Abstract
Analysis of global temporal gene expression by human intestinal cells when exposed to Lactobacillus acidophilus revealed induction of immune-related pathways and NF-κB target genes after a 1-h exposure, compared to a 4- or 8-h exposure. Additionally, an L. acidophilus derivative expressing covalently bound flagellin resulted in increased induction of il8, cxc1, and cxcl2 compared to the parent L. acidophilus.
TEXT
The gastrointestinal tract (GIT) harbors a vast array of microbes, including commensal, pathogenic, and transient bacteria. These microbes can initiate numerous interactions with different cells of the GIT, such as dendritic cells (DCs), M cells, and intestinal epithelial cells (IECs). Receptors on the surface of IECs allow discrimination between pathogenic and gut bacteria, thereby maintaining a vital immunological balance (11). Additionally, this layer of IECs maintains gut barrier functions that are vital to prevent dysfunctions of the intestine that can lead to intestinal diseases, such as irritable bowel syndrome and Crohn's disease. Intestinal epithelial cell lines are important research tools that have long been used to investigate expression of human intestinal markers for studies focused on adhesion, immunomodulation, and infection by both enteric pathogens and beneficial bacteria. More recently, with the availability of global transcriptome arrays these cell line models have been utilized for whole-transcriptome analysis of IECs in response to gut-related bacteria. These studies have identified genes involved in numerous cellular functions, including gut barrier function (2) and immunomodulation (18).
Probiotic bacteria are important components of the food industry, particularly with the increased consumer interest in diet, healthy lifestyles, and functional foods. These bacteria have demonstrated a number of beneficial roles, especially in the GIT (17), including lowering the duration and incidence of diarrhea during antibiotic treatment, protection against pathogens, interaction with dendritic and T cells, and reduced risk of Clostridium difficile-associated disease (3, 5, 10, 13). There are intimate communications between the resident gut microbes and transient microbes with the epithelial layer and immune cells embedded within the GIT. Therefore, there have been increased efforts to investigate and understand the mechanisms of action that occur at this vital interface. Given that Lactobacillus acidophilus NCFM is an important probiotic strain that has been safely consumed for nearly 40 years (19) and recombinant strains of L. acidophilus are also excellent candidates for delivery of therapeutics and treatment of inflammation (9, 14), we investigated the effect of L. acidophilus exposure time on the global gene expression by human intestinal cells. We also determined the effect of exposure with recombinant lactobacilli expressing Salmonella flagellin to IECs on a select group of genes by using quantitative real-time PCR (RT-qPCR).
L. acidophilus NCFM (4) was propagated statically in MRS broth at 37°C to stationary phase (16 h). Cells were harvested by centrifugation (1,717 × g for 10 min at room temperature), washed once in phosphate-buffered saline (PBS, pH 7.4), and then resuspended in PBS to a final concentration of approximately 1 × 108 CFU/ml. The Caco-2 intestinal epithelial cell line was maintained as described previously (16). Prior to exposure assays, Caco-2 monolayers were washed twice with PBS, and 2 ml of minimal essential medium without antibiotics was added per well (16). Two milliliters of triplicate bacterial suspensions or of PBS was added to 21-day Caco-2 monolayers (6-well plates, passage numbers between 23 and 45, seeded at 1.6 × 105 cells per cm2). Plates were centrifuged for 2 min at 429 × g and incubated at 37°C for 1, 4, or 8 h (bacterial cultures) or for 1 h (PBS) for time course gene expression studies.
Total RNA was extracted using Tri reagent (Invitrogen, Grand Island, NY) and purified with the RNAeasy minikit (Qiagen Inc., Valencia, CA). RNA was processed at Expression Analysis (Durham, NC) and hybridized to the humanWG-6 BeadChip (Illumina Inc., San Diego, CA) according to the manufacturer's instructions. Intensity values were captured and normalized via the cubic spline method, and background subtraction was performed. The data were then quantile normalized to uniformly distribute any value of <2 to values between 1 and 2. Transcripts were removed where the detection P value was >0.05 in 95% of the samples. These data were then filtered for genes which displayed a P value of <0.05 and demonstrated at least a +2-fold or −2-fold change. Gene expression results were confirmed by RT-qPCR (see the discussion of materials and methods and Fig. S1 in the supplemental material).
