Abstract
In this study, we tested the hypothesis that the SdiA proteins of Escherichia coli and Salmonella enterica serovar Typhimurium respond to indole. While indole was found to have effects on gene expression and biofilm formation, these effects were not sdiA dependent. However, high concentrations of indole did inhibit N-acyl-l-homoserine lactone (AHL) sensing by SdiA. We conclude that SdiA does not respond to indole but indole can inhibit SdiA activity in E. coli and Salmonella.
TEXT
In prokaryotes, cell-to-cell signaling that allows bacteria to coordinate cellular processes within a larger population, or quorum, is called quorum sensing (9, 24). Bacteria secrete different molecules as intercellular signals, such as N-acyl-l-homoserine lactones (AHLs) and autoinducer 2 (AI-2) in the case of the Proteobacteria and small peptides in the case of the Firmicutes (2, 24). The paradigm for AHL signaling is the LuxR/LuxI system of Vibrio fischeri (23). LuxI, an AHL synthase, produces N-(3-oxo-hexanoyl)-l-homoserine lactone (oxoC6), which can diffuse passively across membranes (14). LuxR binds oxoC6 and responds by activating transcription of the luxICDABEGH operon, which encodes luciferase. Accumulation of AHL signals within a confined environment leads to coordinate activation of light production (9, 10, 27, 28).
Escherichia coli and Salmonella enterica serovar Typhimurium (S. Typhimurium) encode a LuxR homolog, SdiA, but do not encode any type of AHL synthases and do not synthesize AHLs (21). Instead, these organisms respond to the AHLs produced by other species of bacteria, such as Yersinia enterocolitica (7, 21, 32–34). The structure of AHL bound to the N terminus of SdiA has been determined (37). SdiA upregulates two loci in S. Typhimurium, the rck (resistance to complement killing) operon, located on the Salmonella virulence plasmid, and srgE (sdiA-regulated gene), a horizontally acquired gene located on the chromosome (1, 21, 32). The function of SrgE is unknown, but computer algorithms suggest it may be a type III secreted effector (29). Neither of these loci is present in E. coli. In E. coli K-12, sdiA upregulates the acid fitness island (which is not present in S. Typhimurium) while downregulating flagellar genes (8, 13, 18, 25, 35). Enterohemorrhagic E. coli (EHEC) has an additional pathogenicity island, the locus of enterocyte effacement (LEE), which is also downregulated by sdiA (12, 13).
It has been reported that in addition to sensing AHLs, SdiA responds to indole (5, 16–18). Indole is an intermediate product in tryptophan biosynthesis and is produced by the tryptophan degradation enzyme, tryptophanase (TnaA). Similar to AHLs, indole has been shown to be freely diffusible across bacterial membranes (14, 26). While E. coli encodes TnaA and produces indole, S. Typhimurium does not. Indole was found to repress biofilm formation of E. coli (4, 16, 19, 30). Transcription profiling of biofilms indicated that sdiA was upregulated approximately 3-fold in the presence of indole (16). Subsequent work has shown that indole represses biofilm formation of E. coli at 30°C but not 37°C and that this repression is sdiA dependent (18). In this study, we attempted to replicate these findings.
SdiA has no effect on biofilm formation in E. coli K-12 or S. Typhimurium.
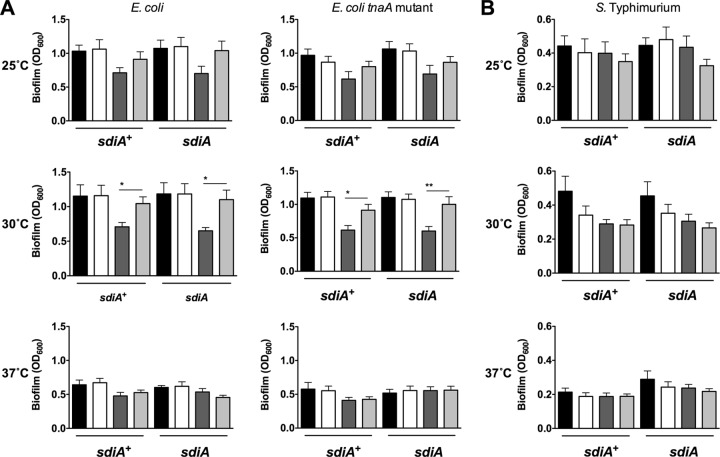
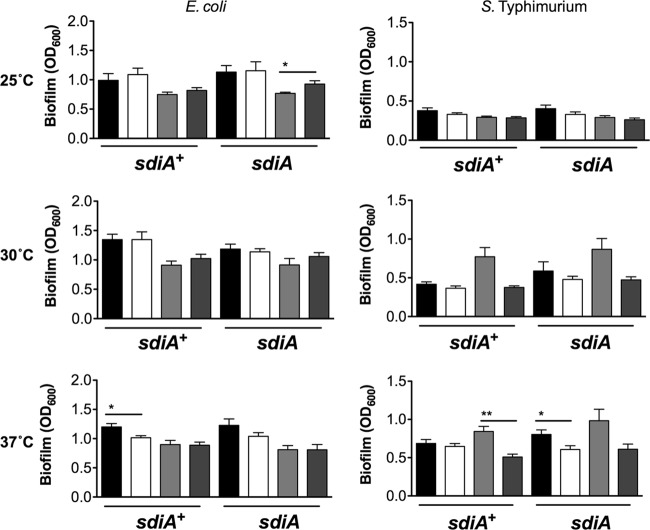
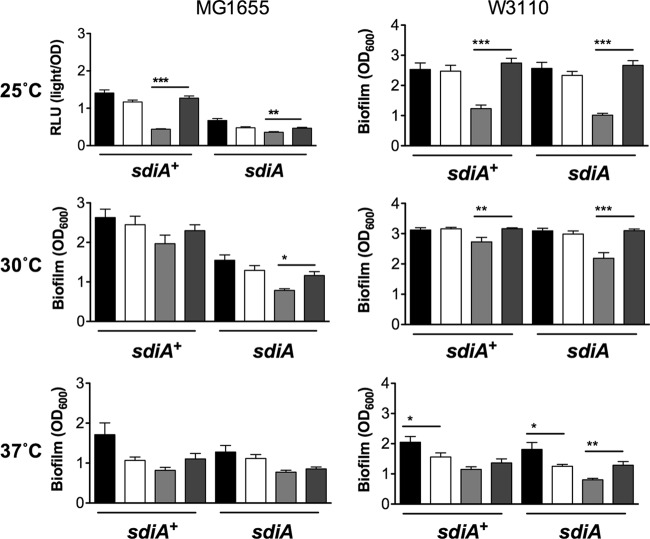
