Abstract
Glutaminase is an enzyme that catalyzes the hydrolysis of l-glutamine to l-glutamate, and it plays an important role in the production of fermented foods by enhancing the umami taste. By using the genome sequence and expressed sequence tag data available for Aspergillus oryzae RIB40, we cloned a novel glutaminase gene (AsgahA) from Aspergillus sojae, which was similar to a previously described gene encoding a salt-tolerant, thermostable glutaminase of Cryptococcus nodaensis (CnGahA). The structural gene was 1,929 bp in length without introns and encoded a glutaminase, AsGahA, which shared 36% identity with CnGahA. The introduction of multiple copies of AsgahA into A. oryzae RIB40 resulted in the overexpression of glutaminase activity. AsGahA was subsequently purified from the overexpressing transformant and characterized. While AsGahA was located at the cell surface in submerged culture, it was secreted extracellularly in solid-state culture. The molecular mass of AsGahA was estimated to be 67 kDa and 135 kDa by SDS-PAGE and gel filtration chromatography, respectively, indicating that the native form of AsGahA was a dimer. The optimal pH of the enzyme was 9.5, and its optimal temperature was 50°C in sodium phosphate buffer (pH 7.0). Analysis of substrate specificity revealed that AsGahA deamidated not only free l-glutamine and l-asparagine but also C-terminal glutaminyl or asparaginyl residues in peptides. Collectively, our results indicate that AsGahA is a novel peptidoglutaminase-asparaginase. Moreover, this is the first report to describe the gene cloning and purification of a peptidoglutaminase-asparaginase.
INTRODUCTION
Glutaminase (glutamine amidohydrolase, EC 3.5.1.2; Gah) catalyzes the hydrolytic deamidation of l-glutamine, resulting in the production of l-glutamate and ammonium. Glutaminases have been identified in bacteria, yeasts, fungi, and mammals (2, 7, 8), and they play an important role in nitrogen metabolism, including glutaminolysis. Since mitochondrial glutaminase is increased in several tumor types and is frequently upregulated in MYC-transformed cells (4), it is thought to represent a potential chemotherapeutic target (30). Moreover, bacterial glutaminase from Achromobacter was shown to exert antileukemic effects in patients with acute lymphoblastic leukemia or acute myeloid leukemia in a preliminary clinical trial (24), and it has attracted significant attention from the pharmaceutical industry due to its potential applications as an anticancer agent.
In addition, l-glutamate (monosodium glutamate) is an important umami taste factor. Thus, the deamidation of glutamine is a key process in the food industry in order to enhance the umami taste. The deamidation of glutamine in proteins found in food also improves functional properties—such as solubility, viscosity, gelation, fat emulsification, and foaming—by increasing the number of negative charges in the protein (5). Although the deamidation of proteins can be performed either enzymatically or nonenzymatically (i.e., chemically), enzymatic protein modification has several advantages over chemical treatment, such as milder reaction conditions, increased reaction rate, and higher specificity. Specifically, transglutaminase, protease, peptidoglutaminase, and protein-glutaminase enzymes have been shown to catalyze protein deamidation (5, 6, 26, 34).
Soy sauce is a traditional Japanese seasoning that is manufactured by fermenting steamed soybeans and roasted wheat with koji molds (Aspergillus sojae or Aspergillus oryzae) along with water and salt. A. sojae is preferentially used to make soy sauce due to its high proteolytic activity. During soy sauce fermentation, glutamate and glutamine are released from proteins by proteases and peptidases that are produced by koji molds. Subsequently, the released glutamine is converted into glutamate by the glutaminase activity of koji molds (23, 31). However, if glutaminase activity is absent or low, most of the glutamine is chemically and irreversibly converted into pyroglutamate, which has no taste (15). Hence, glutaminase is one of the key enzymes responsible for increasing the glutamate concentration in soy sauce fermentation, and it would be desirable to develop a koji mold with high glutaminase activity.
Recently, we cloned the novel salt-tolerant, glutaminase-encoding genes, CngahA and CagahA, from Cryptococcus nodaensis and Cryptococcus albidus, respectively (9). A homology search failed to reveal any characterized gene that was homologous to these glutaminases. Moreover, the glutaminases were shown to belong to an amidase signature superfamily, thus forming a new glutaminase subfamily.
In contrast, the expressed sequence tag (EST) analysis and the genomic sequencing of the A. oryzae RIB40 strain have been completed (1, 16). Thus, we used the EST and genomic data available for A. oryzae RIB40 in order to search for a novel glutaminase present in A. sojae. As a result, we identified a glutaminase gene homologous to CngahA and CagahA, which was named AsgahA. In the present study, we report the cloning of this gene, as well as the purification and characterization of the corresponding enzyme, AsGahA. Our results revealed that this enzyme is a novel peptidoglutaminase-asparaginase.
MATERIALS AND METHODS
Strains and plasmids.
Aspergillus sojae (NBRC 4241) was used as the DNA donor for the gene cloning of glutaminase, and Aspergillus oryzae RIB40 RP-1 (ΔpyrG) (27) was used as the host for gene expression and protein purification. Escherichia coli TOP10 (Invitrogen, Carlsbad, CA) and JM109 strains were employed for recombinant plasmid construction. The pUC18 and pCRTOPO2.1 (Invitrogen) plasmids were utilized for the genomic DNA library and the gene cloning for DNA sequencing, respectively. The expression vector, pMAP, which contains multiple cloning sites under the TAKA-amylase promoter, was constructed from pMAR5 (22) as follows: pMAR5 was digested with SphI, blunted using a DNA blunting kit (TaKaRa, Kyoto, Japan), and subsequently digested with SalI to remove the argB gene. This plasmid was named pMAR5*. The pNr26 plasmid (28) was digested with BamHI, blunted, and subsequently digested with SalI, and the resulting 2.0-kb fragment containing the A. sojae pyrG gene was isolated and ligated into pMAR5* to produce pMAP.
Growth media.
