Abstract
The Greenland ice sheet (GrIS) receives organic carbon (OC) of anthropogenic origin, including pesticides, from the atmosphere and/or local sources, and the fate of these compounds in the ice is currently unknown. The ability of supraglacial heterotrophic microbes to mineralize different types of OC is likely a significant factor determining the fate of anthropogenic OC on the ice sheet. Here we determine the potential of the microbial community from the surface of the GrIS to mineralize the widely used herbicide 2,4-dichlorophenoxyacetic acid (2,4-D). Surface ice cores were collected and incubated for up to 529 days in microcosms simulating in situ conditions. Mineralization of side chain- and ring-labeled [14C]2,4-D was measured in the samples, and quantitative PCR targeting the tfdA genes in total DNA extracted from the ice after the experiment was performed. We show that the supraglacial microbial community on the GrIS contains microbes that are capable of degrading 2,4-D and that they are likely present in very low numbers. They can mineralize 2,4-D at a rate of up to 1 nmol per m2 per day, equivalent to ∼26 ng C m−2 day−1. Thus, the GrIS should not be considered a mere reservoir of all atmospheric contaminants, as it is likely that some deposited compounds will be removed from the system via biodegradation processes before their potential release due to the accelerated melting of the ice sheet.
INTRODUCTION
The Greenland ice sheet (GrIS) is the largest body of ice in the Northern Hemisphere, covering approximately 1.7 million km2. It receives considerable amounts of organic carbon (OC) of anthropogenic origin from the atmosphere and/or local sources (28, 29), including oil-derived hydrocarbons and pesticides (6, 21, 39). The fate of these compounds in the ice is currently unknown. Accelerated melting of the ice sheet (34, 35) may result in an increased release of these chemicals to downstream ecosystems, including soils in the ice-free areas and the coastal waters around Greenland, with potentially harmful consequences.
Microbes associated with the surface ice and debris (supraglacial microbes) on the GrIS are actively involved in carbon cycling processes (16, 41). A great diversity of heterotrophic microbes has been found in the microbial communities of glacier and ice sheet surfaces, with Alpha- and Betaproteobacteria and Bacteroidetes usually being dominant (8, 11, 25, 48). Their ability to mineralize different types of OC substrates is likely a significant factor determining the fate of anthropogenic OC deposition on the GrIS and in the overall carbon cycling in the system.
A wide range of organic compounds has been shown to be degraded by supraglacial heterotrophs, including labile low-molecular-weight compounds such as amino acids and carbohydrates, more complex and recalcitrant compounds such as cellulose, lignin, and some anthropogenic compounds such as polycyclic aromatic hydrocarbons (12, 27). However, a direct link between the diversity of a supraglacial microbial community and its heterotrophic metabolic potential, including degradation of anthropogenic OC, such as pesticides, has not been established to date.
In this study, we determine the potential of the microbial community from the surface of the GrIS to mineralize the herbicide 2,4-dichlorophenoxyacetic acid (2,4-D). Phenoxyacetic acid herbicides have been used extensively for decades (26), resulting in an increased interest in their fate in the environment (31). 2,4-D is one of the most easily degraded compounds among the phenoxyacetic acid herbicides, and the ability to mineralize 2,4-D has been found to be widespread in microbial communities around the globe (19, 22, 45). Functional genes involved in the degradation pathway have also been characterized, including the tfdA gene, encoding an α-ketoglutarate-dependent dioxygenase catalyzing the first step in the degradation pathway (3, 9, 13). Thus, 2,4-D represents a very suitable model compound to use in order to gain an insight into pollutant degradation dynamics in natural environments (2, 14), including those in the rapidly changing Arctic region.
We collected samples of surface ice and debris in the southwestern part of the GrIS and incubated them in custom-built microcosms enabling a simulation of the in situ conditions on the surface of the ice sheet. The diversity of the microbial community in the samples was characterized by cloning and sequencing of 16S rRNA gene amplicons. Mineralization of 2,4-D by the microbial community present was monitored using radiolabeled substrate and quantitative PCR (qPCR) targeting the tfdA genes, involved in the degradation pathway of 2,4-D.
MATERIALS AND METHODS
Sampling.
Samples were collected on the surface of the margin of the GrIS in the Kangerlussuaq, Greenland, area (67°09′N, 50°02′W) during five trips in April, May, July, August, and September 2006. Surface ice cores were collected using a handheld drill and custom-built stainless steel corers (20 cm long, 5 cm in diameter, i.e., an ∼20-cm2 surface area). The ice cores were carefully transferred to custom-built tube-shaped sample containers, avoiding any contact with the ice surface. The corers and sample containers were autoclaved and kept sterile in polypropylene bags prior to use. The collected ice samples were placed into sterile Whirl-Pak bags, and these were placed into insulated boxes and transported to Copenhagen, Denmark. The temperature inside the boxes was between −6 and −14°C during transport. The samples were then kept frozen at −20°C until their placement in microcosms for the experiments.
DNA extraction, amplification, and cloning.
DNA was extracted from surface ice samples collected in April, July, and September 2006. In order to extract DNA from the samples, aliquots were melted at room temperature under sterile conditions. Initially, filtration through 0.22-μm-pore-size filters (Pall, NY) was attempted in order to obtain DNA from the whole community. However, due to DNA contamination from the filters, we decided to use centrifugation and extract DNA from the debris only. This is in line with previous work showing that the majority of microorganisms on glacier surfaces are associated with the surface debris (16, 41). Therefore, 200 ml of the meltwater was centrifuged at maximum speed (22,000 × g) for 10 min, and the supernatant was discarded. DNA was extracted from the pellets using a PowerSoil DNA isolation kit (MO BIO Laboratories, CA) according to the manufacturer's instructions. Approximately 200 mg (wet weight) of pellet was used for each extraction.
