Abstract
The diversity of gene cassette promoters in class 1 integrons was investigated in 47 strains isolated from wastewaters. The weak PcW and PcH1 variants predominated, suggesting that, similar to clinical environments, high rates of gene cassette recombination, rather than high expression of gene cassettes, have been preferentially selected in wastewaters.
TEXT
Integrons are natural gene capture and expression systems, being important players in bacterial adaptation (1). Five classes of resistance integrons have been described, with class 1 being the most prevalent. In class 1 integrons, the gene cassette Pc promoter is located within the integrase intI1 gene (3), and five predominant Pc variants possessing different expression efficiencies, PcW, PcH1, PcWTGN-10, PcH2, and PcS from the weakest to the strongest, have been identified (5). In ∼10% of class 1 integrons, a second promoter (P2) is also present, created by the insertion of a GGG triplet within the attI1 recombination site that optimizes the spacing between the −35 and −10 boxes (from 14 nucleotides [nt] to 17 nt) (5). Usually, the presence of P2 compensates for the weak strength of Pc, enhancing gene cassette expression without interfering with the expression of the integrase gene (4, 5).
Altogether, more than 20 Pc-P2 combinations have been identified in class 1 integrons, regardless of their origin (5, 15). The most frequent configurations are as follows, according to their strength (5): PcW < PcW-P2 < PcH1 < PcH1-P2 < PcWTGN-10-P2 < PcWTGN-10 < PcH2 < PcS-P2 < PcS.
It has also been shown that promoter strength inversely correlates with the integrase gene expression and activity. On one hand, the weaker the promoter, the more active the integrase excision activity (5). On the other hand, the Pc promoter interferes with the level of intI1 transcription, although it depends on the Pc variant: the strong PcS variant prevents intI1 expression, in contrast to the other variants (4). Therefore, the Pc-P2 promoter configurations may give insights not only regarding the level of expression of the gene cassettes but also for the ability of the class 1 integrons to rearrange gene cassette arrays.
Wastewaters have been reported as important reservoirs of integrons and gene cassettes (8–11, 13). In addition, it has been shown that the type of effluent affects both the prevalence and the diversity of gene cassette arrays (10). In particular, a slaughterhouse's effluents have been shown to possess an increased prevalence of integron-carrying bacteria comparing to domestic wastewaters (8, 10). Despite that, the diversity of gene cassette arrays present was found to be higher in bacteria isolated from domestic effluents (10), probably as a result of different selective pressures shaping the diversity of bacterial communities inhabiting those systems (9, 10).
Here we investigated the diversity of gene cassette Pc promoters in 47 class 1 integrons detected in the 46 strains (one strain possessing two integrons) belonging to Enterobacteriaceae and Aeromonadaceae that were isolated from those two distinct wastewater environments: urban wastewaters (n = 23), consisting mostly of domestic discharges, and a slaughterhouse's wastewater (n = 24), consisting of discharges with animal origin. Bacterial isolation and identification, molecular typing, and characterization of integron variable regions have been described in detail in previous studies (8, 10). The characterization of the Pc-P2 region was performed by amplification and sequencing with the primers intI1F (5′-CCTCCCGCACGATGATC-3′) and 5CSrevcompl (5′-CTTGCTGCTTGGATGCC-3′) or by amplification of the complete integron and subsequent sequencing by primer walking as previously described (8, 10).