Soybean powder submerged medium consisted of 3.0% soybean powder and 3.0% KH2PO4 (pH 6.0). DPY medium comprised 2.0% dextrin, 1.0% polypeptone, 0.5% yeast extract, 0.5% KH2PO4, and 0.05% MgSO4 (pH 6.0). Cz-Dox medium (noninduction medium) consisted of 0.1% KH2PO4, 0.05% KCl, 0.2% NaNO3, 3% glucose, 0.05% MgSO4, and 0.001% FeSO4 (pH 6.0). For the Cz-M medium (induction medium), the 2% glucose in the Cz-Dox medium was replaced with 2% maltose. The wheat bran solid medium contained wheat bran with 80% water (wt/wt) and was autoclaved at 121°C for 50 min. The soy sauce koji medium consisted of 2.46 g of defatted soybean meal, 2.7 g of roasted wheat, and 3.6 ml of tap water and was autoclaved at 121°C for 30 min.
DNA and RNA manipulation.
General methods for DNA and RNA manipulation were performed as described by Sambrook et al. (21). Genomic DNA was extracted using the Wizard genomic DNA purification kit (Promega, Madison, WI). Total RNA was extracted from disrupted mycelia using the RNeasy plant minikit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
A glutaminase probe for Southern and Northern blotting was prepared from the cloned genomic AsgahA gene by labeling with a digoxigenin (DIG) labeling mix (Roche Diagnostics, Basel, Switzerland) as per the manufacturer's protocol. Hybridization and detection of signals were performed by using a DIG system in accordance with the manufacturer's instructions (Roche Diagnostics).
All PCRs were performed with Ex Taq polymerase (TaKaRa). Nucleotide sequences were determined by using a Thermo sequenase cycle sequencing kit (USB, Cleveland, OH) and the DNA sequencer LIC-4200L(S)-2 (LI-COR, Lincoln, NE). The universal primers, M13-Forward primer and M13-Reverse primer, were used for sequencing.
Genomic DNA library construction and screening for the gahA gene.
A genomic DNA library was constructed as described below. First, genomic DNA extracted from A. sojae was digested with XbaI, cloned into pUC18, and transformed into E. coli TOP10. Transformants containing the target fragment were screened by colony hybridization using a 0.7-kb PCR fragment as a probe, which corresponded to a region of the EST fragment (AOEST04086). The phage library was constructed using the lambda EMBL3/Gigapack III gold cloning kit (Agilent Technologies, Santa Clara, CA) as previously described (18).
cDNA cloning.
Total RNA was extracted from mycelia grown in soybean powder submerged medium at 30°C for 16 h. To obtain the complete open reading frame (ORF), 5′- and 3′-rapid amplification of cDNA ends (RACE) was performed using the Marathon-Ready cDNA amplification kit (Clontech, Mountain View, CA) according to the manufacturer's instructions. The primers gahA-5R-as (5′-TAGCTATGGTCCCGTACTGTGCAAGCTTGG-3′) and gahA-5R-nas (5′-ATGGCTTGACACACAATCTCCCACCACACC-3′) were used for 5′-RACE, and the primers gahA-3R-s (5′-GCAGCGCAACACTGACCGGCAAATACTCGG-3′) and gahA-3R-ns (5′-AAGAGCGACTTGGAGCAGGAGGCACATCGG-3′) were used for 3′-RACE. The PCR fragments obtained by 5′-RACE and 3′-RACE were cloned into pCR-TOPO2.1 and sequenced. The entire ORF (AsgahA) was obtained by PCR using the first-strand cDNA generated by 3′-RACE as a template and the following primers: gahA-full-s (5′-GGGGAATTCATGTTTCTTAGTACACTCCTCTCACTGG-3′) and gahA-full-as (5′-AAACCCGGGTTAGGAGTATAGGCGAGAGCC-3′). The amplified PCR fragment was cloned into pCR-TOPO2.1 to produce pCR-AsgahA, and the DNA sequence was determined.
Gene expression.
The complete ORF of AsgahA was cloned into pMAP for expression in A. oryzae. pCR-AsgahA was digested with EcoRI and SmaI, and the resulting fragment containing the AsgahA gene was ligated into pMAP, which had been digested with the same enzymes, to produce the pMAP-AsgahA plasmid. A. oryzae RIB40 RP-1 was transformed with pMAP-AsgahA as previously described (28). The transformant was cultured in Cz-Dox or Cz-M medium with shaking at 30°C for 4 days. The mycelia were harvested and washed with 20 mM phosphate buffer (pH 6.0), and 100 mg of the washed mycelia was used to assay enzyme activity.
Enzyme purification.
The transformant was cultured in DPY medium with shaking at 30°C for 4 days. The mycelia were harvested and washed with 20 mM phosphate buffer (pH 6.0). Subsequently, the washed mycelia (100 g) were lysed in 1.0 liter of 20 mM phosphate buffer (pH 6.0) containing 3 mg/ml Yatalase (TaKaRa) at 30°C for 3 h. The lysate was filtered through Miracloth (Merck, Darmstadt, Germany) and centrifuged at 15,000 × g for 15 min. The supernatant was cooled to 5°C, and the following reagents were added to inhibit proteases: phenylmethylsulfonyl fluoride (1.0 mM), EDTA (5 mM), pepstatin A (1.0 μg/ml), leupeptin (0.5 μg/ml), and chymostatin (0.1 μg/ml). Solid ammonium sulfate was added to the cooled supernatant to a final concentration of 1.2 M, and the precipitate was removed by centrifugation at 15,000 × g. The resulting supernatant, which was used as a crude enzyme solution, was applied to a TSKgel phenyl-5PW column (Tosoh Bioscience, Tokyo, Japan) equilibrated with 20 mM phosphate buffer containing 1.2 M ammonium sulfate (pH 7.0). The bound proteins were eluted with a linear gradient of ammonium sulfate from 1.2 M to 0 M and ethylene glycol from 0% to 20% over 60 min at a flow rate of 5.0 ml/min. The active fractions were collected, the buffer was replaced with 20 mM phosphate buffer (pH 7.0) using Centriprep YM-10 (Millipore, Billerica, MA), and the resulting solution was loaded onto a TSKgel DEAE-5PW column (Tosoh Bioscience) equilibrated with the same buffer. The bound proteins were eluted with a linear gradient of NaCl from 0 M to 0.5 M over 60 min at a flow rate of 1.0 ml/min, and the active fractions were collected and concentrated using Centriprep YM-10. The concentrated enzyme solution was loaded onto a TSKgel G3000SW column (Tosoh Bioscience) equilibrated with 100 mM phosphate buffer containing 0.2 M NaCl (pH 7.0) and eluted at a flow rate of 0.7 ml/min.