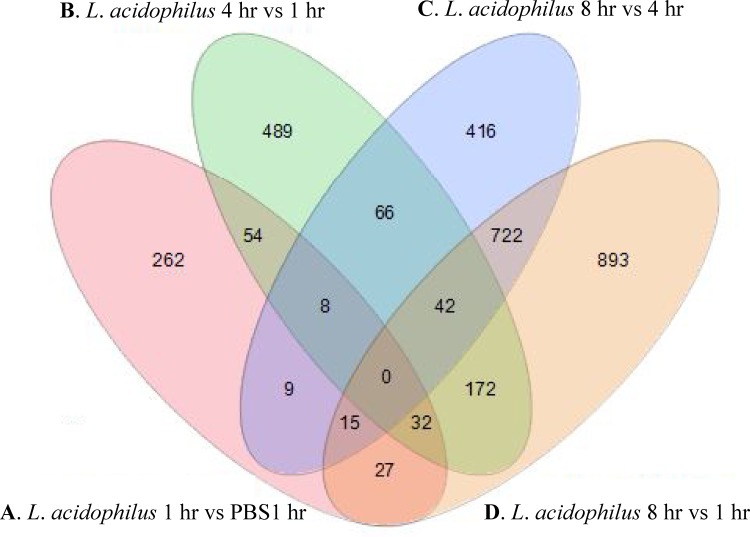
Differentially expressed gene sets were determined between epithelial cells exposed under the following conditions: L. acidophilus versus PBS for 1 h (comparison A), L. acidophilus for 4 versus 1 h (comparison B), L. acidophilus for 8 versus 4 h (comparison C), and L. acidophilus for 8 versus 1 h (comparison D). The number of shared genes among each of the four comparisons was determined (Fig. 1), and the corresponding details of the shared genes between each group are outlined in Table S1 in the supplemental material. Significant differentially expressed gene sets from each comparison were then further analyzed using the GOrilla (Gene Ontology enrichment analysis and visualization) tool to classify their involvement in cellular processes (see the description of GOrilla in the supplemental materials and methods) (8). Pathway analysis revealed that after exposure of IECs to L. acidophilus for 1 h, compared to PBS for 1 h, genes from GO groups related to immune function were enriched (Table 1). These included GO terms associated with response to molecules of bacterial origin, immune system processes, chemokine activity, and chemokine receptor binding. When differentially expressed gene sets after exposure to L. acidophilus for 4 h were compared to those exposed for 1 h, GO terms relating to mitogen-activated protein (MAP) kinase activity, defense responses to viruses, and the Toll-like receptor (TLR) signaling pathway were enriched (Table 1). GO analysis for terms after comparison of 8-h versus the 1- or 4-h exposure, respectively, revealed enrichment terms related to MAP kinase activity and enrichment of genes involved in cellular processes such as amino acid metabolism (see Table S1 in the supplemental material). Depending on the exposure time, epithelial cells generally responded to L. acidophilus by the induction of pathways involved in immune function, cell signaling, and cell processes such as amino acid metabolism.
Fig 1.
Venn diagram illustrating the numbers of shared and unique genes between each comparison. Details of shared and unique genes for each comparison are provided in Table S1 in the supplemental material.
Table 1.