Strains and primers are listed in Tables 1 and 2. The initial report linking sdiA and indole measured biofilm formation of E. coli K-12 strain BW25113 grown in LB broth on polystyrene plates as measured by crystal violet staining (18). Therefore, we measured biofilm formation of E. coli K-12 strain BW25113 grown in LB broth on polystyrene in the presence of AHL (1 μM oxoC6), indole (500 μM), or solvent controls (acidified ethyl acetate for AHL [EA] and dimethyl formamide for indole [DMF]). We tested each of three growth temperatures, 25°C, 30°C, and 37°C. In either the E. coli wild-type or tnaA mutant background, the addition of AHL did not affect biofilm formation compared to results with the solvent control under any of the growth conditions tested (Fig. 1A). The addition of indole significantly repressed biofilm formation at 30°C compared to results with the solvent control. However, this decrease was not dependent upon sdiA (Fig. 1A). With S. Typhimurium we saw no significant effect of sdiA, AHL, or indole on biofilm formation (Fig. 1B). Similar results were observed using 1 mM indole (Fig. 2). Experiments performed using other E. coli K-12 backgrounds also failed to show an sdiA-dependent response to indole (Fig. 3). Interestingly, wild-type MG1655 makes more biofilm than its isogenic sdiA mutant, but this is not dependent upon AHL or indole. This is not seen in the BW25113, W3110, or Salmonella background.
Table 1.
Strains and plasmids used in this work
| Strain or plasmid | Genotypea | Source, reference, or description |
|---|---|---|
| Strains | ||
| S. enterica serovar Typhimurium | ||
| 14028 | Wild type | American Type Culture Collection |
| BA612 | 14028 sdiA1::mTn3 | 1 |
| E. coli | ||
| AL4001 | BA4000 gadW4001::mTn5-lux-kan2 | 8 |
| BA4000 | Nal-resistant mutant of BW25113 | 8 |
| BA760 | MG1655 sdiA::Kanr | P1 transduction of WX2 sdiA::Kanr into MG1655 |
| BA763 | W3110 sdiA::Kanr | P1 transduction of WX2 sdiA::Kanr into W3110 |
| BW25113 | lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78 | 6 |
| JLD800 | BA4000 gadW4001::mTn5-lux-kan2 sdiA271::cam | 8 |
| JNS3003 | BW25113 sdiA+-FRT-cam-FRT | BW25113 with insertion of FRT-cam-FRT cassette using λ Red recombination with primers BA1192 and BA1193 and template pKD3 |
| JNS3212 | BW25113 sdiA+-tnpR-lacZYA | FRT-cam-FRT removed from JNS3003 using Flp recombinase encoded by pCP20; pCE70 inserted at FRT scar using Flp recombinase; pCP20 was subsequently cured by growth at 37°C |
| ME020 | BA4000 gadW4001::mTn5-lux-kan2 sdiA271::cam tnaA::pGP704 | tnaA disrupted by single crossover of pME017 suicide vector into JLD800 chromosome; verified insertion using PCR with primers BA685 and BA2421 |
| ME021 | BA4000 gadW4001::mTn5-lux-kan2 tnaA::pGP704 | tnaA disrupted by single crossover of pME017 suicide vector into AL4001 chromosome; verified insertion using PCR with primers BA685 and BA2421 |
| MG1655 | F− lambda− ilvG rfb-50 rph-1 | E. coli Genetic Stock Center |
| W3110 | F− lambda− IN(rrnD-rrnE)1 rph-1 | 11 |
| WX2 | Δlac sdiA::Kanr | 36 |
| Plasmids | ||
| pCE70 | FRT-tnpR-lacZY oriR6K (Kanr); contains wild-type tnpR Shine-Dalgarno; FRT orientation A | 20 |
| pCP20 | cI857 λPR flp pSC101 oriTS (Ampr Camr) | 6 |
| pGP704 | Suicide vector, oriR6K (Ampr) | 22 |
| pKD3 | FRT-cam-FRT oriR6K (Ampr) | 6 |
| pJNS25 | PsrgE-luxCDABE (Tetr) | 32 |
| pME017 | pGP704 carrying internal portion of tnaA | tnaA fragment amplified with PCR using primers BA2145 and BA2146, BW25113 as template, and Taq DNA polymerase (NEB); fragment cloned into pGEM T-Easy using T4 DNA ligase (Promega); fragment removed with XbaI SphI and ligated into pGP704 cut with XbaI SphI |
Nal, nalidixic acid.