Determination of molecular mass.
The molecular mass of the native enzyme was determined by gel filtration (TSKgel G3000SW) using MW-Marker (Oriental Yeast Co., Tokyo, Japan). SDS-PAGE and native PAGE were carried out using gradient gels (Multigel II mini 10/20; Cosmo Bio Co., Tokyo, Japan) as per the manufacturer's instructions. Gels were stained by using the Quick-CBB kit (Wako Pure Chemical Industries, Osaka, Japan) according to the manufacturer's protocol.
Determination of the N-terminal amino acid sequences of purified AsGahA.
Three polypeptides separated by SDS-PAGE were transferred to an Immun-Blot polyvinylidene difluoride (PVDF) membrane (Bio-Rad Laboratories Inc., Hercules, CA) by the semidry blotting method. The transferred membrane was stained with Coomassie brilliant blue using Quick-CBB staining solution A (Wako Pure Chemical Industries). The stained protein bands were then excised and analyzed with a 492 protein sequencer (Applied Biosystems).
Enzyme assays.
Glutaminase activity using intact mycelia from the transformant grown in submerged culture and the soy sauce koji culture was assayed as previously described (9). A slightly different protocol was used for assaying the glutaminase activity of the purified protein. Briefly, a reaction mixture (380 μl) containing 10 mM glutamine and 0.1 M sodium phosphate buffer (pH 7.0) was preincubated at 37°C for 5 min, and then 20 μl of the purified AsGahA solution (0.2 to 10 μg) was added. Subsequently, this mixture was incubated at 37°C for 30 to 60 min, and the reaction was terminated by adding 100 μl of 0.75 N perchloric acid. After 15 min, the reaction mixture was neutralized with 50 μl of 1.5 M NaOH. Following centrifugation at 18,000 × g for 10 min, the reaction products (l-glutamate and ammonia) in the supernatant were quantified by using glutamate dehydrogenase and F-kit ammonia (Roche Diagnostics), respectively. One unit of enzymatic activity was defined as the amount that produced 1 μmol of l-glutamate or ammonia per minute from l-glutamine under the above-described conditions.
The optimal pH of the enzyme was determined at 37°C in the range of pH 3.5 to 11.0 by using the following buffers: 0.1 M sodium acetate buffer (pH 3.5 to 5.5), 0.1 M sodium phosphate buffer (pH 6.0 to 8.0), 0.1 M Tris-HCl buffer (pH 7.0 to 9.0), and 0.1 M carbonate bicarbonate buffer (pH 9.0 to 11.0). The pH stability was analyzed by preincubating the enzyme at 4°C or 30°C for 24 h in buffers (0.1 M) of various pH values. Residual activity was assayed at 37°C by using the standard assay described above.
The optimal temperature of AsGahA in the range of 30 to 60°C was determined in 0.1 M sodium phosphate buffer (pH 7.0) and/or 0.1 M carbonate bicarbonate buffer (pH 9.5). The thermal stability was measured by preincubating the enzyme at 30 to 60°C for 30 min in 0.1 M sodium phosphate buffer (pH 7.0) and/or 0.1 M carbonate bicarbonate buffer (pH 9.5), and the residual activity was measured at 37°C by using the standard assay.
Substrate specificity was determined in the standard assay solution by using glutamate dehydrogenase and F-kit ammonia. The concentration of substrates utilized in the assay was 10 mM, except for l-γ-Glu-p-nitroanilide (l-γ-Glu-pNA; 1.25 mM). The relative activities against l-Gln, l-γ-Glu-hydrazide, l-γ-Glu-Val-OH, l-γ-Glu-Leu-OH, and l-γ-Glu-Cys-Gly-OH (glutathione) were determined by detecting l-glutamate. The relative activities against carboxybenzoyl (CBZ)-Gln, CBZ-Gln-Gly (Sigma, St. Louis, MO), d-Gln, l-Asn, d-Asn (Fluka), l-Glu, l-Gly-Gln, l-Ala-Gln, l-Gln-Gly (Tokyo Kasei Co., Tokyo, Japan), l-Leu-Gly-Gln, l-Pro-Leu-Gly-Gln, l-Leu-Gly-Asn, l-Leu-Ala-Asn, l-Pro-Leu-Gly-Asn (PH Japan, Hiroshima, Japan), acetamide, acrylamide, and H-Tyr-Gly-NH2 (Kokusan Chemical Co., Tokyo, Japan) were measured by detecting ammonia. The relative activity against l-γ-Glu-pNA was determined by measuring the release of pNA at 405 nm.
Hydroxamate formation of glutaminyl and asparaginyl derivatives was measured by a colorimetric procedure using γ-glutamic acid monohydroxamate as the standard. The reaction mixture (final volume, 200 μl) containing 0.1 M Tris-HCl (pH 9.0), 10 mM the substrates, 0.1 M hydroxylamine, and purified AsGahA (0.1 to 5 μg) was incubated at 37°C for 60 min. The reaction was stopped by adding 35 μl of a stop solution that comprised 28% (wt/vol) trichloroacetic acid, 2.5 M HCl, and 5% FeCl3. After centrifugation at 18,000 × g for 10 min, the absorbance of the supernatant at 540 nm was measured and the hydroxamate content was determined against the standard curve.
The effects of metallic ions were determined in the standard assay containing 2 mM various metallic salts (CuCl2, NaF, CoCl2, CaCl2, HgCl2, SnCl2, MnCl2, MgCl2, NiCl2, ZnCl2, FeCl2, or FeCl3). The effect of NaCl on enzyme activity was determined by using purified AsGahA and intact mycelia in the standard assay in the presence of 0% to 20% NaCl.
The Km and Vmax values were calculated from double reciprocal Lineweaver-Burk plots. The kcat value was calculated based on the molecular mass of dimeric AsGahA (deduced from the gene sequence).
Analysis of the localization of AsGahA in solid-state culture.
