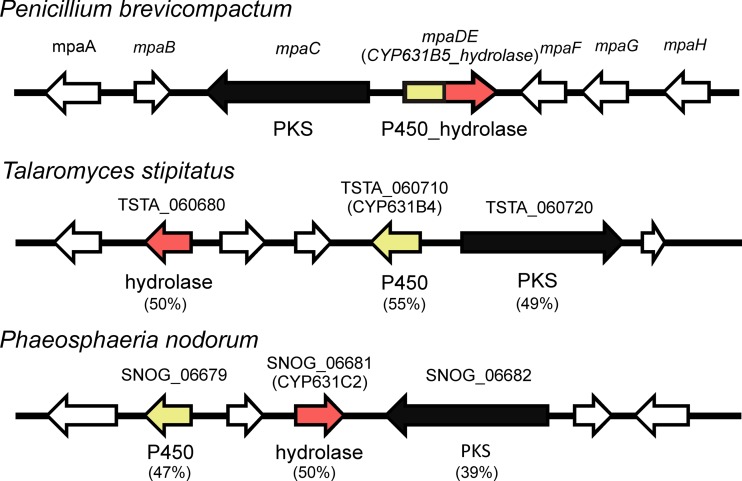
To investigate whether the fusion of MpaDE is widespread in nature, we performed a BLASTP search. This search did not identify any orthologs in any of the organisms within the NCBI database. However, as previously mentioned, BLASTP analysis identified several orthologs of MpaD and MpaE as separate enzymes in a number of fungi. Interestingly, we noticed that in both Talaromyces stipitatus and Phaeosphaeria nodorum, orthologs of MpaD (TSTA_060710 and SNOG_06679) and MpaE (TSTA_060680 and SNOG_06681) are located very close to each other (Fig. 4). In addition, they are placed in the vicinity of a PKS. The two PKSs have ∼40 and ∼50% sequence identity, respectively, and the same domain architecture as MpaC (9) and AusA from A. nidulans that catalyzes the production of 3,5-dimethylorsellinic acid (20). For the remaining members of the P. brevicompactum MPA gene cluster, BLAST hits were found for MpaB and MpaG (see Table S1 in the supplemental material), although not in the vicinity of the corresponding PKSs. Based on the homology and genomic proximity, it can be hypothesized that the PKSs from T. stipitatus and P. nodorum are catalyzing the production of methylorsellinic acid and that the MpaD orthologs are involved in converting this methylorsellinic acid into a lactone. Furthermore, the role of MpaE identified here in the lactonization step of the MPA pathway adds to the list of diverse biochemical functionalities of the polyketide biosynthesis enzymes that belong to metallo-β-lactamase family (2, 13).
Fig 4.
Graphical representation of gene clusters from Talaromyces stipitatus and Phaeosphaeria nodorum with orthologs of MpaC, MpaD, and MpaE. The percent identity to MpaC, MpaD, and MpaE is indicated below each of the orthologs.
In conclusion, our results provide new insights into the biochemical and genetic organization of the MPA biosynthesis pathway. The fusion protein MpaDE may provide an advantage by increasing the rate of reaction due to the close proximity of the two catalytic domains and also by possibly decreasing the side-products from the pathway. As more fungal genomes and polyketide gene clusters get sequenced, it will be of interest to find the extent to which such fusion proteins play a role in shaping the molecular diversity and species-level specificity of fungal polyketides.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to David Nelson (http://drnelson.utmem.edu/CytochromeP450.html) for naming MpaD and its orthologs.
The study was supported by grants 09-064967 and 09-064240 from The Danish Council for Independent Research, Technology, and Production Sciences.
