Abstract
Bacillus methanolicus can utilize methanol as its sole carbon and energy source, and the scientific interest in this thermotolerant bacterium has focused largely on exploring its potential as a biocatalyst for the conversion of methanol into l-lysine and l-glutamate. We present here the genome sequences of the important B. methanolicus model strain MGA3 (ATCC 53907) and the alternative wild-type strain PB1 (NCIMB13113). The physiological diversity of these two strains was demonstrated by a comparative fed-batch methanol cultivation displaying highly different methanol consumption and respiration profiles, as well as major differences in their l-glutamate production levels (406 mmol liter−1 and 11 mmol liter−1, respectively). Both genomes are small (ca 3.4 Mbp) compared to those of other related bacilli, and MGA3 has two plasmids (pBM19 and pBM69), while PB1 has only one (pBM20). In particular, we focus here on genes representing biochemical pathways for methanol oxidation and concomitant formaldehyde assimilation and dissimilation, the important phosphoenol pyruvate/pyruvate anaplerotic node, the tricarboxylic acid cycle including the glyoxylate pathway, and the biosynthetic pathways for l-lysine and l-glutamate. Several unique findings were made, including the discovery of three different methanol dehydrogenase genes in each of the two B. methanolicus strains, and the genomic analyses were accompanied by gene expression studies. Our results provide new insight into a number of peculiar physiological and metabolic traits of B. methanolicus and open up possibilities for system-level metabolic engineering of this bacterium for the production of amino acids and other useful compounds from methanol.
INTRODUCTION
Methylotrophic microorganisms can utilize one-carbon (C1) sources, such as methane and methanol, as their sole sources for energy and biomass generation, and there exist a variety of different enzymes and pathways for C1 metabolism among methylotrophs (14, 15). Bacteria that fix formaldehyde by the ribulose monophosphate (RuMP) pathway belong to three groups: Gram-negative obligate methylotrophs, Gram-positive facultative methylotrophs, and thermotolerant bacilli (3, 4, 19, 38). A number of Gram-positive thermotolerant bacilli with the ability to grow on methanol at temperatures up to 60°C have been isolated, and they were later collectively classified as Bacillus methanolicus (for a review, see reference 11). B. methanolicus is a so-called restricted methylotroph, which means that it can utilize few multicarbon sources for energy and growth. The scientific interest of these organisms has mainly been dedicated to exploring their potential as cell factories for industrial production of l-lysine and l-glutamate from methanol at elevated temperatures. B. methanolicus MGA3 (ATCC 53907) was isolated from soil samples in Minnesota (38), and it has been the major model strain used for metabolic engineering of this bacterium (9, 11, 27).
B. methanolicus has several additional unique traits, including (i) a novel NAD-dependent methanol dehydrogenase (MDH) for methanol oxidation (5, 18, 24), (ii) the ability to secrete up to 60 g liter−1 of l-glutamate (equal to 408 mmol liter−1) in fed-batch methanol cultivation (9, 12), (iii) the ability to grow in cheap seawater-based media (29), and (iv) plasmid-dependent methylotrophy (10, 26). B. methanolicus MGA3 has a natural plasmid, pBM19, carrying mdh and five RuMP pathway genes, and the curing of pBM19 resulted in loss of the ability to grow on methanol. Methanol consumption by this organism involves the concerted recruitment of both plasmid and chromosomal genes, and this discovery represented the first documentation of plasmid-dependent methylotrophy (10, 26). Methanol oxidation is governed by MDH, which is catalytically activated by Act, a member of the Nudix hydrolase family (24, 28). A linear dissimilatory pathway for direct oxidation of formaldehyde to carbon dioxide has been documented based on 13C nuclear magnetic resonance (NMR) data (35), but genetic evidence has been lacking.
Wild-type strains of B. methanolicus produce only about 0.2 g liter−1 l-lysine in fed-batch cultivation, while classical mutants secreting up to 47 g liter−1 of l-lysine (equal to 322 mmol liter−1) have been selected (9, 11, 22). In recent years, several assumed key genes involved in l-lysine biosynthesis have been cloned, and functional expression tools have been developed. Recombinant B. methanolicus strains producing up to 11 g liter−1 of l-lysine have been constructed by manipulation with single genes representing the aspartate pathway (9, 27). We recently showed that multiple genes of the aspartate pathway can play important roles in l-lysine overproduction by B. methanolicus (32). Manipulation with enzymes representing the phosphoenol pyruvate (PEP) and pyruvate anaplerotic node and the entry into the tricarboxylic acid (TCA) cycle has indicated that the oxaloacetate (OAA) precursor supply is not a major bottleneck for l-lysine overproduction by B. methanolicus (9, 12). However, this has to be investigated further with additional relevant genes.
B. methanolicus MGA3 obviously represents a potentially interesting candidate for industrial production of l-glutamate from methanol. In addition, when the goal is to overproduce l-lysine, efficient modes to reduce unwanted l-glutamate production are desirable. Again, genetic insight into the l-glutamate biosynthetic and degradation pathways, as well as the transport capacities, is still lacking and represents a bottleneck for metabolic engineering, aiming at understanding, increasing, and controlling the l-glutamate production by this organism. We present here the genome sequences of B. methanolicus MGA3 and the physiologically very different alternative model strain PB1 (NCIMB13113). The genomes of a number of methylotrophic bacteria have been sequenced during recent years (for a review, see reference 15), and the present report represents the first genome sequences of methylotrophic bacteria belonging to the class of Gram-positive thermotolerant bacilli.
MATERIALS AND METHODS
Bacterial strains, growth media, and recombinant DNA procedures.
B. methanolicus wild-type strains MGA3 (ATCC 53907) and PB1 (NCIMB13113) were generally grown in shake flasks at 50°C in 100 ml of MeOH200 medium containing 200 mM methanol, in Mann10 medium containing 20 g liter−1 mannitol, or in SOBsuc medium (26). When B. methanolicus strains were tested for their ability to grow on alternative C sources, cells were grown in MeOH200 medium but with the methanol replaced with 2% (wt/vol) maltose, raffinose, ribose, fructose, sorbitol, or glucose. Amino acid content was analyzed as previously described (9). Standard recombinant DNA techniques were applied according to reference 37. Escherichia coli DH5α (Invitrogen) was used as the standard cloning host and was generally grown at 37°C in liquid or solid Luria-Bertani (LB) medium supplemented with ampicillin (0.2 g liter−1) when appropriate. Plasmid DNA was isolated by the Wizard Plus SV Minipreps DNA Purification system (Promega), and linear DNA fragments were extracted from agarose gel slabs or PCR mixtures by the QIAquick gel extraction or PCR purification kit (Qiagen). DNA was amplified by the Expand High Fidelity PCR system (Roche), and routine DNA sequencing was performed by Eurofins MWG Operon.
Fed-batch methanol cultivation.
Fed-batch methanol cultivation of B. methanolicus strains MGA3 and PB1 was performed in UMN1 medium containing 150 mM methanol, essentially as described before (27), using the following settings: the initial culture volume was 0.75 liter, and the inoculum was 70 ml divided by the optical density at 600 nm (OD600) of the inoculum at the time of harvest (1.1 to 1.3). The initial and minimum agitation was 450 rpm, and the aeration rate was kept constant at 0.5 vol/vol/min. Agitation was controlled automatically to maintain a dissolved oxygen level of 30%. An automatic enrichment of oxygen up to 60% O2 was applied once the agitation reached 1,300 rpm and continued in combination with the automatic control of agitation until the end of the cultivation, 50 h after inoculation. The initial use of 0.03% (vol/vol) Antifoam 204 (Sigma) and the supply of 4% Antifoam 204 in the methanol feed solution made further batch additions of antifoam agent dispensable during the cultivation. A conversion factor of 0.24 g liter−1 (dry weight) of cells per OD600 unit was used for determination of the dry weight of cells, and the OD600 range for collection of dry weight measurements of cells was between 18 and 53. The correction factors used for the determination of amino acid concentrations were between 1.15 and 1.4 for endpoint samples, based on the volume changes during the cultivation.
Preparation of total DNA and genome sequencing.
Total DNA was isolated from exponentially growing B. methanolicus cells by phenol-chloroform-isoamyl alcohol (25:24:1) extraction and was dissolved in Tris-EDTA (TE) buffer. The quality and concentration of the DNA obtained were verified spectrophotometrically using a NanoDrop ND-1000 spectrophotometer and also by DNA gel electrophoresis. About 300 μl of MGA3 DNA with a concentration of 0.29 g liter−1 was shipped to 454 Life Sciences for pyrosequencing and draft sequence assembly, while 240 μl of PB1 DNA with a concentration of 0.163 g liter−1 was shipped to the Norwegian High-Throughput Sequencing Centre (www.sequencing.uio.no) for 8-kb paired-end GS FLX sequencing. The PB1 draft sequence assembly was built using Newbler at the freely available Bioportal computing service (www.bioportal.uio.no).
Sequence annotation and genome comparison.
The draft MGA3 genome sequence, consisting of 87 large contigs (>500 bp), was submitted to the JCVI Annotation Service, where it was run through JCVI's prokaryotic annotation pipeline (www.jcvi.org/cms/research/projects/annotation-service/) and to the Bacterial Annotation System, BASys (http://basys.ca/basys/cgi/submit.pl) (40). A total of 120 large contigs (9 contigs of 517 to 3,068 bp were found to appear in 2 to 15 near-identical copies) were connected by the use of two different strategies. A total of 16 gaps were closed by connecting the coding sequences of partial genes located at contig ends, or of genes found at contig ends known to be colocalized. The remaining 94 gaps were connected using a primer walking strategy. All contig ends were inspected, and sites for the restriction enzymes EcoRI, BamHI, HindIII, PvuII, AgeI, AflIII, EcoRV, or XhoI were identified. MGA3 genomic DNA was individually digested with each of these enzymes, and the fragmented DNA was circularized by T4 DNA ligase. PCR fragments covering the individual contig ends were amplified from these circularized DNA fragments using primers that annealed close to the contig ends (oriented toward the unknown sequence) in addition to a primer that annealed close to the appropriate restriction site found within the ∼1,000 bp from a contig end (oriented away from the end). The resulting PCR products were then submitted to sequencing directly (if shorter than 1.5 kb) or, alternatively, were subcloned into pGEM-T (Promega) before sequencing. All contig connections were verified by PCR amplification from a genomic template, followed by sequencing. The final genome consists of 10 large contigs and two plasmids and was resubmitted to the JCVI Annotation Service and to the NCBI Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP) (www.ncbi.nlm.nih.gov/genomes/static/Pipeline.html) for final annotation. The PGAAP result formed the basis for the published GenBank data but was manually curated using BLAST (2) before submission.