The transformant and control strain (wild type) were cultured in soy sauce koji medium at 30°C for 42 h. The culture was soaked with 50 ml of 20 mM sodium phosphate buffer (pH 7.0) on ice for 16 h. Subsequently, the culture was homogenized for 1 min with a Polytron homogenizer (Central Scientific Commerce, Inc., Tokyo, Japan) and centrifuged at 28,000 × g for 15 min. The supernatant was collected and used as the extracellular fraction. The precipitate was resuspended in 50 ml of 20 mM sodium phosphate buffer (pH 7.0) and centrifuged at 28,000 × g for 15 min. This washing process was repeated 8 times, and the precipitate was finally resuspended in 30 ml of the same buffer. The resulting suspension containing the precipitate was used as the cell wall-bound fraction. The glutaminase activity of each fraction (1 ml) was assayed for 60 min. Each fraction of the AsGahA-overexpressing transformant was diluted adequately before being assayed.
Analysis of the proteolytic cleavage of AsGahA.
The purified AsGahA and extracellular glutaminase fractions were deglycosylated by incubation with PNGaseF (New England BioLabs, Beverly, MA) at 37°C for 1 h as per the manufacturer's instructions. Subsequently, deglycosylated AsGahA was separated on a gradient gel (Multigel II 10/20 minigel; Cosmo Bio Co.) by SDS-PAGE and transferred to a PVDF membrane (Bio-Rad Laboratories Inc.). Western blot analysis was carried out using the BM chemiluminescence Western blotting kit (Roche Diagnostics). A rabbit antiserum against the purified AsGahA protein was prepared by TaKaRa Bio Inc. (made to order) and employed in Western blotting.
To analyze extracellular proteolytic processing, purified AsGahA was reacted at 30°C for 16 h with the soy sauce koji extract (corresponding to the extracellular glutaminase fraction from the wild-type strain, as described above), which had been incubated for 10 min with or without boiling, and subsequently separated by SDS-PAGE using Multigel II 10/20 minigels (Cosmo Bio Co.). Western blotting was performed as described above. An extract of soy sauce koji medium that was diluted 10-fold was also used in the assay.
Nucleotide sequence accession number.
The nucleotide sequence of AsgahA has been deposited in the DDBJ database under the accession number AB693950.
RESULTS
Cloning and analysis of a glutaminase gene from A. sojae.
In order to find genes in A. sojae that were homologous to CngahA, which encodes a cryptococcal glutaminase, a BLAST search was performed against DOGAN (the genome database of A. oryzae RIB40; http://www.bio.nite.go.jp/dogan/project/view/AO) and against the A. oryzae EST database (http://nribf21.nrib.go.jp/EST2/) by using the nucleotide or amino acid sequence of CnGahA. Four genes homologous to CngahA were identified in the A. oryzae RIB40 genome: AO090003001406 (34% amino acid identity; E value, 1 × 10−90), AO090011000310 (35% amino acid identity; E value, 3 × 10−97), AO090701000634 (28% amino acid identity; E value, 1 × 10−52), and AO090011000138 (31% amino acid identity; E value, 7 × 10−52). These 4 genes were designated AogahA, AogahB, AogahC, and AogahD, respectively. The corresponding gene products were predicted as hypothetical proteins with an amidase signature sequence. However, a search performed against the A. oryzae EST database identified only a single clone (AoEST04086) as the homologue to the 3′ region of CngahA, which was identical to AogahA (AO090003001406).
Thus, we decided to clone the orthologous gene from A. sojae (AsgahA), which is the preferred species for making soy sauce. The EST fragment (AoEST04086) was successfully amplified by PCR using the genome of A. sojae as a template, and it was subsequently used as a probe for Northern and plaque hybridization. Expression of the AsgahA gene was confirmed by Northern blot analysis in A. sojae that had been cultured in soybean powder medium (data not shown). The cDNA was cloned by 3′- and 5′-RACE, and the 7.2-kb genomic DNA was simultaneously cloned by plaque hybridization. The structural gene (ORF) was 1,929 bp in length without introns and encoded a polypeptide of 643 amino acids. This polypeptide, which was termed AsGahA, shared 36% overall amino acid sequence identity with CnGahA. Subsequently, the AsgahA structural gene was overexpressed in A. oryzae RIB40 RP-1 by transformation with the plasmid, pMAP-AsgahA. The introduction of multiple copies of the AsgahA gene in A. oryzae RIB40 was confirmed in the genome of the transformant by Southern hybridization analysis (data not shown).
Glutaminase activity was detected on the cell surface but not in the supernatant. The glutaminase activity of the control strain (wild type) and the transformant in Cz-Dox medium (noninduction medium) was 0.78 and 9.59 mU/mg wet mycelia weight, respectively. However, in Cz-M medium containing maltose as an inducer, the glutaminase activities of the control strain and the transformant were 1.05 and 33.3 mU/mg wet mycelia weight, respectively. Therefore, the glutaminase activity of the transformant in noninduction medium was approximately 10 times higher than that of the wild type and was increased by adding an inducer; in contrast, the activity of the wild type was unaffected by the inducer. This increase in the glutaminase activity of the transformant may be caused by the introduction of the AsgahA gene under the control of the Taka-amylase promoter, which is induced by maltose. These results showed that the AsgahA gene was successfully expressed in A. oryzae and encoded an active glutaminase.
Purification of AsGahA.
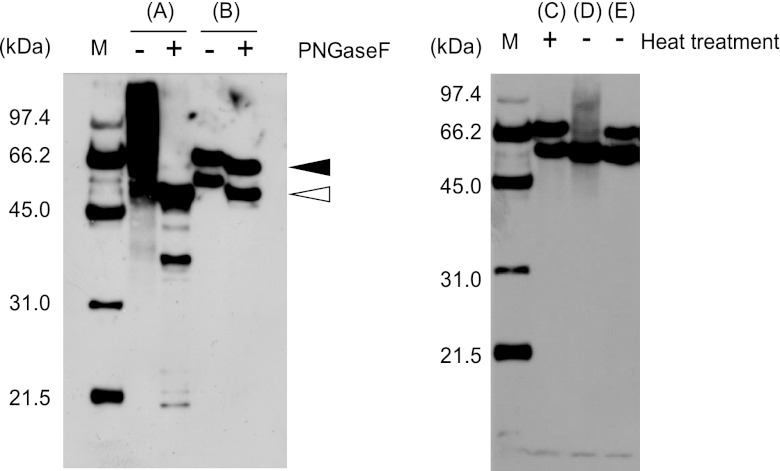
AsGahA was released from the cell surface of the AsGahA-overexpressing transformant by Yatalase treatment and purified by performing 3 chromatography steps (see Table S1 in the supplemental material). Purified AsGahA eluted as a single peak (corresponding to 135 kDa) on gel filtration and produced a single band on native PAGE (see Fig. S1A). In contrast, it separated into 3 polypeptides (A, B, and C) on SDS-PAGE (see Fig. S1B) with the following N-terminal amino acid sequences: polypeptide A, AAIPNGQTLS; polypeptide B, SSDSTITAQL; and polypeptide C, AAIPNGQTLS (see Fig. S2). Thus, the N-terminal sequences of polypeptides A and C were identical. Signal-P (NN) analysis revealed that the signal peptide cleavage site was located between Gly15 and Ala16, and polypeptides A and C contained identical N-terminal amino acid sequences after the removal of the signal peptides. The molecular masses of polypeptides A, B, and C were estimated by SDS-PAGE to be 67 kDa, 59 kDa, and 13 kDa, respectively. Since the sum of the molecular masses of polypeptides B and C was nearly equal to that of polypeptide A, we concluded that polypeptides B and C represent partial degradation products of polypeptide A. The molecular mass of polypeptide A was estimated to be 67 kDa by SDS-PAGE, compared with 135 kDa by gel filtration chromatography, indicating that the native form of AsGahA is a dimer. Furthermore, the mobilities of polypeptides A and B on SDS-PAGE were slightly shifted following deglycosylation by PNGaseF treatment (see Fig. 1B), thus suggesting that AsGahA is a glycoprotein.