GO groups enriched from differentially expressed gene sets of epithelial cells exposed to L. acidophilus for 1 or 4 ha
| Comparison | GO term | Description | GO class | No. of genes differentially expressed |
|---|---|---|---|---|
| A | GO:0002237 | Response to molecule of bacterial origin | BP | 14 |
| A | GO:0032496 | Response to lipopolysaccharide | BP | 13 |
| A | GO:0008009 | Chemokine activity | F | 7 |
| A | GO:0071222 | Cellular response to lipopolysaccharide | BP | 7 |
| A | GO:0042379 | Chemokine receptor binding | F | 7 |
| A | GO:0071219 | Cellular response to molecule of bacterial origin | BP | 7 |
| A | GO:0001894 | Tissue homeostasis | BP | 7 |
| A | GO:0002376 | Immune system process | BP | 29 |
| A | GO:0006954 | Inflammatory response | BP | 13 |
| A | GO:0009611 | Response to wounding | BP | 16 |
| A | GO:0071216 | Cellular response to biotic stimulus | BP | 7 |
| A | GO:0006955 | Immune response | BP | 20 |
| A | GO:0002682 | Regulation of immune system process | BP | 22 |
| A | GO:0060249 | Anatomical structure homeostasis | BP | 8 |
| A | GO:0006952 | Defense response | BP | 21 |
| A | GO:0002684 | Positive regulation of immune system process | BP | 16 |
| A | GO:0002274 | Myeloid leukocyte activation | BP | 5 |
| A | GO:0050896 | Response to stimulus | BP | 100 |
| A | GO:0002687 | Positive regulation of leukocyte migration | BP | 5 |
| A | GO:0009607 | Response to biotic stimulus | BP | 17 |
| A | GO:0050865 | Regulation of cell activation | BP | 12 |
| A | GO:0043069 | Negative regulation of programmed cell death | BP | 16 |
| A | GO:0016337 | Cell-cell adhesion | BP | 12 |
| A | GO:0032101 | Regulation of response to external stimulus | BP | 11 |
| B | GO:0071219 | Cellular response to molecule of bacterial origin | BP | 8 |
| B | GO:0009891 | Positive regulation of biosynthetic process | BP | 50 |
| B | GO:0010557 | Positive regulation of macromolecule biosynthetic process | BP | 46 |
| B | GO:0043406 | Positive regulation of MAP kinase activity | BP | 13 |
| B | GO:0043405 | Regulation of MAP kinase activity | BP | 16 |
| B | GO:0042552 | Myelination | BP | 7 |
| B | GO:0000187 | Activation of MAPK activity | BP | 11 |
| B | GO:0051094 | Positive regulation of developmental process | BP | 29 |
| B | GO:0071902 | Positive regulation of protein serine/threonine kinase activity | BP | 14 |
| B | GO:0008366 | Axon ensheathment | BP | 7 |
| B | GO:0007272 | Ensheathment of neurons | BP | 7 |
| B | GO:0071222 | Cellular response to lipopolysaccharide | BP | 7 |
| B | GO:0009790 | Embryo development | BP | 18 |
| B | GO:0035116 | Embryonic hindlimb morphogenesis | BP | 5 |
| B | GO:0035115 | Embryonic forelimb morphogenesis | BP | 5 |
| B | GO:0051607 | Defense response to virus | BP | 6 |
| B | GO:0002224 | Toll-like receptor signaling pathway | BP | 9 |
| B | GO:0009064 | Glutamine family amino acid metabolic process | BP | 7 |
| B | GO:0010628 | Positive regulation of gene expression | BP | 41 |
BP, biological processes; F, function; A, epithelial cells exposed to L. acidophilus for 1 h compared to epithelial cells exposed to PBS for 1 h; B, epithelial cells exposed to L. acidophilus for 4 h compared to those exposed for 1 h.