The draft PB1 genome sequence, consisting of 82 large contigs organized in 3 scaffolds, was submitted to BASys and the RAST (Rapid Annotation using Subsystem Technology) Annotation server (http://rast.nmpdr.org/) for annotation (6). The scaffolds contained a total of 30 internal gaps, of which 28 were closed by PCR followed by sequencing. All the gaps turned out to contain highly repetitive sequences. The smallest scaffold, scaffold 3, consisted of one large (covering 18,763 bp) and one small (covering 1,424 bp) contig. The large contig covers the main part of the sequence known to represent the pBM19 plasmid in MGA3, while the small part was also found in two of the closed gaps and was later connected to the 5′ end of scaffold 2. Previous experiments have indicated that the PB1 strain contains a pBM19-like plasmid that is slightly larger than that in the MGA3 strain (10). Analysis using the Newbler contig graf viewer (data not shown) suggested that the scaffold 3 sequence is not found in a plasmid context but rather in two copies, interconnected with scaffold 2 and scaffold 1. PCR experiments succeeded in linking the 5′ ends of scaffold 2 with the small contig of scaffold 3; however, all attempts at connecting the large contig of scaffold 3 to scaffold 1 or 2 failed, as did attempts at closing the final gap in scaffold 1 (between contig 1 and contig 2) or the gap between the small and large contigs of scaffold 3. PCR experiments using either PB1 total DNA or isolated PB1 plasmid DNA as the template and primers annealing to the two ends of the large contig of scaffold 3 finally showed that both contigs of scaffold 3 are parts of a large, 20,187-bp plasmid (pBM20) but that the small contig is oriented in the inverse direction from that suggested by Newbler.
Microarray experimental design and microarray construction of strain MGA3.
B. methanolicus MGA3 was grown methylotrophically on methanol and nonmethylotrophically on mannitol, and the gene expression profiles were compared. The experiment was performed in triplicate, from bacterial cultures grown on 3 different days. Custom gene expression microarrays for B. methanolicus (denoted MGA3_1) were designed with the help of eArray software (version 6.0; Agilent Technologies), according to the manufacturer's recommendations. Each customized microarray contained spots in quadruplicate, and the data set was duplicated, representing all the putative 3,641 coding sequences found in the initial draft sequence of B. methanolicus MGA3.
Isolation of total RNA, cDNA labeling, hybridization, and microarray scanning.
Total RNA was isolated using the RNAqueous kit (Ambion). The quality of the RNA preparation was verified by an Agilent Bioanalyzer. Fluorescent Cy3-labeled cDNA was generated using the SuperScript indirect labeling system (Invitrogen), where CyDye fluorescent dyes (GE Healthcare Biosciences) were coupled to the amino allyl cDNA according to the manufacturer's instructions. Microarray hybridizations were carried out using the labeled cDNA together with the In Situ Hybridization Kit Plus (Agilent Technologies). Arrays were incubated at 65°C for 17 h in Agilent microarray hybridization chambers and were subsequently washed according to the Agilent protocol. The washed microarrays were scanned immediately in a DNA Microarray Scanner, model G2567AA (Agilent Technologies). The Feature Extraction software, version 9.5.1.1 (Agilent Technologies), was used for data extraction and quality control. The Agilent standard scenario for normalization was applied to all data sets. Extracted data were analyzed using the GeneSpring GX 10.0.2 software (Silicon Genetics). A 2-fold change (P value log ratio, <0.01) was used as the threshold for targeting genes differently expressed when grown methylotrophically versus nonmethylotrophically.
cDNA synthesis and qPCR.
Quantitative PCR (qPCR) was used to compare the transcription levels of relevant B. methanolicus genes in cells growing methylotrophically on methanol with those in cells growing nonmethylotrophically on mannitol (26), and cDNA was synthesized using the First-Strand cDNA synthesis kit (Amersham Biosciences). qPCR analyses were performed using the ABI Prism 7700 sequence detection system with its default settings (Applied Biosystems) essentially as previously described (9). In brief, the qPCR primers were chosen with the assistance of the Primer Express 2.0 software (Applied Biosystems). The PCR mixture consisted of 12.5 μl of 2× SYBR Universal PCR Master Mix, 600 nM primers, 5 μl cDNA (0.25 g liter−1), and H2O to a final volume of 25 μl. Default cycling conditions were used. Relative quantification of the genes in question was done by normalizing the results, relative to 16S RNA (endogenous control) and a calibrator sample, using a comparative threshold cycle (CT) method (2−ΔΔCT method) according to the PE Applied Biosystems protocol (23, 30).
Primer pair sequences used were as follows: mdh2_f, 5′-GGATACATGTCAAACACTCAAAGTGC-3′; mdh2_r, 5′-TCTAGACACCATCGCATTTTTAATAATTTGG-3′; mdh3_f, 5′-GGATACATGTAAAACACTCAAAGTGC-3′; mdh3_r, 5′-TCTAGACACCATAGCATTTTTAATAATTTGGATG-3′; act_f, 5′-CCTGGTGCTGTAGCTGTAAT-3′; act_r, 5′-GCTTCGTCAAGTGTCAGTTC-3′; hps_f, 5′-GTAGCTGAGGTTCAGGAGTA-3′; hps_r, 5′-CTGCGATCATGTCAACAAGG-3′; phi_f, 5′-GAAGAAGCCGAAGCACTGGT-3′; phi_r, 5′-TGATCGTTACAGCCGCGATG-3′; pfk2_f, 5′-GTTACTGTTCTTGGCCATGTTCAG-3′; pfk2_r, 5′-TTGCAAGCACACGGTCAAA-3′; fba2_f, 5′-GCGAGAGGCGTTTCTGTTG-3′; fba2_r, 5′-CTGCGATTACGTCATCCTCTTG-3′; tkt2_f, 5′-ACGGTCATACACCTGGAGTTGA-3′; tkt2_r, 5′-CCATTCCAACTGCCATAGCA-3′; tal_f, 5′-GCCACCCGCAGCATGT-3′; tal_r, 5′-TGTAAGGCACAGTCGCAATATGA-3′; glpX2_f, 5′-CTGCAGTAGCGCTGAAATGC-3′; glpX2_r, 5′-GCATCATTTTGAGGCAAAAGC-3′; rpe2_f, 5′-CGGCGGAGTGAATAACGAAA-3′; rpe2_r, 5′-CGGCGGATCCTGCTACTAAC-3′; rpiB_f, 5′-GCCGGTTGCGGAAAAAGT-3′; rpiB_r, 5′-CGATACCTGTTCCGCAAATTAAG-3′; pgi_f, 5′-ACGGAACCAGCGATTGCA-3′; pgi_r, 5′-TTCGGATTTTCCGTAAATTGCTTCAGGAAA-3′; zwf1_f, 5′-GGAAACTGGAACGGTTTGCA-3′; zwf1_r, 5′-TATTGGCGTTGCACGAAGGCCA-3′; zwf2_f, 5′-CCACAATCATTTTCCGTTATTGG-3′; zwf2_r, 5′-CTAGGGCGAAGAGATTGGTCGGATAATGA-3′; pgl_f, 5′-TGAGTTAAAAGAAAAGAATTCCTTGTCA-3′; pgl_r, 5′-TAAAGCCGGGAAGCGGACCAAGA-3′; gnd_f, 5′-GCGAACATGGCATCCACTTTA-3′; gnd_r, 5′-CGGTACAGGAGTTTCCGGCGGG-3′; folD_f, 5′-ATGGGATCATGGTTTTACTTAAGGAA-3′; folD_r, 5′-GCTCCGCCCGATCACA-3′; fhs_f, 5′-TTGGCAAAAAGGCTATGATAGCT-3′; fhs_r, 5′-GCTCCTCCTTTTATTCCCATTGTT-3′; fdhA_f, 5′-GTATGGACCGTCGTTTTTTTCAC-3′; fdhA_r, 5′-CCAGCTGCAGAACAGATGGA-3′; fdhD_f, 5′-AGGATTTGCTTACGTGGATTTAGC-3′; fdhD_r, 5′-CAGCACGAGCCAATAAATCG-3′. The primer pair sequences of the pBM19 genes have been published previously (26).
Nucleotide accession numbers.
The Whole Genome Shotgun projects have been deposited at DDBJ/EMBL/GenBank under accession numbers ADWW00000000 for MGA3 and AFEU00000000 for PB1. The versions described in this paper are the first versions, ADWW01000000 and AFEU01000000.
Microarray data accession number.
The complete set of microarray data has been deposited in the Gene Expression Omnibus (GEO) database at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo/) under the accession series GSE35298 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE35298).
RESULTS AND DISCUSSION
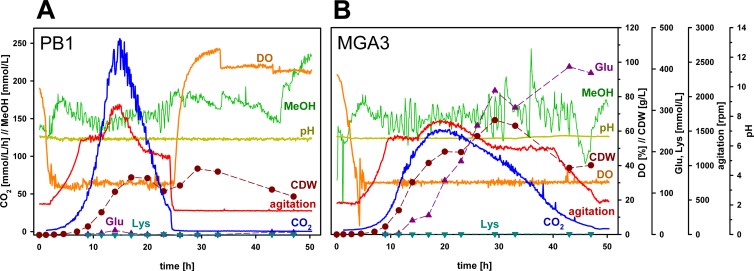
Fed-batch methanol cultivation displayed considerable physiological differences between the B. methanolicus wild-type strains MGA3 (ATCC 53907) and PB1 (NCIMB13113).
