Abstract
Novel markers of fecal pollution in tropical waters are needed since conventional methods recommended for other geographical regions may not apply. To address this, the prevalence of thermotolerant coliforms, enterococci, coliphages, and enterophages was determined by culture methods across a watershed. Additionally, human-, chicken-, and cattle-specific PCR assays were used to identify potential fecal pollution sources in this watershed. An enterococcus quantitative PCR (qPCR) assay was tested and correlated with culture methods at three sites since water quality guidelines could incorporate this technique as a rapid detection method. Various rainfall events reported before sample collection at three sites were considered in the data analyses. Thermotolerant coliforms, enterococci, coliphages, and enterophages were detected across the watershed. Human-specific Bacteroides bacteria, unlike the cattle- and chicken-specific bacteria, were detected mostly at sites with the corresponding fecal impact. Enterococci were detected by qPCR as well, but positive correlations with the culture method were noted at two sites, suggesting that either technique could be used. However, no positive correlations were noted for an inland lake tested, suggesting that qPCR may not be suitable for all water bodies. Concentrations of thermotolerant coliforms and bacteriophages were consistently lower after rainfall events, pointing to a possible dilution effect. Rainfall positively correlated with enterococci detected by culturing and qPCR, but this was not the case for the inland lake. The toolbox of methods and correlations presented here could be potentially applied to assess the microbial quality of various water types.
INTRODUCTION
Monitoring microbial indicators of fecal pollution in tropical waters remains an issue of concern since these may not accurately indicate the presence of microbes associated with fecal matter. Indicators of fecal contamination used in different geographical areas include the thermotolerant coliforms and enterococci (31). These indicator bacteria are present in the intestinal tract of warm-blooded animals, and therefore, their presence in waters may indicate fecal pollution. However, it has been shown elsewhere that thermotolerant coliforms and enterococci may be part of the environmental microbiota of tropical waters (33). In addition, many bacterial indicators cannot be used to indicate the time and source of the fecal contamination since they can replicate outside their host and are present in the feces of different animals (5, 9, 13, 16, 44, 45). These shortcomings with regard to indicator bacteria have prompted the use of alternate indicators, such as bacteriophages, which show promising characteristics. For instance, coliphages, which are normally isolated from feces and fecally contaminated waters, do not appear to replicate outside their host, and certain groups have survival characteristics similar to those of enteric viruses in waters (11, 14, 15, 19). However, certain coliphage groups are present in the feces of different warm-blooded animals and, therefore, may not be used to discriminate the source of the fecal pollution (10).
Recently, isolated phages that infect a specific Enterococcus faecalis type strain (enterophages) have been proposed as good indicators of human-specific fecal contamination. Enterophages have a survival time similar to that of human enteric viruses in marine and fresh waters and sand (8, 32, 36, 43), have been detected in raw and treated domestic sewage in Puerto Rico and Portugal, and have been detected neither in pristine waters nor in animal feces (e.g., pigs, dogs, cattle, and chickens) (8, 36). However, more data are needed in order to accept enterophages as alternate indicators of human fecal contamination. One way to further test enterophages as indicators of fecal pollution is to compare them with currently used indicators and emerging molecular methods in various water types.
Microbial source tracking (MST) methods can complement traditional methods used to assess microbial water quality (37). Specifically, host-specific assays have been tested in various water sources, and several have been successfully used to discriminate among the sources of fecal pollution. Many Bacteroides species make the intestinal microbiota of warm-blooded animals their primary habitat, and some species have shown high levels of host specificity. In addition, Bacteroides bacteria cannot replicate outside the intestinal tract as most species are strictly anaerobic bacteria (3, 24). Quantitative PCR (qPCR) is also perhaps one of the most promising methods that indicate the levels of the target contaminant. However, as with culture-based methods, molecular techniques suffer from shortcomings, such as the inability to distinguish between viable and dead cells or to determine the infectivity status of the target microorganism.
Detection of bacterial and viral indicators by culture or molecular biology-based techniques may be influenced by rainfall events. It has been shown elsewhere that rainfall may lead to higher numbers of indicators and pathogens in surface waters and thus may represent an increased risk to human health (27, 34). Runoff, resuspension of sediments, and sewage overflows, resulting from rainfall events, may contribute to an increase of indicators and pathogens in surface waters (4, 23). Nonetheless, rainfall is often not considered when monitoring the microbial quality of waters. One main question to address is whether sampling under dry conditions alone is sufficient to infer the presence of fecal indicators and pathogens or whether results differ under wet conditions. However, it is relatively difficult to confirm that an increase in microbial indicators and pathogens after precipitation events truly represents a recent input of fecal matter (23). In the present study, we assessed the microbial quality of a tropical watershed using currently used (thermotolerant coliforms, enterococci, and coliphages) and proposed (enterophages) indicators, as well as molecular methods (host-specific assays and qPCR), considering rainfall as a possible influential variable.
MATERIALS AND METHODS
Sample collection and sampling sites.
In all cases, single 1-liter grab samples were collected in sterile plastic bottles with sodium thiosulfate (final concentration of approximately 10.0 mg/liter) (41), up to four times per month from November 2009 to April 2010. A total of 13 samples per site were collected and tested, kept at 6 to 8°C, and processed within 4 to 6 h. Samples were collected from the Rio Grande de Arecibo watershed in Puerto Rico, characterized by sporadic rainfall throughout the year. This watershed, unlike those found in many regions, is composed of small creeks. One of the interesting characteristics of the Rio Grande de Arecibo watershed is that it has very distinct land uses that contribute to human and/or animal fecal matter loadings. The watershed possesses both urban and rural areas inhabited by more than 100,000 people, with waters that are used for recreational activities and as a drinking water source. The watershed is linked to two of the biggest dams on the island, providing approximately 380 million liters of drinking water daily to the north coast (12).