Pathway analysis also identified the involvement of the jun oncogenes (c-jun) in numerous pathways after exposure of epithelial cells to L. acidophilus for 1 h. JUN is a component of the AP-1 transcription factor complex, which is a regulator of numerous cell processes, including tumor necrosis factor (TNF) gene expression (1), and has previously been identified as upregulated by Lactobacillus casei and Lactobacillus rhamnosus (20). Another study identified c-jun as a potential therapeutic target in Crohn's disease treatment, because inhibiting c-jun resulted in the repression of AP-1, thereby suppressing the proinflammatory TNF (12). The expression level of jun was downregulated after a 4- or 8-h exposure compared to the 1-h exposure, indicating a return to baseline expression levels. The induction of c-jun and proinflammatory cytokines by L. acidophilus in this study supports the previously reported non anti-inflammatory properties of this strain (22). However, our results demonstrate that these genes were not upregulated past a 1-h incubation period but returned to expression levels similar to that observed after exposure of epithelial cells to PBS for 1 h.
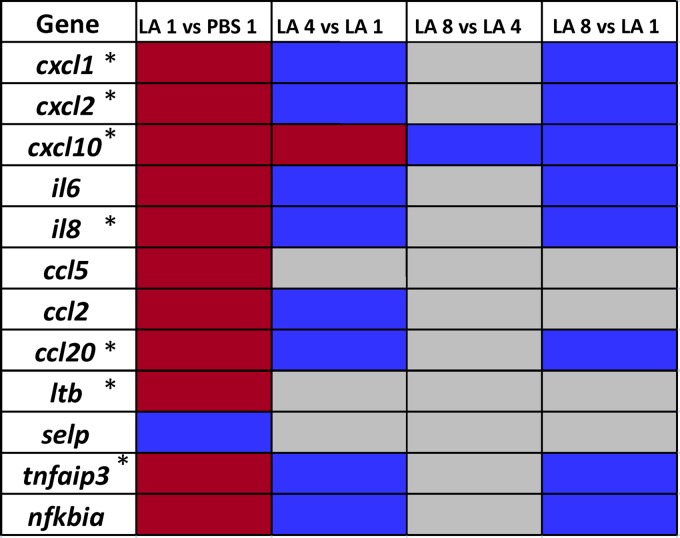
Further investigation of genes identified from GO pathway analysis indicated that immune-related genes (for example, cxcl1, cxcl2, ccl20, il6, and il8) were upregulated mostly after a 1-h exposure of L. acidophilus. Many of these genes were identified as NF-κB target genes (Fig. 2). These genes were upregulated after a 1-h exposure to L. acidophilus compared to a 1-h exposure to PBS, as well as a 4- or 8-h exposure to L. acidophilus (Fig. 2). The chemokine gene cxcl10 was upregulated both at 1 and 4 h, whereas there was no change in the expression profiles of the other genes after the 4- and 8-h exposures. In fact, expression levels with these longer incubation times were similar to those of epithelial cells exposed to PBS for 1 h. These results confirmed the observation that genes related to immunomodulation and also NF-κB target genes are differentially expressed after a 1-h, rather than 4- or 8-h, exposure time. Additionally, the upregulation of the negative regulators of the NF-κB pathway, TNFAIP3 (or A20) and NFKBIA (or IKBA), reinforced this observation and indicated that the downregulation of NF-κB target genes followed an initial upregulation after the 1-h exposure to L. acidophilus (Fig. 2). NF-κB is an important human transcription factor that regulates a large number of genes from different biological processes, including immunomodulation and cell signaling. Activation of receptors on the cell surface, such as TLRs, can induce NF-κB, which is normally sequestered in the cytoplasm, where it is bound by inhibitor proteins (15). Although the subunits of the NF-κB complex were not differentially expressed in this study, the NF-κB antagonists A20 and IKBA and numerous NF-κB-specific target genes were upregulated after the 1-h exposure of L. acidophilus compared to levels in epithelial cells exposed to PBS. These genes were among the most highly expressed genes. In fact, 7 of the 10 most highly upregulated genes after exposure to L. acidophilus compared to PBS for 1 h were NF-κB target genes (Fig. 2). Previous work by van Baarlen and coworkers (20, 21) studied the effect of probiotic bacteria on mucosal gene transcription in vivo. Our results after a 1-h exposure of L. acidophilus to epithelial cells reflect results observed with the other study group's strain of L. acidophilus, Lafti-L10 (20). Interestingly, their study also demonstrated modulation of multiple chemokines, cytokines, and NF-κB target genes, as we observed in our present study. In particular, expression levels of chemokines, such as CXCL2, CXCL10, CCL2, and CCL20 and the interleukin-8 (IL-8) and the NF-κB inhibitors A20 and IKBA, were upregulated in both studies. Considering that in vivo trials are less feasible for many research groups, it was encouraging that results from our in vitro model after a 1-h exposure reflected a similar gene expression trend as the in vivo model in relation to NF-κB target genes, such as chemokines. These data indicate a transition in gene expression from an initial induction after a 1-h exposure to a reduction in gene expression after a 4- or 8-h exposure for immune-related genes. Instead, expression levels of genes involved in cell processes were upregulated after the 4- and 8-h exposures.