Bacterial 16S rRNA genes were amplified from the DNA extracts by PCR with the primers 518F (5′-CC AGC AGC CGC GGT AAT-3′) and 1492R [5′-GG(CT) TAC CTT GTT ACG ACT T-3′]. The reaction mixture (25 μl) for the PCR contained PuReTaq Ready-To-Go beads (GE Healthcare, United Kingdom), 0.125 μl of each primer (10 pmol μl−1), and 1 μl template DNA, representing 2 ml of the original meltwater. The temperature program for PCR was as follows: initial an denaturation step of 10 min at 95°C, followed by 30 cycles of 95°C for 45 s, 55°C for 45 s, and 72°C for 90 s, with a final extension of 72°C for 7 min. Negative controls were included by replacing template DNA with 1 μl of molecular biology-grade water (MO BIO Laboratories, CA). Amplified 16S rRNA genes were purified using an UltraClean PCR cleanup DNA purification kit (MO BIO Laboratories, CA) according to the manufacturer's directions.
Clone libraries were prepared using a TOPO TA cloning kit with the pCR2.1-TOPO vector (Invitrogen, CA) and the manufacturer's recommended protocol. Plasmid DNA from clones from each DNA extract was purified using an UltraClean 6-min miniplasmid prep kit (MO BIO Laboratories, CA). Afterward, 25 μl of the purified plasmid was used for sequencing, which was performed at Eurofins MWG Operon (Ebersberg, Germany).
Phylogenetic analysis.
The bacterial 16S rRNA gene sequences obtained from each site were checked for chimeras using the Bellerophon program (17) and grouped into operational taxonomic units (OTUs) according to their similarity. The OTUs were then used for diversity and phylogenetic analysis. The coverage of the clone libraries (in percent) was calculated as [1 − (n/N)] × 100, where n is the number of unique clones detected in the library, and N is the total number of clones. 16S rRNA gene sequences of the closest relatives were searched for using the BLAST program at the National Center for Biotechnology Information. Additional sequences potentially related to the ones obtained, plus sequences of known 2,4-D degraders (4), were also retrieved. The sequences were aligned using the MUSCLE program (10) and edited manually in the JalView program (47). Phylogenetic analyses were restricted to nucleotide positions that were unambiguously aligned in all sequences. The phylogenetic tree was constructed using the maximum likelihood method and MEGA (version 5) software (43). Bootstrap analysis (1,000 replications) was used to provide confidence estimates for phylogenetic tree topologies.
Mineralization experiments.
Samples collected in May and August were used for 2,4-D mineralization experiments. Individual cores in the original sample containers were put in 750-ml airtight glass containers (Holmegård, Denmark) and placed in a custom-built low-temperature incubator. The incubator was based on a 600-liter chest freezer (Gram AS, Denmark) extended with an 80-cm-high Plexiglas cabinet. Two 35-W high-intensity-discharge (HID) xenon bulbs with reflectors (Bosch, Germany) provided illumination in a 12-h light and 12-h dark regime. Two hundred kilograms of washed and autoclaved quartz pebbles (Bisgaards Akvariegrus, Denmark) was placed at the bottom of the freezer in order to help stabilize the inside temperature. The temperature in the incubator was monitored continuously by a Microlog data logger (Ninasoft, Denmark) frozen halfway down an ice core prepared in a sterilized sample container (described above) by freezing deionized water. The temperature within this ice core fluctuated between −7.5°C when in the dark and −4.0°C when illuminated, and surface melting was observed in all the samples during the light periods. Thus, the incubator simulated the in situ conditions of the surface of an ice sheet in summer. Two turning plates, each capable of holding nine glass containers, were placed on top of the pebble surface, which allowed changing of the position of the samples within the incubator in order to ensure a more even exposure to temperature and light. The samples were repositioned once a week during the whole incubation period.
In the first exploratory experiment, the mineralization rate of side chain-14C-labeled 2,4-D was determined in samples collected in May. 2,4-D (17.1 mCi mmol−1; radiochemical purity, >95%; Izotop, Hungary) dissolved in deionized water (∼500 μl) was added to three ice cores (microcosms 1 to 3) at an initial activity of ∼50,000 dpm, corresponding to ∼1.32 nmol of 2,4-D, so that it covered most of the core surface. The samples were incubated for a total of 529 days, with three additions (marked microcosms 1a to 1c, 2a to 2c, and 3a to 3c) of side chain-labeled 2,4-D made at the beginning (day 0) and then after 137 and 323 days in order to test the reproducibility of the mineralization results (Table 1). In the second experiment, the difference between the mineralization rates of side chain-labeled (microcosms 4 to 6) and ring-labeled (microcosms 7 to 9) 2,4-D was determined for samples collected in August. Side chain-14C-labeled 2,4-D (17.1 mCi mmol−1) and ring-14C-labeled 2,4-D (18.2 mCi mmol−1) (both with a radiochemical purity of >95%; Izotop, Hungary) dissolved in deionized water were added to the ice cores at an initial activity of ∼50,000 dpm, corresponding to ∼1.32 nmol of side chain-labeled and ∼1.24 nmol of ring-labeled 2,4-D. The microcosms were then incubated for 451 days (Table 1). Controls were prepared from frozen deionized water and autoclaved ice samples and incubated alongside the experimental samples. Mineralization of 2,4-D was measured as the amount of 14CO2 trapped in 0.5 M NaOH, as previously described (30). After the experiments, the whole ice cores were melted and DNA was extracted.
