Abstract
Spontaneous cocoa bean fermentations performed under bench- and pilot-scale conditions were studied using an integrated microbiological approach with culture-dependent and culture-independent techniques, as well as analyses of target metabolites from both cocoa pulp and cotyledons. Both fermentation ecosystems reached equilibrium through a two-phase process, starting with the simultaneous growth of the yeasts (with Saccharomyces cerevisiae as the dominant species) and lactic acid bacteria (LAB) (Lactobacillus fermentum and Lactobacillus plantarum were the dominant species), which were gradually replaced by the acetic acid bacteria (AAB) (Acetobacter tropicalis was the dominant species). In both processes, a sequence of substrate consumption (sucrose, glucose, fructose, and citric acid) and metabolite production kinetics (ethanol, lactic acid, and acetic acid) similar to that of previous, larger-scale fermentation experiments was observed. The technological potential of yeast, LAB, and AAB isolates was evaluated using a polyphasic study that included the measurement of stress-tolerant growth and fermentation kinetic parameters in cocoa pulp media. Overall, strains L. fermentum UFLA CHBE8.12 (citric acid fermenting, lactic acid producing, and tolerant to heat, acid, lactic acid, and ethanol), S. cerevisiae UFLA CHYC7.04 (ethanol producing and tolerant to acid, heat, and ethanol), and Acetobacter tropicalis UFLA CHBE16.01 (ethanol and lactic acid oxidizing, acetic acid producing, and tolerant to acid, heat, acetic acid, and ethanol) were selected to form a cocktail starter culture that should lead to better-controlled and more-reliable cocoa bean fermentation processes.
INTRODUCTION
Cocoa fermentation is a key step in the technological transformation of cocoa into chocolate, because the highly bitter, astringent unfermented cocoa beans lack the full chocolate flavor. The fermentation of cocoa beans is therefore the first step of the chocolate-making process, which consists of a natural, 5- to 7-day microbial fermentation of the pectinaceous pulp surrounding the seeds of the tree Theobroma cacao (37, 39). The cocoa pulp is hydrolyzed during fermentation; this aids the drying process by allowing the pulp to be drained. Most importantly, fermentation triggers an array of chemical changes within the cocoa bean that are vital to the development of the complex, beloved flavor of “chocolate.”
The fermentation of cocoa beans occurs at two levels. The first involves reactions that take place in the pulp, in the outer part of the beans, and the second involves several hydrolytic reactions that occur within the cotyledons (39). The microbial activity in the cocoa pulp is a well-defined microbial succession led by yeasts, which dominate the microbial community during the first hours, followed by the lactic acid bacteria (LAB), which decline after 48 h of fermentation, and finally the acetic acid bacteria (AAB) (2, 39). The metabolic diversity of the yeast, LAB, and AAB strains during cocoa fermentation can be interpreted as a natural consequence of the environmental conditions that influence their growth and selection. Changes in the pH, temperature, sugar content, and fermentation products exert selection pressure on the already existing natural biotypes, favoring those strains that are better adapted to this environment (41). An analysis of the bacterial and yeast strains that survive under these stresses could provide useful information concerning the ability of the yeasts and bacteria to initiate growth and complete fermentation. To perform this process, the microbial cells must adapt their own physiology or behavior in response to changing environmental stresses (11).
Chocolate processors require a constant supply of cocoa beans that must conform to an array of criteria. The industrialization of the cocoa fermentation process may allow greater control over the quality of the cocoa beans and the chocolate derived from them (36). The concept of industrializing traditional fermentation processes to enhance their performance and efficiency is not new. For example, wine, beer, cheese, distilled sugar cane beverages, and yogurt were all at one time produced using traditional processes (43). These fermentations have been developed into highly efficient, well-controlled processes in modern fermentor designs often using defined starter cultures (12, 40). However, cocoa fermentation remains an empirical process that does not give rise to beans of consistent quality, which obliges processors to alter their formulations continually (18). The fermentation takes place under uncontrolled environmental conditions that often lead to unsuccessful fermentation, and the variable quality of the product may reflect the vagaries of chance contamination. Although preliminary experiments using defined starter cultures demonstrate satisfying results (8, 19, 35, 36), only one particular study utilized yeast, LAB, and AAB simultaneously as a defined microbial cocktail (36), but without any prior study of the stress tolerance and/or the fermentative kinetic parameters of the individual strains.
The first objective of this study was to investigate the physicochemical changes and the dynamics of the microbial community structure during bench- and pilot-scale spontaneous cocoa bean fermentations. Next, the technological potential of the yeast, LAB, and AAB isolates was evaluated using a polyphasic screening study that measured the isolates' stress tolerance and fermentation kinetic parameters in a cocoa pulp-simulating medium.
MATERIALS AND METHODS
Bench- and pilot-scale cocoa bean fermentations.
Freshly harvested cocoa pods, obtained from a cacao farm located in Itajuípe, Bahia State, Brazil, were broken open manually with a machete, and the beans were immediately transferred to the fermentation site. The beans were mixed in a clean vessel to obtain a homogeneous mixture of 20 kg of wet beans and were transferred to the laboratory. A total of 500 g of cocoa beans was deposited in plastic containers (PCs) (dimensions, 15 cm by 10 cm by 7 cm). The plastic containers were kept in incubators, and the temperature was adjusted every 12 h to simulate the temperature of large-scale fermentations: 28°C at 0 h, 30°C at 12 h, 32°C at 24 h, 35°C at 36 h, 38°C at 48 h, 42°C at 60 h, 46°C at 72 h, and 48°C at 84 h, 96 h, 108 h, 120 h, 132 h, and 144 h. To allow sampling every 12 h, 14 individual plastic containers were prepared for each experiment. The pilot-scale fermentation was performed in a 0.015-m3 double-layer stainless steel conical tank (ST) (10 kg of cocoa beans; noncommercial bioreactors) with temperature control. The stainless steel tank and plastic containers contained holes at the bottom to allow drainage of the sweatings generated during the fermentation. The vessels were partially closed with a steel and plastic lid to ensure adequate insulation. The fermentations were turned every 24 h, and a natural fermentation proceeded for 168 h.
Every 12 h, 200-g samples were randomly collected in sterile bags. The samples for chemical and culture-independent analyses were sealed in plastic bags and were frozen (−20°C). Microbiological analyses were performed immediately after sampling.
Culture-dependent microbiological analysis.
Twenty-five grams of cocoa beans and adhering pulp was added to 225 ml saline-peptone water (0.1% [vol/vol] bacteriological peptone [Himedia], 0.8% [vol/vol] NaCl [Merck, Whitehouse Station, NJ]) and was homogenized in a stomacher at normal speed for 5 min, followed by serial dilutions. LAB were enumerated by pour plate inoculation on MRS agar (Merck) containing 0.2% (vol/vol) sorbic acid (Merck) and 0.1% (vol/vol) cycloheximide (Merck) to inhibit yeast growth and 0.1% (vol/vol) cysteine-HCl to produce anaerobic conditions during incubation. AAB were enumerated by surface inoculation on GYC agar (50 g/liter glucose, 10 g/liter yeast extract, 30 g/liter calcium carbonate, 20 g/liter agar [all from Merck] [pH 5.6]) containing 0.1% cycloheximide to inhibit yeast growth and 50 mg/liter penicillin (Sigma, St. Louis, MO) to inhibit LAB growth. Yeasts were enumerated by surface inoculation on YEPG agar (1% yeast extract [Merck], 2% peptone [Himedia], 2% glucose [Merck] [pH 5.6]) containing 100 mg/liter chloramphenicol (Sigma) and 50 mg/liter chlortetracycline (Sigma) to inhibit bacterial growth. Nutrient agar (NA) containing 0.1% cycloheximide (Merck) was used as a general medium for a viable mesophilic bacterial population and Bacillus spp. Plating was performed in triplicate with 100 μl (surface spread technique) or 1,000 μl (pour plate technique) of each diluted sample. After the spreading, the plates were incubated at 30°C for 3 to 4 days for growth of cultures on MRS agar, YEPG agar, and NA and at 25°C for 5 to 8 days for growth of cultures on GYC agar. Following incubation, the number of CFU was recorded, followed by morphological characterization and counts of each colony type obtained. The square root of the number of colonies of each type was restreaked and purified. The purified isolates originating from GYC agar, YEPG agar, and NA were stored at −80°C in YEPG broth containing 20% (wt/wt) glycerol, and the isolates originating from MRS agar were stored at −80°C in MRS broth containing 20% (wt/wt) glycerol.