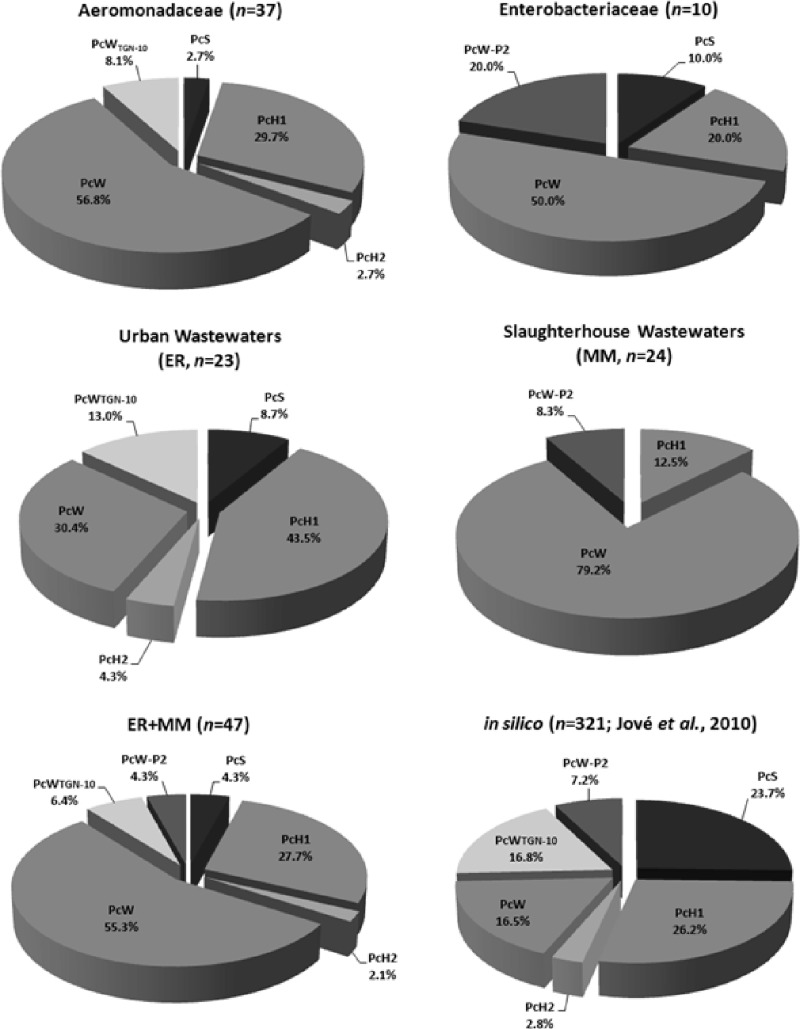
Among 47 class 1 integrons, six different Pc-P2 configurations were detected: PcW, PcW-P2, PcH1, PcWTGN-10, PcH2, and PcS (Fig. 1). The results obtained showed the predominance of weak PcW and PcH1 variants in both types of effluents. Nevertheless, the diversity of the Pc-P2 configurations was dependent on the type of wastewater (χ2 test; P < 0.001): PcW was significantly prevalent in the slaughterhouse's collection, whereas the PcH1 variant prevailed in the urban wastewater (Fig. 1).
Fig 1.
Distribution of the different Pc variants detected in this study among different taxa (Aeromonas spp. and Enterobacteriaceae) and sources (MM, slaughterhouse wastewaters; ER, urban wastewaters); previously reported occurrences based on GenBank in silico analyses (5) are also presented.
A higher diversity of Pc variants was observed in integrons from urban wastewater isolates: five different Pc-P2 configurations were detected, namely, PcH1 (44%), PcW (30%), PcWTGN-10 (13%), PcS (9%), and PcH2 (4%). In integrons from the slaughterhouse's wastewaters, gene cassette promoters were limited to three configurations: PcW (79%), PcH1 (13%), and PcW-P2 (8%). Occurrence of the PcH1 variant in integrons from urban wastewaters, as well as the PcW variant in animal wastewaters, was much higher than that previously reported from in silico studies (26.2% and 16.5%, respectively), though differences may be related to the exclusion of identical arrays made in the in silico analysis (5).
Previous studies concerning clinical environments identified a high predominance of weak PcW variants in Salmonella (6, 14), and PcW and PcH1 variants also prevailed in Escherichia coli strains from human and animal origins (2), although the authors did not discriminate between PcW and PcWTGN-10 (15). In this study, the control of gene cassette expression was also associated with weak PcW and PcH1 variants in both wastewaters, suggesting the existence of a dynamic gene cassette pool in these environments that is similar to what has been described for clinical settings (2, 6, 14).
As shown in Table 1, different Pc variants were not species specific, as previously reported (12, 15). Also, different promoters were found in integrons with identical gene cassette arrays (e.g., aacA4-cr-blaOXA-1-catB3-arr3; aadA2). Nevertheless, the dfrA1-aadA1 gene cassette array was always associated with the PcW promoter. As reported in previous studies, it is thought that this array is considerably stable and that its transfer occurs by the mobilization of the complete integron structure in larger elements rather than by individual resistance gene cassettes (7, 8).
Table 1.