FIG 1.
Differences in the glycosylation or the specific proteolysis and molecular mass of AsGahA between solid-state culture and submerged culture. The soy sauce koji extracts of the AsGahA-overexpressing transformant (A) and purified AsGahA (B) were deglycosylated by treatment with or without PNGase F. Purified AsGahA was reacted with the soy sauce koji extract of A. oryzae RIB40 (wild type) with (C) or without (D) heat treatment (boiling for 10 min) or with a 10-fold dilution of the soy sauce koji extract without heat treatment (E). Incubation with the soy sauce koji extract was carried out at 30°C for 16 h. Proteins were subjected to SDS-PAGE with a gradient gel (10% to 20%), electroblotted onto a PVDF membrane, and immunodetected using antiserum against the purified AsGahA protein. The peptides A and B are indicated by the closed and open triangles, respectively.
Characterization of AsGahA.
The properties of AsGahA were investigated by using the purified enzyme. The optimal pH of the enzyme was 9.5 (see Fig. S3A). At 30°C, the pH stability was decreased at pH values above 9.0, whereas it was not affected at 4°C (see Fig. S3B). The optimal temperatures for enzyme activity at pH 9.5 and at pH 7.0 were 45°C and 50°C, respectively (see Fig. S3C). Following the incubation of purified AsGahA at 50°C in 0.1 M phosphate buffer (pH 7.0), 85% of the enzyme activity was retained, whereas AsGahA was completely inactivated by incubation in 0.1 M carbonate buffer (pH 9.5) (see Fig. S3D). These results demonstrate that AsGahA is unstable at high temperatures under alkaline pH conditions.
The glutaminase activity of AsGahA was inhibited by approximately 85% in the presence of 18% sodium chloride. In order to investigate the effect of metallic ions on AsGahA, the glutaminase activity of purified AsGahA was measured in the presence of various metallic ions. While partial inhibition of glutaminase activity by HgCl2 was observed (45.0% ± 5.2%), other metallic ions (FeCl2, FeCl3, CuCl2, NaF, CoCl2, CaCl2, SnCl2, MnCl2, MgCl2, NiCl2, and ZnCl2) did not exert any significant effect (data not shown).
As shown in Tables 1 and 2, analysis of substrate specificity showed that AsGahA deamidated not only free glutamine but also C-terminal glutaminyl residues in peptides. l-Glutamate was not detected in the reaction solution of each peptide substrate. Since AsGahA did not deamidate l-glutamate and γ-glutaminyl derivatives—as indicated by detecting the liberation of ammonia—we concluded that AsGahA hydrolyzes the γ-amide bond of glutamine or glutaminyl residues in peptides rather than the α-amide bond. The Km values for each peptide were 10 times lower than those for free glutamine (Table 2). The kcat/Km value of Cbz-Gln was the highest among all the substrates tested. However, Cbz-Gln-Gly was hardly deamidated (Table 2), suggesting that AsGahA specifically deamidates the γ-amide bond of glutamine residues located at the C-terminal end of peptides. Moreover, AsGahA also deamidated free asparagine and C-terminal asparaginyl residues in peptides (Tables 1 and 2), demonstrating that AsGahA represents a novel peptidoglutaminase-asparaginase.
TABLE 1.
Substrate specificities of AsGahA
| Substrate | Specific activitya (μmol · min−1 · mg−1) |
|
|---|---|---|
| Deamidation | Transferring | |
| l-Gln | 75.2 ± 25 | 55.9 ± 8.3 |
| l-Gly-l-Gln | 212.3 ± 45 | 119.6 ± 16.3 |
| l-Ala-l-Gln | 317.3 ± 78 | 119.2 ± 21.3 |
| l-Leu-Gly-Gln | 185.9 ± 20 | 106.2 ± 14.1 |
| l-Pro-Leu-Gly-Gln | 160.5 ± 23 | 70.6 ± 7.8 |
| Cbz-Gln | 236.3 ± 67 | 72.1 ± 3.1 |
| Cbz-Gln-Gly | 0.09 ± 0.01 | 0.16 ± 0.03 |
| d-Gln | 23.1 ± 2.3 | 9.6 ± 2.9 |
| l-Gln-Gly | ND | ND |
| l-Asn | 84.0 ± 26 | 47.4 ± 5.5 |
| l-Gly-l-Asn | 113.5 ± 35 | 98.5 ± 2.1 |
| l-Leu-Ala-Asn | 70.4 ± 26 | 36.9 ± 1.8 |
| l-Leu-Gly-Asn | 119.9 ± 5.0 | 80.2 ± 7.6 |
| l-Pro-Leu-Gly-Asn | 207.1 ± 38 | 87.9 ± 5.6 |
| d-Asn | 0.25 ± 0.03 | 0.17 ± 0.03 |
| Cbz-Asn | 24.7 ± 8.9 | 22.2 ± 3.4 |
| l-Glu | ND | 0.3 ± 0.1 |
| Acrylamide | 0.18 ± 0.04 | 1.1 ± 0.1 |
| Acetoamide | ND | 0.1 ± 0.02 |
| l-Gly-Tyr-NH2 | ND | ND |
| l-γ-Glu-Leu-OH | ND | ND |
| l-γ-Glu-Val-OH | ND | ND |
| l-γ-Glu-Cys-Gly-OH | ND | ND |
The deamidation activity was determined by measuring ammonia liberated from various substrates. The transferring activity was determined by hydroxamate formation between hydroxylamine and various substrates. ND, not detectable; defined as specific activity of less than 0.05 μmol · min−1 · mg−1. Assays were carried out in triplicate and the results represent the means ± standard deviations.