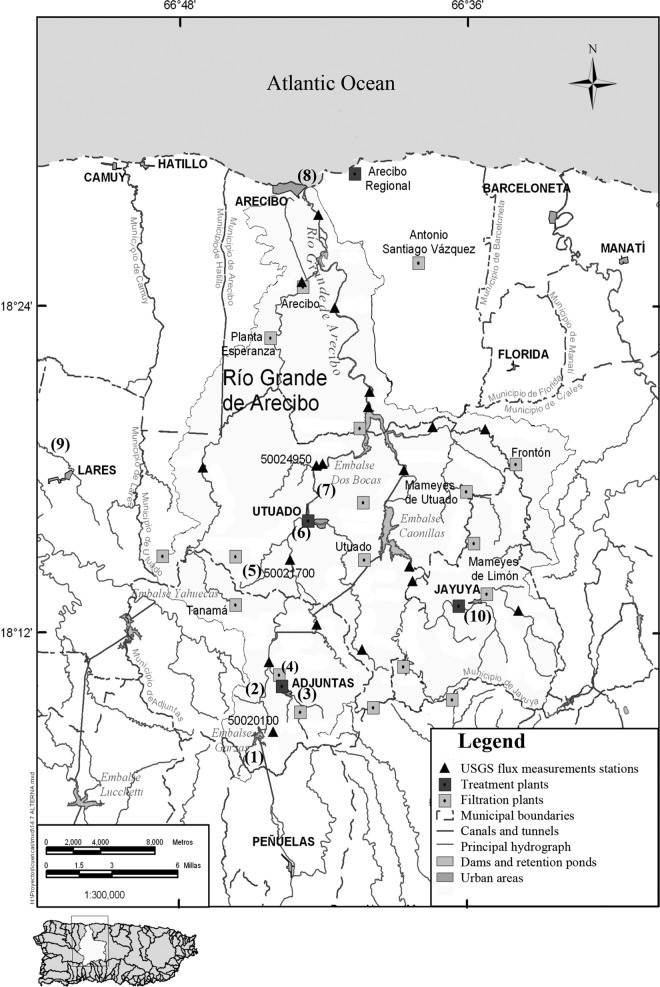
Ten sampling sites were selected based on the potential different fecal pollution sources associated with them. Site 1 (Lago Garza) is bordered on the southwestern part by the Reserva Forestal de Toro Negro; thus, the number of households is limited and farm animals, such as poultry and cattle, are scarce or absent. Site 2 (Rio Vaca) is located downstream from site 1, and residences, poultry, and cattle are also limited. Sites 3 and 4 (Rio Cidra) are located downstream from the urban nucleus of the municipality of Adjuntas. Therefore, both sites are presumably influenced by urban runoff and local poultry. Site 3 is located upstream of the domestic wastewater treatment plant (WTP) at Adjuntas, and site 4 is located downstream from this WTP. Site 5 (Rio Grande de Arecibo) is lined by houses that drain their wastewaters directly into the river, and poultry and horses were also frequently observed nearby or in the river. Site 6 is located at the southern entrance of the urban nucleus of the municipality of Utuado. It is bordered by houses that manage their wastewater through septic tanks and by grass fields, where cattle were frequently observed. Site 7 is located 200 m downstream from the domestic WTP at Utuado, and poultry and cattle were frequently observed. Site 8 is located where the Rio Grande de Arecibo discharges into the Atlantic Ocean, is impacted by cattle grazing, and is used for recreational activities. The Rio Criminales (site 9) is located between a fenced farm with more than 100 cows and human residences with septic tanks. Site 10 is located at the Rio Caguana and receives the input of a WTP. This place is also impacted by cattle and horses (Fig. 1).
FIG 1.
Rio Grande de Arecibo watershed in Puerto Rico. Water samples were collected from Lago Garza (site 1), Rio Vaca (2), upstream from the WTP at Adjuntas (3), downstream from the WTP at Adjuntas (4), Rio Grande de Arecibo (5), upstream from the WTP at Utuado (6), downstream from the WTP at Utuado (7), the estuary (8), Rio Criminales (9), and Rio Caguana (10). (Reproduced from reference 13 with permission.)
Enumeration of indicators by culture methods.
Thermotolerant coliforms were enumerated using m-FC agar incubated at 45°C for 24 h, and enterococci were enumerated using m-Enterococcus agar incubated at 37°C for up to 48 h (40). Coliphages and enterophages were quantified using the single-layer method as described previously (8). The type strains used for phage enumeration were Escherichia coli ATCC 15597 and E. faecalis ATCC 19433. Plates were incubated at 22, 37, 41, and 45°C to detect bacteriophages that replicate at different temperatures. However, data presented in this study correspond to those phages that replicate at 22°C since preliminary analyses suggested that this may be the optimal temperature.
DNA extraction and PCR conditions.
Water samples (100 ml) were filtered through polycarbonate membranes (0.4-μm pore size, 47-mm diameter) (GE Water and Process Technologies, Trevose, PA). Total DNA was extracted from the membranes using Mo Bio PowerSoil kits (Mo Bio Laboratories, Carlsbad, CA) according to the manufacturer's protocol. DNA concentration was estimated using a NanoDrop ND-1000 UV spectrophotometer (NanoDrop Technologies, Wilmington, DE). DNA extracts were stored at −20°C until further processing and analyzed using host-specific PCR assays commonly used in fecal source tracking studies and the qPCR assay for general enterococci (29) (Table 1). PCR was performed using cattle- and human-specific Bacteroides assays targeting 16S rRNA genes and two chicken-specific assays targeting functional genes (6, 7, 28). For convenience, cattle- and human-specific Bacteroides will be referred as CSB and HSB, respectively. PCR amplifications were performed in 25 μl using the polymerase TaKaRa Ex Taq (TaKaRa Bio Inc.) in a Bio-Rad Tetrad2 Peltier thermal cycler (Bio-Rad, Hercules, CA) under the following cycling conditions: an initial denaturation step at 95°C for 5 min, followed by 35 cycles of 1 min at 95°C, 1 min at optimum annealing temperature, and 1 min at 72°C. PCR products were visualized in 1.5% agarose gels using GelStar nucleic acid gel stain (Lonza, Rockland, ME).
TABLE 1.
Summary of oligonucleotide primers and probes for PCR and TaqMan qPCR
| Assay | Primer or probe name and sequence (5′ to 3′)a | Ta (°C)b | Size (bp) | Reference |
|---|---|---|---|---|
| General Bacteroides | Bac32F, AACGCTAGCTACAGGCTT | 53 | 694 | 6 |
| Bac708R, CAATCGGAGTTCTTCGTG | ||||
| Human-specific Bacteroides | HF183, ATCATGAGTTCACATGTCCG | 63 | 543 | 7 |
| Cattle-specific Bacteroides | CF128, CCAACYTTCCCGWTACTC | 62 | 598 | 7 |
| Chicken-specific Bacteroides | CP2-9F, GTAAGACAGCAACCCCATGTA | 56 | 245 | 28 |
| CP2-9R, ACCTATGGTTCAACACGCTTTA | ||||
| Chicken-specific Clostridium | CP3-49F, GTCCAGCGCCTCATTGAT | 57 | 329 | 28 |
| CP3-49R, TGGTGATCGACTTTTCCAAT | ||||
| General Enterococcus qPCR (Entero1) | ECST748F, AGAAATTCCAAACGAACTTG | 60 | 92 | 29 |
| ENC854R, CAGTGCTCTACCTCCATCATT | ||||
| GPL813, FAM-TGGTTCTCTCCGAAATAGCTTTAGGGCTA-TAMRAc | ||||
| GPL813TQ, FAM-TGGTTCTCTCCGAAATAGCTTTAGGGCTA-TAMRA |
The Bac708R primer was used for all 16S Bacteroides assays as a reverse primer.