Phenotypic characterization of bacterial colonies originating from MRS, GYC, and NA plates was performed by conventional microbiological methods: Gram staining, microscopic examination, catalase and oxidase activity, a motility test, spore formation, acid and gas production from glucose, and acid and gas production from lactate and acetate (only for isolates originating from GYC agar plates showing a clear zone around the colony [presumptive AAB]). Yeast colonies were physiologically characterized by determining their morphology, spore formation, and fermentation of different carbon sources according to the method of Kurtzman et al. (17).
Molecular identification of representative microbial strains was performed by sequence analysis of the full-length 16S rRNA gene or the internal transcribed spacer (ITS) region for bacteria or yeasts, respectively. Bacterial or yeast cultures were grown under appropriate conditions, collected from agar plates with a sterile pipette tip, and resuspended in 40 μl of PCR buffer. The suspension was heated for 10 min at 95°C, and 1 μl was used as a DNA template in PCR experiments. For bacterial isolates originating from MRS agar and NA, a 16S rRNA PCR was carried out using primers 27-F and 1512-R (23). For bacterial isolates originating from GYC agar plates showing a clear zone around the colony, primers 16Sd and 16Sr were used for the amplification of the 16S rRNA gene region conserved among AAB (34). For yeast isolates, ITS-PCR was carried out using primers ITS1 and ITS4 (26). Isolates identified as Saccharomyces cerevisiae had their identity confirmed through a species-specific PCR assay with HO gene-derived primers (32). The rRNA gene region was amplified in a Thermo PCYL220 thermal cycler (Thermo Fisher Scientific Inc., Waltham, MA). The PCR products were sequenced using an ABI3730 XL automatic DNA sequencer. The sequences were aligned using the BioEdit 7.7 sequence alignment editor and were compared to the GenBank database using the BLAST algorithm (National Center for Biotechnology Information, Bethesda, MD).
For the differentiation of AAB species closely related by their rRNA gene sequences, some specific biochemical tests were performed in order to validate the data obtained by 16S rRNA sequencing. Growth on 30% d-glucose, 0.3% maltose, 0.3% methanol, and 10% ethanol was examined by using a basal medium (0.05% yeast extract, 0.3% [wt/vol] vitamin-free Casamino Acids [Difco], and 2.5% agar) and appropriate concentrations of carbon sources. The medium without the carbon source was used as a control. Growth was checked after 7 days of incubation at 28°C. The utilization of ammonium as the sole nitrogen source in the presence of ethanol as a carbon source was tested using Frateur's modified Hoyer ethanol/vitamin medium containing 2.5% agar. The acid production from d- and l-arabitol was tested in phenol red broth with the carbon source added at a final concentration of 1%. The results were assessed with reference to the control after incubation at 28°C for 48 h. All the tests were performed as described previously (15, 34).
Total-community DNA isolation.
Beans and pulp were physically separated by adding 100 ml of sterile distilled water to 100 g of the cocoa beans and adhering pulp in a plastic bag and homogenizing in a stomacher at normal speed for 5 min. The pulp fraction was recovered by decanting. Forty milliliters of the pulp fraction was lyophilized, and the freeze-dried cocoa pulp was ground thoroughly with a sterile pestle. Thirty milligrams of freeze-dried pulp was mixed by homogenization twice in 1.5 ml of phosphate-buffered saline. The combined fluids were mixed by vortexing for a further 10 min and were subsequently centrifuged at 100 × g and 4°C for 10 min to remove large particles. The supernatant was further centrifuged at 8,000 × g and 4°C for 20 min to pellet the yeast and bacterial cells, which were subsequently frozen at −20°C for at least 1 h. This procedure was performed twice for the preparation of yeast and bacterial cells separately. Bacterial cells were lysed as described by Pereira et al. (31). For the lysis of yeast cells, the pellet was resuspended in 600 μl sorbitol buffer (1 M sorbitol, 100 mM EDTA, 14 mM β-mercaptoethanol) with 10 mg/ml of lysing enzymes and was incubated at 30°C for 1 h. A pellet of the total cells was obtained by centrifugation for 5 min at 5,000 × g and were resuspended in 180 μl of buffer ATL (supplied in the QiAamp DNA Mini Kit). After cell lysis, the DNA in the supernatant was further purified by using step 4 (bacterial procedure) and step 2 (yeast procedure) of the protocol for DNA purification from tissues (supplied in the QIAamp DNA Mini Kit; Qiagen, Hilden, Germany). The final samples were stored at −20°C until further use.
Nested PCR–DGGE strategy.
In order to increase the sensitivity of denaturing gradient gel electrophoresis (DGGE) and to facilitate the DGGE by analyzing fragments of the same length, a two-step nested-PCR technique was applied. For analysis of bacterial diversity, primers 27F and 1512R were used to amplify the nearly complete gene encoding 16S rRNA under conventional PCR conditions in the first PCR step (23). Subsequently, the product of the first PCR was used as a template for a nested PCR targeting the V3 region of the 16S rRNA gene using primers GC-338f and 518r (29) to create a DNA fragment suitable for DGGE analysis. For analysis of yeast diversity, PCR amplification of the ITS regions was performed using primers ITS1-F and ITS4 in the first step, followed by nested PCR using the DGGE primers GC-ITS1-F and ITS2 (42). Reactions were performed in a Mastercycler (Eppendorf, Hamburg, Germany). The PCR products were analyzed by DGGE using a Bio-Rad DCode Universal Mutation Detection system (Bio-Rad, Richmond, CA). The PCR products of the second step were loaded onto 8% (wt/vol) polyacrylamide gels in a running buffer containing 1× TAE (20 mM Tris, 10 mM acetate, 0.5 mM EDTA [pH 8.0]). Optimal separation was achieved with a 30 to 55% urea-formamide denaturing gradient for the bacterial community and a 12 to 60% gradient for the yeast community (100% corresponded to 7 M urea and 40% [vol/vol] formamide).
The DGGE bands of interest were excised from the gel with a sterile scalpel, disrupted in 60 μl of sterile Milli-Q water, and left overnight at 4°C to let the DNA diffuse out of the bands. Ten microliters of the eluted DNA of each DGGE band was reamplified by using the appropriate primers and the conditions described above. The PCR products for sequencing were purified using the QIAquick PCR purification kit (Qiagen). The samples were analyzed with an automated DNA sequencer (Applied Biosystems, Foster City, CA). Searches in GenBank with BLAST were performed to determine the closest known sequences obtained.
Physical-chemical analysis.
The temperature of the fermenting cocoa pulp-bean mass was determined every 12 h with a Delta Ohm portable data logger, model HD 2105.2. For analysis of target metabolites, aqueous extracts from fermentation samples were obtained as described previously (26, 36). The amounts (mg g–1) of alcohols (ethanol and methanol), organic acids (lactic, acetic, and citric acids), and carbohydrates (glucose, sucrose, and fructose) were determined from pulp and bean extracts with a high-performance liquid chromatography (HPLC) apparatus (HP series 1200; Hewlett-Packard Company), equipped with an Aminex HPX-87H column (Bio-Rad Laboratories, Hercules, CA) connected to a refractive index (RI) detector (HPG1362A; Hewlett-Packard Company). All samples were analyzed in triplicate, and the average values and standard deviations are presented. The column was eluted with a degassed mobile phase containing 4 mM H2SO4 at 30°C at a flow rate of 0.6 ml/min.
Screening on agar plates for stress tolerance.
For the analysis of tolerance to ethanol, lactic acid, acetic acid, glucose, and fructose, approximately 106 CFU ml−1 of the yeast or AAB isolates was plated on basal medium (0.05% yeast extract, 0.3% [wt/vol] vitamin-free Casamino Acids [Difco], and 2.5% agar), and the LAB isolates were plated on MRS agar. The plates were supplemented with either 6, 10, or 12% (vol/wt) ethanol; 1, 2, 3, or 5% (vol/wt) lactic acid; 1, 2, 3, or 5% (vol/wt) acetic acid; 5, 15, or 30% (wt/wt) glucose; or 5, 15, or 30% (wt/wt) fructose. A medium without a carbon source was used as a control. Growth was observed after 7 days at 28°C. To evaluate heat tolerance, the isolates were grown at 30, 37, and 45°C on YEPG agar (for yeast and AAB isolates) or MRS agar (for LAB isolates). To evaluate pH tolerance, each medium was adjusted to pH 2, 3, or 5.
Evaluation of the fermentation performance of yeasts, LAB, and AAB.