Fig 2.
Modulation of NF-κB activity. Target genes of NF-κB were upregulated (red boxes), downregulated (blue boxes), or not significantly altered in expression (gray boxes). The columns, from left to right, show results for exposure to L. acidophilus versus PBS for 1 h, exposure to L. acidophilus for 4 versus 1 h, exposure to L. acidophilus for 8 versus 4 h, and exposure to L. acidophilus for 8 versus 1 h. Genes marked with asterisks were among the 10 most highly upregulated genes from the LA 1 versus PBS 1 comparison. LA, L. acidophilus.
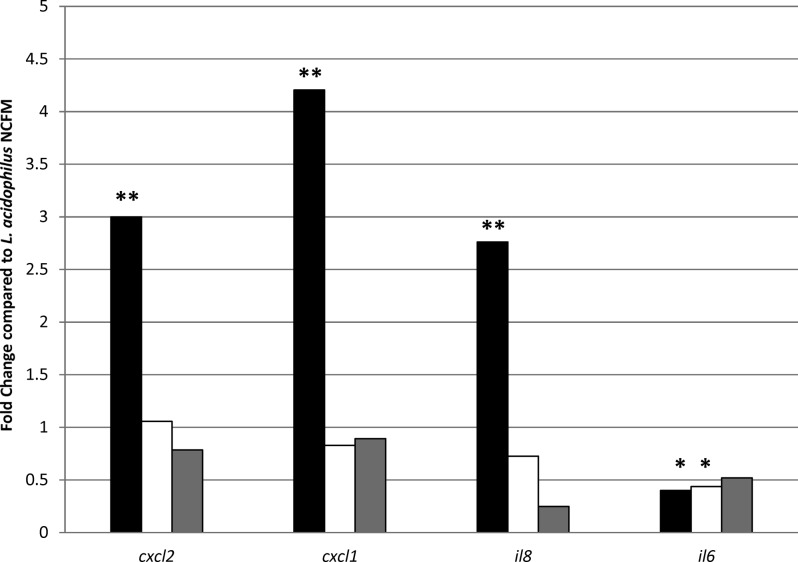
In addition to their probiotic properties, some lactic acid bacteria have potential for delivery of vaccines and biotherapeutics to the intestinal mucosa. L. acidophilus NCFM is an ideal candidate for these applications, as it can survive stomach acids and intestinal bile for transit to the mucosa of the GIT, an important site of mucosal immunity. Additionally, genetic tools are available and have been utilized to genetically alter this strain for the purpose of vaccine delivery (6, 7). In the present study, recombinant lactobacilli expressing flagellin from Salmonella were exposed to Caco-2 epithelial cells for 1 h. A 1-h exposure time was chosen based on the results of the whole-genome array described above, as this exposure time resulted in the induction of immune-related genes. Exposure of bacteria to epithelial cells and RNA isolation were performed as described above. These recombinant strains were previously constructed using two different anchor motifs to display flagellin on the cell surface either by covalent (LPXTG motif, NCK2158) or noncovalent (NCK2160) bonds (9). Previous results with these strains demonstrated different immunological responses in relation to cytokine production from DCs and DC maturation (9). In particular, NCK2158 (covalently bound flagellin) induced more IL-12-producing and CD83+ cells than the controls. RNA was isolated after exposure of epithelial cells to 2 × 108 CFU/ml of the parent strain L. acidophilus (NCK56), a control strain containing the empty vector (NCK1985) (7), and both recombinant strains, NCK2158 and NCK2160. RT-qPCR was used to monitor expression levels of the chemokine genes cxcl1, cxcl2, il6, and il8 between epithelial cells exposed to the parent strain L. acidophilus, the control strain NCK1985, or the recombinant strains (Fig. 3). A marked difference in expression levels of cxcl1, cxcl2, and il8 was observed between the recombinant