Table 2.
Primers used in this work
| Primer | Sequencea | Description |
|---|---|---|
| BA685 | AGATCTCTGGCGCGTCGTCGCCACCTACAGGC | tnaA insertion verification |
| BA1192 | TGTTACGCGGCCGCTACTGGCTTAATTTGAgtgtaggctggagctgcttc | sdiA+-FRT-cam-FRT |
| BA1193 | TTGCATCTGGCACGCAGGACAGAAAAGAGAcatatgaatatcctccttag | sdiA+-FRT-cam-FRT |
| BA2145 | TCTAGACTGATTAAAAAACGCGAGCAGGAAAAAG | Internal tnaA portion |
| BA2146 | GCATGCCATCGACCAGATACTGTACCTGCGCGATAC | Internal tnaA portion |
| BA2421 | GTGGCACTTTTCGGGGAAATGTGCGCGGAACCCC | tnaA insertion verification |
Lowercase letters indicate the portion of the primer that binds the template pKD3.
Fig 1.
Biofilm formation in the presence of AHL and 500 μM indole in E. coli K-12 and S. Typhimurium. (A) Biofilm formation of E. coli K-12 BW25113 in either the wild-type (AL4001), sdiA mutant (JLD800), tnaA mutant (ME021), or tnaA sdiA double mutant (ME020) background. (B) Biofilm formation of S. Typhimurium 14028 or the isogenic sdiA mutant (BA612). Crystal violet absorbance at the optical density at 600 nm (OD600) is indicated by bars for 1 μM AHL (black), 0.1% ethyl acetate (EA) (white), 500 μM indole (dark gray), and 0.1% dimethylformamide (DMF) (light gray). All data points are the averages for nine biological replicates, and error bars indicate SEM. Statistical significance in comparison to results for the solvent control is denoted by asterisks representing t test P values: ∗, <0.05; ∗∗, <0.005.
Fig 2.
Biofilm formation in the presence of AHL and 1 mM indole in E. coli K-12 and S. Typhimurium. Biofilm formation of E. coli K-12 in either the wild type (AL4001) or the sdiA mutant (JLD800) and of S. Typhimurium 14028 or the isogenic sdiA mutant (BA612) was analyzed. Crystal violet absorbance at 600 nm is indicated by bars for 1 μM AHL (black), 0.1% EA (white), 1 mM indole (light gray), and 0.1% DMF (dark gray). All data points are the averages for nine biological replicates, and error bars indicate SEM. Statistical significance in comparison to results with the solvent control is denoted by asterisks representing t test P values: ∗, <0.05; ∗∗, <0.005.
Fig 3.
Biofilm formation in the presence of AHL and indole in other E. coli backgrounds. Biofilm formation of E. coli MG1655 and W3110 or the isogenic sdiA mutants, BA763 and BA760, is shown. Crystal violet absorbance at 600 nm is indicated by bars for 1 μM AHL (black), 0.1% EA (white), 1 mM indole (light gray), and 0.1% DMF (dark gray). All data points are the averages for nine biological replicates, and error bars indicate SEM. Statistical significance in comparison to results with the solvent control is denoted by asterisks representing t test P values: ∗, <0.05; ∗∗, <0.005; ∗∗∗, <0.0005.
SdiA reporter strains show little or no response to indole.