Promoter configurations determined in this study
| Promoter configuration | Gene cassette arraya | Organism | Strain designationb | Resistance phenotypec | Putative integron locationd | GenBank accession no. |
|---|---|---|---|---|---|---|
| PcS | ND | Klebsiella oxytoca | ER.1.13 | AMP, ERY, NAL, STR | C | JN837680 |
| orfER.1.7::ISAs12-aadA13 | Aeromonas salmonicida | ER.1.7 | AMP, CEF, NAL, STR | C | HQ170513 | |
| PcH2 | blaGES-7-aacA4 | Aeromonas media | ER.1.8 | AMP, CEF, KAN, NAL, STR (CAZ, GEN) | C | HQ170511 |
| PcWTGN-10 | aacA4-CR-blaOXA-1-catB3-arr3 | Aeromonas allosaccharophila | ER.1.4 | AMP, CEF, ERY, GEN, KAN, NAL, STR (CIP) | C, P | HQ170510 |
| aacA4-CR-blaOXA-1-catB3-arr3 | Aeromonas media | ER.1.25 | AMP, CEF, CIP, ERY, NAL | C | HQ170516 | |
| dfrA12-orfF-aadA2 | Aeromonas media | ER.1.1 | AMP, CEF, ERY, KAN, NAL, STR, STX | C, P | FJ460175 | |
| PcH1 | aacA4-CR-blaOXA-1-catB3-arr3 | Aeromonas allosaccharophila | ER.1.16 | AMP, CEF, ERY, GEN, KAN, NAL, STR, TET | C | HQ170517 |
| ND | Enterobacter cloacae | ER.1.10 | AMP, CEF, ERY (STR) | C, P | JN837679 | |
| catB8-aadA1 | Aeromonas allosaccharophila | ER.1.16 | AMP, CEF, ERY, GEN, KAN, NAL, STR, TET | C | HQ170518 | |
| blaOXA-2-aadA1-blaOXA-2-gcuD | Aeromonas caviae | ER.1.26 | AMP, CEF, NAL, STR (ERY) | C | HQ170515 | |
| aadA1 | Aeromonas jandaei | MM.1.24 | AMP, CEF, ERY, IPM, STR | C | JQ326986 | |
| aadA1 | Aeromonas media | ER.1.5 | AMP, CEF, ERY, KAN, NAL, STR, STX (CHL) | C | FJ460176 | |
| aadA2 | Aeromonas media | ER.1.11 | AMP, CEF, NAL, STR (ATM, STX, ERY) | C | FJ460179 | |
| aacA4-catB3-blaOXA-10-aadA1 | Aeromonas media | ER.1.17 | AMP, CEF, KAN, NAL, STR, TET (STX, ERY) | C | HQ170514 | |
| catB8-aadA17 | Aeromonas media | ER.1.18 | AMP, CAZ, CEF, NAL, STR | C | FJ460181 | |
| catB3-aadA1 | Aeromonas sp. | MM.1.6 | AMP, ERY, IPM, STR, TET (CEF, STX) | C | JQ326975 | |
| aadA1 | Aeromonas veronii | ER.1.24 | AMP, CEF, ERY, KAN, NAL, STR | C | FJ460183 | |
| aadA2 | Aeromonas veronii | MM.1.10 | AMP (ERY, STR, TET) | C | EU089667 | |
| dfrA17-aadA5 | Shigella sp. | ER.1.23 | AMP, CEF, CIP, ERY, NAL, STR, STX, TET | C | FJ460182 | |
| PcW-P2 | aadA1 | Escherichia coli | MM.1.15 | ERY, STR, TET | C | JQ326978 |
| aadA1 | Escherichia coli | MM.1.9 | CEF, ERY, STR, TET | C | EU089666 | |
| PcW | dfrA1-aadA1 | Aeromonas salmonicida | MM.1.19 | AMP, CEF, STR, STX, TET (ERY) | C | JQ326982 |
| dfrA1-aadA1 | Aeromonas salmonicida | MM.1.3 | AMP, CEF, STR, STX (ERY) | C, P | JQ326972 | |
| dfrA1-aadA1 | Aeromonas salmonicida | MM.1.4 | AMP, CEF, STR, STX, TET (ERY) | C | JQ326973 | |
| dfrA1-aadA1 | Aeromonas salmonicida | MM.1.23 | AMP, CEF, STR, STX, TET (ERY) | C | JQ326985 | |
| dfrA1-aadA1 | Aeromonas salmonicida | MM.1.29 | AMP, CAZ, CHL, CEF, ERY, GEN, IPM, STR, STX, TET | C | JQ326989 | |
| ND | Aeromonas sp. | ER.1.21 | AMP, CAZ, CEF, ERY, NAL | C, P | JN837681 | |
| dfrA1-aadA1 | Aeromonas sp. | MM.1.8 | AMP, CEF, STR, STX, TET (ERY) | C | JQ326976 | |