Optimum annealing temperature (Ta) determined using temperature gradient PCR.
TAMRA, 6-carboxytetramethylrhodamine.
The TaqMan qPCR assay targeting the 23S rRNA gene of Enterococcus spp. (Entero1) was performed in 25-μl reaction mixtures containing 1× TaqMan universal PCR master mix with AmpErase uracil-N-glycosylase (Applied Biosystems, Foster City, CA), 0.2 mg/ml bovine serum albumin (Sigma), 200 nM (each) primer, and 6-carboxyfluorescein (FAM)-labeled TaqMan probe. The qPCR assays were performed using a 7900 HT Fast real-time sequence detector (Applied Biosystems). All reaction mixtures were prepared in triplicate in MicroAmp Optical 96-well reaction plates with MicroAmp Optical Caps (Applied Biosystems). The amplification protocol involved incubation at 50°C for 2 min to activate uracil-N-glycosylase, followed by 10 min of incubation at 95°C and 40 cycles of 95°C for 15 s and 60°C for 1 min. PCR data were analyzed using ABI's Sequence Detector software (version 2.2.2). Duplicate serial dilutions of E. faecalis genomic DNA (10−8 to 10−12 g/reaction mixture) were used to generate standard curves. Percent amplification efficiencies were calculated according to the instrument manufacturer's instructions (Applied Biosystems). No-template controls were used to check for cross-contamination (two per PCR plate). Assays were performed with 2-μl DNA extracts in a total volume of 100 μl, and 10-fold dilutions of each DNA extract were used to test for PCR inhibition. Based on the standard curve, qPCR intensities (QI) were expressed as a unit of pg/2 μl (i.e., genomic DNA mass/reaction volume). Subsequently, the concentrations of the target gene (C) in water samples were calculated by the following equation: C (pg/100 ml) = QI (pg/2 μl) × CF × DF, where CF (μl/ml) is a conversion factor of 1 and DF is a dilution factor of 50 (35).
USGS precipitation data.
Precipitation data reported 24 and 48 h and 1 week before sample collection were obtained from the U.S. Geological Survey (USGS) Caribbean Water Science Center (http://pr.water.usgs.gov/) for sites 1, 6, and 7 (USGS stations 50020100, 50021700, and 50024950, respectively) (Table 2). Precipitation data were collected from these sites due to their proximity to a USGS station.
TABLE 2.
Reported rainfall during the sampling period (November 2009 to April 2010)a
| Site | USGS station | Rainfall (mm; mean ± SD) for indicated period before sample collection |
||
|---|---|---|---|---|
| 24 h | 48 h | 1 wk | ||
| Lago Garza (site 1) | 50020100 | 1.6 ± 0.4 | 5.7 ± 0.4 | 64.3 ± 40.3 |
| Upstream from WTP at Utuado (site 6) | 50021700 | 13.3 ± 15.3 | 18.5 ± 15.7 | 60.2 ± 35.6 |
| Downstream from WTP at Utuado (site 7) | 50024950 | 8.3 ± 10.2 | 8.6 ± 10.3 | 40.6 ± 25.6 |
Data were collected from Lago Garza (site 1), upstream from the WTP at Utuado (site 6), and downstream from the WTP at Utuado (site 7) for 24 h, 48 h, and 1 week before collection of the samples. Results represent the mean precipitation during the sampling period.
Statistical analyses.
The mean concentrations of cultivable thermotolerant coliforms, enterococci, coliphages, and enterophages and enterococci detected by qPCR were calculated using a Bayesian approach as described previously (30). Estimates were calculated assuming normal distribution of response. Comparisons among concentrations at different temperatures, on sample dates, and at sample sites were analyzed using a one-way or two-way analysis of variance (ANOVA) (30). Interaction effects were not considered in these analyses, and in all Bayesian analyses, credible intervals (CIs) were calculated. In addition, to determine correlations between rainfall and the concentrations of the indicators (24 h, 48 h, and 1 week) before sample collection, a cross-correlation analysis was used. However, prediction of the response to or effect of rainfall on microbial concentrations was estimated using nonlinear regression as described in references 20, 21, 26, 30, and 39. In all cases, the error around the point estimate, the 95% CI, is calculated. The 95% CI is the area under the curve of the posterior distribution, compared to the 95% confidence interval, i.e., the probability that the point estimate would be between the lower and upper bounds (21). All analyses were performed with the software package IBM SPSStatistics v.19.
RESULTS
Detection of indicators by culture-based methods.
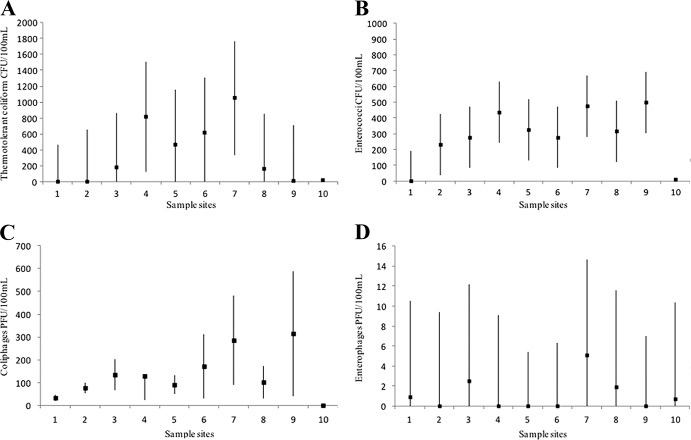
Thermotolerant coliforms showed the lowest counts at sites 1 and 9, with mean concentrations of 0.0 and 11.6 CFU/100 ml, respectively. The highest concentrations of thermotolerant coliforms correspond to sites 4 and 7, with mean concentrations of 817.8 and 1,053.0 CFU/100 ml, respectively (Fig. 2A). The variances of thermotolerant coliforms did not differ across the sampled points (354.0 to 356.0 CFU/100 ml), except for site 10, in which the standard deviation (SD) was 32.3 CFU/100 ml. Enterococci exhibited lower counts at sites 1 and 10, with mean concentrations of 0.0 and 9.6 CFU/100 ml, respectively, and higher counts at site 9 (499.2 CFU/100 ml) (Fig. 2B). The variances of enterococci did not differ across most of the sampled points, exhibiting SDs of 99.23 to 99.67 CFU/100 ml, except for site 10, which showed an SD of 21.2 CFU/100 ml. In terms of the phages, these were detected in lower concentrations than were the bacterial indicators. The lowest and highest means for coliphages correspond to sites 1 and 9 (32.7 PFU/100 ml and 314.0 PFU/100 ml, respectively) (Fig. 2C). The variances for the coliphages differed across the sampling sites, in which the lowest and highest SDs correspond to sites 1 (6.3 PFU/100 ml) and 9 (139.1 PFU/100 ml), respectively. In terms of the enterophages, the highest mean concentrations correspond to sites 3 and 7 (Fig. 2D) and variances did not differ across the sampling sites (SD = 4.9). CIs for the bacterial and viral indicators are represented in Fig. 2.