The LAB and AAB strain fermentations were performed in 2-liter Erlenmeyer flasks containing 500 ml of cocoa pulp simulation medium for LAB (PSM-LAB) (13 g/liter fructose, 25 g/liter glucose, 10 g/liter citric acid, 5 g/liter yeast extract, 5 g/liter soya peptone, 0.5 g/liter magnesium sulfate-heptahydrate, 0.2 g/liter manganese sulfate-monohydrate, 1 ml/liter Tween 80) or cocoa pulp simulation medium for AAB (PSM-AAB) (10 g/liter calcium lactate-pentahydrate, 10 ml/liter ethanol, 10 g/liter yeast extract, 5 g/liter soya peptone), as described by Lefeber et al. (20). PSM-LAB and PSM-AAB were supplemented with 20% (vol/vol) fresh cocoa pulp. A cocoa pulp simulation medium for yeast (PSM-yeast) was formulated containing 17 g/liter fructose, 25 g/liter glucose, 10 g/liter citric acid, 5 g/liter yeast extract, 5 g/liter soya peptone, and 20% (vol/vol) fresh cocoa pulp. The precultures were grown in the same substrate at 30°C for 48 h and were then used to inoculate each of the fermentations (106 CFU ml−1). The fermentations were performed in triplicate for each strain at 30°C for 24 h under static conditions. The alcohol (ethanol and methanol), organic acid (lactic, acetic, and citric acids), and carbohydrate (glucose, sucrose, and fructose) contents were quantified in triplicate at 0, 12, and 24 h of fermentation using HPLC with a model HP series 1200 system (Hewlett-Packard Company) equipped with an Aminex HPX-87H column (Bio-Rad Laboratories, Hercules, CA) connected to an RI detector (HPG1362A; Hewlett-Packard Company). The column was eluted with a degassed mobile phase containing 4 mM H2SO4 at 30°C at a flow rate of 0.6 ml/min.
RESULTS
Microbial counts.
The results of the culture-dependent microbiological approach demonstrated that the yeasts, LAB, and AAB were able to grow in bench- and pilot-scale cocoa fermentations (Fig. 1a). The LAB and yeasts developed simultaneously and reached a maximum population of 8 log CFU g−1 after 12 h of fermentation. The LAB counts remained high throughout the fermentations, while the yeasts progressively decreased in number and became undetectable after 72 and 108 h in the PC and ST fermentations, respectively. The AAB population started to develop after 12 h (5.20 and 6.36 log CFU g−1 in the ST and PC, respectively) and was present up to 96 h after the start of fermentation. Although nutrient agar medium was used to monitor the growth of aerobic mesophilic bacteria and Bacillus spp., several Gram-negative and aerobic rods that produced catalase and were non-spore forming (later identified as AAB) grew on this medium (data not shown).
Fig 1.
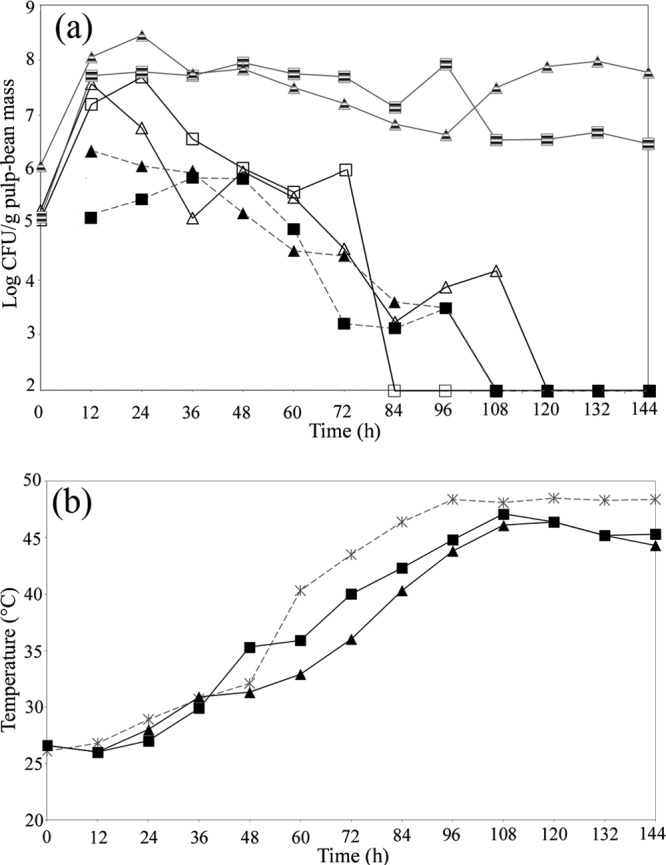
(a) Evolution of LAB in MRS medium (hatched symbols), yeasts in YEPG medium (open symbols), and AAB in GYC medium (filled symbols). Squares, ST; triangles, PC. (b) Temperatures inside the ST ▲ and the PC ■ and the incubation temperature (*).
Identification and distribution of the isolates.
A total of 96 MRS, 68 GYC, 98 YEPG, and 71 NA isolates were randomly picked up at different times during the fermentation processes and were identified via biochemical and molecular methods (Tables 1 and 2). The 16S rRNA gene sequence analysis identified all of the homofermentative or facultatively heterofermentative MRS isolates as Lactobacillus plantarum (n = 39). The other 57 isolates were obligately heterofermentative organisms, and most of them were identified as Lactobacillus fermentum (n = 52); the remaining 5 obligately heterofermentative isolates were closely related to Lactobacillus vaccinostercus. At the onset of fermentation, L. plantarum was the dominant species of the LAB, with a population of approximately 4 log CFU g−1. After 12 to 24 h, L. fermentum was the dominant species isolated and reached a maximum population of 8.01 log CFU g−1 at 24 h and 8.09 log CFU g−1 at 60 h in the PC and ST, respectively.
Table 1.
Phylogenetic affiliationsa and estimated average levels of yeast and bacterial isolates from PC fermentation
| Isolate identification | GenBank accession no. of closest relative | Estimated average level (log CFU g−1)b of the isolate at the following fermentation time (h): |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 12 | 24 | 36 | 48 | 60 | 72 | 84 | 96 | 108 | 120 | 132 | 144 | ||
| Yeasts | ||||||||||||||
| S. cerevisiae | AM711362.1 | 5.32 | 7.08 | 8.29 | 6.17 | 5.87 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| P. kluyveri | FM199971.1 | <1 | <1 | <1 | 6.09 | 6.01 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| H. uvarum | FJ515178.1 | <1 | <1 | <1 | <1 | 6.33 | 5.93 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| I. orientalis | EU315767.1 | <1 | <1 | <1 | 6.13 | 5.92 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| LAB | ||||||||||||||
| L. plantarum | HQ293084.1 | 4.11 | 7.23 | 7.87 | 6.96 | 6.84 | 6.23 | 6.76 | <10 | <10 | 5.98 | 5.76 | 5.59 | <10 |
| L. fermentum | HQ293040.2 | <10 | 7.01 | 8.01 | 7.02 | 6.92 | 6.77 | 6.08 | 6.60 | 6.52 | 6.17 | 6.02 | 5.91 | 6.01 |
| L. vaccinostercus | AB218801.1 | <10 | <10 | 7.96 | 7.11 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
| AAB | ||||||||||||||
| A. malorum | FJ831444.1 | <1 | <1 | 5.23 | <1 | <1 | 4.31 | 3.76 | <1 | 3.49 | <1 | <1 | <1 | <1 |
| A. cerevisiae | HM562995.1 | <1 | 4.00 | <1 | <1 | 5.00 | 4.00 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| A. tropicalis | DQ523494.1 | <1 | <1 | 4.00 | 5.74 | 5.52 | 4.74 | 3.84 | 3.12 | <1 | <1 | <1 | <1 | <1 |
| A. ghanensis | HM562984.1 | <1 | 5.10 | <1 | 5.00 | <1 | <1 | 3.87 | <1 | <1 | <1 | <1 | <1 | <1 |
| Species isolated on NA | ||||||||||||||
| B. subtilis | HQ286641.1 | 5.91 | 6.73 | 6.98 | 5.76 | 4.00 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| B. megaterium | FJ174651.1 | 5.32 | 5.00 | 6.32 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| S. pasteuri | HM130543.1 | 5.17 | 5.00 | 6.21 | <1 | 6.00 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| S. xylosus | HM854231.1 | <1 | 6.19 | 5.00 | <1 | 6.00 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| S. saprophyticus | HM130543.1 | <1 | 5.00 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| T. saanichensis | EU215774.1 | 5.00 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
BLAST searches were based on sequences of type and cultured strains at GenBank (National Center for Biotechnology Information). An isolate was assumed to belong to a given species if the similarity between the query rRNA gene sequence and the sequences in the databases was higher than 97%.
Obtained after morphological characterization and molecular identification of each colony type at the sampling time. A value of <1 log CFU g−1 means that no colonies were found.
Table 2.