TABLE 2.
Kinetic properties of AsGahAa
| Substrate | Km (mM) | kcat (s−1) | kcat/Km (s−1 · mM−1) |
|---|---|---|---|
| l-Gln | 5.22 ± 0.10 | 489 ± 103 | 93 ± 21 |
| l-Gly-Gln | 0.66 ± 0.12 | 711 ± 84 | 1085 ± 74 |
| l-Leu-Gly-Gln | 0.43 ± 0.13 | 751 ± 67 | 1870 ± 666 |
| l-Pro-Leu-Gly-Gln | 0.33 ± 0.08 | 833 ± 20 | 2662 ± 786 |
| Cbz-Gln | 0.07 ± 0.02 | 833 ± 63 | 11986 ± 3014 |
| l-Asn | 8.03 ± 0.18 | 1023 ± 39 | 127 ± 7.0 |
| l-Gly-Asn | 0.63 ± 0.03 | 439 ± 46 | 692 ± 36 |
| l-Leu-Gly-Asn | 1.06 ± 0.06 | 408 ± 56 | 388 ± 71 |
| l-Pro-Leu-Gly-Asn | 0.74 ± 0.19 | 715 ± 77 | 991 ± 144 |
Assays were carried out in triplicate and the results represent the means ± standard deviations.
As indicated in Table 1, AsGahA also catalyzed the incorporation of hydroxylamine into glutamine and asparagine residues in peptides. The substrate specificities of this transferring activity approximately corresponded to those of the deamidation activity detected by ammonia liberation. Analysis of glutamate or p-nitroanilide liberation revealed that AsGahA did not show any activity against gamma-glutamyl derivatives (l-γ-Glu-Val-OH, l-γ-Glu-Leu-OH, l-γ-Glu-Cys-Gly-OH [glutathione], and l-γ-Glu-pNA) (Table 1). In addition, γ-glutamyl-valine and γ-glutamyl-leucine could not be detected by liquid chromatography-mass spectrometry among the reaction products of AsGahA in the presence of l-glutamine and l-valine or l-leucine (data not shown). Thus, these results indicate that AsGahA does not possess γ-glutamyl transpeptidase activity. Other substrates, such as acrylamide, acetoamide (a substrate for amidase), and H-Tyr-Gly-NH2 (a substrate for peptide amidase) were hardly deamidated (Table 1).
Localization and characterization of AsGahA in solid-state culture.
The extracellular and cell wall-bound glutaminase activity of the AsGahA-overexpressing transformant cultured in soy sauce koji medium was 17.9 ± 3.77 U/g koji and 2.98 ± 0.93 U/g koji, respectively, indicating the extracellular glutaminase activity was 6 times higher than that of the cell wall-bound glutaminase activity. This result suggests that AsGahA is localized in the extracellular fraction in solid-state culture but that it is localized at the cell surface under submerged culture conditions. We found that the extracellular and cell wall-bound glutaminase activities of the control strain were 0.47 ± 0.01 U/g koji and 0.11 ± 0.07 U/g koji, respectively. However, the cell wall-bound glutaminase activity of the AsGahA-overexpressing transformant was much higher under solid-state culture conditions, indicating that a proportion of the overproduced glutaminase was trapped on the cell surface.
Figures 1A and B show the altered mobility of AsGahA on SDS-PAGE after deglycosylation. While AsGahA extracted from solid-state culture produced a smeared band by Western blot analysis using antiserum against AsGahA, one major band and some minor bands were observed after deglycosylation (Fig. 1A). We concluded that the smeared band resulted from variation in the glycosylation of AsGahA, and AsGahA extracted from solid-state culture was more glycosylated than that from submerged culture (Fig. 1B). The molecular mass of the major band was similar to that of the lower band seen for purified AsGahA (corresponding to peptide B) after deglycosylation.
Degradation of the purified AsGahA by treatment with the soy sauce koji extract in vitro is shown in Fig. 1C to E. The upper band seen for purified AsGahA (corresponding to peptide A) disappeared following treatment with the soy sauce koji extract, which contained proteases and peptidases, but the lower band remained and increased slightly in intensity. This proteolytic cleavage did not occur in the reaction mixture containing 3 mg/ml Yatalase, which was used for the release of AsGahA from cells (data not shown). These results suggest that AsGahA was produced as a large protein (corresponding to peptide A), which was then cleaved specifically by a protease produced by A. oryzae in solid-state culture.
DISCUSSION
The glutaminase activity of koji molds is known to be important for soy sauce production. In the present study, we cloned a novel glutaminase gene from A. sojae, which was discovered by performing a BLAST search against EST and genomic sequences of A. oryzae RIB40 to search for homologues to the salt-tolerant and thermostable glutaminase, CnGahA. Substrate specificity analysis revealed that this protein, termed AsGahA, was a unique enzyme that deamidated not only free glutamine and asparagine but also C-terminal glutaminyl or asparaginyl residues in peptides. Furthermore, AsGahA displayed a substrate preference for C-terminal glutaminyl or asparaginyl residues over free glutamine or asparagine.
Several proteins indicating glutaminase activity (e.g., GtaA [14, 29], GgtA [13], and Gls [17]) have been reported in A. oryzae. The gtaA gene was originally cloned as a glutaminase gene from a koji mold. GtaA protein is a monomer enzyme and is able to hydrolyze l-γ-Glu-pNA, which is not a substrate for AsGahA. GgtA protein is a γ-glutamyltranspeptidase, hydrolyzing not only l-Gln but also γ-glutamyl compounds such as a glutathione and γ-Glu-Lys, which cannot be hydrolyzed by AsGahA. Unlike AsGahA, Gls is a salt-tolerant glutaminase that has no transferring activity in the presence of l-Gln and hydroxylamine. In addition, AsGahA does not show amino acid similarity to these proteins, suggesting that AsGahA differs from the characterized glutaminases of A. oryzae.