We previously presented a total of 14 different wild-type isolates of B. methanolicus (10), and among these, MGA3 has been the major model strain used for metabolic engineering (9, 11, 12, 26, 27, 32). The majority of these strains—including MGA3—have been isolated from soil samples in the Minnesota area, while one strain, PB1, has been isolated from a sugar beet factory wastewater treatment system in Europe (20). We made a physiological comparison of the two B. methanolicus wild-type strains MGA3 and PB1 by analyzing both strains in fed-batch methanol cultivation. These two strains displayed fundamental differences with respect to growth, amino acid production, and respiration profiles (Fig. 1). The maximum biomass yield of PB1 is substantially lower than that of MGA3, and PB1 produces very little l-glutamate (11 mmol liter−1) compared to MGA3 (406 mmol liter−1). Also, the CO2 evolution profiles of the two strains were highly different. Maximum l-lysine production levels were below 2 mmol liter−1 for both strains. Whether the observed differences in agitation between the two strains had any effect on these data remained unknown. The data for MGA3 were similar to analogous data presented previously under such conditions (12, 26), and together these data indicate physiological differences between these two wild-type strains. In order to generate a solid basis for understanding the similarities and differences in cell biology and physiology of B. methanolicus strains at a genome-scale level, we decided to include both wild-type strains as models for genome sequencing.
FIG 1.
Summary of on-line and off-line analyses from fed-batch methanol cultivation of PB1 (A) and MGA3 (B) in UMN1 medium. CO2, CO2 evolution rate as a function of time (mmol liter−1 h−1); CDW, dry weight (g liter−1) of cells (filled circles), calculated from OD600 measurements by correlation to selected measurements of dry weights of cells; Glu, glutamate concentration in the medium (mmol liter−1) (filled triangles); Lys, lysine concentration in the medium (mmol liter−1) (filled inverted triangles); MeOH, methanol concentration (mmol liter−1), determined by in-line mass spectrometric measurement in the exhaust gas and automatically held constant at 150 mmol liter−1; DO, dissolved oxygen (%). Values for CO2, dry weight of cells, Glu, and Lys are corrected for volume changes during cultivation.
Assembly, annotation, and general features of the B. methanolicus MGA3 and PB1 genomes.
The B. methanolicus MGA3 genome was sequenced with 20-fold coverage, and the direct outcome was 87 large contigs totally encoding about 3.4 Mb of DNA sequence. After manual gap filling using sequence alignments and PCR, the total number of large contigs was reduced to 12, including the previously described plasmid pBM19 (10) and the 69-kb plasmid pBM69 discovered here (see below). Nine contigs of 517 to 3,068 bp were found to appear in 2 to 15 near-identical copies mainly associated with genes encoding recombinases, transposases, and the 16S-23S-5S rRNA clusters. All attempts to experimentally close these remaining gaps failed due to highly repetitive DNA sequences in these regions. The average size of the 110 manually filled gaps was 251 bp, with the majority of the gap sequences being represented by the original 31 small contigs (101 to 449 bp), while the average length of new sequence was 79 bp. The size of the genome represented within the remaining 12 contigs is 3,398,992 bp (3,399 kb), and based on the average size of the manually filled gaps (249 bp), we estimated that this should represent at least 99.93% of the total MGA3 genome sequence.
The B. methanolicus PB1 genome was sequenced in 8-kb paired ends with 23-fold coverage, and the direct outcome from the Newbler assembly was 30 large contigs organized in 3 scaffolds, totally encoding about 3.4 Mb of DNA sequence. After manual gap filling using sequence alignments and PCR, the total number of large contigs was reduced to 4, including a pBM19-like plasmid (see below). In addition, 3 smaller contigs (contigs 5, 6, and 7, of 1,484 bp, 759 bp, and 304 bp, respectively), which had not been included in the original scaffolds and were not connected in the manual gap filling, were included in the final genome, since they were found to encode missing tRNA and rRNA genes organized in the same order as in the MGA3 strain. The size of the genome represented within the remaining 7 contigs is 3,426,513 bp (3,426 kb), and based on the average size of the manually filled gaps (123 bp), we estimate that this should represent at least 99.98% of the total PB1 genome sequence.
Both genome sequences were automatically annotated by using the services of JCVI (formerly TIGR), BASys (40), and PGAAP (from NCBI). In addition, genes of particular interest were all manually annotated using BLAST (2). The overall GC contents of both genomes were low, 38.5% (MGA3) and 39.0% (PB1), well below the GC contents of Bacillus subtilis and Bacillus halodurans (Table 1). Both the genome size and the number of open reading frames (ORF) were low compared to those of B. subtilis, and about 75% of all ORFs identified encoded proteins with annotated functions.
TABLE 1.
Genome features of selected Bacillus spp.
| Characteristic |
B. methanolicus |
B. subtilis 168 | B. halodurans C-125 | |
|---|---|---|---|---|
| MGA3 | PB1 | |||
| No. of large contigsa | 10 (+2)b | 3 (+1) | 1 | 1 |
| No. of plasmids | 2 (19 kb, 69 kb) | 1 (20 kb) | 0 | 0 |
| Avg contig size (bp) | 283,249 | 856,628 | ||
| Total size (bp) | 3,398,992 | 3,426,513 | 4,215,606 | 4,202,353 |
| GC content (%) | 38.5 | 39.0 | 43 | 43 |
| No. of predicted protein coding sequences | 3,435 | 3,410 | 4,100 | 4,066 |
| Status | Partially assembled | Partially assembled | Complete | Complete |
| GenBank accession no. | ADWW00000000 | AFEU00000000 | AL009126 | BA000004 |
>5 kb.
Numbers in parentheses refer to the plasmids.
B. methanolicus plasmids.
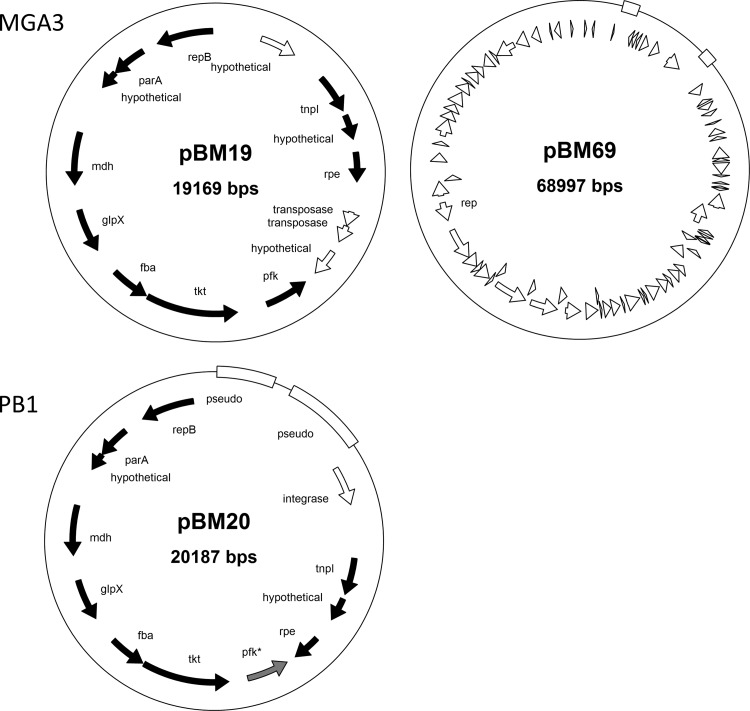
In contrast to B. subtilis and B. halodorans, which carry no natural plasmids (Table 1), B. methanolicus strains harbor plasmids. The pivotal role of pBM19 for B. methanolicus MGA3 methylotrophy has been well documented by us (10, 26). We show here that the corresponding PB1 plasmid, designated pBM20, is 20 kb and contains mdh and RuMP pathway genes, analogously to pBM19. The phosphofructokinase gene of pBM20, pfk, contained a 2-nucleotide insertion leading to a frameshift mutation and a concomitant translational stop codon that should render this gene nonfunctional. This implies that this strain must recruit Pfk activity needed for the RuMP pathway from an alternative gene located on the chromosome (see below). We also noticed that pBM19 and pBM20 are somewhat different with respect to the remaining genes and their organization (Fig. 2). Nonetheless, the biological impact of these discrepancies remains unknown.
FIG 2.
Physical maps of plasmids pBM19 and pBM69 from MGA3 and of plasmid pBM20 from PB1. Filled arrows show genes that are conserved between pBM19 and pBM20, while open arrows and boxes indicate coding regions that differ between the plasmids. The pfk gene in pBM20 is nonfunctional due to a frameshift mutation. For more detailed information about the coding sequences in pBM69, see GenBank accession code ADWW01000012 (locus tags MGA3_17472 to MGA3_17882). For the pBM19 and pBM20 genes, see the text.
We also discovered that MGA3 has one additional, circular, 69-kb large plasmid designated pBM69, with 79 predicted coding sequences (CDSs), including a putative replication initiation protein gene. Its deduced gene product shared 52% overall primary sequence identity to the replication initiation protein of plasmid pBMB67 in Bacillus thuringiensis (13). The remaining CDSs of pBM69 encoded a number of hypothetical and unknown proteins with no assigned roles in cell metabolism. No plasmid analogous to pBM69 was identified in PB1, and the relevant CDSs were also not found on the PB1 chromosome.
B. methanolicus carries three mdh genes and one act gene.
The methanol dehydrogenase gene mdh was originally cloned and characterized from B. methanolicus strain C1 (21), and later we showed that the corresponding mdh gene of B. methanolicus MGA3 was located on plasmid pBM19 (10). Inspection of the MGA3 genome sequence unraveled two additional chromosomal genes, denoted mdh2 and mdh3, both encoding putative MDH proteins. Still, only one chromosomal act gene, encoding the MDH activator protein (28), was found (Table 2). The deduced Mdh2 and Mdh3 proteins were 96% identical to each other, and they shared only 61% and 62% overall primary sequence identity to the pBM19-encoded Mdh protein.
TABLE 2.