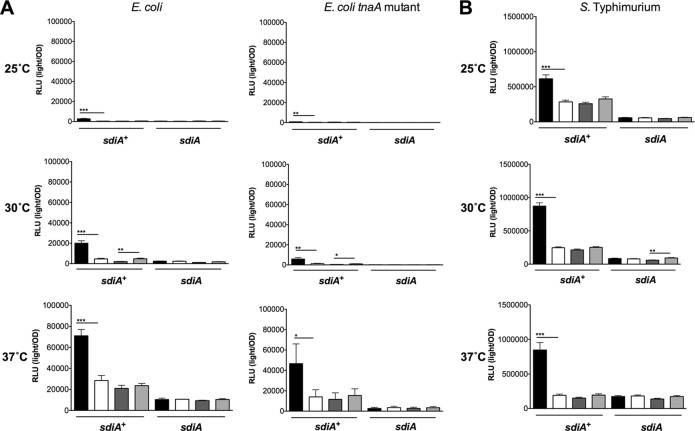
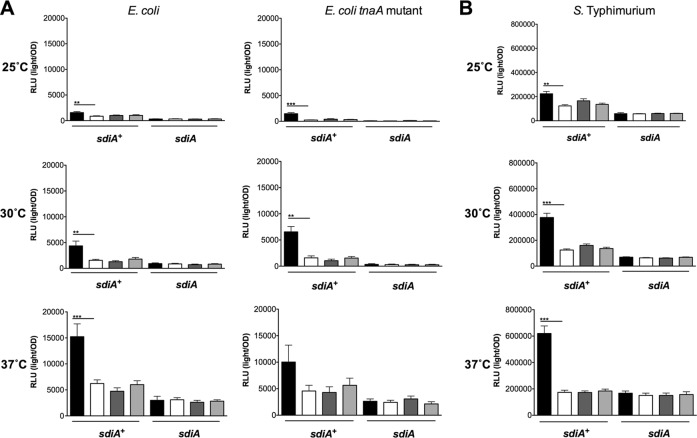
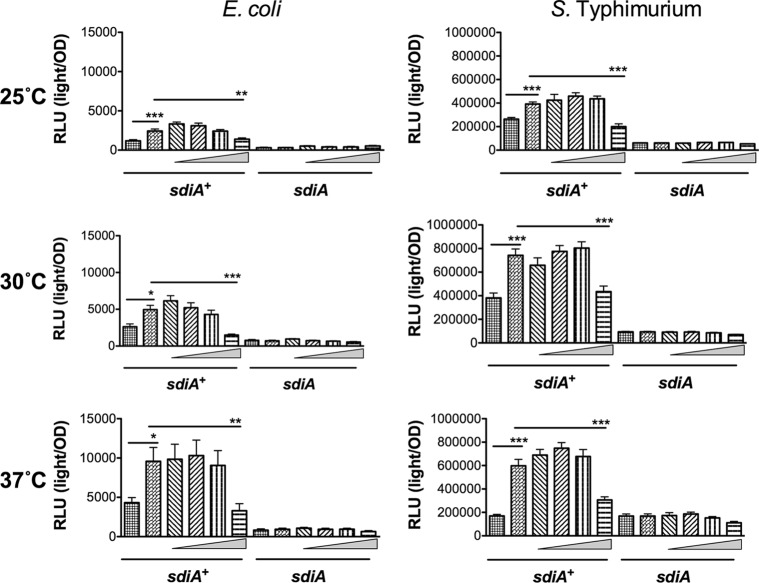
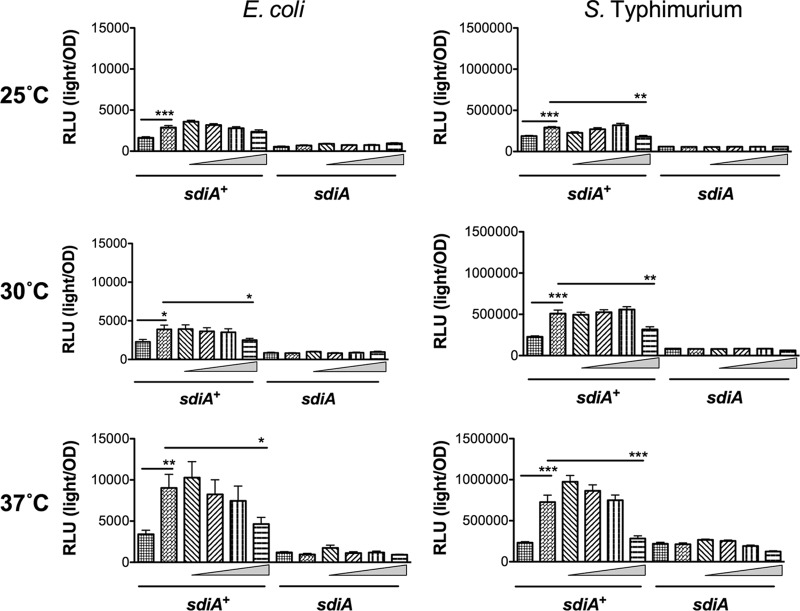
To more thoroughly investigate SdiA activity in the presence of indole, we utilized previously described sdiA-dependent reporter strains of E. coli and S. Typhimurium (8, 32). In E. coli K-12, the most sensitive reporter of SdiA activity is a chromosomal gadW::Tn5-luxCDABE fusion (8). GadW is a transcription factor encoded within the acid fitness island. For S. Typhimurium, the most sensitive reporter is a plasmid-based srgE-luxCDABE fusion (32). We tested the E. coli K-12 and S. Typhimurium reporter strains grown in LB in the presence of AHL (1 μM oxoC6), indole (500 μM), or solvent controls during standing or shaking growth at each of three temperatures, 25°C, 30°C, and 37°C. Both fusions show sdiA-dependent activation only in the presence of AHL (Fig. 4 and 5). Indole had repressive effects on the fusions under some conditions, but in most instances this was not sdiA dependent. We believe the overall trend throughout the experiments is not sdiA dependent, although statistical significance is achieved with only the wild type or only with the sdiA mutant in some experiments. Experiments using 0.1 mM and 1 mM indole yielded similar conclusions (Fig. 6 and 7 and data not shown).