| dfrA1-aadA1 | Escherichia coli | MM.1.5 | AMP, CEF, ERY, STR, STX, TET | P | JQ326974 | |
| aadA2 | Aeromonas caviae | ER.1.9 | ATM, CAZ, NAL (STR) | C | EU089666 | |
| blaOXA-1-aadA1 | Escherichia coli | MM.1.12 | AMP, ERY, STR, TET (CEF) | C | EU089669 | |
| dfrA1-aadA1 | Escherichia coli | MM.1.11 | ERY, TET, STX (CEF, STR) | C | EU089668 | |
| dfrA1-aadA1 | Escherichia coli | MM.1.13 | AMP, CEF, CHL, ERY, STR, STX, TET | C | JQ326977 | |
| dfrA1-aadA1 | Aeromonas allosaccharophila | ER.1.6 | AMP, CEF, NAL, STR, STX (IPM, ERY) | C | FJ460177 | |
| Empty integron | Aeromonas caviae | ER.1.2 | AMP, CAZ, CEF, ERY, KAN, NAL, STR (CHL, CIP) | C | JN837678 | |
| dcyA | Aeromonas caviae | ER.1.20 | AMP, CEF, NAL | C, P | HQ170512 | |
| Empty integron | Aeromonas media | ER.1.22 | AMP, CEF, CHL, CIP, ERY, NAL, TET (STR, KAN) | C, P | JN837682 | |
| dfrA1-aadA1 | Aeromonas salmonicida | MM.1.2 | AMP, CEF, STR, STX, TET (ERY) | C | JQ326971 | |
| dfrA1-aadA1 | Aeromonas salmonicida | MM.1.16 | AMP, CEF, STR, STX, TET (ERY) | C | JQ326979 | |
| dfrA1-aadA1 | Aeromonas salmonicida | MM.1.17 | AMP, CEF, STR, STX, TET (ERY) | C | JQ326980 | |
| dfrA1-aadA1 | Aeromonas salmonicida | MM.1.18 | AMP, CEF, STR, STX (ERY, TET) | C | JQ326981 | |
| dfrA1-aadA1 | Aeromonas salmonicida | MM.1.20 | AMP, CEF, STR, STX, TET (ERY) | C | JQ326983 | |
| dfrA1-aadA1 | Aeromonas salmonicida | MM.1.22 | AMP, CEF, STR, STX, TET (ERY) | C | JQ326984 | |
| dfrA1-aadA1 | Aeromonas salmonicida | MM.1.26 | AMP, CEF, STR, STX, TET (ERY) | C | JQ326987 | |
| aadA2 | Aeromonas sp. | MM.1.1a | AMP, CEF, CHL, STR, TET (ERY) | C | JQ326970 | |
| ND | Aeromonas veronii | MM.1.27 | AMP (ERY) | C | JQ326988 | |
| Empty integron | Kluyvera cryocrescens | ER.1.27 | AMP, CEF, CIP, ERY, NAL | C, P | JN837683 |
ND, not detected.
MM strains were obtained from the slaughterhouse's wastewaters, whereas ER strains refer to urban wastewaters.
Intermediary susceptibility phenotypes are shown in parentheses. AMP, ampicillin; ATM, aztreonam; CAZ, ceftazidime; CEF, cephalothin; CIP, ciprofloxacin; CHL, chloramphenicol; ERY, erythromycin; GEN, gentamicin; IPM, imipenem; KAN, kanamycin; NAL, nalidixic acid; STR, streptomycin; TET, tetracycline; STX, trimethoprim-sulfamethoxazole.
In summary, the results obtained showed the predominance of weak promoter variants in both types of wastewaters. Although some differences emerged in the distribution of the Pc variants between the two types of wastewaters, they converged in a trend that favors a high rate of recombination of gene cassettes, contributing to genome plasticity. Moreover, in this study, the trend of the Pc variants was more pronounced than those previously reported, which concerned mostly integrons from clinical origin. Nevertheless, we are aware that the number of integrons analyzed in this study is limited and that different methodologies applied among clinical studies may vary.