Phylogenetic affiliationsa and estimated average levels of yeast and bacterial isolates from ST fermentation
| Isolate identification | GenBank accession no. of closest relative | Estimated average level (log CFU g−1)b of the isolate at the following fermentation time (h): |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 12 | 24 | 36 | 48 | 60 | 72 | 84 | 96 | 108 | 120 | 132 | 144 | ||
| Yeasts | ||||||||||||||
| S. cerevisiae | AM711362.1 | 5.30 | 7.14 | 8.07 | 6.27 | 6.45 | 5.71 | 5.93 | 4.45 | 3.95 | <1 | <1 | <1 | <1 |
| P. kluyveri | FM199971.1 | <1 | 6.00 | <1 | 6.34 | 6.56 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| H. uvarum | FJ515178.1 | <1 | <1 | <1 | <1 | 5.00 | 5.65 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| I. orientalis | EU315767.1 | <1 | <1 | <1 | 6.56 | 5.00 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| D. etchellsii | AJ586528.1 | <1 | 6.00 | 7.00 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| K. ohmeri | EF196811.1 | <1 | 6.00 | 7.00 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| LAB | ||||||||||||||
| L. plantarum | HQ293084.1 | 4.55 | 7.30 | 7.97 | <10 | <10 | 8.05 | 8.08 | <10 | <10 | <10 | <10 | <10 | <10 |
| L. fermentum | HQ293040.2 | <10 | <10 | 8.01 | 7.16 | 7.72 | 8.09 | <10 | 7.07 | 7.96 | 6.48 | 6.37 | 6.21 | 6.09 |
| AAB | ||||||||||||||
| A. malorum | FJ831444.1 | <1 | <1 | 6.08 | 5.51 | <1 | 4.32 | 3.00 | <1 | <1 | <1 | <1 | <1 | <1 |
| A. cerevisiae | HM562995.1 | <1 | 5.00 | <1 | <1 | 5.00 | 4.61 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| A. tropicalis | DQ523494.1 | <1 | 6.22 | 5.00 | 5.17 | 5.10 | 4.37 | 3.84 | 3.61 | 3.50 | <1 | <1 | <1 | <1 |
| A. ghanensis | HM562984.1 | <1 | <1 | <1 | <1 | 5.00 | 4.00 | 3.87 | <1 | <1 | <1 | <1 | <1 | <1 |
| Species isolated on NA | ||||||||||||||
| B. subtilis | HQ286641.1 | 6.81 | 6.92 | 7.42 | 5.11 | 4.28 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| B. megaterium | FJ174651.1 | 5.00 | 6.31 | 7.00 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| T. saanichensis | EU215774.1 | <1 | 6.00 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| S. aureus | FJ899095.1 | 5.00 | 6.00 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| S. equorum | AM945662.1 | <1 | <1 | <1 | 6.00 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| P. agglomerans | EU596536.1 | 6.20 | <1 | 7.00 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| P. terrea | HM562993.1 | <1 | 6.00 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
BLAST searches were based on sequences of type and cultured strains at GenBank (National Center for Biotechnology Information). An isolate was assumed to belong to a given species if the similarity between the query rRNA gene sequence and the sequences in the databases was higher than 97%.
Obtained after morphological characterization and molecular identification of each colony type at the sampling time. A value of <1 log CFU g−1 means that no colonies were found.
All of the isolates from the GYC agar plates oxidized acetate and lactate to CO2 and H2O and were assigned to the genus Acetobacter. The sequencing of the 16S rRNA genes and the specific biochemical tests supported the identification of these isolates as four different species of Acetobacter, namely, Acetobacter tropicalis (n = 39), A. malorum (n = 13), A. cerevisiae (n = 9), and A. ghanensis (n = 7). A. tropicalis was the dominant species and reached a maximum population after 36 h (5.74 log CFU g−1) and 12 h (6.22 log CFU g−1) in the PC and ST, respectively.
The 16S rRNA gene sequence analysis demonstrated that a range of other bacterial species grew on NA, including members of the Gram-positive order Bacillales and the Gram-negative families Enterobacteriaceae and Acetobacteraceae. The members of the order Bacillales were subdivided into the endospore-forming bacteria Bacillus subtilis (n = 29) and Bacillus megaterium (n = 11) and the non-spore-forming bacteria Staphylococcus pasteuri (n = 7), Staphylococcus aureus (n = 3), Staphylococcus saprophyticus (n = 3), Staphylococcus equorum (n = 1), and Staphylococcus xylosus (n = 1). The family Enterobacteriaceae was represented by the species Tatumella saanichensis (n = 3), Pantoea agglomerans (n = 3), and Pantoea terrea (n = 2), while the family Acetobacteraceae was represented by the species A. malorum (n = 6), A. cerevisiae (n = 6), A. pomorum (n = 2), and A. pasteurianus (n = 2) (data not shown). In this medium, B. subtilis was the dominant species, with a maximum population of 6.98 log CFU g−1 at 24 h and 6.92 log CFU g−1 at 12 h in the PC and ST, respectively.
The yeast isolates from the YEPG agar plates were Saccharomyces cerevisiae (n = 63), Pichia kluyveri (n = 13), Hanseniaspora uvarum (n = 9), Issatchenkia orientalis (n = 7), Debaryomyces etchellsii (n = 5), and Kodamaea ohmeri (n = 1). S. cerevisiae was the most prevalent yeast found at the start of fermentation and grew to a maximum population of 8.29 log CFU g−1 and 8.07 log CFU g−1 at 12 h in the PC and ST, respectively. Occasionally, Pichia kudriavzevii, H. uvarum, and I. orientalis were found in both fermentation processes, while D. etchellsii and K. ohmeri were isolated only at 12 h and 24 h in the ST fermentation.
Culture-independent microbiological analysis using a nested-PCR-DGGE strategy.
As displayed in Fig. 2, the bacterial and yeast DGGE profiles revealed a stable microbial composition independent of the fermentation time. The PCR-DGGE revealed that Lactobacillus fermentum, L. plantarum, and Acetobacter spp. (A. tropicalis and Acetobacter senegalensis were the closest relatives found using sequence comparisons) were the dominant bacterial species, as revealed through sequencing of the most intense bands (Fig. 2A). Members of the Enterobacteriaceae family (Tatumella ptyseos and Pantoea terrea) and species of Staphylococcus (S. saprophyticus and a Staphylococcus sp.) were identified at different times in both fermentation processes, while a Bacillus sp. and B. subtilis were detected at the end of the ST fermentation.
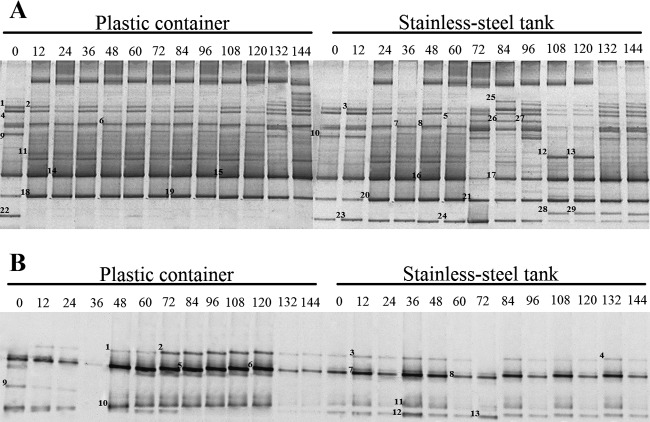
Fig 2.
16S rRNA gene PCR-DGGE profiles of the bacterial communities (A) and ITS PCR-DGGE profiles of the yeast communities (B) associated with the cocoa bean fermentation samples from the PC and the ST. (A) The closest relatives of the fragments sequenced, based on a search of GenBank (>97% similarity), were L. plantarum (bands 1, 2, and 3), L. fermentum (bands 4, 5, 6, 7, and 8), P. terrea (bands 9 and 10), a Staphylococcus sp. (band 11), B. subtilis (bands 12 and 13), an Acetobacter sp. (bands 14, 15, 16, 17, 18, 19, 20, and 21), S. saprophyticus (bands 22, 23, and 24), an uncultured Lactobacillus sp. (band 25), T. ptyseos (bands 26 and 27), and a Bacillus sp. (bands 28 and 29). (B) The closest relatives of the fragments sequenced, based on a search of GenBank (>97% similarity), were a Hanseniaspora sp. (bands 1, 2, 3, 4, 5, 6, 7, and 8), a Debaryomyces sp. (band 9), S. cerevisiae (bands 10 and 11), and P. kluyveri (bands 12 and 13).
The PCR-DGGE revealed that the yeast population was composed of a few species (Fig. 2B). The most intense bands corresponded to the Hanseniaspora species (>99% identity with H. opuntiae, H. uvarum, and H. guilliermondii). Saccharomyces cerevisiae could also be detected in almost all of the samples, but its band density was typically weak. Pichia kluyveri was detected throughout the ST fermentation, while a Debaryomyces sp. was observed at the beginning of the PC fermentation.