Fig 4.
Response of SdiA to AHL and 500 μM indole in E. coli K-12 and S. Typhimurium during shaking growth conditions. (A) Expression of the gadW::Tn5-luxCDABE fusion in E. coli K-12 in either the wild-type (AL4001), sdiA mutant (JLD800), tnaA mutant (ME021), or tnaA sdiA double mutant (ME020) background.(B) Expression of the srgE-luxCDABE fusion in S. Typhimurium 14028 or the isogenic sdiA mutant (BA612). Relative light units (light/OD590) after 9 h of growth with shaking are indicated by bars for 1 μM AHL (black), 0.1% ethyl acetate (EA) (white), 500 μM indole (dark gray), and 0.1% dimethylformamide (DMF) (light gray). All data points are the averages for nine biological replicates, and error bars indicate SEM. Statistical significance in comparison to results with the solvent control is denoted by asterisks representing t test P values: ∗, <0.05; ∗∗, <0.005; ∗∗∗, <0.0005.
Fig 5.
Response of SdiA to AHL and 500 μM indole in E. coli K-12 and S. Typhimurium during standing growth conditions. (A) Expression of the gadW::Tn5-luxCDABE fusion in E. coli K-12 in either the wild-type (AL4001), sdiA mutant (JLD800), tnaA mutant (ME021), or tnaA sdiA double mutant (ME020) background. (B) Expression of the srgE-luxCDABE fusion in S. Typhimurium 14028 or the isogenic sdiA mutant (BA612). Relative light units (light/OD590) after 9 h of standing growth are indicated by bars for 1 μM AHL (black), 0.1% ethyl acetate (EA) (white), 500 μM indole (dark gray), and 0.1% dimethylformamide (DMF) (light gray). All data points are the averages for nine biological replicates, and error bars indicate SEM. Statistical significance in comparison to results with the solvent control is denoted by asterisks representing t test P values: ∗∗, <0.005; ∗∗∗, <0.0005.
Fig 6.
Response of SdiA to AHL and 1 mM indole in E. coli K-12 and S. Typhimurium during shaking growth conditions. (A) Expression of the gadW::Tn5-luxCDABE fusion in E. coli K-12 in either the wild-type (AL4001), sdiA mutant (JLD800), tnaA mutant (ME021), or tnaA sdiA double mutant (ME020) backgrounds. (B) Expression of the srgE-luxCDABE fusion in S. Typhimurium 14028 or the isogenic sdiA mutant (BA612). Relative light units (light/OD590) after 9 h of growth with shaking are indicated by bars for 1 μM AHL (black), 0.1% ethyl acetate (EA) (white), 1 mM indole (dark gray), and 0.1% dimethylformamide (DMF) (light gray). All data points are the averages for nine biological replicates, and error bars indicate SEM. Statistical significance in comparison to results with the solvent control is denoted by asterisks representing t test P values: ∗, <0.05; ∗∗, <0.005.
Fig 7.
Response of SdiA to AHL and 1 mM indole in E. coli K-12 and S. Typhimurium during standing growth conditions. (A) Expression of the gadW::Tn5-luxCDABE fusion in E. coli K-12 in either the wild-type (AL4001), sdiA mutant (JLD800), tnaA mutant (ME021), or tnaA sdiA double mutant (ME020) background. (B) Expression of the srgE-luxCDABE fusion in S. Typhimurium 14028 or the isogenic sdiA mutant (BA612). Relative light units (light/OD590) after 9 h of standing growth are indicated by bars for 1 μM AHL (black), 0.1% ethyl acetate (EA) (white), 1 mM indole (dark gray), and 0.1% dimethylformamide (DMF) (light gray). All data points are the averages for nine biological replicates, and error bars indicate SEM. Statistical significance in comparison to results with the solvent control is denoted by asterisks representing t test P values: ∗, <0.05; ∗∗, <0.005.