FIG 2.
Credible intervals (CIs) for thermotolerant coliforms (A), enterococci (B), coliphages (C), and enterophages (D) in the Rio Grande de Arecibo watershed. Results represent the means of the sampling period (n = 13), and credible intervals, representing positive values, were drawn as well. Except for enterophages, CIs could not be calculated for site 10 since most data are zeros.
Detection of host-specific Bacteroides by PCR and enterococci by qPCR.
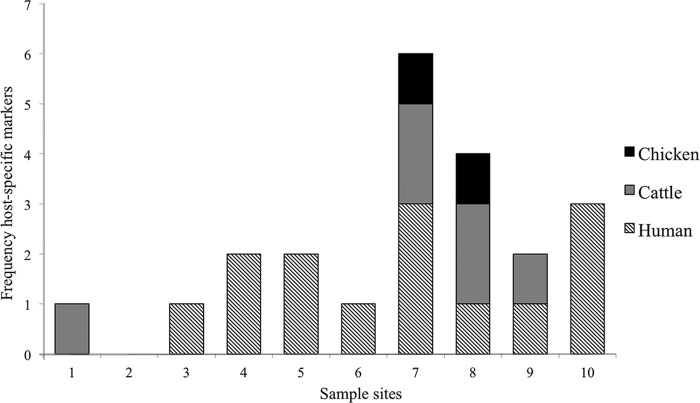
Of the 13 samples tested per site, HSB were not detected at sites 1 and 2 and were detected once at sites 3, 6, 8, and 9; twice at sites 4 and 5; and three times at sites 7 and 10. CSB were detected once at sites 1 and 9 and twice at sites 7 and 8 but were not detected in samples collected from sites 2 to 6 and 10. Chicken-specific bacterial markers were detected once with both CP2-9 and CP3-49 at sites 7 and 8, respectively (Fig. 3).
FIG 3.
Presence of human-, cattle-, and chicken-specific markers in the Rio Grande de Arecibo watershed. Numbers represent the frequency of detection of the HF183, CF128, CP2-9, and CP3-40 markers during the sampling period (n = 13). Both chicken-specific markers were detected only once during the sampling period.
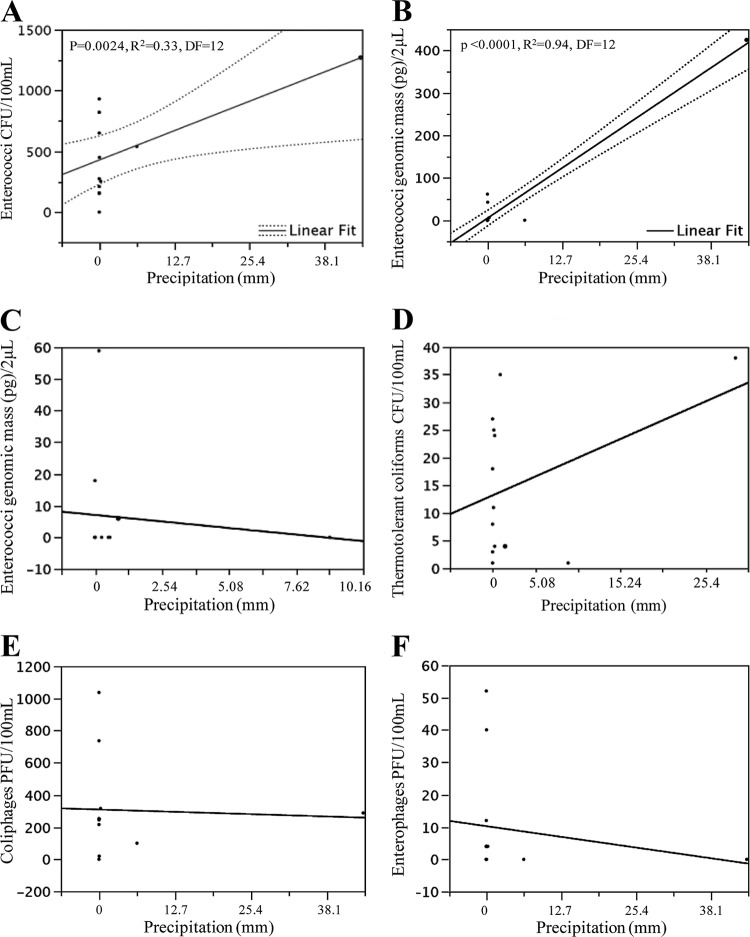
The range of quantification (ROQ) for the Enterococcus qPCR was 10−8 to 10−12 g of genomic DNA per reaction. The qPCR amplification efficiency ranged from 94.2 to 98.4%, with R2 values of ≥0.994. No signals were detected in the negative controls (i.e., no-template reactions), indicating the absence of cross-contamination in this study. None of the samples showed increases of signal intensity compared to the undiluted DNA templates, suggesting that PCR inhibition did not interfere with the amplification efficiency. Enterococci were detected by qPCR across all sites (Fig. 4A). The lowest means correspond to sites 1, 2, and 3 (0.0 pg/100 ml), and the highest correspond to site 7 (30.3 pg/100 ml). Variance was the highest at site 1 (SD, 275.9 pg/100 ml) but remained constant throughout the rest of the sampled sites (approximately 98.0 pg/100 ml). In terms of possible correlations between qPCR and culture techniques for the detection of enterococci, a positive correlation between the two methods was found in this study (Fig. 4B). This was the case for sites 6 and 7 (P = <0.0001, R2 = 0.78, DF = 12; P = 0.013, R2 = 0.39, DF = 12), but no correlation was noted for samples collected from site 1.