Temperature of the cocoa mass.
The temperature inside the PC and ST fermentations ranged from an initial 26.6°C to a maximum 46 to 47°C obtained at 108 h of fermentation (Fig. 1b). After 108 h of fermentation, a slight decrease to 45 to 44°C was observed.
Substrate consumption and fermentation products.
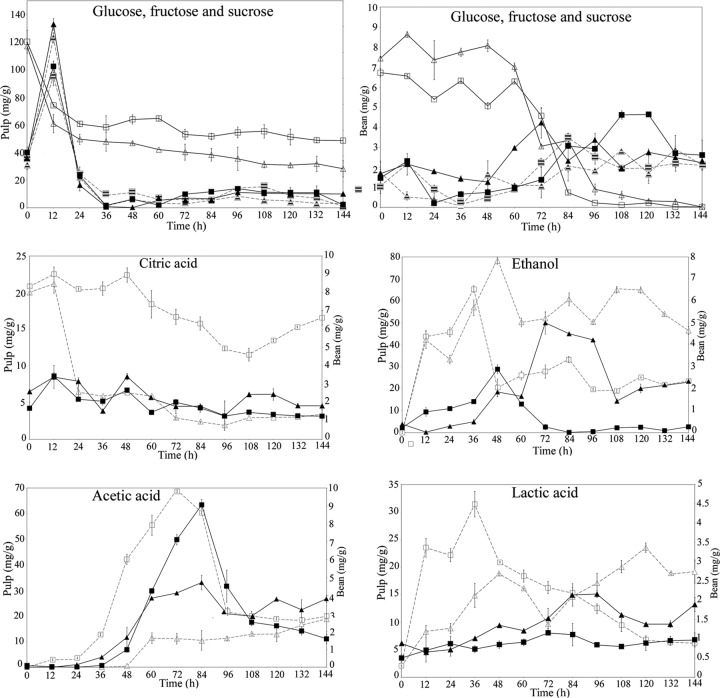
The substrate consumption and the metabolite production of the spontaneous cocoa bean fermentations are shown in Fig. 3. During the first 12 h of fermentation, the hydrolysis of nearly 50% of the pulp sucrose increases the levels of glucose and fructose over 90 mg/g up to 130 mg/g. Glucose and fructose were consumed simultaneously and rapidly up to 36 to 48 h, and afterwards, no significant changes were observed. The major carbohydrate inside the beans was sucrose (6.73 mg/g and 7.44 mg/g in the PC and ST, respectively), which was continuously hydrolyzed into fructose and glucose.
Fig 3.
(Top) Residual glucose, fructose, and sucrose levels. Samples were taken from the ST (triangles) and the PC (squares). Open symbols, sucrose; filled symbols, glucose; hatched symbols, fructose. (Center and bottom) Production of ethanol, methanol, and organic acids during cocoa bean fermentation. Ethanol and organic acid contents in the pulp (open symbols) and in the beans (filled symbols) are shown. Error bars represent standard deviations.
Ethanol was produced and was subsequently consumed concomitantly with the growth of the yeast and the AAB (maximum concentrations, 65.37 mg/g at 36 h and 79 mg/g at 48 h in the PC and ST, respectively) (Fig. 3). The ethanol produced in the pulp diffused into the beans and reached maximum concentrations of 2.89 mg/g at 36 h and 4.51 mg/g at 84 h in the PC and ST, respectively, after which it began evaporating.
The ST fermentation exhibited an unexpected profile, with little acetic acid production; this acid was not oxidized during the first 48 h of fermentation, and, with a linear increase, peaked only at the end of the fermentation process (18.86 mg/g). During the PC fermentation, acetic acid quickly appeared within 48 h (from 24 to 72 h). After 72 h of fermentation, the acetic acid was likely oxidized to CO2 and H2O, dropping below 20 mg g−1 at the end of the fermentation. A portion of the acetic acid content produced in the pulp diffused into the beans after 36 h of fermentation. The final concentrations of acetic acid in the cotyledon were 1.58 mg/g and 3.82 mg/g in the PC and ST, respectively. The concentration of lactic acid in the pulp increased in the first 36 to 48 h, followed by a drop coinciding with the appearance of the AAB; there was again an increase in the concentration during the last 3 days of the ST fermentation. The final lactic acid content in the pulp was 6.2 mg/g in the PC fermentation, and the concentration in the ST was approximately 3-fold higher (19.22 mg/g). The lactic acid content in the cotyledons was nearly 2-fold higher at the end of the ST fermentation process (1.89 mg/g and 0.97 mg/g in the ST and PC, respectively). Rapid decreases in the citric acid content in the pulp were observed after 12 h and 48 h for the ST and PC fermentations, respectively, while inside the beans, the citric acid content was stable during both fermentation processes, with final concentrations of 1.26 and 1.82 mg/g in the PC and ST, respectively.
Polyphasic selection study.
To investigate the physiological adaptation of the yeasts (184 isolates), the LAB (156 isolates), and the AAB (112 isolates) to the cocoa fermentation conditions, and to select for strains better adapted to this environment, several growth parameters were evaluated. Initially, the isolates were screened for their abilities to withstand the stressors imposed by cocoa fermentation, including variations in the pH, the temperature, and the concentrations of ethanol, lactic acid, acetic acid, glucose, and fructose. Isolates from traditional cocoa fermentations performed in wooden boxes in Brazil (G. V. M. Pereira et al., submitted for publication) were also included in this step. The best-adapted strains were selected and were subjected to additional kinetic analysis performed in cocoa pulp simulation media. Tables S3, S4, and S5 in the supplemental material summarize the kinetic analysis data.
Yeasts.
All of the cocoa yeast isolates tolerated the pH range of a typical cocoa fermentation process (from pH 2.0 to pH 5.0). The maximum growth temperature was 30 to 37°C, except for four I. orientalis and three S. cerevisiae isolates, which were able to grow up to 45°C. Although all of the isolates exhibited a remarkable ability to grow in the presence of 15% glucose and fructose, the best growth was observed in the presence of 5% carbohydrates. The majority of the yeasts identified as S. cerevisiae, P. kluyveri, and D. etchellsii were tolerant of media amended with as much as 12% ethanol, while the other yeast species tolerated only 6% ethanol (except for Pichia fermentans UFLA CHYA4.0 and UFLA CHYD31.07, I. orientalis UFLA CHYB6.02 and UFLA CHYC6.02, and Candida humilis UFLA CHD14.32) (data not shown). From these results, 15 well-adapted isolates of S. cerevisiae, P. fermentans, P. kluyveri, D. etchellsii, and I. orientalis were selected for simulated cocoa fermentation in PSM-yeast. Isolates of K. ohmeri, Candida orthopsilosis, C. humilis, Candida intermedia, and Schizosaccharomyces pombe, which were not resistant to the stressors employed in this study, were also included in this step. The fermentation kinetics of the pure cultures is reported in Table S3 in the supplemental material. S. cerevisiae strains UFLA CHYC7.04 and UFLA CHYB4.03 exhibited the highest conversion rate of substrate to ethanol (49.33 and 49.81% of the theoretical value, respectively). Within the non-Saccharomyces species, D. etchellsii UFLA CHYB5.56, C. humilis UFLA CHYD14.32, P. kluyveri UFLA CHYC2.02, and I. orientalis UFLA CHYB6.02 and UFLA CHYC6.02 exhibited the highest fermentation efficiencies (>35% conversion of the substrate into ethanol), while K. ohmeri BM2.01, P. fermentans UFLA CHYA4.01 and UFLA CHYD31.07, C. orthopsilosis UFLA CHYC5.02, C. intermedia UFLA CHYE21.01, and Schizosaccharomyces pombe UFLA CHYE5.39 exhibited the lowest values (<20% conversion). None of the organic acid contents changed significantly in PSM-yeast during 24 h of fermentation.
LAB.
The results demonstrated the overall ability of the LAB isolates to grow over a wide range of pH values and lactic acid concentrations. In general, the majority of the L. plantarum and L. fermentum strains shared the ability to grow at 45°C and 12% ethanol, while Weissella sp. isolates did not (data not shown). Six strains of L. fermentum, five of L. plantarum, and one of Weissella ghanensis were selected on the basis of their growth properties for further kinetic analysis in PSM-LAB (see Table S4 in the supplemental material). The strictly heterofermentative LAB strains (L. fermentum and the Weissella sp.) fermented glucose but not fructose, converted citric acid, and accumulated lactic acid and acetic acid as the main products. Interestingly, the conversion of citric acid by the strictly heterofermentative LAB strains was more efficient than the conversion of carbohydrates (glucose or fructose) into these products. All the citric acid content was consumed in 12 h of fermentation, while a residual content of carbohydrates was observed at the end of fermentations. The production of lactic acid was higher for L. fermentum UFLA CHBE8.12 (7.72 g liter−1), while the production of acetic acid was higher for Weissella ghanensis UFLA CHBE8.23 and L. fermentum UFLA CHBE6.01 (6.25 g liter−1). None of the strains of L. plantarum tested were able to produce lactic or acetic acid when inoculated in the synthetic medium used in this study (see Table S4).