Kikuchi et al. identified 2 types of peptidoglutaminases that hydrolyze the γ-amide bond of glutamine in peptides from Bacillus circulans (11, 12). While peptidoglutaminase I deamidates free glutamine and C-terminal glutaminyl residues, thereby displaying similar substrate specificity to AsGahA, peptidoglutaminase II deamidates N-terminal and internal glutaminyl residues in peptides. However, both peptidoglutaminases are unable to deamidate free asparagine and/or C-terminal asparaginyl residues.
A peptide amidase is an enzyme that hydrolyzes the C-terminal amide bond in peptide amides and is a member of the amidase signature superfamily (19, 25). AsGahA showed 35% amino acid sequence identity with a characterized peptide amidase (Q8RJN5). However, this peptide amidase does not deamidate the amide group in the side chain of free glutamine or asparagine nor does it deamidate C-terminal glutaminyl or asparaginyl residues in peptides (25). Moreover, AsGahA did not deamidate H-Tyr-Gly-NH2, which is a typical substrate for the peptide amidase. Thus, AsGahA does not appear to possess peptide amidase activity. Alternatively, protein-glutaminase and transglutaminase are enzymes that deamidate internal glutaminyl residues in proteins and/or peptides, respectively (3, 32). However, AsGahA hardly deamidated Cbz-Gln-Gly—a good substrate of these enzymes—in the presence or absence of NH2OH. Collectively, these results suggest that AsGahA is a novel type of peptidoglutaminase-asparaginase. To our knowledge, this is the first report to clone a gene encoding an enzyme that deamidates C-terminal glutaminyl or asparaginyl residues in peptides.
We previously showed that the cryptococcal glutaminases form a new subclass (glutamine amidohydrolase [GAH] subclass) in the amidase signature superfamily (9), in which AsGahA was also classified. Iwasa et al. reported that while CaGahA deamidated free glutamine and Cbz-Gln, it did not deamidate Gly-Gln (10); therefore, they concluded that CaGahA is not a peptidoglutaminase. In the present study, we used the purified enzyme to confirm that CaGahA is able to deamidate not only Cbz-Gln but also C-terminal glutaminyl residues, including Gly-Gln (see Table S2). This suggests that the GAH subclass of the amidase signature superfamily deamidates C-terminal glutaminyl or asparaginyl residues in peptides. The different substrate specificities of CaGahA observed between the distinct studies may result from the detection method employed or the detection sensitivity. While Iwasa et al. determined the deamidation activity of CaGahA by detecting ammonia via Nesslerization, we measured ammonia liberated from various substrates by an enzymatic assay using l-glutamate dehydrogenase.
The AsGahA protein purified from submerged culture existed as a complex of 3 polypeptides (A, B, and C). We concluded that polypeptides B and C were partially cleaved from polypeptide A during purification. Polypeptide A disappeared from the Western blot following incubation of purified AsGahA with an extract containing proteases from solid-state culture, whereas polypeptide B was still visible and slightly increased in intensity. In contrast, a significant amount of the extracellular AsGahA from solid-state culture was present as polypeptide B. Since the cell wall-bound glutaminase activity of the AsGahA-overexpressing transformant was higher than that of the control strain, we hypothesized that a proportion of AsGahA was trapped on the cell surface under conditions of solid-state culture. Yano et al. purified the intracellular and extracellular glutaminase enzymes independently and showed that they were identical (33). Moreover, Koibuchi et al. suggested that the cell wall-bound glutaminase is released from mycelia by self-digestion at a late growth phase (14). Therefore, it is possible that AsGahA (polypeptide A) is initially located at the cell surface and is subsequently cleaved by extracellular proteases around the mycelia, resulting in its release from the cell surface. The polypeptide A of AsGahA may be less likely to be attacked by proteases in submerged culture than by those in solid-state culture, because the extracellular proteases diffuse more easily in submerged culture conditions than in solid-state culture conditions. Notably, proteins from the GAH subfamily differed with regard to the length and sequence of the N-terminal region (9). Thus, the N-terminal region of AsGahA may be involved in attachment to the cell surface.
It is of interest to determine which glutaminases of koji molds are involved in glutamate production during soy sauce fermentation. GtaA and GgtA are localized extracellularly, whereas Gls is predicted to be localized in the cytoplasm by the PSORT-II program (http://psort.hgc.jp/). AsGahA was secreted under solid-state culture conditions; however, a proportion of AsGahA was located on the cell surface. Therefore, since the concentration of glutamate in soy sauce moromi is positively correlated with the cell wall-bound glutaminase activity of Aspergillus species (23), this glutaminase may be involved in glutamate production during the fermentation of soy sauce. Furthermore, several short acidic peptides containing l-Glu or l-Asp residues at the C terminus have been isolated from soy sauce (20). Thus, these peptides might be deamidated by AsGahA during soy sauce fermentation. To investigate this possibility, experiments to manufacture soy sauce using disruptants and/or overexpressing transformants of the AsgahA gene are in progress.
Supplementary Material
ACKNOWLEDGMENT
We thank Kuniko Shiraishi for experimental assistance.