FIG 4.
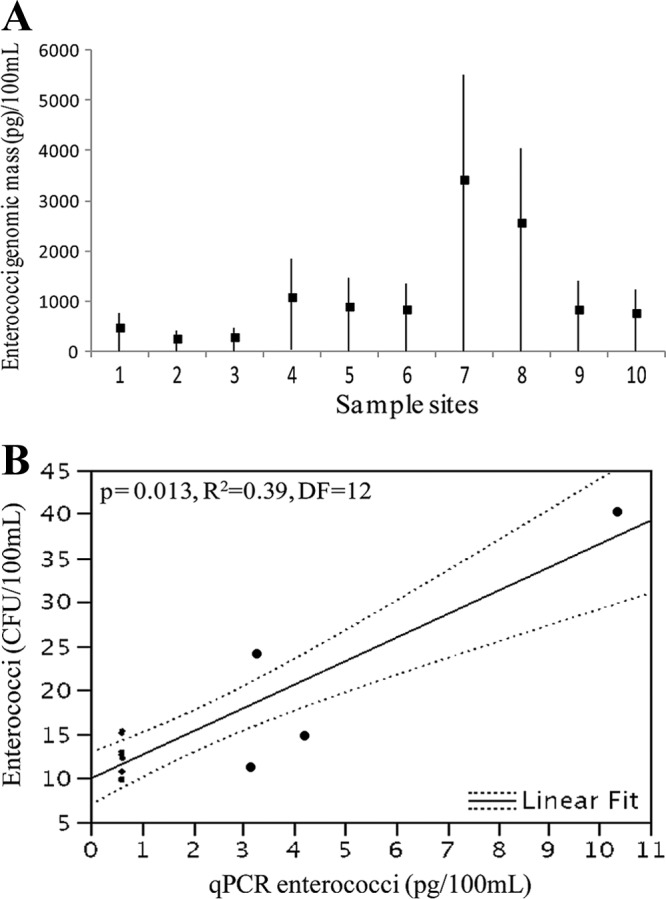
qPCR for enterococci and correlation between qPCR and culture-based techniques for enterococci. (A) Results show the mean genomic mass (pg)/100 ml (n = 13) for enterococci across the Rio Grande de Arecibo watershed and the corresponding credible intervals. (B) Correlation between qPCR and culture-based methods for the detection of enterococci (site 7).
Correlations of indicators with rainfall and detection methods.
At site 1, a positive correlation was noted with enterococci detected by culture methods and rainfall reported for 24 h and for 48 h, but not for 1 week, before sample collection. At sites 6 and 7, a positive correlation was noted with the precipitation reported for 24 h (Fig. 5A), for 48 h, and for 1 week before sample collection. For Enterococcus qPCR data from sites 6 and 7, a positive correlation was found at 24 h (Fig. 5B), for 48 h, and for 1 week before sample collection, but this was not the case for site 1 (Fig. 5C). Results from the correlation analyses between enterococci detected by culture methods and those detected by qPCR are shown in Table 3. No correlation was found with thermotolerant coliforms and precipitation in any of the study sites (Fig. 5D). Similarly, no correlations were found with coliphages or enterophages and precipitation (Fig. 5E and F, respectively). In addition, positive correlations between thermotolerant coliforms and coliphages were found at sites 1 and 7 (nonparametric Spearman's test: ρ = 0.45, P = 0.014, and ρ = 0.85, P = 0.049, respectively). No other correlations between any of the microbial indicators detected by culture methods were noted.
FIG 5.
Correlation between rainfall and microbial indicators. Precipitation data were collected from sites 1, 6, and 7 due to the proximity of a USGS station. Results show the possible effect of precipitation reported 24 h before collection of the samples on enterococci detected by culture methods (site 7) (A), 24 h before collection of the samples on enterococci detected by qPCR (site 7) (B), 24 h before collection of the samples on enterococci detected by qPCR (site 1) (C), 48 h before collection of the samples on thermotolerant coliforms (site 1) (D), 24 h before collection of the samples on coliphages (site 7) (E), and 24 h before collection of the samples on enterophages (site 7) (F).
TABLE 3.
Correlations between enterococci (detected by culture methods and qPCR) and rainfall reported for 24 and 48 h and 1 week before sample collectiona
| Method | Site | Value for indicated time before sample collection |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 24 h |
48 h |
1 wk |
||||||||
| P | R2 | DF | P | R2 | DF | P | R2 | DF | ||
| Culture | 1 | 0.013 | 0.39 | 12 | 0.042 | 0.26 | 12 | NC | NC | NC |
| 6 | <0.0001 | 0.82 | 12 | <0.0001 | 0.82 | 12 | <0.0001 | 0.45 | 12 | |
| 7 | 0.024 | 0.33 | 12 | 0.023 | 0.33 | 12 | 0.0042 | 0.5 | 12 | |
| qPCR | 1 | NC | NC | NC | NC | NC | NC | NC | NC | NC |
| 6 | <0.001 | 0.79 | 12 | <0.0001 | 0.78 | 12 | <0.0001 | 0.45 | 12 | |
| 7 | <0.001 | 0.94 | 12 | <0.0001 | 0.93 | 12 | 0.0089 | 0.42 | 12 | |
Enterococci were not always correlated with specific rainfall events.
NC, not correlated.
DISCUSSION
Prevalence of the bacterial and viral indicators detected by culture methods.
Detection of thermotolerant coliforms throughout the watershed suggests an input of fecal matter, although it is relatively difficult to identify the possible sources. High concentrations of these indicator bacteria at sites 4 and 7 may suggest an inefficient removal by sewage treatment or an additional input of fecal matter, but future studies are needed to confirm this. Moreover, rainfall may play an important role in the loading dynamics. For instance, when no precipitation was reported at site 1, thermotolerant coliforms ranged between 0 and ∼40 CFU/100 ml and 1 CFU/100 ml was detected when 10.2 mm of rain was reported 24 h before collection of the samples (data not shown). This may suggest that rainfall may have a possible dilution effect on the thermotolerant coliforms. Interestingly, 0 to ∼35 CFU/100 ml was detected when no precipitation was reported, but ∼40 CFU/100 ml was detected when >25.4 mm of rain was reported for 48 h before sample collection at site 1. This may suggest that tropical sediments may be a source of thermotolerant coliforms and that these may have been resuspended due to the rainfall reported in the previous 48 h. However, future studies are still needed to determine the prevalence of these indicator bacteria in tropical sediments and other possible loading mechanisms after rainfall events. A similar pattern was noted with thermotolerant coliforms and rainfall reported for 24 and 48 h before collection of the samples at sites 6 and 7, but unlike site 1, high numbers of these bacteria could also be associated with their transport from higher sites of the watershed. Additional studies are needed to determine the possible transport mechanisms of these indicator bacteria in tropical watersheds. No differences in the concentrations of thermotolerant coliforms were noted when correlated with the rainfall reported for 1 week before sample collection. This suggests that different periods of precipitation may differently influence the numbers of these bacteria when detected by culture methods.