Table 1.
Substrate additions and 2,4-D mineralization rates in long-term laboratory incubations of surface ice cores from GrIS and estimated 2,4-D degradation potential on the surface of GrIS
| Microcosma | No. of days | Amt of 2,4-D added |
Amt of 2,4-D mineralized (nmol) | tfdA gene copy no. (104) | 2,4-D degradation potential |
||
|---|---|---|---|---|---|---|---|
| dpm | nmol | nmol day−1 m−2 | ng C day−1 m−2 | ||||
| 1a | 133 | 49,234 | 1.30 | 0.17 | <1 | 0.65 | 16 |
| 1b | 166 | 50,338 | 1.33 | 0.12 | <1 | 0.38 | 9.0 |
| 1c | 206 | 46,802 | 1.23 | 0.13 | <1 | 0.31 | 7.4 |
| 2a | 133 | 49,234 | 1.30 | 0.82 | <1 | 3.07 | 74 |
| 2b | 166 | 50,338 | 1.33 | 0.53 | <1 | 1.60 | 38 |
| 2c | 206 | 46,802 | 1.23 | 0.46 | <1 | 1.12 | 27 |
| 3a | 133 | 49,234 | 1.30 | 0.56 | <1 | 2.09 | 50 |
| 3b | 166 | 48,308 | 1.27 | 0.35 | <1 | 1.05 | 25 |
| 3c | 206 | 46,802 | 1.23 | 0.48 | <1 | 1.16 | 28 |
| Control a | 133 | 49,234 | 1.30 | 0.13 | NDb | 0.49 | 12 |
| Control b | 166 | 48,308 | 1.27 | 0.14 | ND | 0.42 | 10 |
| Control c | 206 | 46,802 | 1.23 | 0.11 | ND | 0.27 | 6.5 |
| 4 | 451 | 48,252 | 1.27 | 0.22 | <1 | 0.24 | 5.8 |
| 5 | 451 | 48,252 | 1.27 | 0.51 | <1 | 0.57 | 14 |
| 6 | 451 | 48,252 | 1.27 | 0.91 | <1-1.7 | 1.00 | 24 |
| Control (4–6) | 451 | 48,252 | 1.27 | 0.25 | ND | 0.28 | 6.8 |
| 7 | 451 | 53,358 | 1.32 | 0.29 | 1.5-3.1 | 0.32 | 31 |
| 8 | 451 | 53,358 | 1.32 | 0.13 | <1 | 0.14 | 13 |
| 9 | 451 | 53,358 | 1.32 | 0.23 | <1 | 0.26 | 25 |
| Control (7–9) | 451 | 53,358 | 1.32 | 0.13 | ND | 0.15 | 14 |
a, b, and c, 1st, 2nd, and 3rd additions, respectively.
ND, not determined.
Quantitative PCR.
DNA was extracted from the ice cores used in both experiments. The whole ice cores were melted under sterile conditions, the meltwater was centrifuged at 22,000 × g for 10 min, and the supernatant was discarded. DNA was then extracted from the pellet as described above. Real-time PCR quantification of the entire known diversity of the tfdA gene and melting curve analysis were performed with the tfdA 215-bp primer set as previously described (3). Standards with known quantities of the bacterium Cupriavidus necator JMP134 possessing the class I tfdA gene were prepared by addition of 100 μl of washed 10-fold dilutions of C. necator JMP134 cells in 0.015 M phosphate buffer (pH 7.4) to produce final cell concentrations of 3 × 107, 3 × 106, 3 × 105, 3 × 104, and 3 × 103 CFU ml−1. qPCRs were performed in triplicate on all DNA samples, and confirmatory melting curve analysis was performed on all qPCR products. All sample and reagent handling was done in a separate clean lab in order to prevent contamination. The detection limit was 1 × 104 of gene copies per sample (a whole melted ice core of ∼400 ml).
Nucleotide sequence accession numbers.
The sequences of the OTUs have been deposited in GenBank under accession numbers JQ407490 to JQ407508.