strains NCK2158 and NCK2160 expressing flagellin. The interleukin gene il6 was downregulated after exposure to the recombinant strains, whereas cxcl1, cxcl2, and il8 were significantly induced after exposure to NCK2158 compared to NCK2160, NCK1985, and L. acidophilus (Fig. 3). In fact, there was no significant difference in the expression levels of cxcl1, cxcl2, and il8 when epithelial cells were exposed to NCK2160 compared to NCK56 and NCK1985. These results reinforced the previous observation (9) that prior determination of cell surface display methods is vital for further vaccine delivery research, and they demonstrate that the epithelial cell line is a useful tool to monitor the effects of recombinant lactobacilli on gene expression by human intestinal cells.
Fig 3.
RT-qPCR results for genes from epithelial cells after exposure to recombinant lactobacilli. A 1-fold change indicates no difference in expression compared to the parent strain, L. acidophilus NCFM (NCK56). Results for NCK2158 (covalently bound flagellin; black bars), NCK2160 (noncovalently bound flagellin; white bars), and NCK1985 (empty vector control; gray bars) are shown. **, P < 0.01; *, P < 0.05 compared to L. acidophilus (NCK56).
In conclusion, this study demonstrated that exposure time with L. acidophilus impacted the gene expression profiles of IECs. In our study, a 1-h, rather than 4- or 8-h exposure time, resulted in maximal differential expression of immune-related genes and genes targeted by the NF-κB complex. These results showed that after an initial exposure to L. acidophilus, expression of the immune-related genes returned toward a baseline level. Additionally, the epithelial cell line proved to be a useful model to determine the expression of various immune markers after exposure to recombinant lactobacilli expressing different cell surface properties.
Microarray data accession number.
Microarray data were deposited in the NCBI Gene Expression Omnibus (GEO) database under accession number GSE34175.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by The Southeast Dairy Foods Research Center and by the National Dairy Council and Dairy Management Inc. as administered by the Dairy Research Institute.
We thank Rosemary Dawes for her technical assistance with Caco-2 cell maintenance and Yong Jun Goh, Evelyn Durmaz, and Steve Silferd for helpful discussions. We thank Akinobu Kajikawa for providing the recombinant strains. We also thank Yong Jun Goh for critical reading of the manuscript.