It has been suggested elsewhere that some enterococci are naturally occurring in tropical waters (33). Results presented here suggest that many Enterococcus spp. detected by culture methods are of fecal origin. The reason for this is that site 1 is one of the highest sites of the watershed and possibly one of the sites less impacted by human or animal activities, and very low numbers were detected. These results are consistent with previous studies in which lower numbers of indicator bacteria have been detected in inland lakes (22). Higher numbers of enterococci at sites 4 and 7 suggest that these sites are point sources of fecal pollution. Interestingly, site 10 represents a point source of fecal contamination as well, but low numbers may be due to an efficient removal by sewage treatment. Also, the possibility remains that rainfall may also contribute to the input of enterococci into tropical surface waters. Even though rainfall did not correlate with enterococci detected at site 1, a positive correlation was noted with rainfall reported for 1 week before sample collection at site 6. In addition, a positive correlation between enterococci and rainfall reported for 24 h and for 48 h and 1 week before sample collection at site 7 suggests that loading of enterococci into surface waters may be influenced by precipitation events. Based on previous studies, it is possible that sediments and runoff could contribute to the input of these bacteria into surface waters, but future studies are still needed in order to confirm if this is the case for waters in Puerto Rico. Results presented here are consistent with previous studies in which higher numbers of enterococci are detected after rainfall events in Hawaiian marine waters (46). However, previous studies have not considered the effect of various precipitation periods prior to sample collection. The present study suggests that various rainfall episodes may be considered before inferring a fecal input in tropical surface waters. However, the possibility remains that, although positive correlations were noted, rainfall may not always result in an increase in numbers of enterococci.
Both coliphages and enterophages were detected throughout the watershed, indicating that the sampled sites may be impacted by human or animal fecal matter to some extent. The high concentrations of coliphages (compared to enterophages) suggest that more than one source could be contributing to their input and/or that they are present in higher numbers in feces. The low concentrations of enterophages throughout the watershed may suggest that (i) these are present in low concentrations in human feces, (ii) few sources of human fecal pollution are contributing to the input of these bacteriophages (e.g., septic tanks or domestic WTP), and (iii) enterophages are able to survive for short periods in tropical fresh waters and/or (iv) may be diluted as a result of precipitation events. Even though enterophage concentrations were relatively low, further studies need to compare their numbers with those of enteric viruses under similar conditions since a reliable indicator should be at least as abundant as the pathogen of concern. Also, the possibility remains that rainfall may have a possible dilution effect on both phages. The reason for this is that up to 1,000 and 50 coliphage and enterophage PFU/100 ml, respectively, were detected when no rainfall was reported and approximately 300 and 0 PFU/100 ml, respectively, were detected when >38.1 mm of rain was reported.
Monitoring indicator bacteria by molecular biology-based techniques.
Human and animal fecal contaminations are among the major concerns for public health since pathogens could be present (25, 42, 47). Therefore, it is important to validate source-specific markers, particularly in the tropics, as most studies were done in temperate regions. One promising tool for indicating human fecal contamination is the Bacteroides marker HF183 (1, 38). In the present study, HSB were detected mostly in samples collected downstream of all three domestic WTPs, suggesting that their presence is the result of the input of human fecal matter. This supports the specificity of the HF183 marker in tropical inland waters with human fecal impact. In terms of the cattle-specific markers, these have been shown to be a promising assay to detect ruminant pollution in environmental waters. However, some studies have shown that CF128 cross-reacts with DNA extracts from various farm animals (2). In the present study, CSB were detected at sites 1, 2, and 3, in which cattle are not present, but the assay was expected to detect CSB at site 6 since cattle grazing takes place there. Site 5, which could be impacted by horse fecal matter, tested positive for CSB by using the CF128 marker, and this is comparable with previous studies in which CF128 amplified CSB DNA in horse fecal matter (24). Similarly, chicken-specific bacteria were not detected using CP2-9 or CP3-49 in waters impacted by chicken fecal matter. Sample sites 4, 5, and 7 were considered to be impacted by chicken fecal matter, but chicken-specific Bacteroides bacteria were detected once at site 7 during the sampling period and chicken-specific Clostridium bacteria were detected once in samples collected from site 8. However, it should be noted that the numbers of chickens at these sites are relatively low, suggesting that the assays may not be sensitive enough to detect low levels of poultry pollution.
qPCR is currently being considered as a possible rapid detection method to be incorporated into U.S. Environmental Protection Agency guidelines; therefore, it is important to test this technique in different water types prior to implementation. Positive correlations between qPCR and culture methods in temperate waters suggest that detection of enterococci by qPCR could also be used as a tool for determining health-related risks and that both techniques may be reliable for the detection of enterococci (18). In the present study, enterococci genomic mass detected by qPCR was found to be higher at sites with known point sources of fecal pollution. However, detection of the multiple copies of the 23S rRNA gene present in the bacterial genome and viable-noncultivable bacteria, as well as DNA from dead cells, may add to the apparent increase in enterococcal genomic mass (17). Rainfall may also contribute to the increase of enterococci detected by qPCR. At sites 6 and 7, a positive correlation with rainfall may suggest that it may have an indirect contribution to the input of Enterococcus into tropical surface waters (e.g., septic tank overflows and animal droppings). However, as with culture methods, it is difficult to predict that rainfall may always have a positive correlation with enterococci detected by qPCR.