RESULTS
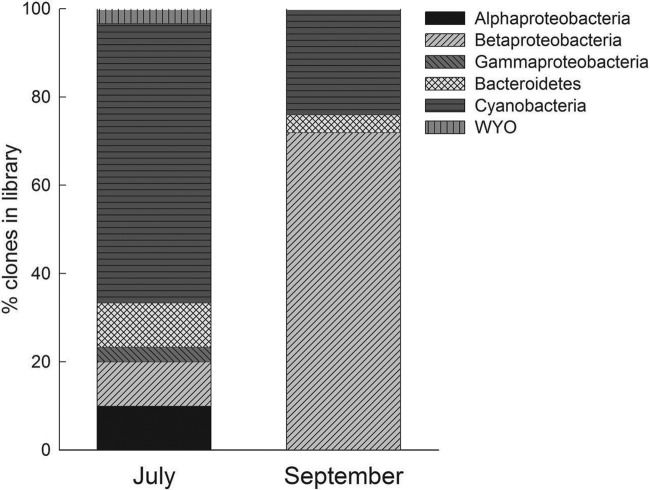
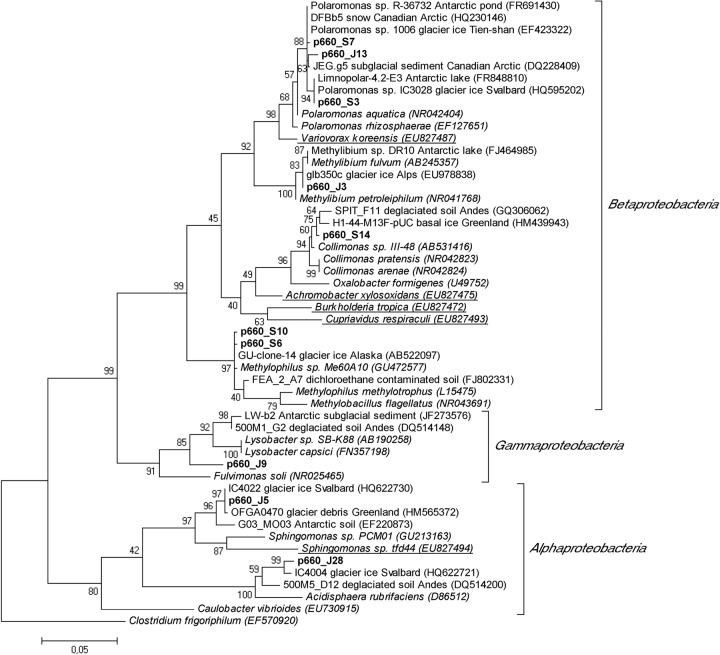
No DNA could be extracted from the particles in surface ice samples collected in April 2006, as evidenced by the complete lack of PCR amplification for these samples. Clone libraries constructed from the July and September samples are shown in Fig. 1. Eleven unique OTUs were found in the July library. Cyanobacteria, including the genera Nostoc and Phormidium, were the dominant component of the supraglacial microbial community (3 OTUs, ∼60% of clones in the library). Alpha- and Betaproteobacteria and Bacteroidetes (Hymenobacter) were also well represented, and single clones from Gammaproteobacteria and the candidate division WYO were detected. In the September clone library, 10 unique OTUs were found, with Betaproteobacteria being dominant (5 OTUs, ∼70% of clones in the library). Cyanobacteria and Bacteroidetes were also found in the September library. All the detected taxa, with the exception of clone J8, were most closely related to taxa from ice and/or snow environments in polar or alpine regions. Figure 2 shows a phylogenetic analysis of all proteobacteria detected. The detected Betaproteobacteria clustered within the families Comamonadaceae (associated with the genera Polaromonas and Methylibium) and Oxalobacteraceae (Collimonas) within the Burkholderiales and in the order Methylophilales (Methylophilus). None of the clones appeared to cluster within the genera of Betaproteobacteria containing known 2,4-D degraders, such as Variovorax, Burkholderia, Cupriavidus, or Achromobacter (Fig. 2). The one gammaproteobacterium detected clustered into the order Xanthomonadales and thus was distant from other known 2,4-D-degrading Gammaproteobacteria (e.g., Pseudomonas spp.). In the Alphaproteobacteria, one clone (J28) clustered within the Acetobacteraceae, while the phylogenetic position of the other clone (J5) was not clear; however, both appeared to be distant from any known 2,4-D degraders in the genus Sphingomonas (Fig. 2).
Fig 1.
Composition of the 16S rRNA gene clone libraries prepared from the July (30 clones; coverage, 63%) and September (25 clones; coverage, 60%) ice samples.
Fig 2.
Phylogenetic analysis of proteobacterial clones from the surface ice on the GrIS (beginning with “p660,” in bold). The maximum likelihood tree was constructed on the basis of 640-base-pair-long 16S rRNA gene sequences and rooted with Clostridium frigoriphilum. Underlined sequences denote known 2,4-D degraders (from Bælum et al. [4]).
Figures 3 and 4 show the mineralization of 14C-2,4-D in the surface ice cores, quantified as the percentage of the initial activity (in dpm) accumulated as trapped 14CO2. The percent dpm values are converted into nmol of 2,4-D in Table 1. In the first experiment (microcosms 1 to 3), the maximum cumulative mineralization of side chain-labeled 2,4-D in the samples collected in May 2006 was 63%, corresponding to 0.82 nmol, after 133 days during the first addition. The average cumulative mineralization rates were 40% ± 25% (mean ± standard deviation [SD], n = 3) in the first incubation (after 133 days of incubation), 26% ± 15% in the second incubation (166 days), and 29% ± 16% in the third incubation (206 days). The cumulative mineralization rates in the deionized water control incubation were 10%, 11%, and 9% in the three consecutive incubations, respectively, and the cumulative mineralization of the autoclaved ice control was 11% during the last 206-day incubation (Fig. 3). The great differences in mineralization of 2,4-D between the individual ice cores were reproduced in the repeated additions, with the mineralization of one of the ice cores not being significantly different from that for the blank incubations (Fig. 3).
Fig 3.
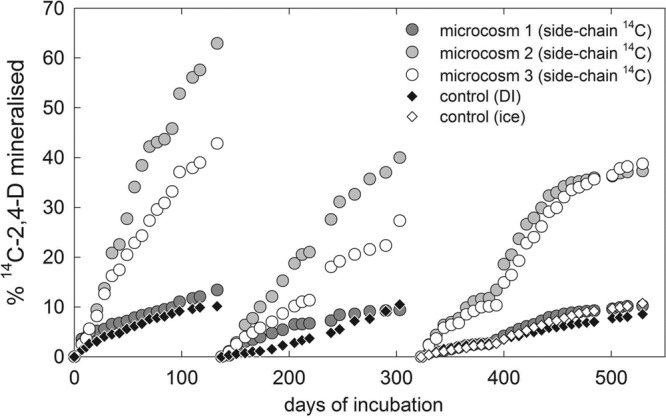
Mineralization of side chain-14C-labeled 2,4-D in samples collected in May. 2,4-D was added to three ice cores (microcosms 1 to 3) at an initial activity of ∼50,000 dpm, corresponding to ∼1.32 nmol of 2,4-D. The control consisted of frozen autoclaved deionized water (DI) in all three incubations, with an additional control of refrozen autoclaved ice (ice) in the last incubation.