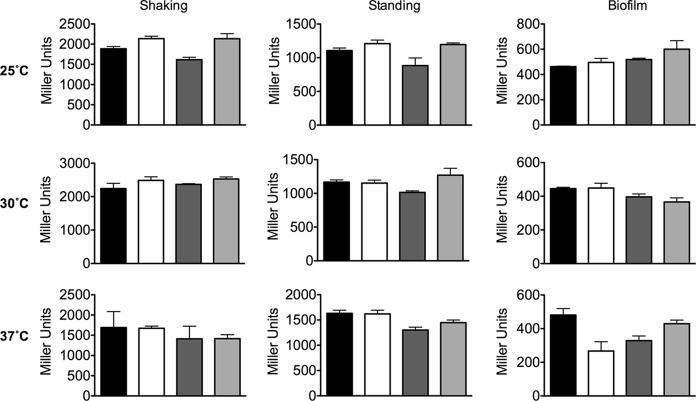
Indole does not regulate sdiA expression.
Transcription profiling of E. coli biofilms grown in LB glucose in the presence and absence of indole has indicated that the sdiA gene is upregulated in the presence of indole (16). We constructed an sdiA-tnpR-lacZY chromosomal fusion in E. coli K-12 strain BW25113 to test the regulation of sdiA by indole. We saw no significant effect of AHL or indole on sdiA expression compared to results with the solvent control at 25°C, 30°C, or 37°C during shaking or standing growth conditions in broth cultures or during growth in biofilms (Fig. 8).
Fig 8.
Expression of E. coli sdiA during growth in shaking or standing liquid cultures or in biofilms. Expression of the sdiA-tnpR-lacZY fusion in E. coli K-12 (JNS3212) is shown. Miller units after 9 h of growth in standing or shaking liquid, or 16 h of growth in biofilm, are indicated by bars for 1 μM AHL (black), 0.1% ethyl acetate (EA) (white), 500 μM indole (dark gray), and 0.1% dimethylformamide (DMF) (light gray). All data points are the averages for three biological replicates, and error bars indicate SEM.
Indole inhibits detection of AHL by SdiA.
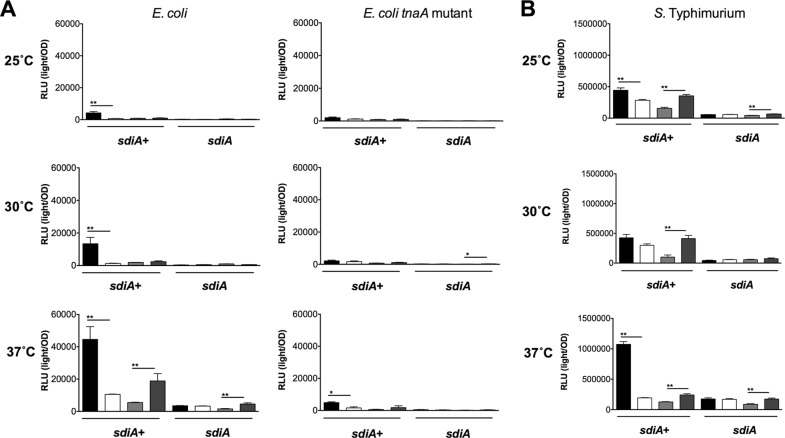
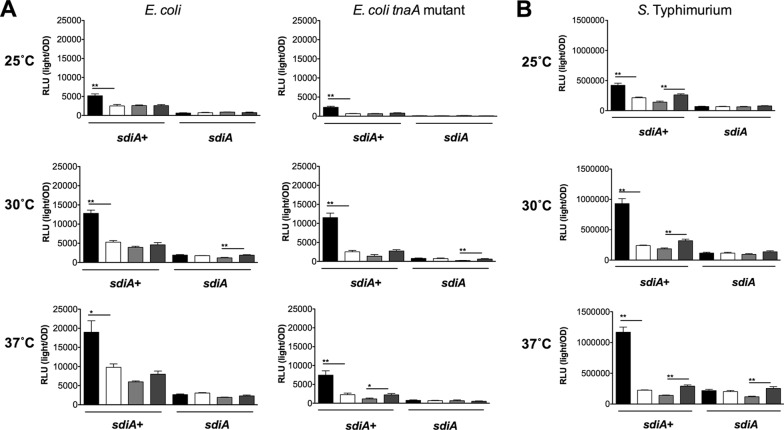
In order to determine whether indole could alter AHL sensing by SdiA, we measured gadW::Tn5-luxCDABE and srgE-luxCDABE expression in E. coli and S. Typhimurium, respectively, in the presence of 100 nM AHL and increasing indole concentrations (1 μM, 10 μM, 100 μM, and 1 mM) during growth at 25°C, 30°C, and 37°C under shaking and standing conditions. Interestingly, at high concentrations, indole inhibited the detection of AHL by E. coli and Salmonella (Fig. 9 and 10). While SdiA activity was never reduced to the level of that of an sdiA mutant, the inhibition was significant at the highest concentration of 1 mM indole.
Fig 9.