Footnotes
Published ahead of print 4 May 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ameyar M, Wisniewska M, Weitzman JB. 2003. A role for AP-1 in apoptosis: the case for and against. Biochimie 85: 747–752 [DOI] [PubMed] [Google Scholar]
- 2.Anderson RC, et al. 2010. Lactobacillus plantarum MB452 enhances the function of the intestinal barrier by increasing the expression levels of genes involved in tight junction formation. BMC Microbiol. 10: 316 doi:10.1186/1471-2180-10-316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avadhani A, Miley H. 2011. Probiotics for prevention of antibiotic-associated diarrhea and Clostridium difficile-associated disease in hospitalized adults: a meta-analysis. J. Am. Acad. Nurse Pract. 23: 269–274 [DOI] [PubMed] [Google Scholar]
- 4.Barefoot SF, Klaenhammer TR. 1983. Detection and activity of lactacin B, a bacteriocin produced by Lactobacillus acidophilus. Appl. Environ. Microbiol. 45: 1808–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corr SC, et al. 2007. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl. Acad. Sci. U. S. A. 104: 7617–7621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Douglas GL, Klaenhammer TR. 2011. Directed chromosomal integration and expression of the reporter gene gusA3 in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 77: 7365–7371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duong T, Miller MJ, Barrangou R, Azcarate-Peril MA, Klaenhammer TR. 2011. Construction of vectors for inducible and constitutive gene expression in Lactobacillus. Microb. Biotechnol. 4: 357–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48 doi:10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kajikawa A, et al. 2011. Dissimilar properties of two recombinant Lactobacillus acidophilus strains displaying Salmonella FliC with different anchoring motifs. Appl. Environ. Microbiol. 77: 6587–6596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konstantinov SR, et al. 2008. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc. Natl. Acad. Sci. U. S. A. 105: 19474–19479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lebeer S, Vanderleyden J, De Keersmaecker SC. 2010. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat. Rev. Microbiol. 8: 171–184 [DOI] [PubMed] [Google Scholar]
- 12.Lin YP, Thibodeaux CH, Pena JA, Ferry GD, Versalovic J. 2008. Probiotic Lactobacillus reuteri suppress proinflammatory cytokines via c-Jun. Inflamm. Bowel Dis. 14: 1068–1083 [DOI] [PubMed] [Google Scholar]
- 13.Lonnermark E, et al. 2010. Intake of Lactobacillus plantarum reduces certain gastrointestinal symptoms during treatment with antibiotics. J. Clin. Gastroenterol. 44: 106–112 [DOI] [PubMed] [Google Scholar]
- 14.Mohamadzadeh M, Duong T, Sandwick SJ, Hoover T, Klaenhammer TR. 2009. Dendritic cell targeting of Bacillus anthracis protective antigen expressed by Lactobacillus acidophilus protects mice from lethal challenge. Proc. Natl. Acad. Sci. U. S. A. 106: 4331–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oeckinghaus A, Hayden MS, Ghosh S. 2011. Crosstalk in NF-κB signaling pathways. Nat. Immunol. 12: 695–708 [DOI] [PubMed] [Google Scholar]
- 16.O'Flaherty SJ, Klaenhammer TR. 2010. Functional and phenotypic characterization of a protein from Lactobacillus acidophilus involved in cell morphology, stress tolerance and adherence to intestinal cells. Microbiology 156: 3360–3367 [DOI] [PubMed] [Google Scholar]
- 17.O'Flaherty S, Klaenhammer TR. 2010. The role and potential of probiotic bacteria in the gut, and the communication between gut microflora and gut/host. Int. Dairy J. 20: 262–268 [Google Scholar]
- 18.Putaala H, et al. 2010. Analysis of the human intestinal transcriptional response to Lactobacillus acidophilus, Lactobacillus salivarius, Bifidobacterium lactis and Escherichia coli. Benef. Microbes 1: 283–295 [DOI] [PubMed] [Google Scholar]
- 19.Sanders ME, Klaenhammer TR. 2001. Invited review: the scientific basis of Lactobacillus acidophilus NCFM functionality as a probiotic. J. Dairy Sci. 84: 319–331 [DOI] [PubMed] [Google Scholar]
- 20.van Baarlen P, et al. 2011. Human mucosal in vivo transcriptome responses to three lactobacilli indicate how probiotics may modulate human cellular pathways. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl. 1): 4562–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Baarlen P, et al. 2009. Differential NF-κB pathways induction by Lactobacillus plantarum in the duodenum of healthy humans correlating with immune tolerance. Proc. Natl. Acad. Sci. U. S. A. 106: 2371–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zoumpopoulou G, Tsakalidou E, Dewulf J, Pot B, Grangette C. 2009. Differential crosstalk between epithelial cells, dendritic cells and bacteria in a co-culture model. Int. J. Food Microbiol. 131: 40–51 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.