Fig 4.
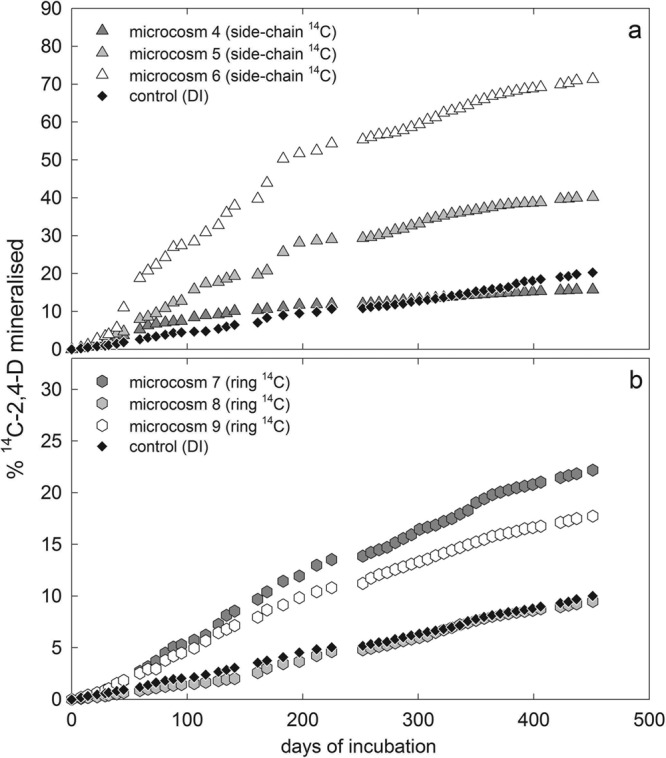
Mineralization of side chain-14C-labeled (microcosms 4 to 6) and ring-14C-labeled (microcosms 7 to 9) 2,4-D in samples collected in August. 2,4-D was added to the ice cores at an initial activity of ∼50,000 dpm, corresponding to ∼1.32 nmol of side chain-labeled 2,4-D and ∼1.24 nmol 2,4-D of ring-labeled 2,4-D. The control consisted of frozen autoclaved deionized water (DI) in all three incubations.
Figure 4 shows a comparison between microbial mineralization of side chain- and ring-labeled 2,4-D in the ice cores collected in August 2006 after 451 days of incubation (microcosms 4 to 9). The maximum cumulative mineralization of the side chain-labeled 2,4-D was 71%, corresponding to 0.91 nmol, with an average of 43% ± 27%. Two out of the three incubations showed a mineralization rate significantly higher than that of the control, which reached ∼20% (Fig. 4a). Mineralization rates of ring-labeled 2,4-D were lower than those of side chain-labeled substrate during incubation, with a maximum of 22% (0.29 nmol) and an average of 16% ± 6% of the initial activity. Two out of the three incubations showed a mineralization rate significantly higher than that of the control (∼10%; Fig. 4b).
The tfdA gene was detected by qPCR only in microcosms 6 (side chain-labeled 2,4-D) and 7 (ring-labeled 2,4-D). In microcosm 6, the gene was detected in only 2 out of 3 replications, and the gene copy numbers were 1.7 × 104 and 1.1 × 104 copies per sample, respectively, which are near the limit of detection of the qPCR assay. In microcosm 7, the tfdA gene was detected in all three replications, with gene copy numbers of 2.9 × 104, 3.1 × 104, and 1.5 ×104 per sample, respectively (Table 1).
The estimated potential for 2,4-D degradation rate per unit area of the ice surface is shown in Table 1, expressed as nmol of 2,4-D per unit area (m2) per day and as ng C m−2 s−1. We used the following assumptions for our estimates. First, the surface area of the incubated cores was 0.002 m2. Second, the degradation rates were linear over time. This is a simplification; however, it would be difficult to apply nonlinear fits derived from closed-system incubations on in situ conditions. Third, for the C estimates, we assume that 2 atoms of C are metabolized when side chain-labeled 2,4-D is mineralized, while we assume that all 8 atoms are metabolized when ring-labeled 2,4-D is mineralized. The degradation potential of 2,4-D based on side chain-labeled 2,4-D data is then estimated to be 1.1 ± 0.83 nmol m−2 day−1 (mean ± SD; n = 9), corresponding to 26 ± 20 ng C m−2 day−1; the degradation potential based on ring-labeled 2,4-D mineralization is 0.24 ± 0.09 nmol m−2 day−1 (mean ± SD; n = 3), corresponding to 23 ± 9.1 ng C m−2 day−1. The mean degradation potential using all the data (n = 12) is estimated to be 0.93 ± 0.81 nmol of 2,4-D per m2 per day or 26 ± 18 ng C m−2 day−1.
DISCUSSION
The surface of the GrIS is known to harbor metabolically active microbial communities that are involved in a number of carbon cycling processes (16, 41). Our results suggest that the supraglacial community on the GrIS contains low numbers of active microbes capable of degradation of the herbicide 2,4-dichlorophenoxyacetic acid in situ.