Indole inhibits AHL detection by SdiA in E. coli K-12 and S. Typhimurium during shaking growth conditions. (A) Expression of the gadW::Tn5-luxCDABE fusion in E. coli K-12 in either the wild-type (AL4001), sdiA mutant (JLD800), tnaA mutant (ME021), or tnaA sdiA double mutant (ME020) background. (B) Expression of the srgE-luxCDABE fusion in S. Typhimurium 14028 or sdiA mutant (BA612). Relative light units (light/OD590) after 9 h of growth with shaking are indicated by bars for 0.1% ethyl acetate (EA) and 0.5% dimethylformamide (DMF) (hatched), 100 nM AHL + 0.5% DMF (bricked lines), 100 nM AHL + 1 μM indole (diagonal downward-slanting lines), 100 nM AHL + 10 μM indole (diagonal upward-slanting lines), 100 nM AHL + 100 μM indole (vertical lines), and 100 nM AHL + 1 mM indole (horizontal lines). All data points are the averages for nine biological replicates, and error bars indicate SEM. Statistical significance in comparison to results with the solvent control or the AHL + solvent control is denoted by asterisks representing t test P values: ∗, <0.05; ∗∗, <0.005; ∗∗∗, <0.0005.
Fig 10.
Indole inhibits AHL detection by SdiA of E. coli K-12 and S. Typhimurium during standing growth conditions. (A) Expression of the gadW::Tn5-luxCDABE fusion in E. coli K-12 in either the wild-type (AL4001), sdiA mutant (JLD800), tnaA mutant (ME021), or tnaA sdiA double mutant (ME020) background. (B) Expression of the srgE-luxCDABE fusion in S. Typhimurium 14028 or the sdiA mutant (BA612). Relative light units (light/OD590) after 9 h of growth with shaking are indicated by bars for 0.1% ethyl acetate (EA) and 0.5% dimethylformamide (DMF) (hatched), 100 nM AHL + 0.5% DMF (bricked lines), 100 nM AHL + 1 μM indole (diagonal downward-slanting lines), 100 nM AHL + 10 μM indole (diagonal upward-slanting lines), 100 nM AHL + 100 μM indole (vertical lines), 100 nM AHL + 1 mM indole (horizontal lines). Each data point is the average for nine biological replicates, and error bars indicate SEM. Statistical significance in comparison to results with the solvent control or the AHL + solvent control is denoted by asterisks representing t test P values: ∗, <0.05; ∗∗, <0.005; ∗∗∗, <0.0005.
Based on the results obtained in this work, SdiA does not respond to indole. Indole has repressive effects on reporter gene expression in some instances, but these are not sdiA dependent. Indole also represses biofilm formation at lower temperatures, but this is not sdiA dependent. We do not know why our result differs from previously published results. We obtained our results with three different E. coli backgrounds and with S. Typhimurium, suggesting that strain background is not the issue. However, we did find that high concentrations of indole inhibit the detection of AHL by SdiA. We see a gradation of inhibition between 100 μM and 1 mM indole, which may be physiologically relevant since indole is reported to be present in the mouse, rat, and human gut at ∼140 μM, ∼68 μM, and ∼300 to 1,074 μM concentrations, respectively (3, 15, 31, 38).
ACKNOWLEDGMENTS
The project described was supported by awards R01AI073971 and R01AI097116 from the National Institute of Allergy and Infectious Diseases.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
Published ahead of print 18 May 2012
REFERENCES
- 1. Ahmer BM, Reeuwijk JV, Timmers CD, Valentine PJ, Heffron F. 1998. Salmonella typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J. Bacteriol. 180:1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Atkinson S, Williams P. 2009. Quorum sensing and social networking in the microbial world. J. R. Soc. Interface 6:959–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Botsford JL, Demoss RD. 1972. Escherichia coli tryptophanase in the enteric environment. J. Bacteriol. 109:74–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bunders CA, et al. 2011. Intercepting bacterial indole signaling with flustramine derivatives. J. Am. Chem. Soc. 133:20160–20163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chu W, et al. 2012. Indole production promotes Escherichia coli mixed-culture growth with Pseudomonas aeruginosa by inhibiting quorum signaling. Appl. Environ. Microbiol. 78:411–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dyszel JL, et al. 2010. Salmonella enterica serovar Typhimurium can detect acyl homoserine lactone production by Yersinia enterocolitica in mice. J. Bacteriol. 192:29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dyszel JL, et al. 2010. E. coli K-12 and EHEC genes regulated by SdiA. PLoS One 5:e8946 doi:10.1371/journal.pone.0008946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fuqua WC, Winans SC, Greenberg EP. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hense BA, et al. 2007. Does efficiency sensing unify diffusion and quorum sensing? Nat. Rev. Microbiol. 5:230–239 [DOI] [PubMed] [Google Scholar]
- 11. Hill CW, Harnish BW. 1981. Inversions between ribosomal RNA genes of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 78:7069–7072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hughes DT, et al. 2010. Chemical sensing in mammalian host-bacterial commensal associations. Proc. Natl. Acad. Sci. U. S. A. 107:9831–9836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kanamaru K, Kanamaru K, Tatsuno I, Tobe T, Sasakawa C. 2000. SdiA, an Escherichia coli homologue of quorum-sensing regulators, controls the expression of virulence factors in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 38:805–816 [DOI] [PubMed] [Google Scholar]