Footnotes
Published ahead of print 18 May 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Akao T, et al. 2007. Analysis of expressed sequence tags from the fungus Aspergillus oryzae cultured under different conditions. DNA Res. 14:47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Durá AM, Flores M, Toldrá F. 2002. Purification and characterization of a glutaminase from Debaryomyces spp. Int. J. Food Microbiol. 76:117–126 [DOI] [PubMed] [Google Scholar]
- 3. Folk JE, Chung SI. 1985. Transglutaminases. Methods Enzymol. 113:358–375 [DOI] [PubMed] [Google Scholar]
- 4. Gao P, et al. 2009. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458:762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hamada JS. 1994. Deamidation of food proteins to improve functionality. Crit. Rev. Food Sci. Nutr. 34:283–292 [DOI] [PubMed] [Google Scholar]
- 6. Hamada JS, Mashall WE. 1988. Enhancement of peptidoglutaminase deamidation of soy protein by heat treatment and/or proteolysis. J. Food Sci. 53:1132–1134 [Google Scholar]
- 7. Heini HG, Gebhardt R, Brecht A, Mecke D. 1987. Purification and characterization of rat liver glutaminase. Eur. J. Biochem. 162:541–546 [DOI] [PubMed] [Google Scholar]
- 8. Imada A, Igarasi S, Nakahama K, Isono M. 1973. Asparaginase and glutaminase activities of micro-organisms. J. Gen. Microbiol. 76:85–99 [DOI] [PubMed] [Google Scholar]
- 9. Ito K, Umitsuki G, Oguma T, Koyama Y. 2011. Salt-tolerant and thermostable glutaminase of Cryptococcus species form a new glutaminase family. Biosci. Biotechnol. Biochem. 75:1317–1324 [DOI] [PubMed] [Google Scholar]
- 10. Iwasa T, Fujii S, Yokotsuka T. 1987. On the glutaminase produced by Cryptococcus albidus ATCC 20293. J. Jpn. Soy Sauce Res. Inst. 13:205–210 [Google Scholar]
- 11. Kikuchi M, Hayashi H, Nakano E, Sakaguchi K. 1971. Peptidoglutaminase. Enzyme for selective deamidation of γ-amide of peptide-bond glutamine. Biochemistry 10:1222–1229 [DOI] [PubMed] [Google Scholar]
- 12. Kikuchi M, Sakaguchi K. 1973. Some enzymatic properties and substrate specidicities of peptidoglutaminase-I and II. Agr. Biol. Chem. 37:1813–1821 [Google Scholar]
- 13. Kitamoto N, Yasuda S, Ito H, Azeyanagi T. 2011. New glutaminase and method for producing same. Japanese patent JP 4651203 (B2)
- 14. Koibuchi K, Nagasaki H, Yuasa A, Kataoka J, Kitamoto K. 2000. Molecular cloning and characterization of a gene encoding glutaminase from Aspergillus oryzae. Appl. Microbiol. Biotechnol. 54:59–68 [DOI] [PubMed] [Google Scholar]
- 15. Kurose E, Oyama Y, Matuo T, Sugimori T. 1969. Biosynthesis and degradation of glutamic acid in microorganisms relating to the soy sauce brewing. J. Ferment. Technol. 47:693–700 [Google Scholar]
- 16. Machida M, et al. 2005. Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157–1161 [DOI] [PubMed] [Google Scholar]
- 17. Masuo N, et al. 2004. Molecular cloning, overexpression, and purification of Micrococcus luteus K-3-type glutaminase from Aspergillus oryzae RIB40. Protein Expr. Purif. 38:272–278 [DOI] [PubMed] [Google Scholar]
- 18. Matsushima K, et al. 2001. Pre-termination in aflR of Aspergillus sojae inhibits aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 55:585–589 [DOI] [PubMed] [Google Scholar]
- 19. Neumann S, Kula MR. 2002. Gene cloning, overexpression and biochemical characterization of the peptide amidase from Stenotrophomonas maltophilia. Appl. Microbiol. Biotechnol. 58:772–780 [DOI] [PubMed] [Google Scholar]
- 20. Oka S, Nagata K. 1973. Isolation and characterization of acidic peptide in soy sauce. Agr. Biol. Chem. 38:1195–1202 [Google Scholar]
- 21. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 22. Shibuya I, Tsuchiya K, Tamura G, Ishikawa T, Hara S. 1992. Overproduction of an alpha-amylase/glucoamylase fusion protein in Aspergillus oryzae using a high expression vector. Biosci. Biotech. Biochem. 56:1674–1675 [DOI] [PubMed] [Google Scholar]
- 23. Shikata H, Yasui T, Ishigami Y, Oomori K. 1979. Studies on the glutaminase of Shoyu Koji (part 2). J. Jpn. Soy Sauce Res. Inst. 5:21–25 [Google Scholar]
- 24. Spiers AS, Wade HE. 1976. Bacterial glutaminase in treatment of acute leukaemia. Br. Med. J. 1:1317–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stelkes-Ritter U, Wyzgol K, Kula MR. 1995. Purification and characterization of a newly screened microbial peptide amidase. Appl. Microbiol. Biotechnol. 44:393–398 [DOI] [PubMed] [Google Scholar]
- 26. Suppavorasatit I, De Mejia EG, Cadwallader KR. 2011. Optimization of the enzymatic deamidation of soy protein by protein-glutaminase and its effect on the functional properties of the protein. J. Agric. Food Chem. 59:11621–11628 [DOI] [PubMed] [Google Scholar]
- 27. Takahashi T, Masuda T, Koyama Y. 2006. Enhanced gene targeting frequency in ku70 and ku80 disruption mutants of Aspergillus sojae and Aspergillus oryzae. Mol. Genet. Genomics 275:460–470 [DOI] [PubMed] [Google Scholar]
- 28. Takahashi T, Hatamoto T, Koyama Y, Abe K. 2004. Efficient gene disruption in the koji-mold Aspergillus sojae using a novel variation of the positive-negative method. Mol. Genet. Genomics 272:344–352 [DOI] [PubMed] [Google Scholar]
- 29. Thammarongtham C, Turner G, Moir AJ, Tanticharoen M, Cheevadhanarak S. 2001. A new class of glutaminase from Aspergillus oryzae. J. Mol. Microbiol. Biotechnol. 3:611–617 [PubMed] [Google Scholar]
- 30. Wang JB, et al. 2010. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell 18:207–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yamamoto S, Hirooka H. 1974. Production of glutaminase by Aspergillus sojae. J. Ferment. Technol. 52:564–569 [Google Scholar]
- 32. Yamaguchi S, Jeenes DJ, Archer DB. 2001. Protein-glutaminase from Chryseobacterium proteolyticum, an enzyme that deamidates glutaminyl residues in proteins. Purification, characterization and gene cloning. Eur. J. Biochem. 268:1410–1421 [DOI] [PubMed] [Google Scholar]
- 33. Yano T, Ito M, Tomita K, Kumagai H, Tochikura T. 1988. Purification and properties of glutaminase from Aspergillus oryzae. J. Ferment. Technol. 66:137–143 [Google Scholar]
- 34. Yong YH, Yamaguchi S, Matsumura Y. 2006. Effects of enzymatic deamidation by protein-glutaminase on structure and functional properties of wheat gluten. J. Agric. Food Chem. 54:6034–6040 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.