Footnotes
Published ahead of print 27 April 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Annalora AJ, et al. 2010. Crystal structure of CYP24A1, a mitochondrial cytochrome P450 involved in vitamin D metabolism. J. Mol. Biol. 396: 441– 451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Awakawa T, et al. 2009. Physically discrete β-lactamase-type thioesterase catalyzes product release in atrochrysone synthesis by iterative type I polyketide synthase. Chem. Biol. 16: 613– 623 [DOI] [PubMed] [Google Scholar]
- 3.Bentley R. 2000. Mycophenolic acid: a one hundred year odyssey from antibiotic to immunosuppressant. Chem. Rev. 100: 3801– 3826 [DOI] [PubMed] [Google Scholar]
- 4.Burge C, Karlin S. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78– 94 [DOI] [PubMed] [Google Scholar]
- 5.Cove DJ. 1966. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 113: 51. [DOI] [PubMed] [Google Scholar]
- 6.Fujimoto H, Fujimaki T, Okuyama E, Yamazaki M. 1999. Immunomodulatory constituents from an ascomycete, Microascus tardifaciens. Chem. Pharm. Bull. (Tokyo) 47: 1426– 1432 [DOI] [PubMed] [Google Scholar]
- 7.Girvan HM, et al. 2007. Structural and spectroscopic characterization of P450 BM3 mutants with unprecedented P450 heme iron ligand sets. New heme ligation states influence conformational equilibria in P450 BM3. J. Biol. Chem. 282: 564– 572 [DOI] [PubMed] [Google Scholar]
- 8.Hansen BG, et al. 2011. A new class of IMP dehydrogenase with a role in self-resistance of mycophenolic acid producing fungi. BMC Microbiol. 11: 202 doi:10.1186/1471-2180-11-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen BG, et al. 2011. Versatile enzyme expression and characterization system for Aspergillus nidulans, with the Penicillium brevicompactum polyketide synthase gene from the mycophenolic acid gene cluster as a test case. Appl. Environ. Microbiol. 77: 3044– 3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen BG, et al. 2012. Adaptive evolution of drug targets in producer and non-producer organisms. Biochem. J. 441: 219– 226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hedstrom L. 2009. IMP dehydrogenase: structure, mechanism, and inhibition. Chem. Rev. 109: 2903– 2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitazume T, Takaya N, Nakayama N, Shoun H. 2000. Fusarium oxysporum fatty-acid subterminal hydroxylase (CYP505) is a membrane-bound eukaryotic counterpart of Bacillus megaterium cytochrome P450BM3. J. Biol. Chem. 275: 39734– 39740 [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Chooi YH, Sheng Y, Valentine JS, Tang Y. 2011. Comparative characterization of fungal anthracenone and naphthacenedione biosynthetic pathways reveals an α-hydroxylation-dependent Claisen-like cyclization catalyzed by a dimanganese thioesterase. J. Am. Chem. Soc. 133: 15773– 15785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu D, et al. 2008. Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures. Biochemistry 47: 7706– 7714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu D, et al. 2007. Structure and specificity of a quorum-quenching lactonase (AiiB) from Agrobacterium tumefaciens. Biochemistry 46: 11789– 11799 [DOI] [PubMed] [Google Scholar]
- 16.Meyer V, Wu B, Ram AF. 2011. Aspergillus as a multi-purpose cell factory: current status and perspectives. Biotechnol. Lett. 33: 469– 476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nielsen JB, Nielsen ML, Mortensen UH. 2008. Transient disruption of non-homologous end-joining facilitates targeted genome manipulations in the filamentous fungus Aspergillus nidulans. Fungal Genet. Biol. 45: 165– 170 [DOI] [PubMed] [Google Scholar]
- 18.Nielsen KF, Månsson M, Rank C, Frisvad JC, Larsen TO. 2011. Dereplication of microbial natural products by LC-DAD-TOFMS. J. Nat. Prod. 74: 2338– 2348 [DOI] [PubMed] [Google Scholar]
- 19.Nielsen ML, Albertsen L, Lettier G, Nielsen JB, Mortensen UH. 2006. Efficient PCR-based gene targeting with a recyclable marker for Aspergillus nidulans. Fungal Genet. Biol. 43: 54– 64 [DOI] [PubMed] [Google Scholar]
- 20.Nielsen ML, et al. 2011. A genome-wide polyketide synthase deletion library uncovers novel genetic links to polyketides and meroterpenoids in Aspergillus nidulans. FEMS Microbiol. Lett. 321: 157– 166 [DOI] [PubMed] [Google Scholar]
- 21.Nørholm MH. 2010. A mutant Pfu DNA polymerase designed for advanced uracil-excision DNA engineering. BMC Biotechnol. 10: 21 doi:10.1186/1472-6750-10-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nour-Eldin HH, Hansen BG, Nørholm MH, Jensen JK, Halkier BA. 2006. Advancing uracil-excision based cloning toward an ideal technique for cloning PCR fragments. Nucleic Acids Res. 34: e122 doi:10.1093/nar/gkl635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Regueira TB, et al. 2011. Molecular basis for mycophenolic acid biosynthesis in Penicillium brevicompactum. Appl. Environ. Microbiol. 77: 3035– 3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rowland P, et al. 2006. Crystal structure of human cytochrome P450 2D6. J. Biol. Chem. 281: 7614– 7622 [DOI] [PubMed] [Google Scholar]
- 25.Salamov AA, Solovyev VV. 2000. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10: 516– 522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Söding J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21: 951– 960 [DOI] [PubMed] [Google Scholar]
- 27.Stanke M, Morgenstern B. 2005. AUGUSTUS: a Web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33: W465– W467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Todd RB, Davis MA, Hynes MJ. 2007. Genetic manipulation of Aspergillus nidulans: meiotic progeny for genetic analysis and strain construction. Nat. Protoc. 2: 811– 821 [DOI] [PubMed] [Google Scholar]
- 29.Ullah JH, et al. 1998. The crystal structure of the L1 metallo-β-lactamase from Stenotrophomonas maltophilia at 1.7 Å resolution. J. Mol. Biol. 284: 125– 136 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.