- 14. Kaplan HB, Greenberg EP. 1985. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J. Bacteriol. 163:1210–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Karlin DA, Mastromarino AJ, Jones RD, Stroehlein JR, Lorentz O. 1985. Fecal skatole and indole and breath methane and hydrogen in patients with large bowel polyps or cancer. J. Cancer Res. Clin. Oncol. 109:135–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee J, Jayaraman A, Wood TK. 2007. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 7:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee J, Maeda T, Hong SH, Wood TK. 2009. Reconfiguring the quorum-sensing regulator SdiA of Escherichia coli to control biofilm formation via indole and N-acylhomoserine lactones. Appl. Environ. Microbiol. 75:1703–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee J, et al. 2008. Indole cell signaling occurs primarily at low temperatures in Escherichia coli. ISME J. 2:1007–1023 [DOI] [PubMed] [Google Scholar]
- 19. Martino PD, Fursy R, Bret L, Sundararaju B, Phillips RS. 2003. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can. J. Microbiol. 49:443–449 [DOI] [PubMed] [Google Scholar]
- 20. Merighi M, Ellermeier CD, Slauch JM, Gunn JS. 2005. Resolvase-in vivo expression technology analysis of the Salmonella enterica serovar Typhimurium PhoP and PmrA regulons in BALB/c mice. J. Bacteriol. 187:7407–7416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Michael B, Smith JN, Swift S, Heffron F, Ahmer BM. 2001. SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J. Bacteriol. 183:5733–5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miyashiro T, Ruby EG. 2012. Shedding light on bioluminescence regulation in Vibrio fischeri. Mol. Microbiol. doi:10.1111/j.1365–2958.2012.08065.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ng W-L, Bassler BL. 2009. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43:197–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nikaido E, Yamaguchi A, Nishino K. 2008. AcrAB multidrug efflux pump regulation in Salmonella enterica serovar Typhimurium by RamA in response to environmental signals. J. Biol. Chem. 283:24245–24253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Piñero-Fernandez S, Chimerel C, Keyser UF, Summers DK. 2011. Indole transport across Escherichia coli membranes. J. Bacteriol. 193:1793–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Redfield RJ. 2002. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 10:365–370 [DOI] [PubMed] [Google Scholar]
- 28. Roux A, Payne SM, Gilmore MS. 2009. Microbial telesensing: probing the environment for friends, foes, and food. Cell Host Microbe 6:115–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Samudrala R, Heffron F, McDermott JE. 2009. Accurate prediction of secreted substrates and identification of a conserved putative secretion signal for type III secretion systems. PLoS Pathog. 5:e1000375 doi:10.1371/journal.ppat.1000375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scherzer R, et al. 2009. New tryptophanase inhibitors: towards prevention of bacterial biofilm formation. J. Enzyme Inhib. Med. Chem. 24:350–355 [DOI] [PubMed] [Google Scholar]
- 31. Sims J, Renwick AG. 1983. The effects of saccharin on the metabolism of dietary tryptophan to indole, a known cocarcinogen for the urinary bladder of the rat. Toxicol. Appl. Pharmacol. 67:132–151 [DOI] [PubMed] [Google Scholar]
- 32. Smith JN, Ahmer BMM. 2003. Detection of other microbial species by Salmonella: expression of the SdiA regulon. J. Bacteriol. 185:1357–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smith JN, et al. 2008. SdiA, an N-acylhomoserine lactone receptor, becomes active during the transit of Salmonella enterica through the gastrointestinal tract of turtles. PLoS One 3:e2826 doi:10.1371/journal.pone.0002826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Soares JA, Ahmer BM. 2011. Detection of acyl-homoserine lactones by Escherichia and Salmonella. Curr. Opin. Microbiol. 14:188–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Van Houdt R, Aertsen A, Moons P, Vanoirbeek K, Michiels CW. 2006. N-Acyl-l-homoserine lactone signal interception by Escherichia coli. FEMS Microbiol. Lett. 256:83–89 [DOI] [PubMed] [Google Scholar]
- 36. Wang XD, de Boer PA, Rothfield LI. 1991. A factor that positively regulates cell division by activating transcription of the major cluster of essential cell division genes of Escherichia coli. EMBO J. 10:3363–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yao Y, et al. 2006. Structure of the Escherichia coli quorum sensing protein SdiA: activation of the folding switch by acyl homoserine lactones. J. Mol. Biol. 355:262–273 [DOI] [PubMed] [Google Scholar]
- 38. Zuccato E, et al. 1993. Role of bile acids and metabolic activity of colonic bacteria in increased risk of colon cancer after cholecystectomy. Dig. Dis. Sci. 38:514–519 [DOI] [PubMed] [Google Scholar]