To the best of our knowledge, this constitutes the first investigation concerning the configuration of gene cassette promoters in class 1 integrons from wastewater environments. More epidemiological studies focusing on the analysis of promoter configurations is necessary to refine data on their distributions and determine to what extent the class 1 integron promoters' ecologies differ according to the wastewater type.
ACKNOWLEDGMENTS
This work was supported by Fundação para a Ciência e a Tecnologia (FCT) through project POCTI/BME/45881/2002 and grants SFRH/BD/19443/2004 and SFRH/BPD/72256/2010 (A. Moura) and SFRH/BPD/63487/2009 (I. Henriques).
We thank António Nogueira (Department of Biology, University of Aveiro) for his assistance in statistical analysis.
Footnotes
Published ahead of print 11 May 2012
REFERENCES
- 1. Cambray G, Guerout AM, Mazel D. 2010. Integrons. Annu. Rev. Genet. 44:141–156 [DOI] [PubMed] [Google Scholar]
- 2. Cocchi S, et al. 2007. Distribution and characterization of integrons in Escherichia coli strains of animal and human origin. FEMS Immunol. Med. Microbiol. 50:126–132 [DOI] [PubMed] [Google Scholar]
- 3. Collis CM, Hall RM. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39:155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guérin E, JovÉ T, Tabesse A, Mazel D, Ploy MC. 2011. High-level gene cassette transcription prevents integrase expression in class 1 integrons. J. Bacteriol. 193:5675–5682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jové T, Da Ré S, Denis F, Mazel D, Ploy MC. 2010. Inverse correlation between promoter strength and excision activity in class 1 integrons. PLoS Genet. 6:e1000793 doi:10.1371/journal.pgen.1000793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lindstedt BA, Heir E, Nygård I, Kapperud G. 2003. Characterization of class I integrons in clinical strains of Salmonella enterica subsp. enterica serovars Typhimurium and Enteritidis from Norwegian hospitals. J. Med. Microbiol. 52:141–149 [DOI] [PubMed] [Google Scholar]
- 7. Martinez-Freijo P, Fluit AC, Schmitz FJ, Verhoef J, Jones ME. 1999. Many class 1 integrons comprise distinct stable structures occurring in different species of Enterobacteriaceae isolated from widespread geographic regions in Europe. Antimicrob. Agents Chemother. 43:686–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moura A, Henriques I, Ribeiro R, Correia A. 2007. Prevalence and characterization of integrons from bacteria isolated from a slaughterhouse wastewater treatment plant. J. Antimicrob. Chemother. 60:1243–1250 [DOI] [PubMed] [Google Scholar]
- 9. Moura A, Henriques I, Smalla K, Correia A. 2010. Wastewater bacterial communities bring together broad-host range plasmids, integrons and a wide diversity of uncharacterized gene cassettes. Res. Microbiol. 161:58–66 [DOI] [PubMed] [Google Scholar]
- 10. Moura A, Pereira P, Henriques I, Correia A. 2012. Novel gene cassettes and integrons in antibiotic resistant bacteria isolated from urban wastewaters. Res. Microbiol. 163:92–100 [DOI] [PubMed] [Google Scholar]
- 11. Pellegrini C, et al. 2011. Occurrence of class 1 and 2 integrons in resistant enterobacteriaceae collected from a urban wastewater treatment plant: first report from central Italy. Microb. Drug Resist. 17:229–234 [DOI] [PubMed] [Google Scholar]
- 12. Peters EDJ, Leverstein-van Hall MA, Box ATA, Verhoef J, Fluit AC. 2001. Novel gene cassettes and integrons. Antimicrob. Agents Chemother. 45:2961–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pignato S, Coniglio MA, Faro G, Weill FX, Giammanco G. 2009. Plasmid-mediated multiple antibiotic resistance of Escherichia coli in crude and treated wastewater used in agriculture. J. Water Health 7:251–258 [DOI] [PubMed] [Google Scholar]
- 14. Schmitz FJ, et al. 2001. Increased prevalence of class I integrons in Escherichia coli, Klebsiella species, and Enterobacter species isolates over a 7-year period in a German University Hospital. J. Clin. Microbiol. 39:3724–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vinué L, Jové T, Torres C, Ploy MC. 2011. Diversity of class 1 integron gene cassette Pc promoter variants in clinical Escherichia coli strains and description of a new P2 promoter variant. Int. J. Antimicrob. Agents 38:526–529 [DOI] [PubMed] [Google Scholar]