Correlations between culture-based and qPCR methods in this study may suggest that either of the techniques may be used to detect enterococci in tropical inland waters. Correlation results in this study are among the few obtained for tropical inland waters, but similar outcomes have been reported across the United States (22). Specifically, both methods were positively correlated in Hawaiian marine waters, but this correlation was not noted for fresh waters and estuaries (46). Results suggest that qPCR may not be suitable for all water types since different outcomes are obtained by the two methods and a correlation may not be expected in various water types at all times. Future studies need to determine if correlations between qPCR and culture methods in tropical inland waters exist throughout the year. Correlations between molecular and culture techniques and between thermotolerant coliforms and coliphages suggest that the bacteriological and virological quality of tropical inland waters should be evaluated using diverse MST tools. This opens up the possibility of using a toolbox whenever determining the microbiological quality of tropical inland waters.
Conclusions.
Few studies have evaluated the microbial quality of tropical inland waters. One problem is the need to develop appropriate water quality standards for tropical waters since guidelines used in other areas may not be applicable. The present study tested combinations of markers to assess the microbial quality of a tropical watershed in Puerto Rico, but results presented here need to be further tested in other tropical watersheds. Even though a different combination of MST techniques was tested in this study, there is still a need for developing a more robust toolbox to infer fecal pollution and the possible sources in various tropical water types. In terms of the bacterial indicators, these alone may not be suitable when inferring fecal contamination or its source in tropical inland waters. Additional studies are needed to further examine the loading dynamics of indicator bacteria for tropical fresh waters as a result of rainfall events. Enumeration of coliphages and enterophages may be a more appropriate way to infer fecal contamination since these are not ubiquitous in tropical waters and their numbers do not increase after rainfall events. In terms of the host-specific markers in this study, only HF183 seemed a reliable means of inferring the targeted source of fecal contamination in tropical waters. Cattle- and chicken-specific Bacteroides and Clostridium markers may not distinguish the source of the fecal pollution in tropical waters; therefore, additional markers that could more reliably detect cattle- and chicken-specific sources of fecal pollution in Puerto Rico remain an important need. Positive correlations between culture and molecular methods in this study should be considered before implementing qPCR as a rapid method for inferring fecal pollution in the tropics since it may not apply to all water bodies, specifically to inland lakes. qPCR assays targeting enterococci in tropical waters must be carefully considered and further tested in other tropical areas since rainfall may cause overestimation of the numbers of enterococci detected by this method. The methods and results presented here could be potentially useful in other water types.
ACKNOWLEDGMENTS
We thank the USGS for the precipitation data, the Department of Natural Resources of Puerto Rico for information about the Rio Grande de Arecibo watershed, and Gwendolyn Argüello and Jean F. Ruiz for processing the samples. We also thank Pablo A. Ortiz-Pineda for managing the figures at the modification stage.
This research was partially supported by MBRS-RISE (NIH grant no. 2R25GM061151-09) and the U.S. Environmental Protection Agency.
Footnotes
Published ahead of print 18 May 2012
REFERENCES
- 1. Ahmed W, Goonetilleke A, Powell D, Gardner T. 2009. Evaluation of multiple sewage-associated Bacteroides PCR markers for sewage pollution tracking. Water Res. 43:4872–4877 [DOI] [PubMed] [Google Scholar]
- 2. Ahmed W, Powell D, Goonetilleke A, Gardner T. 2008. Detection and source identification of faecal pollution in non-sewered catchment by means of host-specific molecular markers. Water Sci. Technol. 58:579–586 [DOI] [PubMed] [Google Scholar]
- 3. Allsop K, Stickler DJ. 1985. An assessment of Bacteroides fragilis group organisms as indicators of human faecal pollution. J. Appl. Bacteriol. 58:95–99 [DOI] [PubMed] [Google Scholar]
- 4. Arnone RD, Walling JP. 2007. Waterborne pathogens in urban watersheds. J. Water Health 5:149–162 [DOI] [PubMed] [Google Scholar]
- 5. Bahirathan M, Puente L, Seyfried P. 1998. Use of yellow-pigmented enterococci as a specific indicator of human and nonhuman sources of fecal pollution. Can. J. Microbiol. 44:1066–1071 [DOI] [PubMed] [Google Scholar]
- 6. Bernhard AE, Field KG. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 66:1587–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bernhard AE, Field KG. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bonilla N, et al. 2010. Enterophages, a group of phages infecting Enterococcus faecalis, and their potential as alternate indicators of human faecal contamination. Water Sci. Technol. 61:293–300 [DOI] [PubMed] [Google Scholar]
- 9. Byappanahalli MN, Shively DA, Nevers MB, Sadowsky MJ, Whitman RL. 2003. Growth and survival of Escherichia coli and enterococci populations in the macro-alga Cladophora (Chlorophyta). FEMS Microbiol. Ecol. 46:203–211 [DOI] [PubMed] [Google Scholar]
- 10. Calci KR, Burkhardt W, III, Watkins WD, Rippey SR. 1998. Occurrence of male-specific bacteriophage in feral and domestic animal wastes, human feces, and human-associated wastewaters. Appl. Environ. Microbiol. 64:5027–5029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cole D, Long SC, Sobsey MD. 2003. Evaluation of F+ RNA and DNA coliphages as source-specific indicators of fecal contamination in surface waters. Appl. Environ. Microbiol. 69:6507–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Department of Natural Resources 2005. Inventario de recursos de agua de Puerto Rico (Borrador). Oficina del Plan de Aguas, Department of Natural Resources, San Juan, PR [Google Scholar]
- 13. Devriese LA, Van de Kerckhove A, Kilpper-Balz R, Schleifer KH. 1987. Characterization and identification of enterococcus species isolated from the intestines of animals. Int. J. Syst. Bacteriol. 37:257–259 [Google Scholar]
- 14. Furuse K, et al. 1983. Bacteriophage distribution in human faeces: continuous survey of healthy subjects and patients with internal and leukaemic diseases. J. Gen. Virol. 64:2039–2043 [DOI] [PubMed] [Google Scholar]