Genes involved in methanol oxidation and concomitant formaldehyde metabolism in the B. methanolicus MGA3 genome sequence and their counterparts in PB1
| Locus tag in MGA3 | Locus tag in PB1 | % Amino acid identity | Common name | Gene | EC no. | Localizationa | Pathwayb | Upregulated by methanolc |
|---|---|---|---|---|---|---|---|---|
| MGA3_17392 | PB1_17533 | 93 | Methanol dehydrogenase | mdh | 1.1.1.244 | pBM19 | MO | Yes |
| PB1_14569 | 89d | Methanol dehydrogenase 1 | mdh1d | 1.1.1.244 | NA | |||
| MGA3_07340 | PB1_12584e | 91e | Methanol dehydrogenase 2 | mdh2 | 1.1.1.244 | C | MO | No |
| MGA3_10725 | Methanol dehydrogenase 3 | mdh3 | 1.1.1.244 | C | MO | No | ||
| MGA3_09170 | PB1_14394 | 97 | Methanol dehydrogenase activator protein | act | C | MO | No | |
| MGA3_15306 | PB1_00480 | 98 | 3-Hexulose-6-phosphate synthase | hps | 4.1.2.43 | C | FA | Yes |
| MGA3_15301 | PB1_00485 | 98 | 6-Phospho-3-hexuloisomerase | phi | 5.3.1.27 | C | FA | Yes |
| MGA3_17457 | 6-Phosphofructokinase | pfk | 2.7.1.11 | pBM19 | FA | Yes | ||
| MGA3_03000 | PB1_16819 | 96 | 6-Phosphofructokinase-2 | pfk2 | 2.7.1.11 | C | FA | No |
| MGA3_17377 | PB1_17523 | 96 | Fructose-bisphosphate aldolase | fba | 4.1.2.13 | pBM19 | FA | Yes |
| MGA3_01355 | PB1_07992 | 94 | Fructose-bisphosphate aldolase-2 | fba2 | 4.1.2.13 | C | FA | No |
| MGA3_17462 | PB1_17518 | 97 | Transketolase | tkt | 2.2.1.1 | pBM19 | FA | Yes |
| MGA3_15171 | PB1_00595 | 94 | Transketolase-2 | tkt2 | 2.2.1.1 | C | FA | No |
| MGA3_01350 | PB1_07997 | 90 | Transaldolase | tal | 2.2.1.2 | C | FA | No |
| MGA3_17382 | PB1_17528 | 96 | Fructose-bisphosphatase | glpX | 3.1.3.11 | pBM19 | FA | Yes |
| MGA3_01340 | PB1_08007 | 97 | Fructose-bisphosphatase-2 | glpX2 | 3.1.3.11 | C | FA | No |
| MGA3_17437 | PB1_17503 | 94 | Ribulose-phosphate 3-epimerase | rpe | 5.1.3.1 | pBM19 | FA | Yes |
| MGA3_14311 | Ribulose-phosphate 3-epimerase-2 | rpe2 | 5.1.3.1 | C | FA | No | ||
| MGA3_01280 | PB1_08082 | 95 | Ribose 5-phosphate isomerase | rpiB | 5.3.1.6 | C | FA | No |
| MGA3_16421 | PB1_10192 | 95 | Glucose-6-phosphate isomerase | pgi | 5.3.1.9 | C | FOI | No |
| MGA3_09280 | PB1_14524 | 94 | Glucose-6-phosphate dehydrogenase | zwf1 | 1.1.1.49 | C | FOI | ND |
| MGA3_15311 | PB1_00475 | 91 | Glucose-6-phosphate dehydrogenase | zwf2 | 1.1.1.49 | C | FOI | Yes |
| MGA3_11305 | PB1_04625 | 93 | 6-Phosphogluconolactonase | pgl | 3.1.1.31 | C | FOI | No |
| MGA3_09290 | PB1_14534 | 96 | 6-Phosphogluconate dehydrogenase, decarboxylating | gnd | 1.1.1.44 | C | FOI | No |
| MGA3_09460 | PB1_14689 | 88 | 5,10-Methylene-tetrahydrofolate dehydrogenase/5,10-methylene-tetrahydrofolate cyclohydrolase | folD | 1.5.1.5/3.5.4.9 | C | FOV | No |
| MGA3_08300 | PB1_13509 | 94 | Formate-tetrahydrofolate ligase | fhs | 6.3.4.3 | C | FOV | No |
| MGA3_06625 | PB1_11719 | 94 | Formate dehydrogenase, alpha chain | fdhA | 1.2.1.2 | C | FOV | No |
| MGA3_06630 | PB1_11724 | 86 | Formate dehydrogenase family accessory protein | fdhD | C | FOV | No |
C, chromosome.
MO, methanol oxidation; FA, formaldehyde assimilation II (RuMP cycle); FOI, formaldehyde oxidation I (cyclic pentose phosphate pathway); FOV, formaldehyde oxidation V (tetrahydrofolate pathway).
Yes or No, upregulated transcription or not upregulated transcription, respectively, on methanol versus mannitol in MGA3. NA, not analyzed; ND, not detectable.
mdh1 is found only in PB1 and is compared to the MGA3 mdh gene.
The PB1 counterpart of mdh2/mdh3 has been given the gene designation mdh2 and is 92% identical to mdh3.
Inspection of the PB1 genome sequence confirmed the presence of three methanol dehydrogenase genes, one located on pBM20 and two on the chromosome, and one chromosomal act gene (Table 2). The mdh gene located on pBM20 was 92% identical to the MGA3 mdh gene located on pBM19, and the respective deduced gene products displayed 93% primary sequence identity. One of the chromosomal PB1 genes, denoted mdh1, encoded an Mdh1 protein with 92% and 89% primary sequence identity to the PB1 and MGA3 Mdh proteins, respectively. The second chromosomal PB1 gene, denoted mdh2, encoded a putative Mdh2 protein with 91% and 92% primary sequence identity to the MGA3 Mdh2 and Mdh3 proteins, respectively, and only 59% and 60% identity to the PB1 Mdh and Mdh1 proteins, respectively. We are currently in the process of recombinant production, purification, and biochemical characterization of all six MDH proteins.
B. methanolicus encodes chromosomal homologues to all the RuMP pathway genes of pBM19 and pBM20.
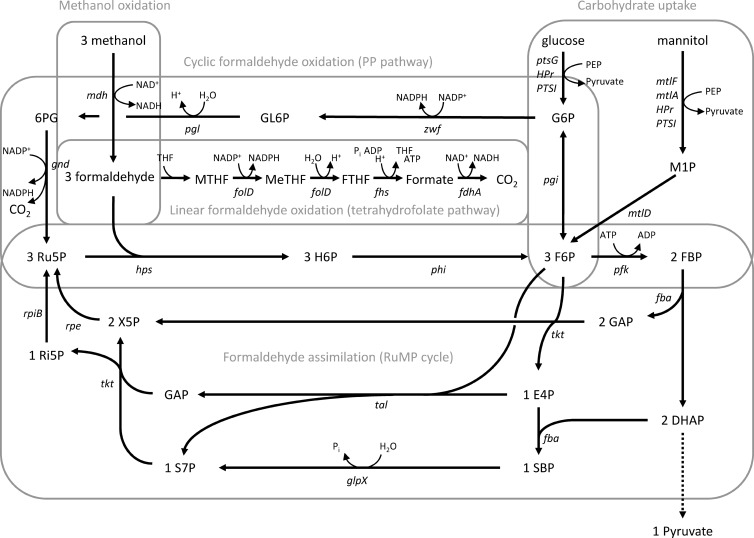
Inspection of the B. methanolicus MGA3 genome sequence revealed a number of hitherto unknown, putative RuMP pathway genes (Table 2; Fig. 3). A tal gene encoding transaldolase was identified, suggesting that this bacterium has all the genes putatively representing both a fructose bisphosphate aldolase/seduheptulose bisphosphatase (FBPA/SBPase) variant (26) and a FBPA/transaldolase (TA) regeneration variant of the RuMP pathway (see reference 10). Whether both of these variants are biologically significant during the methylotrophic growth of B. methanolicus remains unknown. Moreover, a rpiB gene (encoding ribose-5-phosphate isomerase) was found, which likely represents the previously missing step of the RuMP pathway regeneration phase, the conversion of ribose-5-phosphate to ribulose-5-phosphate (10). The rpiB gene would play a role in the RuMP pathway irrespective of the variant of the regeneration phase being used. Homologues representing all five RuMP pathway genes located on pBM19 were found on the chromosome (pfk2, encoding 6-phosphofructokinase-2; fba2, encoding fructose-bisphosphate aldolase-2; tkt2, encoding transketolase-2; glpX2, encoding fructose bisphosphatase-2; rpe2, encoding ribulose-5-phosphate 3-epimerase-2). Taken together with hps, phi, rpiB, and possibly also tal, these findings imply that B. methanolicus has a complete assimilatory RuMP pathway solely encoded by chromosomal genes. The same observations overall were made in the PB1 genome sequence (Table 2), except that this strain has no chromosomal rpe2 homologue, and pBM20 carries no functional pfk gene (see above).
FIG 3.
Proposed methylotrophic and important carbohydrate assimilation pathways in B. methanolicus. Genes: mdh, methanol dehydrogenase (EC 1.1.1.244); hps, 3-hexulose-6-phosphate synthase (EC 4.1.2.43); phi, 6-phospho-3-hexuloisomerase (EC 5.3.1.27); pfk, 6-phosphofructokinase, (EC 2.7.1.11); fba, fructose-bisphosphate aldolase (EC 4.1.2.13); tkt, transketolase (EC 2.2.1.1); glpX, fructose-bisphosphatase (EC 3.1.3.1); tal, transaldolase (EC 2.2.1.2); rpe, ribulose-phosphate 3-epimerase (EC 5.1.3.1); rpiB, ribose-5-phosphate isomerase (EC 5.3.1.6); folD, 5,10-methylene-tetrahydrofolate dehydrogenase/5,10-methenyl-tetrahydrofolate cyclohydrolase (EC 1.5.1.5/EC 3.5.4.9); fhs, formate-tetrahydrofolate ligase (EC 6.3.4.3); fdhA, formate dehydrogenase (EC 1.2.1.2); pgi, glucose-6-phosphate isomerase (EC 5.3.1.9); zwf, glucose-6-phosphate dehydrogenase (EC 1.1.1.49); gnd, 6-phosphogluconate dehydrogenase (EC 1.1.1.44); pgl, 6-phosphogluconolactonase (EC 3.1.1.31); PTSI, phosphoenolpyruvate–protein phosphotransferase (EC 2.7.3.9); HPr, phosphocarrier protein; ptsG, PTS-glucose-specific transporter subunit IICBA (EC 2.7.1.69); mtlF, mannitol-specific phosphotransferase enzyme IIA component (EC 2.7.1.69); mtlA, PTS system mannitol-specific enzyme IIBC component (EC 2.7.1.69); mtlD, mannitol-1-phosphate 5-dehydrogenase (EC 1.1.1.17). Metabolites: H6P, 3-hexulose 6-phosphate; F6P, fructose-6-phosphate; FBP, fructose-1,6-bisphosphate; GAP, glyceraldehyde 3-phosphate; DHAP, dihydroxyacetone phosphate; E4P, erythrose-4-phosphate; SBP, sedoheptulose-1,7-bisphosphate; S7P, sedoheptulose-7-phosphate; Ri5P, ribose-5-phosphate; X5P, xylulose-5-phosphate; Ru5P, ribulose-5-phosphate; MTHF, methylene tetrahydrofolate; MeTHF, methenyl tetrahydrofolate; FTHF, formate tetrahydrofolate; G6P, glucose-6-phosphate; GL6P, gluconolactone 6-phosphate; 6PG, 6-phosphogluconate; M1P, mannitol-1-phosphate.