AAB.
All of the cocoa AAB isolates grew in a basal medium supplemented with as much as 5% acetic acid and a pH adjusted to 2.0, 3.0, or 5.0, while these isolates failed to grow at 15 and 30% glucose and fructose concentrations. All of the isolates grew at 30 and 37°C, as well as in 6% ethanol. Only the A. tropicalis isolates grew at 45°C and in 12% ethanol (data not shown). Five stress-tolerant A. tropicalis strains and representative isolates of the other AAB species were subjected to simulated cocoa fermentation in PSM-AAB (see Table S5 in the supplemental material). The AAB species oxidized almost all of the ethanol into acetic acid within 24 h of fermentation. These species also oxidized lactic acid during their growth. The production of acetic acid was higher for A. tropicalis UFLA CHBE16.01, A. senegalensis UFLA CHBE6.16, A. ghanensis UFLA CHBA3.01, A. malorum UFLA CHBA1.01, and Gluconobacter oxydans UFLA CHBD7.06, which produced 14.08, 13.49, 13.25, 11.45, and 10.73 g/liter, respectively, than for the other isolates, which produced <10 g/liter.
DISCUSSION
Microbiological and physicochemical performance of cocoa bean fermentations.
The microbiological communities in the cocoa bean fermentations performed under bench- and pilot-scale conditions (Fig. 1a; Tables 1 and 2) were similar to those observed in previous spontaneous, larger-scale fermentations (2, 14, 16, 18, 25, 27, 30, 39). The most common LAB (L. fermentum and L. plantarum), AAB (A. tropicalis), and yeast (S. cerevisiae and Hanseniaspora spp.) cocoa species appear to dominate the cocoa fermentations. The composition and metabolic activity of these well-adapted cocoa species remained unchanged from those in larger cocoa fermentations once a stable ecosystem was achieved. These similarities suggested that the small-scale fermentations provided a suitable model system for larger-scale fermentations, at least with regard to microbial ecology. The use of a suitable model system may allow the evaluation of a starter culture under controlled conditions prior to its use in the field.
Several studies have attempted to cure small quantities of cacao beans (<20 kg) rapidly and effectively in order to research aspects of the cocoa fermentations, such as the effects of the cultivar, pod storage, pulp removal, and acid production (5, 10, 33). However, in these studies, the temperature of the cocoa bean mass did not increase normally, often reaching only 35 to 37°C. Therefore, in the present study, the bench- and pilot-scale fermentations were placed in a temperature-controlled incubator. The program chosen was designed to mimic the increases observed in a traditional fermentation, which allowed the successive growth of the yeasts and the LAB (25 to 32°C; 0 to 48 h), followed by the AAB (40 to 48°C; 60 to 144 h). For both fermentation processes, the temperature of the cocoa bean mass closely followed the temperature of the incubator (Fig. 1b).
The culture-based approach demonstrated that the LAB species L. fermentum and L. plantarum and the yeast S. cerevisiae dominated the early stages of fermentation. The richness in fermentable carbohydrates and the low oxygen content of the cocoa mass favored the growth of these microbial groups, which were hypothesized to rapidly metabolize reducing sugars and citric acid and to produce mainly ethanol and lactic acid (Fig. 3). In addition, the simultaneous growth of both the yeast and the LAB can be explained by additional modes of LAB-yeast interaction. For example, the death and autolysis of the yeast cells releases vitamins and other nutrients, and/or the CO2 produced by the yeast creates microaerophilic conditions, which favor LAB growth (9). Moreover, in the beginning, L. plantarum (facultatively heterofermentative) dominated the LAB community, but after 36 h, L. fermentum (strictly heterofermentative) became the dominant LAB species (Tables 1 and 2). A similar growth dynamic between these two LAB species was observed during large-scale cocoa fermentations (26). This result demonstrated that the yeasts were most adaptable when they were associated with the facultatively heterofermentative LAB rather than the strictly heterofermentative LAB, as observed previously during sourdough fermentations (13). Dircks (7) observed that the increased ethanol content during a controlled inoculation of cacao bean fermentations using different yeast species inhibited the growth of L. fermentum. Despite the complex microbial ecology of cocoa bean fermentations, these types of interactions have not been considered.
Higher concentrations of citric and lactic acid were recovered during PC fermentation, in which L. plantarum populations were higher, and amounts of L. fermentum were smaller, than those for the ST fermentation. The homolactic metabolism of L. plantarum allows this species to achieve a high cell density within a reasonable fermentation time and, in contrast to L. fermentum, to produce large amounts of lactic acid. In contrast, the heterolactic metabolism of L. fermentum leads to the rapid conversion of citric acid and the production of almost equal amounts (on a mass basis) of lactic acid and acetic acid (3). This led to higher final concentrations of lactic acid (but not of citric acid) in the cocoa beans from the PC fermentation than in those from the ST fermentation (Fig. 3). Although the diffusion of lactic acid into the cocoa beans contributes to the breakdown of the seed cell structure, because it is nonvolatile, such an excess is not reduced during drying, and high concentrations of the residual lactic acid will impart a sour flavor to the chocolate (1).
The kinetics of ethanol production in the pulp of cocoa fermentations corresponded to yeast growth (specifically the growth of the yeast S. cerevisiae [Tables 1 and 2]) and the utilization of the sugars. However, over time, the ethanol concentration in each fermentation varied with the lowest content recovered from the PC at 48 h of fermentation. This result may have been due to the higher yeast population observed in the ST than in the PC fermentation. The metabolic activity of the yeasts and the LAB (temperature increase; ethanol and lactic acid accumulation [Fig. 1b and 3]) favored AAB growth (Fig. 1a), which was accompanied by the production of acetic acid and a reduction in the concentration of ethanol (Fig. 3). This finding agreed with the classic description of these bacteria in the literature (22, 39). Both culture-independent and culture-based approaches suggested a major role for the genus Acetobacter in bench- and pilot-scale cocoa fermentations. The presence of Gluconobacter species, previously reported in larger cocoa fermentations (22, 39), was not confirmed in this study. A. tropicalis was the dominant species in both fermentations. The dominance of A. tropicalis was observed previously during larger cocoa fermentations in Australia (7). The A. tropicalis species is associated mainly with fruits and fermented foods and has been selected to produce artisanal vinegar (24). The dominance of this species during cocoa bean fermentation may be explained by its acid and heat resistance (24). The levels of acetic acid produced in the pulp from the PC fermentation were much lower than those produced during ST fermentation. This difference might be due to the low availability of oxygen inside the tank; consequently, less ethanol may be oxidized to acetic acid.
Bacillus, Staphylococcus, and Enterobacteria were isolated during the last 0 to 48 h. The PCR-DGGE profile confirmed the presence of these three bacterial groups in both fermentation processes (Fig. 2). The main species isolated were Bacillus subtilis, B. megaterium, and Staphylococcus pasteuri. Their growth appeared to be inhibited by the high population of the LAB and/or yeast, and no growth was observed after 48 h of fermentation (Tables 1 and 2). The appearance of Bacillus spp. during cocoa bean fermentations is less predictable than that of other microbial groups. Their role during cocoa fermentation is not well known, and this bacterium has never been involved as a starter in attempts to control the fermentation process. It is believed that the growth of a low population of certain Bacillus strains might have a beneficial action, e.g., by acting as a complementary partner to the yeasts in the pulp depectinization process during the advanced stage of cocoa fermentation (28). However, a high population may contribute to the acidity and perhaps to the undesirable flavors of fermented cocoa beans (38). In this sense, cocoa fermentations conducted with a defined inoculum of LAB and yeasts may prove useful for the control of Bacillus spp. during industrialized fermentations. Although the conditions in the cocoa mass do not favor the development of Enterobacteria and Staphylococcus species (low pH and high temperatures), their presence was reported using culture-independent methods during Ghanaian, Brazilian, and Ivorian cocoa bean fermentations (30). The appearance of these bacterial species underlines the importance of monitoring the hygiene of the fermentation process to ensure that these species do not become dominant and spoil the beans (2). Their presence may indicate human contact with the beans or may also be associated with pod surfaces and the material used.