The bacterial taxa detected in the surface ice samples were similar to those from other icy environments, including glacier ice (23, 37, 50), subglacial sediment (38, 42, 49), snow (15, 24), and recently deglaciated soil (32, 36). The low diversity and the similarity to other icy environment clones suggest that the bacterial community on the GrIS is selected from a pool of propagules deposited on the surface of the ice sheet, based on the level of adaptation to the conditions in the surface ice. The dominance of cyanobacteria in the July sample is consistent with their high numbers and activity during the peak summer season on the GrIS (16, 41). The shift from the dominance of cyanobacterial to that of mostly heterotrophic Betaproteobacteria over the course of the season may suggest accumulation of OC from autochthonous and/or allochthonous sources (40, 41), thus making the environment conducive for heterotrophic growth. The dominance of Betaproteobacteria, especially the family Comamonadaceae, has also been reported from some glacial environments (25, 38, 42, 48, 49). Bacteroidetes, particularly the genus Hymenobacter, are also often found in glacial environments (23, 25).
The fact that no taxa closely related to known 2,4-D degraders were detected in the clone libraries has two possible explanations. First, some of the bacteria detected in the GrIS samples are novel 2,4-D degraders, and second, the detected clones represent only the dominant members of the community and so the actual 2,4-D degraders, though present in the community, are missing in the clone libraries. While we cannot exclude the former explanation, the latter is more likely, for reasons further discussed below.
The cumulative mineralization of ring-labeled 2,4-D in the surface ice (∼15 to 20% of the initial 14C mineralized after >400 days) was lower than that in other environments where 2,4-D degradation has been investigated, such as agricultural soil (2, 20, 33), although the incubation period was at least an order of magnitude longer than that in those studies. Several factors are likely responsible for this result. First, the low temperature on the glacier surface, simulated in the incubations, may slow down the metabolism of supraglacial microbes compared to the rate for their counterparts in temperate regions. We assume that most mineralization activity in our experiments took place in the melting surface of the ice core, where the temperature was ∼0°C, thus mimicking the conditions on the very surface of the ice sheet during summer (1). It is also possible that some degradation occurred within the ice where the temperature was between −7.5 and −4°C, since microbial activity at subzero temperatures in glacier ice has been reported on the basis of chemical analysis of ice cores and laboratory experiments (5, 7, 46). Second, the concentration of substrate (2,4-D) in the system was very low compared to the concentrations typically used in previous studies, and so a first-order kinetic may be expected (20). Third, active microbes possessing the required degradation pathway are likely present in the surface ice on the GrIS in very low numbers. This is supported by the absence of clones similar to known 2,4-D degraders in the clone libraries and by the fact that the functional gene tdfA involved in the degradation pathway was detected only in incubations showing the highest 2,4-D degradation rates (Fig. 3). We suggest that the numbers of copies of tdfA, and thus the number of likely 2,4-D degraders in the ice cores, were around the detection limit of the qPCR method, but due to the high sensitivity of the 14C substrate method, their activity could still be observed and quantified. However, the presence of another gene involved in 2,4-D degradation that is not detected by the primer set used cannot be ruled out. The low numbers of 2,4-D degraders are also likely responsible for the great differences in degradation rates between replicates, due to the stochastic distribution of their cells in the ice cores. This is also supported by the results of a recent study in which high spatial heterogeneity in the degradation rates of xenobiotics as opposed to easily degradable compounds, such as glucose, was detected (18).
The difference in the mineralization rates of side chain- and ring-labeled 2,4-D may be the result of the mechanism of 2,4-D degradation. It is possible that the acid side chain can be attacked and metabolized by a wider spectrum of microbes and that fewer microorganisms can attack the aromatic ring. Thus, the difference in degradation of side chain- and ring-labeled 2,4-D may reflect the proportion of 2,4-D degraders that can also attack the ring.
Using the incubation data, we can also crudely estimate the potential 2,4-D degradation rate per unit area of the ice surface (Table 1). As the melting season can be up to 130 days on the surface of the GrIS margin (44), the maximum potential for the degradation of 2,4-D on the surface of the GrIS can be estimated to be 120 nmol 2,4-D per m2 per year, or ∼3 μg Cm−2 year−1. Although 2-methyl-4-chlorophenoxyacetic acid (MCPA), a phenoxyacetic acid herbicide similar to 2,4-D, has been detected in the snow on the surface of the GrIS at a concentration of 0.12 μg liter−1 (21), no consistent data on rates of deposition of 2,4-D or similar compounds on the GrIS are available. Therefore, it is not yet possible to quantitatively evaluate the role of the supraglacial microbes in the removal of this sort of pollutant from the Greenland supraglacial environment.
In summary, we show that the supraglacial microbial community on the Greenland ice sheet contains microbes capable of degrading 2,4-D during long-term incubation simulating in situ conditions. These degraders are likely present in very low numbers but nonetheless can mineralize 2,4-D at a rate of up to 1 nmol m−2 day−1, equivalent to ∼26 ng C m−2 day−1. Thus, the ice sheet should not be considered a mere reservoir of all atmospheric contaminants. Indeed, it is likely that some deposited organic compounds (xenobiotic and otherwise) will be removed from the system by mineralization or other biodegradative activities before their potential release to soils and waters due to the accelerated melting of the ice sheet.
ACKNOWLEDGMENTS
This research project was funded by Danish Research Council (FNU 10-085274).
We thank Jens Bisgaard (GEUS) for technical help and three anonymous reviewers for their comments.