B. methanolicus has genes representing both a cyclic dissimilatory RuMP pathway and a linear tetrahydrofolate pathway for formaldehyde dissimilation.
A linear dissimilatory pathway for formaldehyde oxidation via formate and to CO2 in B. methanolicus MGA3 has been experimentally documented by using 13C labeling experiments (35). This pathway was proposed to include formaldehyde dehydrogenase (FADH) and formate dehydrogenase (FDH) activities, as in B. subtilis. The B. methanolicus genome sequences contained no fadh gene. However, a putative fdh gene and a formate dehydrogenase family accessory protein gene, fdhD, were identified, as well as folD (encoding bifunctional methylene tetrahydrofolate dehydrogenase/methylene/methenyl tetrahydrofolate cyclohydrolase) and fhs (encoding formate tetrahydrofolate ligase) (Table 2). Together, these genes represent a putative linear dissimilatory tetrahydrofolate pathway for the oxidation of formaldehyde into CO2 (see Fig. 3). Such pathways are widely distributed among methylotrophic bacteria, and in addition to playing a role in formaldehyde detoxification and energy generation, they are also important for providing C1 units for biosynthetic reactions in the cells (41).
A set of genes, including pgi (encoding glucose-6-phosphate isomerase), zwf1, and zwf2 (encoding glucose-6-phosphate dehydrogenases), pgl (encoding 6-phosphogluconolactonase), and gnd (encoding 6-phosphogluconate dehydrogenase), were found that should represent—together with hps and phi—an alternative cyclic dissimilatory RuMP pathway for the oxidation of formaldehyde into CO2 and the regeneration of Ru5P (Fig. 3). This pathway is analogous to the oxidative pentose phosphate (PP) pathway widely distributed in bacteria. The zwf2 gene was colocalized with hps and phi as a putative operon on the chromosome (see below), while zwf1 was located elsewhere. The overall findings made here were similar in MGA3 and PB1 (Table 2), although the organization of the respective genes differed slightly. The cyclic dissimilatory RuMP pathway plays an important energetic role in generating reducing power [NAD(P)H], and it has been argued that it represents a substitute for the lack of a complete TCA cycle for such purposes in many methylotrophic bacteria (15).
Gene expression analysis of B. methanolicus MGA3 cells growing on methanol versus on mannitol.
Our findings here imply that B. methanolicus MGA3 has chromosomal genes presumably constituting complete pathways for methanol oxidation (mdh2, mdh3, and act), formaldehyde dissimilation (one linear tetrahydrofolate and one cyclic RuMP pathway), and formaldehyde assimilation (assimilatory RuMP pathway). We demonstrated previously that the pBM19-encoded RuMP pathway genes are transcriptionally upregulated in MGA3 cells growing on methanol (26), and it was of interest to extend our understanding regarding the regulation of gene expression upon methylotrophic versus nonmethylotrophic growth. Initially, we implemented a whole-genome gene expression microarray to compare the transcription levels of all genes in MGA3 cells growing on methanol versus on mannitol. In total, 102 genes were significantly upregulated in cells growing on methanol, while 64 genes were upregulated in cells growing on mannitol (for details, see http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE35298).
We then applied qPCR for targeted analyses of all genes with assigned putative roles in methanol consumption as listed in Table 2. In agreement with previous data, all five RuMP pathway genes positioned on pBM19 were transcriptionally upregulated by methanol (Table 2). In contrast, the transcription levels of the respective chromosomal homologues, as well as those of rpiB and tal, were not significantly affected by the carbon source. Taken together with the higher copy number of all pBM19 genes (10), it was tempting to assume that these chromosomal genes do not play important roles for methylotrophy in B. methanolicus. Considering that the pfk gene located on pBM20 in PB1 is inactive (see Fig. 2), it is plausible to believe that the overall regulation and role of RuMP pathway genes might be different in these two wild-type strains. The transcription level of zwf2 was 3-fold upregulated in cells growing on methanol compared to that in cells growing on mannitol, indicating that this gene is cotranscribed and is accordingly coregulated as part of an operon together with the RuMP pathway genes hps and phi. In contrast, zwf1 transcripts were hardly detectable under any growth conditions, suggesting that zwf1 is a pseudogene with no biological function. The transcription levels of the remaining putative cyclic dissimilatory RuMP pathway genes gnd, pgi, and pgl were 2- to 3-fold lower in cells growing on methanol than in cells growing on mannitol. It should be kept in mind that these genes most likely play important roles in the generation of reducing power and energy in cells during growth on mannitol (see below). Transcription of the putative dissimilatory tetrahydrofolate pathway genes folD, fhs, fdhA, and fdhD was similar on methanol and mannitol, suggesting that these genes are important for cell metabolism under both growth conditions tested. In total there was good agreement between the qPCR data and the corresponding microarray data.
B. methanolicus mannitol and glucose uptake systems.
B. methanolicus strains can grow on few multicarbon sources, and they are therefore collectively designated restricted methylotrophs (11). Testing of MGA3 and PB1 for growth in minimal medium with a number of different C sources (20 g liter−1) in this study revealed that both wild-type strains grew well on methanol, mannitol, and glucose, while they could not grow on maltose, raffinose, ribose, and sorbitol under the conditions tested. These results are somewhat different from those of previous reports stating that PB1 can grow on all the C sources mentioned above (4), and the reason for this discrepancy is unknown. The ability to grow rapidly on the alternative C source mannitol is a very important and useful trait, enabling the study of genetic regulation and physiology related to methylotrophy. In bacteria, mannitol and glucose enter the cells via mannitol-1-phosphate and glucose-6-phosphate, respectively, and are converted to fructose-6-phosphate, which can either be further metabolized via the Entner-Doudoroff pathway or, alternatively, oxidized via the PP pathway (Fig. 3). Gene clusters putatively involved in mannitol uptake and assimilation were identified in the B. methanolicus MGA3 and PB1 genomes (Table 3), including the genes mtlD (mannitol-1-phosphate 5-dehydrogenase), mtlF (mannitol-specific phosphotransferase [PTS] enzyme IIA component), mtlR (transcriptional regulator), and mtlA (mannitol-specific PTS enzyme IIBC component). These four genes are organized as an operon on the chromosome, as in Geobacillus stearothermophilus (GenBank accession no. BSU18943) (25) and similar to the pattern in B. subtilis (GenBank accession no. AL009126). Also, a ptsG gene putatively encoding a glucose uptake system was identified in both genome sequences (Table 3). The gene expression data (see above) showed that transcript levels of all the four mannitol uptake and assimilation genes are significantly higher in MGA3 cells growing on mannitol than in cells growing on methanol (Table 3). These data confirm that this gene cluster is active during growth on mannitol.
TABLE 3.
Genes involved in mannitol and glucose uptake and metabolism
| Locus tag in MGA3 | Locus tag in PB1 | % Amino acid identity | Common name | Gene | EC no. | Upregulated by mannitola |
|---|---|---|---|---|---|---|
| MGA3_12915 | PB1_06237 | 88 | Mannitol-1-phosphate 5-dehydrogenase | mtlD | 1.1.1.17 | Yes |
| MGA3_12920 | PB1_06242 | 97 | Mannitol-specific phosphotransferase enzyme IIA component | mtlF | 2.7.1.69 | Yes |
| MGA3_12925 | PB1_06247 | 88 | Transcriptional regulator | mtlR | Yes | |
| MGA3_12930 | PB1_06252 | 93 | PTS system mannitol-specific enzyme IIBC component | mtlA | 2.7.1.69 | Yes |
| MGA3_16763 | PB1_05647 | 92 | PTS-glucose-specific transporter subunit IICBA | ptsG | 2.7.1.69 | No |
| MGA3_16768 | PB1_05652 | 88 | BglG family transcriptional antiterminator | glcT | No | |
| MGA3_09280 | PB1_14524 | 94 | Glucose-6-phosphate dehydrogenase | zwf1 | 1.1.1.49 | ND |
| MGA3_15311 | PB1_00475 | 91 | Glucose-6-phosphate dehydrogenase | zwf2 | 1.1.1.49 | No |
| MGA3_13541 | PB1_02110 | 97 | Phosphocarrier protein HPr | hpr | No | |
| MGA3_13546 | PB1_02105 | 90 | PtsI—phosphoenolpyruvate-protein phosphotransferase | ptsP | 2.7.3.9 | No |
Yes or No, upregulated transcription or not upregulated transcription, respectively, on mannitol versus on methanol in MGA3.
ND, Not detectable.
The anaplerotic node around PEP and pyruvate.