- 15. Gantzer C, Henny J, Schwartzbrod L. 2002. Bacteroides fragilis and Escherichia coli bacteriophages in human faeces. Int. J. Hyg. Environ. Health 205:325–328 [DOI] [PubMed] [Google Scholar]
- 16. Hardina CM, Fujioka RS. 1991. Soil: the environmental source of Escherichia coli and enterococci in Hawaii's streams. Environ. Toxicol. 6:185–195 [Google Scholar]
- 17. Harwood VJ, Gordon KV, Staley C. 2011. Validation of rapid methods for enumeration of markers for human sewage contamination in recreational waters. WERF report PATH3C09. Water Environment Research Foundation, Alexandria, VA [Google Scholar]
- 18. Haugland RA, Siefring SC, Wymer LJ, Brenner KP, Dufour AP. 2005. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res. 39:559–568 [DOI] [PubMed] [Google Scholar]
- 19. Hernandez-Delgado EA, Toranzos GA. 1995. In situ replication studies of somatic and male-specific coliphages in a tropical pristine river. Water Sci. Technol. 31:247–250 [Google Scholar]
- 20. Hoff PD. 2010. A first course in Bayesian statistical methods. Springer Science Business Media, New York, NY [Google Scholar]
- 21. Kéry M. 2010. Introduction to WinBugs for ecologist: a Bayesian approach to regression, ANOVA, mixed models and related analysis. Academic Press, London, United Kingdom [Google Scholar]
- 22. Kinzelman JL, Bushon RN, Dorevitch S, Noble RT. 2011. Comparative evaluation of molecular and culture methods for fecal indicator bacteria for use in inland recreational waters. Water Environment Research Foundation, Alexandria, VA [Google Scholar]
- 23. Kistemann T, et al. 2002. Microbial load of drinking water reservoir tributaries during extreme rainfall and runoff. Appl. Environ. Microbiol. 68:2188–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lamendella R, Domingo JW, Oerther DB, Vogel JR, Stoeckel DM. 2007. Assessment of fecal pollution sources in a small northern-plains watershed using PCR and phylogenetic analyses of Bacteroidetes 16S rRNA gene. FEMS Microbiol. Ecol. 59:651–660 [DOI] [PubMed] [Google Scholar]
- 25. Licence K, Oates KR, Synge BA, Reid TM. 2001. An outbreak of E. coli O157 infection with evidence of spread from animals to man through contamination of a private water supply. Epidemiol. Infect. 126:135–138 [PMC free article] [PubMed] [Google Scholar]
- 26. Link WA, Barker RJ. 2010. Bayesian inference with ecological application. Academic Press, London, United Kingdom [Google Scholar]
- 27. Lipp EK, et al. 2001. The effects of seasonal variability and weather on microbial fecal pollution and enteric pathogens in a subtropical estuary. Estuaries 24:266–276 [Google Scholar]
- 28. Lu J, Santo Domingo J, Shanks OC. 2007. Identification of chicken-specific fecal microbial sequences using a metagenomic approach. Water Res. 41:3561–3574 [DOI] [PubMed] [Google Scholar]
- 29. Ludwig W, Schleifer KH. 2000. How quantitative is quantitative PCR with respect to cell counts? Syst. Appl. Microbiol. 23:556–562 [DOI] [PubMed] [Google Scholar]
- 30. McCarthy MA. 2007. Bayesian methods for ecology. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 31. Muñiz I, Jimenez L, Toranzos GA, Hazen TC. 1989. Survival and activity of Streptococcus faecalis and Escherichia coli in tropical fresh water. Microb. Ecol. 18:125–134 [DOI] [PubMed] [Google Scholar]
- 32. Rao VC, Seidel KM, Goyal SM, Metcalf TG, Melnick JL. 1984. Isolation of enteroviruses from water, suspended solids, and sediments from Galveston Bay: survival of poliovirus and rotavirus adsorbed to sediments. Appl. Environ. Microbiol. 48:404–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rivera SC, Hazen TC, Toranzos GA. 1988. Isolation of fecal coliforms from pristine sites in a tropical rain forest. Appl. Environ. Microbiol. 54:513–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rose JB, et al. 2001. Climate variability and change in the United States: potential impacts on water- and foodborne diseases caused by microbiologic agents. Environ. Health Perspect. 109(Suppl. 2):211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ryu H, et al. 2012. Development and evaluation of a quantitative PCR assay targeting of sandhill crane (Grus canadensis) fecal pollution. Appl. Environ. Microbiol. 78:4338–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Santiago-Rodriguez TM, et al. 2010. Characterization of Enterococcus faecalis-infecting phages (enterophages) as markers of human fecal pollution in recreational waters. Water Res. 44:4716–4725 [DOI] [PubMed] [Google Scholar]
- 37. Santo-Domingo JW, Bambic DG, Edge TA, Wuertz S. 2007. Quo vadis source tracking? Towards a strategic framework for environmental monitoring of fecal pollution. Water Res. 41:3539–3552 [DOI] [PubMed] [Google Scholar]
- 38. Sercu B, Van De Werfhorst LC, Murray J, Holden PA. 2009. Storm drains are sources of human fecal pollution during dry weather in three urban southern California watersheds. Environ. Sci. Technol. 43:293–298 [DOI] [PubMed] [Google Scholar]
- 39. Sivia DS, Skilling J. 2011. Data analysis: a Bayesian tutorial, vol 2 Oxford University Press, Oxford, England, United Kingdom [Google Scholar]
- 40.US Environmental Protection Agency 2002. Method 1600: membrane filter test method for enterococci in water, EPA-821-R- 02-022 Office of Water, US Environmental Protection Agency, Washington, DC [Google Scholar]
- 41.US Environmental Protection Agency 1978. Microbiological methods for monitoring the environment: water and wastes. US Environmental Protection Agency, Washington, DC [Google Scholar]
- 42. Walters SP, Gannon VP, Field KG. 2007. Detection of Bacteroidales fecal indicators and the zoonotic pathogens E. coli O157:H7, Salmonella, and Campylobacter in river water. Environ. Sci. Technol. 41:1856–1862 [DOI] [PubMed] [Google Scholar]
- 43. Ward RL, Knowlton DR, Winston PE. 1986. Mechanism of inactivation of enteric viruses in fresh water. Appl. Environ. Microbiol. 52:450–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Whitlock JE, Jones DT, Harwood VJ. 2002. Identification of the sources of fecal coliforms in an urban watershed using antibiotic resistance analysis. Water Res. 36:4273–4282 [DOI] [PubMed] [Google Scholar]
- 45. Whitman RL, Shively DA, Pawlik H, Nevers MB, Byappanahalli MN. 2003. Occurrence of Escherichia coli and enterococci in Cladophora (Chlorophyta) in nearshore water and beach sand of Lake Michigan. Appl. Environ. Microbiol. 69:4714–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yan T, Goto DK, Feng F. 2011. Concentration dynamics of fecal indicators in Hawaii's coastal and inland sand, soil, and water during rainfall events. PATH6R09. Water Environment Research Foundation, Alexandria, VA [Google Scholar]
- 47. Yoder JS, et al. 2004. Surveillance for waterborne-disease outbreaks associated with recreational water—United States, 2001–2002. MMWR Surveill. Summ. 53(8):1–22 [PubMed] [Google Scholar]