Footnotes
Published ahead of print 11 May 2012
REFERENCES
- 1. Anesio AM, Laybourn-Parry J. 2012. Glaciers and ice sheets as a biome. Trends Ecol. Evol. 27:219–225 [DOI] [PubMed] [Google Scholar]
- 2. Bælum J, et al. 2008. Direct analysis of tfdA gene expression by indigenous bacteria in phenoxy acid amended agricultural soil. ISME J. 2:677–687 [DOI] [PubMed] [Google Scholar]
- 3. Bælum J, Jacobsen CS. 2009. TaqMan probe-based real-time PCR assay for detection and discrimination of class I, II, and III tfdA genes in soils treated with phenoxy acid herbicides. Appl. Environ. Microbiol. 75:2969–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bælum J, Jacobsen CS, Holben WE. 2010. Comparison of 16S rRNA gene phylogeny and functional tfdA gene distribution in thirty-one different 2,4-dichlorophenoxyacetic acid and 4-chloro-2-methylphenoxyacetic acid degraders. Syst. Appl. Microbiol. 33:67–70 [DOI] [PubMed] [Google Scholar]
- 5. Bakermans C, Skidmore ML. 2011. Microbial metabolism in ice and brine at −5°C. Environ. Microbiol. 13:2269–2278 [DOI] [PubMed] [Google Scholar]
- 6. Bhatia MP, Das SB, Longnecker K, Charette MA, Kujawinski EB. 2010. Molecular characterization of dissolved organic matter associated with the Greenland ice sheet. Geochim. Cosmochim. Acta 74:3768–3784 [Google Scholar]
- 7. Campen RK, Sowers T, Alley RB. 2003. Evidence of microbial consortia metabolizing within a low-latitude mountain glacier. Geology 31:231–234 [Google Scholar]
- 8. Christner BC, Kvitko BH, Reeve JN. 2003. Molecular identification of Bacteria and Eukarya inhabiting an Antarctic cryoconite hole. Extremophiles 7:177–183 [DOI] [PubMed] [Google Scholar]
- 9. Don RH, Weightman AJ, Knackmuss HJ, Timmis KN. 1985. Transposon mutagenesis and cloning analysis of the pathways for degradation of 2,4-dichlorophenoxyacetic acid and 3-chlorobenzoate in Alcaligenes eutrophus JMP134(pJP4). J. Bacteriol. 161:85–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Edwards A, et al. 2011. Possible interactions between bacterial diversity, microbial activity and supraglacial hydrology of cryoconite holes in Svalbard. ISME J. 5:150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Foreman CM, Sattler B, Mikucki JA, Porazinska DL, Priscu JC. 2007. Metabolic activity and diversity of cryoconites in the Taylor Valley, Antarctica. J. Geophys. Res. 112:G04S32. [Google Scholar]
- 13. Fukumori F, Hausinger RP. 1993. Purification and characterization of 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase. J. Biol. Chem. 268:24311–24317 [PubMed] [Google Scholar]
- 14. Greer CW, Hawari J, Samson R. 1990. Influence of environmental factors on 2,4-dichlorophenoxyacetic acid degradation by Pseudomonas cepacia isolated from peat. Arch. Microbiol. 154:317–322 [DOI] [PubMed] [Google Scholar]
- 15. Harding T, Jungblut AD, Lovejoy C, Vincent WF. 2011. Microbes in high Arctic snow and implications for the cold biosphere. Appl. Environ. Microbiol. 77:3234–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hodson A, et al. 2010. The cryoconite ecosystem on the Greenland ice sheet. Ann. Glaciol. 51(56):123–129 [Google Scholar]
- 17. Huber T, Faulkner G, Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319 [DOI] [PubMed] [Google Scholar]
- 18. Hybholt TK, Aamand J, Johnsen AR. 2011. Quantification of centimeter-scale spatial variation in PAH, glucose and benzoic acid mineralization and soil organic matter in road-side soil. Environ. Pollut. 159:1085–1091 [DOI] [PubMed] [Google Scholar]
- 19. Itoh K, et al. 2000. Presence of 2,4-D-catabolizing bacteria in a Japanese arable soil that belong to BANA (Bradyrhizobium-Agromonas-Nitrobacter-Afipia) cluster in α-Proteobacteria. Microb. Environ. 15:113–117 [Google Scholar]
- 20. Jacobsen CS, Pedersen JC. 1992. Mineralization of 2,4-dichlorophenoxyacetic acid (2,4-D) in soil inoculated with Pseudomonas cepacia DBO1(pRO101), Alcaligenes eutrophus AEO106(pRO101) and Alcaligenes eutrophus JMP134(pJP4): effects of inoculation level and substrate concentration. Biodegradation 2:253–263 [DOI] [PubMed] [Google Scholar]
- 21. Jacobsen CS, et al. 2012. Global contamination of the Greenlandic icecap with organic contaminants—distribution of contaminants and studies of the microbiological and photochemical degradation. Danmarks og Grønlands Geologiske Undersøgelse Rapport 2012-14. Geological Survey of Denmark and Greenland, Copenhagen, Denmark. [Google Scholar]
- 22. Kamagata Y, et al. 1998. Pristine environments harbor a new group of oligotrophic 2,4-dichlorophenoxy acid-degrading bacteria. Appl. Environ. Microbiol. 63:2266–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Klassen JL, Foght JM. 2011. Characterization of Hymenobacter isolates from Victoria Upper Glacier, Antarctica reveals five new species and substantial non-vertical evolution within this genus. Extremophiles 15:45–57 [DOI] [PubMed] [Google Scholar]
- 24. Larose C, et al. 2010. Microbial sequences retrieved from environmental samples from seasonal Arctic snow and meltwater from Svalbard, Norway. Extremophiles 14:205–212 [DOI] [PubMed] [Google Scholar]