Pyruvate and phosphoenol pyruvate (PEP) represent key precursor metabolites in all living cells, together with acetyl coenzyme A (acetyl-CoA), oxaloacetate (OAA), and α-ketoglutarate (1). The supply of these metabolites in the cells has a strong impact on the biotechnological production of amino acids and other useful compounds. For example, l-lysine overproduction relies on an efficient supply of the precursor metabolite OAA, while l-glutamate production relies on high entry of carbon into the TCA cycle. This implies that regulating the carbon flow at the anaplerotic node represented by PEP and pyruvate might be of crucial importance when the goal is to overproduce either l-lysine or l-glutamate. Inspection of the MGA3 and PB1 genome sequences confirmed that B. methanolicus has one single pyruvate carboxylase (PC)-encoding gene represented by the recently cloned pyc gene of MGA3 (9) and one putative PEP carboxykinase gene (pckA), but no PEP carboxylase gene. We also identified the pdhA and pdhB genes putatively encoding the large and small subunits, respectively, of pyruvate dehydrogenase (Pdh) (Table 4). For example, we believe that the reduced l-glutamate production in MGA3 cells upon PC overexpression (9) implies that there is an imbalance in the concentrations of OAA versus acetyl-CoA, eventually resulting in reduced entry of PEP and pyruvate into the TCA cycle. Possibly, pdhA and pdhB may represent two additional targets, together with pyc, to redirect and control carbon flow at this point. Moreover, alanine dehydrogenase can convert pyruvate into the amino acid l-alanine, thus representing an alternative route. B. methanolicus MGA3 cells produce substantial levels of this amino acid (13 g liter−1) in fed-batch methanol cultivation (9), and the ald gene (Table 4) might therefore represent another mutational target to maximize carbon flow from pyruvate toward l-lysine or l-glutamate.
TABLE 4.
Genes involved in the TCA cycle, glyoxylate shunt, and anaplerotic reactions
| Locus tag in MGA3 | Locus tag in PB1 | % Amino acid identity | Common name | Gene | EC no. | Pathwaya |
|---|---|---|---|---|---|---|
| MGA3_03025 | PB1_16794 | 97 | Citrate (Si)-synthase II | citY | 2.3.3.1 | TCA |
| MGA3_16693 | PB1_00055 | 96 | Aconitate hydratase | acnA | 4.2.1.3 | TCA |
| MGA3_03030 | PB1_16789 | 97 | Isocitrate dehydrogenase [NADP(+)] | icd | 1.1.1.42 | TCA |
| MGA3_06345 | PB1_11459 | 83 | Dihydrolipoyllysine-residue succinyltransferase | odhB | 2.3.1.61 | TCA |
| MGA3_06350 | PB1_11464 | 89 | Oxoglutarate dehydrogenase (succinyl transferring) | odhA | 1.2.4.2 | TCA |
| MGA3_14936 | PB1_00700 | 94 | 2-Oxoglutarate synthase, alpha subunit | korA | 1.2.7.3 | TCA |
| MGA3_14941 | PB1_00695 | 96 | 2-Oxoglutarate synthase, beta subunit | korB | 1.2.7.3 | TCA |
| MGA3_14471 | PB1_01175 | 98 | Succinyl-CoA synthetase, subunit beta | sucC | 6.2.1.5 | TCA |
| MGA3_14476 | PB1_01170 | 97 | Succinyl-CoA synthetase, subunit alpha | sucD | 6.2.1.5 | TCA |
| MGA3_03260 | PB1_16529 | 96 | Succinate dehydrogenase, cytochrome b558 subunit | sdhC | 1.3.99.1 | TCA |
| MGA3_03265 | PB1_16524 | 96 | Succinate dehydrogenase, flavoprotein subunit | sdhA | 1.3.99.1 | TCA |
| MGA3_03270 | PB1_16519 | 98 | Succinate dehydrogenase, iron-sulfur subunit | sdhB | 1.3.99.1 | TCA |
| MGA3_04115 | PB1_03575 | 92 | Fumarate hydratase, class II | fumC | 4.2.1.2 | TCA |
| MGA3_10765 | PB1_04250 | 94 | Fumarate hydratase, class I | fumA | 4.2.1.2 | TCA |
| MGA3_03035 | PB1_16784 | 94 | Malate dehydrogenase | citH | 1.1.1.37 | TCA |
| MGA3_12305 | PB1_05300 | 95 | Malate:quinone oxidoreductase | mqo | 1.1.5.4 | TCA |
| MGA3_12485 | PB1_05532 | 97 | Isocitrate lyase | aceA | 4.1.3.1 | GxS |
| MGA3_12490 | PB1_05537 | 95 | Malate synthase | aceB | 2.3.3.9 | GxS |
| MGA3_13826 | PB1_01810 | 96 | Pyruvate carboxylase | pyc | 6.4.1.1 | AN |
| MGA3_08600 | PB1_13864 | 94 | Aspartate transaminase | aspB | 2.6.1.1 | AN |
| MGA3_09380 | PB1_14609 | 90 | Dihydrolipoyl dehydrogenase (E3) | bfmbC | 1.8.1.4 | AN |
| MGA3_13696 | PB1_01955 | 94 | Pyruvate dehydrogenase (acetyl transferring) (E1), α subunit | pdhA | 1.2.4.1 | AN |
| MGA3_13701 | PB1_01950 | 98 | Pyruvate dehydrogenase (acetyl transferring) (E1), β subunit | pdhB | 1.2.4.1 | AN |
| MGA3_13706 | PB1_01945 | 86 | Dihydrolipoyllysine-residue acetyltransferase (E2) | pdhC | 2.3.1.12 | AN |
| MGA3_13711 | PB1_01940 | 95 | Dihydrolipoyl dehydrogenase (E3) | pdhD | 1.8.1.4 | AN |
| MGA3_08930 | PB1_14164 | 96 | Glutamate dehydrogenase (NAD+) | yweB | 1.4.1.2 | AN |
| MGA3_09355 | PB1_14584 | 96 | Methylmalonyl-CoA mutase, large subunit | mutB | 5.4.99.2 | AN |
| MGA3_01385 | PB1_07957 | 94 | Methylmalonyl-CoA mutase, large subunit | mutB2 | 5.4.99.2 | AN |
| MGA3_09360 | PB1_14589 | 85 | Methylmalonyl-CoA mutase, small subunit | mutA | 5.4.99.2 | AN |
| MGA3_02480 | PB1_17299 | 96 | Phosphoenolpyruvate carboxykinase (ATP) | pckA | 4.1.1.49 | AN |
| MGA3_02925 | PB1_16924 | 96 | Alanine dehydrogenase | ald | 1.4.1.1 | AN |
Determined by the ExPASy Proteomics Server (http://expasy.org/).
TCA, tricarboxylic acid cycle; GxS, glyoxylate shunt; AN, anaplerotic.
B. methanolicus has genes representing a complete TCA cycle and a glyoxylate shunt, but no ethylmalonyl-CoA pathway.
It is well known that many methylotrophic bacteria lack a complete TCA cycle and that the formaldehyde assimilatory and dissimilatory routes play major roles in the generation of energy and reducing power (15). Some methylotrophs also lack a glyoxylate shunt for acetyl-CoA assimilation but instead possess an alternative ethylmalonyl-CoA pathway for such purposes (1, 34). B. methanolicus MGA3 cells presumably have a high entry of methanol-derived carbon into the first three steps of the TCA cycle catalyzed by citrate synthase (CS), aconitate hydratase (Acn), and isocitrate dehydrogenase (Icd), leading to the generation of α-ketoglutarate. A substantial portion of this precursor metabolite is then directly converted to l-glutamate (9, 12). Together with the lack of any significant 2-oxoglutarate dehydrogenase (OGDH) activity in cell extracts (12), this has led to the assumption that B. methanolicus also lacks a complete TCA cycle. However, genes representing all biochemical steps of a complete TCA cycle were identified in both B. methanolicus genomes, including the odhAB operon encoding a putative OGDH enzyme (Table 4). B. subtilis encodes two CS genes, but we identified only one CS gene, citY, in both B. methanolicus strains MGA3 and PB1. We demonstrated previously that citY encoded an active CSII protein, and a frameshift mutation in this gene resulted in dramatically reduced l-glutamate production in a MGA3 genetic background (12).
Genes representing a glyoxylate cycle, aceA and aceB, encoding isocitrate lyase and malate synthase, respectively, were also found, while no genes representing any ethylmalonyl-CoA pathway were identified. The latter pathway was recently documented in the Gram-negative methylotroph Methylobacterium extorquens AM1 (1, 34), and to our knowledge, no analogous pathways have been identified in any Gram-positive methylotrophic bacterium.
Genes and regulators putatively involved in l-glutamate biosynthesis.
The biosynthesis of l-glutamate represents a major part of cellular metabolism linking carbon metabolism and nitrogen metabolism. Also included here are the biosynthetic pathways for the amino acids l-glutamine and l-arginine. In contrast to the closely related B. subtilis, B. methanolicus MGA3 synthesizes and secretes large amounts of l-glutamate, and it was of interest to explore whether this difference can be directly attributed to the genes and regulators involved in l-glutamate metabolism. In B. subtilis, the central enzymes involved are glutamate synthase (GOGAT; encoded by the gltAB operon), two different glutamate dehydrogenases (GDH; encoded by rocG and gudB), and glutamine synthetase (GS; encoded by glnA). In this bacterium, l-glutamate is reported to be synthesized exclusively by the reductive amination of α-ketoglutarate catalyzed by GOGAT, and mutants deficient in this enzyme are l-glutamate auxotrophs (17). GDH, on the other hand, is only devoted to glutamate catabolism into α-ketoglutarate and further degradation in the TCA cycle by OGDH. Overexpression of rocG in B. subtilis resulted in an ability to grow on l-glutamate (17). l-Glutamate can be aminated to l-glutamine, catalyzed by GS (17).
Putative B. methanolicus genes involved in l-glutamate metabolism are listed in Table 5. In contrast to B. subtilis, which has two GDH genes, rocG and gudB, B. methanolicus has only one GDH gene, yweB. B. methanolicus has a gltAB operon putatively encoding a GOGAT similar to that reported for B. subtilis (7), and it has one additional gltA2 gene located elsewhere on the chromosome. One putative GS gene, glnA, was also identified and is analogous to a corresponding gene in B. subtilis. The MGA3 and PB1 genome sequences were similar with respect to all these genes, and still these two strains are very different with respect to l-glutamate production levels in fed-batch methanol cultivation (see above). Biochemical characterization and regulation of all these gene products are in progress in our research group, and this work will provide new insight into l-glutamate biosynthesis in B. methanolicus strains.