The PCR-DGGE method has some potential limitations that made it necessary to use this technique in combination with a culture-dependent method. Species present in low concentrations could occasionally be grown from agar plates but in many cases did not generate a detectable DGGE band. Thus, it is possible that the DGGE fingerprint might mask the perturbations of low-abundance community members in our study. This finding illustrates the intrinsic limitation of DGGE analysis in visualizing only the predominant species of a microbial community (31). Interestingly, S. cerevisiae yielded only a relatively weak band in the denaturing gels compared to Hanseniaspora spp. (Fig. 2b), even though S. cerevisiae dominated the yeast species isolated by using the plating method from both fermentation processes. A possible explanation may be that the S. cerevisiae ITS fragment is less efficiently amplified than that of the Hanseniaspora species present during the cocoa fermentation by using the protocol described here (25). An accurate assessment of the microbial ecologies using PCR-DGGE requires appropriately designed primers, and the use of poorly targeted primers will skew estimates of the microbial diversity present (4).
Polyphasic selection study.
To our knowledge, this study represents the first assessment of the growth and stress tolerance of yeasts, LAB, and AAB under cocoa-fermenting conditions. The criteria used were based on the physical and chemical changes that each microbial group faces in the cocoa pulp substrate when their metabolisms are most active. At the beginning of the process, the yeast and LAB cells are affected by osmotic stress due to the high sugar content in the cocoa pulp (39). As the fermentation progresses, other stressors become relevant as ethanol and lactic acid accumulate and the environment acidifies. After 2 days of fermentation, the AAB become the dominant group and are faced with an ethanol-rich environment, followed by the accumulation of acetic acid and an increase in the temperature to a mean value of 45°C (39). In the present study, the results indicated that the stressors of the early fermentation phase influenced the prevalence of the best-adapted yeast, S. cerevisiae. In addition, some strains of the non-Saccharomyces yeasts, such as P. fermentans, P. kluyveri, D. etchellsii, and I. orientalis, were also well adapted to the conditions imposed. The limited number of yeasts capable of growth at 45°C explains the decreasing yeast population (Fig. 1a) after AAB-facilitated exothermic ethanol oxidation raises the temperature to 48°C. Similarly, the abilities of the L. fermentum and L. plantarum strains to grow over a wide range of pH values, temperatures, and ethanol and lactic acid concentrations correlated with the stable population of the LAB during the entire fermentation process (Fig. 1a). Finally, the ability of A. tropicalis isolates to tolerate the stressors imposed during the second stage of the fermentation might be considered an advantage of this species, which led to its dominance in the fermentations performed.
As expected, the fermenting capacity of S. cerevisiae strains was generally higher than that of the non-Saccharomyces species in PSM-yeast (see Tables S3 to S5 in the supplemental material). The production of ethanol by the yeasts could affect the course of the cocoa fermentation in several ways, including the inhibition of certain microbial species, the inability of the cocoa beans to germinate and their decompartmentalization, and the use of ethanol as a precursor for the formation of acetic acid. However, some non-Saccharomyces strains (D. etchellsii UFLA CHYB5.56, C. humilis UFLA CHYD14.32, P. kluyveri UFLA CHYC2.02, and I. orientalis UFLA CHYB6.02 and UFLA CHYC6.02) also produced significant levels of ethanol. This result, together with the ability of these non-Saccharomyces strains to tolerate cocoa-fermenting conditions, indicated that these strains could be used in a controlled cocoa fermentation multiculture starter with S. cerevisiae. The secondary products of their metabolism (e.g., organic acids, aldehydes, ketones, higher alcohols, and esters) and their glycosidase production are likely to be significant and should impact bean and chocolate quality (2). These potentially important influences have been overlooked in previous work on cocoa fermentation.
The strictly heterofermentative L. fermentum strains were characterized by rapid citric acid conversion, and in the consumption of sugars, glucose was preferentially metabolized over fructose. Citric acid is used for the oxidation of NADH plus H+ to bypass the energy-limiting ethanol pathway and to maximize the growth rate on glucose, thereby producing mannitol and lactic acid plus acetic acid (21). The metabolism of citric acid by cocoa-specific L. fermentum has been observed previously (20). The consumption of citric acid resulted in the production of organic acids with a higher pKa value, which increased the pH of the environment and allowed better bacterial growth and microbiological control of the environment (39).
All of the AAB strains oxidized lactic acid (through lactate dehydrogenase, pyruvate decarboxylase, and acetaldehyde dehydrogenase activities) and ethanol (via the sequential action of pyrroquinoline-quinone-dependent alcohol dehydrogenase and acetaldehyde dehydrogenase activities) into acetic acid in PSM-AAB. Thus, the simultaneous metabolism of the LAB and yeasts stimulates the growth of the AAB during a natural cocoa fermentation. In agreement with the ability to tolerate cocoa fermentation stress, the oxidation process of A. tropicalis UFLA CBE16.01 was the most effective, followed by A. senegalensis UFLA CBE6.16 and A. ghanensis UFLA CBA3.01. Interestingly, the species A. senegalensis and A. ghanensis were recently described during Ghanaian cocoa bean fermentation processes (6), which likely explains their adaptation to the cocoa pulp habitat.
In conclusion, this study demonstrated that the bench- and pilot-scale cocoa fermentation ecosystems reach equilibrium starting with the simultaneous growth of the yeasts and LAB, which are gradually replaced by the AAB. Overall, the dominant species largely overlapped with those commonly associated with larger cocoa bean fermentations (S. cerevisiae and Hanseniaspora spp. in the yeast group, L. fermentum and L. plantarum in the LAB group, and A. tropicalis in the AAB group), proving their adaptation to the cocoa environment under the conditions applied. In both processes, the course of substrate consumption and metabolite production was similar to that in the larger, spontaneous cocoa bean fermentation processes. However, further studies should evaluate the impact of excessive production of these metabolites in the pulp on the technological and sensorial quality of the resultant chocolate.
The polyphasic selection study allowed us to construct a better picture of the physiology and ecology of the indigenous yeast, LAB, and AAB strains. As a result, some strains from these three major groups were selected as potential starter cultures. In particular, L. fermentum UFLA CBE8.12 (citric acid fermenting, lactic acid producing, and tolerant to heat, acid, lactic acid, and ethanol), S. cerevisiae UFLA CYC7.04 (ethanol producing and acid, heat, and ethanol tolerant), and A. tropicalis UFLA CBE16.01 (ethanol and lactic acid oxidizing, acetic acid producing, and tolerant to acid, heat, acetic acid, and ethanol) were selected as candidates for a mixed-strain starter cocktail that should lead to better-controlled and more-reliable cocoa bean fermentation processes. In addition, analysis of the non-Saccharomyces strains (D. etchellsii UFLA CYB5.56, C. humilis UFLA CYD14.32, P. kluyveri UFLA CYC2.02, and I. orientalis UFLA CYB6.02 and UFLA CYC6.02) indicated that these should be tested in association with S. cerevisiae for cocoa starter cultures in future studies.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Brazilian agencies Conselho Nacional de Desenvolvimento Científico e Tecnológico of Brasil (CNPQ), Fundação de Amparo a Pesquisa do Estado de Minas Gerais (FAPEMIG), and CAPES for scholarships, and Almirante Cacau Agrícola (Masterfoods) for financial support.