- 25. Liu Y, et al. 2009. Bacterial diversity in the snow over Tibetan Plateau glaciers. Extremophiles 13:411–423 [DOI] [PubMed] [Google Scholar]
- 26. Loos MA. 1975. Phenoxyalkanoic acids, p 1–128. In Kearney PC, Kaufmann DD. (ed), Herbicides: chemistry, degradation and mode of action, vol 1 Marcel Dekker, New York, NY [Google Scholar]
- 27. Margesin R, Zacke G, Schinner F. 2002. Characterization of heterotrophic microorganisms in alpine glacier cryoconite. Arct. Antarct. Alp. Res. 34:88–93 [Google Scholar]
- 28. Masclet P, Hoyau V, Jaffrezo JL, Cachier H. 2000. Polycyclic aromatic hydrocarbon deposition on the ice sheet of Greenland. Part I. Superficial snow. Atmos. Environ. 34:3195–3207 [Google Scholar]
- 29. McConnell JR, et al. 2007. 20th-century industrial black carbon emissions altered Arctic climate forcing. Science 317:1381–1384 [DOI] [PubMed] [Google Scholar]
- 30. Mortensen SK, Jacobsen CS. 2004. Influence of frozen storage on herbicide degradation capacity in surface and subsurface sandy soils. Environ. Sci. Technol. 38:6625–6632 [DOI] [PubMed] [Google Scholar]
- 31. Müller MD, Buser H-R. 1997. Conversion reactions of various phenoxyalkanoic acid herbicides in soil. 1. Enantiomerization and enantioselective degradation of the chiral 2-phenoxypropionic acid herbicides. Environ. Sci. Technol. 31:1953–1959 [Google Scholar]
- 32. Nemergut DR, et al. 2007. Microbial community succession in an unvegetated, recently deglaciated soil. Microb. Ecol. 53:110–122 [DOI] [PubMed] [Google Scholar]
- 33. Ou L-T. 1984. 2,4-D degradation and 2,4-D degrading microorganisms in soils. Soil Sci. 137:100–107 [Google Scholar]
- 34. Pritchard HD, Arthern RJ, Vaughan DG, Edwards LA. 2009. Extensive dynamic thinning on the margins of the Greenland and Antarctic ice sheets. Nature 461:971–975 [DOI] [PubMed] [Google Scholar]
- 35. Rignot E, Velicogna I, van den Broeke MR, Monaghan A, Lenaerts J. 2011. Acceleration of the contribution of the Greenland and Antarctic ice sheets to sea level rise. Geophys. Res. Lett. 38:05503 doi:10.1029/2011GL046583. [Google Scholar]
- 36. Schmidt SK, et al. 2009. Microbial activity and diversity during extreme freeze-thaw cycles in periglacial soils, 5400 m elevation, Cordillera Vilcanota, Peru. Extremophiles 13:807–816 [DOI] [PubMed] [Google Scholar]
- 37. Simon C, Wiezer A, Strittmatter AW, Daniel R. 2009. Phylogenetic diversity and metabolic potential revealed in a glacier ice metagenome. Appl. Environ. Microbiol. 75:7519–7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Skidmore M, Anderson SP, Sharp M, Foght J, Lanoil BD. 2005. Comparison of microbial community composition in two subglacial environments reveals a possible role for microbes in chemical weathering processes. Appl. Environ. Microbiol. 71:6986–6997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Slater JF, Currie LA, Dibb JE, Benner BA., Jr 2002. Distinguishing the relative contribution of fossil fuel and biomass combustion aerosols deposited at Summit, Greenland through isotopic and molecular characterization of insoluble carbon. Atmos. Environ. 36:4463–4477 [Google Scholar]
- 40. Stibal M, et al. 2010. Organic matter content and quality in supraglacial debris across the ablation zone of the Greenland ice sheet. Ann. Glaciol. 51(56):1–8 [Google Scholar]
- 41. Stibal M, et al. 2012. Environmental controls on microbial abundance and activity on the Greenland ice sheet: a multivariate analysis approach. Microb. Ecol. 63:74–84 [DOI] [PubMed] [Google Scholar]
- 42. Stibal M, Hasan F, Wadham JL, Sharp MJ, Anesio AM. 2012. Prokaryotic diversity in sediments beneath two polar glaciers with contrasting organic carbon substrates. Extremophiles 16:255–265 [DOI] [PubMed] [Google Scholar]
- 43. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tedesco M, et al. 2011. The role of albedo and accumulation in the 2010 melting record in Greenland. Environ. Res. Lett. 6:014005 doi:10.1088/1748-9326/6/1/014005. [Google Scholar]
- 45. Tonso NL, Matheson VG, Holben WE. 1994. Polyphasic characterization of a suite of bacterial isolates capable of degrading 2,4-D. Microb. Ecol. 30:3–24 [DOI] [PubMed] [Google Scholar]
- 46. Tung HC, Price PB, Bramall NE, Vrdoljak G. 2006. Microorganisms metabolizing on clay grains in 3-km-deep Greenland basal ice. Astrobiology 6:69–86 [DOI] [PubMed] [Google Scholar]
- 47. Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xiang S-R, Shang T-C, Chen Y, Jing Z-F, Yao T. 2009. Dominant bacteria and biomass in the Kuytun 51 Glacier. Appl. Environ. Microbiol. 75:7287–7290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yde JC, et al. 2010. Basal ice microbiology at the margin of the Greenland ice sheet. Ann. Glaciol. 51(56):71–79 [Google Scholar]
- 50. Zhang XF, Yao TD, Tian LD, Xu SJ, An LZ. 2008. Phylogenetic and physiological diversity of bacteria isolated from Puruogangri ice core. Microb. Ecol. 55:476–488 [DOI] [PubMed] [Google Scholar]