TABLE 5.
Genes involved in glutamate synthesis and metabolism
| Locus tag in MGA3 | Locus tag in PB1 | % Amino acid identity | Common name | Gene | EC no. |
|---|---|---|---|---|---|
| MGA3_02745 | PB1_17089 | 95 | Carbon catabolite repression transcriptional regulator | ccpA | |
| MGA3_06010 | PB1_11134 | 90 | Glutaminase | glsA | 3.5.1.2 |
| MGA3_08135 | PB1_13334 | 85 | Transcriptional activator of arginine utilization operons | rocR | |
| MGA3_08930 | PB1_14164 | 96 | Glutamate dehydrogenase | yweB | 1.4.1.2 |
| MGA3_09430 | PB1_14659 | 98 | Arginine repressor | ahrC | |
| MGA3_10580 | PB1_03940 | 92 | Glutamate synthase (NADPH) | gltA2 | 1.4.1.13 |
| MGA3_11185 | PB1_04590 | 93 | Transcriptional pleiotropic regulator involved in global nitrogen regulation | tnrA | |
| MGA3_12495 | PB1_05542 | 96 | 1-Pyrroline-5-carboxylate dehydrogenase | rocA | 1.5.1.12 |
| MGA3_12285 | PB1_05275 | 96 | Glutamate synthase (NADPH) | gltB | 1.4.1.13 |
| MGA3_12290 | PB1_05280 | 96 | Glutamate synthase (NADPH) | gltA | 1.4.1.13 |
| MGA3_12295 | PB1_05285 | 96 | Transcriptional activator of the glutamate synthase operon | gltC | |
| MGA3_13010 | PB1_06417 | 97 | Arginase | rocF | 3.5.3.1 |
| MGA3_14511 | PB1_01135 | 99 | Transcriptional pleiotropic repressor | codY | |
| MGA3_15006 | PB1_00630 | 94 | Transcriptional repressor of glutamine synthetase | glnR | |
| MGA3_15011 | PB1_00625 | 98 | Glutamine synthetase, type I | glnA | 6.3.1.2 |
| MGA3_16406 | PB1_10177 | 92 | Ornithine aminotransferase | rocD | 2.6.1.13 |
In Corynebacterium glutamicum, it has been reported that the NCgl1221 gene encodes an active l-glutamate exporter (33, 42). Putative counterparts to this gene, originally annotated as a small-conductance mechanosensitive channel, were found in the MGA3 and PB1 genomes (locus tags MGA3_01925 and PB1_07357, here annotated as mechanosensitive ion channel family proteins). The B. methanolicus genes are considerably shorter than NClg1221, and the primary sequences of the deduced B. methanolicus gene products are 93% identical to each other and about 28% identical to the NClg1221 gene product. Whether these genes play any role in l-glutamate secretion in B. methanolicus remains to be experimentally tested.
l-Glutamate metabolism in B. subtilis is tightly controlled, and several transcriptional regulators are involved. Transcription of the gltAB operon is repressed by the global regulator TnrA in the absence of ammonium, while GltC activates gltAB transcription in the presence of sugars and in the absence of arginine. Transcription of rocG is subject to carbon catabolite repression exerted by CcpA in the presence of arginine, and rocG transcription is mediated by the activators RocR and AhrC. The gudB gene, on the other hand, is constitutively expressed in B. subtilis. Moreover, the global transcriptional regulators CodY and GlnR play important roles in regulating nitrogen metabolism in B. subtilis (16). GS, in addition to being feedback regulated, also plays a role in regulating the activity of the transcription factors GlnR and TnrA. Genes putatively encoding all the assumed important transcriptional regulators AhrC, CcpA, CodY, GlnR, GltC, RocR, and TnrA were also identified in the B. methanolicus MGA3 and PB1 genome sequences (Table 5). The role and impact of these regulatory genes for controlling l-glutamate and nitrogen metabolism in B. methanolicus remain to be tested experimentally.
B. methanolicus presumably uses the acetylase variant of the l-lysine biosynthetic pathway.
Some B. methanolicus genes encoding enzymes of the aspartate pathway have previously been cloned and characterized, including lysA (encoding diaminopimelate decarboxylase [LysA]) (31), the three aspartokinase (AK) genes dapG (AKI), lysC (AKII), and yclM (AKIII) (27, 39), and hom1 and hom2 (encoding homoserine dehydrogenase [HD I and II, respectively]) (9). The asd (encoding aspartate-semialdehyde dehydrogenase) and dapA (encoding dihydrodipicolinate synthase) genes were also cloned by us (27), and we recently demonstrated that they both encode active enzymes (32). Overexpression of dapG, lysC, and yclM increased l-lysine production in wild-type B. methanolicus MGA3 up to 60-fold (corresponding to 11 g liter−1) in fed-batch methanol cultivation (27), and we showed that hom1 is the major target for S-(2-aminoethyl)-l-cysteine (AEC) resistance in B. methanolicus (9). The B. methanolicus genome sequences provided the remaining genes representing a complete aspartate pathway (Table 6). In particular, the finding of dapH (encoding tetrahydrodipicolinate N-acetyltransferase), patA (encoding acetyl-diaminopimelate aminotransferase), and dapL (encoding N-acetyl-diaminopimelate deacetylase) indicates that this organism uses the acetylase variant of the l-lysine biosynthetic pathway, as previously proposed by us (11). The dapH and dapL genes are colocalized as an operon. Also, the asd, dapG, and dapA genes are localized as a dap operon, together with dpaA and dpaB (encoding dipicolinate synthase subunits A and B, respectively) and presumably under coordinated transcriptional control, analogous to the situation in B. subtilis. The two l-threonine pathway genes thrB (encoding homoserine kinase) and thrC (encoding homoserine synthase) are also localized as an operon together with hom1. PB1 has no hom2 gene, and the biological impact of this remains unknown.
TABLE 6.
Genes involved in the aspartate pathway
| Step | Locus tag in MGA3 | Locus tag in PB1 | % Amino acid identity | Common name | Gene | EC no. |
|---|---|---|---|---|---|---|
| −1 | MGA3_08600 | PB1_13864 | 94 | Aspartate transaminase | aspB | 2.6.1.1 |
| 1 | MGA3_14816 | PB1_00835 | 95 | Aspartate kinase I | dapG | 2.7.2.4 |
| 1 | MGA3_03250 | PB1_16544 | 91 | Aspartate kinase II | lysC | 2.7.2.4 |
| 1 | MGA3_06765 | PB1_11794 | 92 | Aspartate kinase III | yclM | 2.7.2.4 |
| 2 | MGA3_14811 | PB1_00840 | 92 | Aspartate-semialdehyde dehydrogenase | asd | 1.2.1.11 |
| 3 | MGA3_14821 | PB1_00830 | 95 | Dihydrodipicolinate synthase | dapA | 4.2.1.52 |
| 4 | MGA3_08665 | PB1_13924 | 95 | Dihydrodipicolinate reductase | dapB | 1.3.1.26 |
| 5 | MGA3_13651 | PB1_02000 | 95 | Tetrahydrodipicolinate N-acetyltransferase | dapH | 2.3.1.89 |
| 6 | MGA3_13566 | PB1_02085 | 89 | Acetyl-diaminopimelate aminotransferase | patA | 2.6.1.- |
| 7 | MGA3_13656 | PB1_01995 | 90 | N-Acetyl-diaminopimelate deacetylase | dapL | 3.5.1.47 |
| 8 | MGA3_07875 | PB1_13099 | 88 | Diaminopimelate epimerase | dapF | 5.1.1.7 |
| 9 | MGA3_09090 | PB1_14319 | 92 | Diaminopimelate decarboxylase | lysA | 4.1.1.20 |
| 10 | MGA3_07720 | PB1_05235 | 62 | Putative l-lysine exporter | lysE | |
| 10 | PB1_00380 | Putative l-lysine exporter | lysE2 | |||
| 3a | MGA3_16241 | PB1_10002 | 95 | Homoserine dehydrogenase 1 | hom1a | 1.1.1.3 |
| 3a | MGA3_11045 | Homoserine dehydrogenase 2 | hom2 | 1.1.1.3 | ||
| 3b | MGA3_16251 | PB1_10012 | 93 | Homoserine kinase | thrB | 2.7.1.39 |
| 3c | MGA3_16246 | PB1_10007 | 96 | Threonine synthase | thrC | 4.2.3.1 |
| 4a | MGA3_14801 | PB1_00850 | 94 | Dipicolinate synthase, subunit A | dpaA | |
| 4a | MGA3_14806 | PB1_00845 | 95 | Dipicolinate synthase, subunit B | dpaB |
Only one hom gene is found in PB1.
l-Lysine export in B. methanolicus strains.
A putative lysine exporter gene, lysE, was identified in the MGA3 genome, and the deduced LysE primary sequence showed highest similarity (53%) to the yisU gene-encoded protein from B. subtilis 168, annotated as a not yet characterized putative lysine exporter (36). Both the YisU and LysE amino acid sequences shared only a low similarity of 20% with the C. glutamicum LysECg primary sequence. In contrast to C. glutamicum (8), no putative regulator genes were found associated with any of the putative lysine exporter genes in the B. methanolicus strains. Genome mining revealed two putative lysine exporter genes in the PB1 genome sequence, denoted lysE and lysE2, and the deduced amino acid sequences of the respective gene products share 20% identity with each other. The deduced LysE protein of MGA3 shared 62% and 44% primary sequence identity to the deduced LysE and LysE2 proteins of PB1, respectively. We are currently in the process of investigating the biological function and roles of all these genes for l-lysine export in B. methanolicus strains.
Concluding remarks.
The first genome sequencing of bacteria belonging to thermotolerant bacilli is presented. Our results include a genomewide comparison between two Bacillus methanolicus wild-type strains, MGA3 and PB1, as well as a comparison of this species with other nonmethylotrophic bacilli. MGA3 and PB1 displayed major physiological differences in fed-batch methanol cultivation, and together these two strains represent a valuable model system for understanding plasmid-dependent methylotrophy. The differences in methanol consumption and respiration rate between these two strains were accompanied by different organization of the methanol dehydrogenase genes and of certain RuMP pathway genes. Our results open up possibilities for systems-level metabolic engineering of B. methanolicus for the overproduction of amino acids and other useful compounds from methanol. For example, PB1 produced very little l-glutamate compared to MGA3, and this could not be explained genetically. Ongoing analyses of the regulation and biochemistry of l-glutamate biosynthesis genes and enzymes should provide new insight into key bottlenecks for l-glutamate overproduction from methanol by B. methanolicus.