Footnotes
Published ahead of print 25 May 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Afoakwa EO, Paterson A, Fowler M, Ryan A. 2008. Flavour formation and character in cocoa and chocolate: a critical review. Crit. Rev. Food Sci. Nutr. 48:840–857 [DOI] [PubMed] [Google Scholar]
- 2. Ardhana M, Fleet G. 2003. The microbial ecology of cocoa bean fermentations in Indonesia. Int. J. Food Microbiol. 86:87–99 [DOI] [PubMed] [Google Scholar]
- 3. Axelsson L. 2004. Lactic acid bacteria: classification and physiology, p 1–66 In Salminen S, von Wright A, Ouwehand A. (ed), Lactic acid bacteria: microbiological and functional aspects, 3rd ed Marcel Dekker, New York, NY [Google Scholar]
- 4. Beh AL, Fleet GH, Prakitchaiwattana C, Heard GM. 2006. Evaluation of molecular methods for the analysis of yeast in food and beverages. Adv. Exp. Med. Biol. 571:69–106 [DOI] [PubMed] [Google Scholar]
- 5. Clapperton JF, Lockwood G, Yow STK, Um OHK. 1994. Effects of planting materiais on flavour. Cocoa Growers Bull. 48:47–63 [Google Scholar]
- 6. Cleenwerck I, et al. 2007. Acetobacter ghanensis sp. nov., a novel acetic acid bacterium isolated from traditional heap fermentations of Ghanaian cocoa beans. Int. J. Syst. Evol. Microbiol. 57:1647–1652 [DOI] [PubMed] [Google Scholar]
- 7. Dircks HD. 2009. Ph. D. thesis. University of New South Wales, Sydney, Australia [Google Scholar]
- 8. Dzogbefia VP, Buamah R, Oldham JH. 1999. The controlled fermentation of cocoa (Theobroma cacao L.) using yeasts: enzymatic process and associated physico-chemical changes in cocoa sweatings. Food Biotechnol. 13:1–12 [Google Scholar]
- 9. Fleet GH. 2007. Yeast in foods and beverages: impact on product quality and safety. Curr. Opinion. Biotechnol. 18:170–175 [DOI] [PubMed] [Google Scholar]
- 10. Frederick H. 1960. Cocoa manual. English ed Inter-American Institute of Agricultural Sciences; Turrialba, Costa Rica [Google Scholar]
- 11. Gasch AP, Werner-Washburne M. 2002. The genomics of yeast responses to environmental stress and starvation. Funct. Integr. Genomics 2:181–192 [DOI] [PubMed] [Google Scholar]
- 12. Holzapfel WH. 2002. Appropriate starter culture technologies for small scale fermentation in developing countries. Int. J. Food Microbiol. 75:197–212 [DOI] [PubMed] [Google Scholar]
- 13. Iacumin L, et al. 2009. Description of the microflora of sourdoughs by culture-dependent and culture-independent methods. Food Microbiol. 26:128–135 [DOI] [PubMed] [Google Scholar]
- 14. Jespersen L, Nielsen DS, Hønholt S, Jakobsen M. 2005. Occurrence and diversity of yeasts involved in fermentation of West African cocoa beans. FEMS Yeast Res. 5:441–453 [DOI] [PubMed] [Google Scholar]
- 15. Joyeux A, Lafon-Lafourcade S, Ribereau-Gayon P. 1984. Evolution of acetic acid bacteria during fermentation and storage of wine. Appl. Environ. Microbiol. 48:153–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kostinek M, et al. 2008. Diversity of predominant lactic acid bacteria associated with cocoa fermentation in Nigeria. Curr. Microbiol. 56:306–314 [DOI] [PubMed] [Google Scholar]
- 17. Kurtzman CP, Fell JW, Boekhout T. 2011. The yeasts, a taxonomic study, 4th ed Elsevier, Amsterdam, Netherlands [Google Scholar]
- 18. Lagunes-Gálvez S, Loiseau G, Paredes JL, Barel M, Guiraud J-P. 2007. Study on the microflora and biochemistry of cocoa fermentation in the Dominican Republic. Int. J. Food Microbiol. 114:124–130 [DOI] [PubMed] [Google Scholar]
- 19. Leal GA, Jr, Gomes LH, Efraim P, Tavares FC, Figueira A. 2008. Fermentation of cocoa (Theobroma cacao L.) seeds with a hybrid Kluyveromyces marxianus strain improved product quality attributes. FEMS Yeast Res. 8:788–798 [DOI] [PubMed] [Google Scholar]
- 20. Lefeber T, Janssens M, Camu N, De Vuyst L. 2010. Kinetic analysis of strains of lactic acid bacteria and acetic acid bacteria in cocoa pulp simulation media to compose a starter culture for cocoa bean fermentation. Appl. Environ. Microbiol. 76:7708–7716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li X, Jiang B, Beilei P, Wanmeng M, Zhang T. 2008. Purification and partial characterization of Lactobacillus species SK007 lactate dehydrogenase (LDH) catalyzing phenylpyruvic acid (PPA) conversion into phenyllactic acid (PLA). J. Agric. Food Chem. 56:2392–2399 [DOI] [PubMed] [Google Scholar]
- 22. Lima LJR, Almeida MH, Nout MJ, Zwietering MH. 2011. Theobroma cacao L., “the food of the gods”: quality determinants of commercial cocoa beans, with particular reference to the impact of fermentation. Crit. Rev. Food Sci. Nutr. 51:731–761 [DOI] [PubMed] [Google Scholar]
- 23. Magalhães KT, Pereira GVM, Dias DR, Schwan RF. 2010. Microbial communities and chemical changes during fermentation of sugary Brazilian kefir. World J. Microbiol. Biotechnol. 26:1241–1250 [DOI] [PubMed] [Google Scholar]
- 24. Ndoye B, et al. 2006. Thermoresistant properties of acetic acid bacteria isolated from tropical products of sub-Saharan Africa and destined to industrial vinegar. Enzyme Microb. Technol. 39:916–923 [Google Scholar]
- 25. Nielsen DS, Honholt S, Tano-Debrah K, Jespersen L. 2005. Yeast populations associated with Ghanaian cocoa fermentations analysed using denaturing gradient gel electrophoresis (DGGE). Yeast. 22:271–284 [DOI] [PubMed] [Google Scholar]
- 26. Nielsen DS, et al. 2007. The microbiology of Ghanaian cocoa fermentations analysed using culture-dependent and culture independent methods. Int. J. Food Microbiol. 114:168–186 [DOI] [PubMed] [Google Scholar]
- 27. Ostovar K, Keeney PG. 1973. Isolation and characterization of microorganisms involved in the fermentation of Trinidad's cacao beans. J. Food Sci. 38:611–617 [Google Scholar]
- 28. Ouattara HG, et al. 2008. Implication of Bacillus sp. in the production of pectinolytic enzymes during cocoa fermentation. World J. Microbiol. Biotechnol. 24:1753–1760 [Google Scholar]
- 29. Ovreas L, Forney L, Daae FL, Torsvik V. 1997. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 63:3367–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Papalexandratou Z, Camu N, Falony G, De Vuyst L. 2011. Comparison of the bacterial species diversity of spontaneous cocoa bean fermentations carried out at selected farms in Ivory Coast and Brazil. Food Microbiol. 28:964–973 [DOI] [PubMed] [Google Scholar]
- 31. Pereira GVM, Magalhães KT, Lorenzetii ER, Souza TP, Schwan RS. 2012. A multiphasic approach for the identification of endophytic bacteria in strawberry fruit and their potential for plant growth promotion. Microb. Ecol. 63:405–417 [DOI] [PubMed] [Google Scholar]
- 32. Pereira GVM, Ramos CL, Galvão C, Dias ES, Schwan RF. 2010. Use of specific PCR primers to identify three important industrial species of Saccharomyces genus: Saccharomyces cerevisiae, Saccharomyces bayanus and Saccharomyces pastorianus. Lett. Appl. Microbiol. 5:131–137 [DOI] [PubMed] [Google Scholar]
- 33. Quesnel VC, Lopez A. 1975. A sweat-box for fermenting small samples of cocoa. Trop. Agric. 52:309–316 [Google Scholar]
- 34. Ruiz A, Poblet M, Mas A, Guillamón JM. 2000. Identification of acetic acid bacteria by RFLP of PCR-amplified 16S rDNA and 16S–23S rDNA intergenic spacer. Int. J. Syst. Evol. Microbiol. 50:1981–1987 [DOI] [PubMed] [Google Scholar]
- 35. Samah OA, Ptih MF, Selamat J. 1992. Biochemical changes during fermentation of cocoa beans inoculated with Saccharomyces cerevisiae (wild strain). J. Food Sci. Technol. 29:341–343 [Google Scholar]
- 36. Schwan RF. 1998. Cocoa fermentations conducted with a defined microbial cocktail inoculum. Appl. Environ. Microbiol. 64:1477–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schwan RF, Rose AH, Board RG. 1995. Microbial fermentation of cocoa beans, with emphasis on enzymatic degradation of the pulp. J. Appl. Bacteriol. Symp. Suppl. 79:96S–107S [Google Scholar]
- 38. Schwan RF, Vanetti MCD, Silva DO, Lopez A, Moraes CA. 1986. Characterization and distribution of aerobic, spore-forming bacteria from cacao fermentations in Bahia. J. Food Sci. 51:1583–1584 [Google Scholar]
- 39. Schwan RF, Wheals AE. 2004. The microbiology of cocoa fermentation and its role in chocolate quality. Crit. Rev. Food Sci. Nutr. 44:205–221 [DOI] [PubMed] [Google Scholar]
- 40. Steinkraus KH. 2004. Industrialization of indigenous fermented foods, 2nd ed CRC Press, Boca Raton, FL [Google Scholar]
- 41. Vandenbergh PA. 1993. Lactic acid bacteria, their metabolic products and interference with microbial growth. FEMS Microbiol. Rev. 12:221–238 [Google Scholar]
- 42. White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322 In Innis MA, Gelfand DH, Sninsky JJ, White TJ. (ed), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA [Google Scholar]
- 43. Wood BJB. 1998. Microbiology of fermented foods, 2nd ed Thompson Science, London, United Kingdom [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.