ACKNOWLEDGMENTS
This work was supported by a research grant from The Research Council of Norway.
We thank Bård Karsten Gustavsen and Lihua Yu for valuable contributions during the MGA3 genome gap-filling process and Roman Netzer for extracting the B. methanolicus genomic DNA and for valuable discussions. We also thank Ave Tooming-Klunderud at the Norwegian High-Throughput Sequencing Centre for valuable discussions during the PB1 assembly process.
Footnotes
Published ahead of print 18 May 2012
REFERENCES
- 1. Alber BE. 2011. Biotechnological potential of the ethylmalonyl-CoA pathway. Appl. Microbiol. Biotechnol. 89:17–25 [DOI] [PubMed] [Google Scholar]
- 2. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anthony C. 1982. The biochemistry of methylotrophs. Academic Press, London, United Kingdom [Google Scholar]
- 4. Arfman N, et al. 1992. Environmental regulation of alcohol metabolism in thermotolerant methylotrophic Bacillus strains. Arch. Microbiol. 157:272–278 [DOI] [PubMed] [Google Scholar]
- 5. Arfman N, et al. 1997. Properties of an NAD(H)-containing methanol dehydrogenase and its activator protein from Bacillus methanolicus. Eur. J. Biochem. 244:426–433 [DOI] [PubMed] [Google Scholar]
- 6. Aziz RK, et al. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75 doi: 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Belitsky BR. 2002. Biosynthesis of amino acids of the glutamate and aspartate families, alanine, and polyamines, p 203–231 In Sonenshein AL, Hoch JA, Losick L. (ed), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, DC [Google Scholar]
- 8. Bellmann A, et al. 2001. Expression control and specificity of the basic amino acid exporter LysE of Corynebacterium glutamicum. Microbiology 147:1765–1774 [DOI] [PubMed] [Google Scholar]
- 9. Brautaset T, et al. 2010. Bacillus methanolicus pyruvate carboxylase and homoserine dehydrogenase I and II and their roles for l-lysine production from methanol at 50 degrees C. Appl. Microbiol. Biotechnol. 87:951–964 [DOI] [PubMed] [Google Scholar]
- 10. Brautaset T, Jakobsen M, Flickinger MC, Valla S, Ellingsen TE. 2004. Plasmid-dependent methylotrophy in thermotolerant Bacillus methanolicus. J. Bacteriol. 186:1229–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brautaset T, Jakobsen M, Josefsen KD, Flickinger MC, Ellingsen TE. 2007. Bacillus methanolicus: a candidate for industrial production of amino acids from methanol at 50 degrees C. Appl. Microbiol. Biotechnol. 74:22–34 [DOI] [PubMed] [Google Scholar]
- 12. Brautaset T, et al. 2003. Role of the Bacillus methanolicus citrate synthase II gene, citY, in regulating the secretion of glutamate in l-lysine-secreting mutants. Appl. Environ. Microbiol. 69:3986–3995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chao L, et al. 2007. Complete nucleotide sequence of pBMB67, a 67-kb plasmid from Bacillus thuringiensis strain YBT-1520. Plasmid 57:44–54 [DOI] [PubMed] [Google Scholar]
- 14. Chistoserdova L. 2011. Modularity of methylotrophy, revisited. Environ. Microbiol. 13:2603–2622 [DOI] [PubMed] [Google Scholar]
- 15. Chistoserdova L, Kalyuzhnaya MG, Lidstrom ME. 2009. The expanding world of methylotrophic metabolism. Annu. Rev. Microbiol. 63:477–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Commichau FM, Forchhammer K, Stülke J. 2006. Regulatory links between carbon and nitrogen metabolism. Curr. Opin. Microbiol. 9:167–172 [DOI] [PubMed] [Google Scholar]
- 17. Commichau FM, Gunka K, Landmann JJ, Stülke J. 2008. Glutamate metabolism in Bacillus subtilis: gene expression and enzyme activities evolved to avoid futile cycles and to allow rapid responses to perturbations of the system. J. Bacteriol. 190:3557–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de Vries GE, Arfman N, Terpstra P, Dijkhuizen L. 1992. Cloning, expression, and sequence analysis of the Bacillus methanolicus C1 methanol dehydrogenase gene. J. Bacteriol. 174:5346–5353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Vries GE, Kües U, Stahl U. 1990. Physiology and genetics of methylotrophic bacteria. FEMS Microbiol. Rev. 6:57–101 [DOI] [PubMed] [Google Scholar]
- 20. Dijkhuizen L, et al. 1988. Isolation and initial characterization of thermotolerant methylotrophic Bacillus strains. FEMs Microbiol. Lett. 52:209–214 [Google Scholar]
- 21. Dijkhuizen L, Levering PR, De Vries GE. 1992. The physiology and biochemistry of aerobic methanol-utilizing Gram-negative and Gram-positive bacteria, p 149–181 In Murrel JC, Dalton H. (ed), Methane and methanol utilizers. Plenum Press, New York, NY [Google Scholar]
- 22. Hanson RS, Dillingham R, Olson P. 1996. Production of l-lysine and some other amino acids by mutants of B. methanolicus, p 227–236 In Lindstrom ME, Tabita FR. (ed), Microbial growth on C1 compounds. Kluwer Academic Publishers, Dordrecht, Netherlands [Google Scholar]
- 23. Heid CA, Stevens J, Livak KJ, Williams PM. 1996. Real time quantitative PCR. Genome Res. 6:986–994 [DOI] [PubMed] [Google Scholar]
- 24. Hektor HJ, Kloosterman H, Dijkhuizen L. 2002. Identification of a magnesium-dependent NAD(P)(H)-binding domain in the nicotinoprotein methanol dehydrogenase from Bacillus methanolicus. J. Biol. Chem. 277:46966–46973 [DOI] [PubMed] [Google Scholar]
- 25. Henstra SA, Tolner B, Duurkens RHT, Konings WN, Robillard GT. 1996. Cloning, expression, and isolation of the mannitol transport protein from the thermophilic bacterium Bacillus stearothermophilus. J. Bacteriol. 178:5586–5591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jakobsen ØM, et al. 2006. Upregulated transcription of plasmid and chromosomal ribulose monophosphate pathway genes is critical for methanol assimilation rate and methanol tolerance in the methylotrophic bacterium Bacillus methanolicus. J. Bacteriol. 188:3063–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jakobsen ØM, et al. 2009. Overexpression of wild-type aspartokinase increases l-lysine production in the thermotolerant methylotrophic bacterium Bacillus methanolicus. Appl. Environ. Microbiol. 75:652–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kloosterman H, Vrijbloed JW, Dijkhuizen L. 2002. Molecular, biochemical, and functional characterization of a Nudix hydrolase protein that stimulates the activity of a nicotinoprotein alcohol dehydrogenase. J. Biol. Chem. 277:34785–34792 [DOI] [PubMed] [Google Scholar]
- 29. Komives CF, Cheung LY, Pluschkell SB, Flickinger MC. 2005. Growth of Bacillus methanolicus in seawater-based media. J. Ind. Microbiol. Biotechnol. 32:61–66 [DOI] [PubMed] [Google Scholar]
- 30. Livak KJ. 1997. Comparative CT method. User bulletin no. 2. PE Applied Biosystems, Foster City, CA [Google Scholar]
- 31. Mills DA, Flickinger MC. 1993. Cloning and sequence analysis of the meso-diaminopimelate decarboxylase gene from Bacillus methanolicus MGA3 and comparison to other decarboxylase genes. Appl. Environ. Microbiol. 59:2927–2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nærdal I, Netzer R, Ellingsen TE, Brautaset T. 2011. Analysis and manipulation of aspartate pathway genes for l-lysine overproduction from methanol by Bacillus methanolicus. Appl. Environ. Microbiol. 77:6020–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nakamura J, Hirano S, Ito H, Wachi M. 2007. Mutations of the Corynebacterium glutamicum NCgl1221 gene, encoding a mechanosensitive channel homolog, induce l-glutamic acid production. Appl. Environ. Microbiol. 73:4491–4498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Peyraud R, et al. 2009. Demonstration of the ethylmalonyl-CoA pathway by using 13C metabolomics. Proc. Natl. Acad. Sci. U. S. A. 106:4846–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pluschkell SB, Flickinger MC. 2002. Dissimilation of [13C]methanol by continuous cultures of Bacillus methanolicus MGA3 at 50 degrees C studied by 13C NMR and isotope-ratio mass spectrometry. Microbiology 148:3223–3233 [DOI] [PubMed] [Google Scholar]
- 36. Saier MH, et al. 2002. Transport capabilities encoded within the Bacillus subtilis genome. J. Mol. Microbiol. Biotechnol. 4:37–67 [PubMed] [Google Scholar]
- 37. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 38. Schendel FJ, Bremmon CE, Flickinger MC, Guettler M, Hanson RS. 1990. l-Lysine production at 50 degrees C by mutants of a newly isolated and characterized methylotrophic Bacillus sp. Appl. Environ. Microbiol. 56:963–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schendel FJ, Flickinger MC. 1992. Cloning and nucleotide sequence of the gene coding for aspartokinase II from a thermophilic methylotrophic Bacillus sp. Appl. Environ. Microbiol. 58:2806–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Van Domselaar GH, et al. 2005. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res. 33:W455–W459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vorholt JA. 2002. Cofactor-dependent pathways of formaldehyde oxidation in methylotrophic bacteria. Arch. Microbiol. 178:239–249 [DOI] [PubMed] [Google Scholar]
- 42. Yao W, et al. 2009. Expression and localization of the Corynebacterium glutamicum NCgl1221 protein encoding an l-glutamic acid exporter. Microbiol. Res. 164:680–687 [DOI] [PubMed] [Google Scholar]