Abstract
FK506 is a potent immunosuppressant that has a wide range of clinical applications. Its 23-member macrocyclic scaffold, mainly with a polyketide origin, features two methoxy groups at C-13 and C-15 and one allyl side chain at C-21, due to the region-specific incorporation of two unusual extender units derived from methoxymalonyl-acyl carrier protein (ACP) and allylmalonyl-coenzyme A (CoA), respectively. Whether their intracellular formations can be a bottleneck for FK506 production remains elusive. In this study, we report the improvement of FK506 yield in the producing strain Streptomyces tsukubaensis by the duplication of two sets of pathway-specific genes individually encoding the biosyntheses of these two extender units, thereby providing a promising approach to generate high-FK506-producing strains via genetic manipulation. Taking advantage of the fact that S. tsukubaensis is amenable to two actinophage (ΦC31 and VWB) integrase-mediated recombination systems, we genetically enhanced the biosyntheses of methoxymalonyl-ACP and allylmalonyl-CoA, as indicated by transcriptional analysis. Together with the optimization of glucose supplementation, the maximal FK506 titer eventually increased by approximately 150% in comparison with that of the original strain. The strategy of engineering the biosynthesis of unusual extender units described here may be applicable to improving the production of other polyketide or nonribosomal peptide natural products that contain pathway-specific building blocks.
INTRODUCTION
FK506 (tacrolimus), isolated from a variety of soil Streptomyces species, is a natural product with potent immunosuppressive activity. FK506 interacts with a receptor known as FK506-binding protein 12 (FKBP12); the FK506-FKBP12 complex then acts on a target protein, calcineurin, by inhibiting its Ser/Thr phosphatase activity. This leads to the arrest of T cell proliferation at the G0-G1 stage (18). As a consequence, FK506 has been clinically approved as an immunosuppressant to prevent the rejection of transplanted organs and for the treatment of various inflammatory diseases. Recently, other promising biological activities, such as neuroprotection and regeneration, were reported (9, 31), creating significant interest in the further development of FK506 for different medicinal uses.
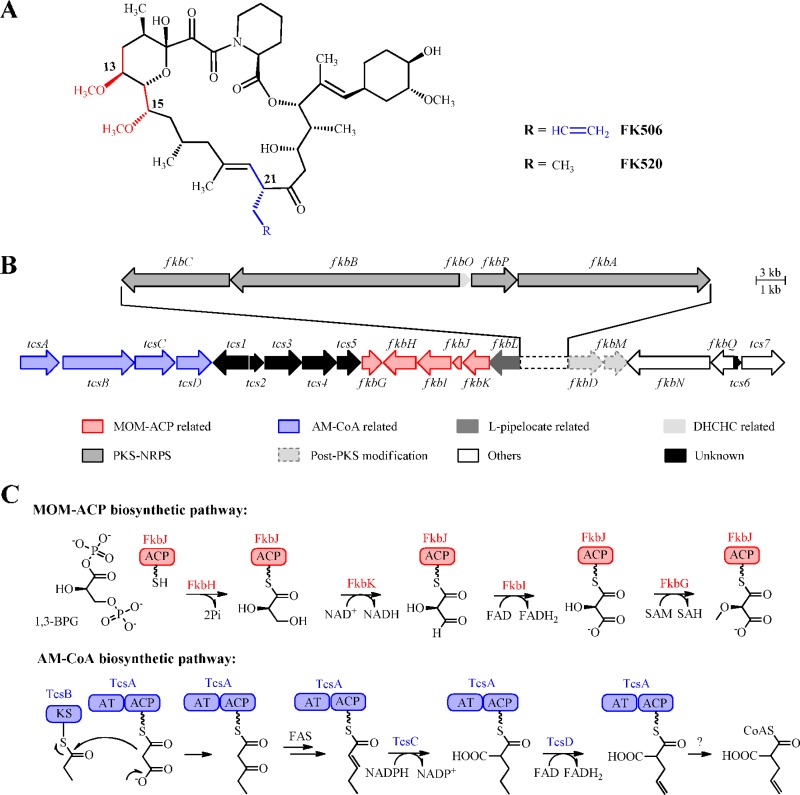
FK506 biosynthesis involves a hybrid polyketide synthase (PKS)-nonribosomal peptide synthetase (NRPS) system, in line with the fact that the corresponding structure belongs to an amide bond-containing macrolide family with members that include its naturally occurring analogues FK520 and rapamycin (21, 22, 28, 35). FkbA to FkbC, constituting a typical type I PKS system, have functional domains that are organized colinearly with their activities in the biosynthetic assembly process: each module normally consists of ketoacylsynthase (KS), acyltransferase (AT), and acyl carrier protein (ACP) for chain elongation and optionally contains dehydratase (DH), enoylreductase (ER), and ketoreductase (KR) for reductively processing β-keto (oxidation state of beta-C in the growing polyketide chain) functionality. These PKSs are responsible for the construction of the main backbone of FK506 by catalyzing 10 two-carbon extensions from a chorismate-derived starter unit (2). An unnatural amino acid residue, l-pipecolate, terminates the resulting polyketide intermediate with an amide bond (7), whose formation is catalyzed by the NRPS FkbP, which subsequently closes the 23-member macrocyclic core via intramolecular lactonization (8). Besides malonyl-coenzyme A (M-CoA) and methylmalonyl (MM)-CoA, which often serve as common substrates of typical type I PKSs for elongation, the PKSs in FK506 biosynthesis feature the incorporation of two unusual extender units derived from methoxymalonyl (MOM)-ACP and allylmalonyl (AM)-CoA into the polyketide skeleton to produce two methoxy groups at C-13 and C-15 and one allyl side chain at C-21, respectively (Fig. 1A).
Fig 1.
Chemical structures of FK506 and FK520, the FK506 biosynthetic gene cluster in S. tsukubaensis, and the biosynthetic pathways of MOM-ACP and AM-CoA. (A) Structures of FK506 and its analogue, FK520. The polyketide skeletons consist of two methoxy groups at C-13 and C-15, while the allyl side chain at C-21 of FK506 is replaced by an ethyl group in FK520. The FK506 macrocyclic scaffolds derived from MOM-ACP and AM-CoA are shown in red and blue, respectively. (B) Schematic representation of the FK506 biosynthetic gene cluster in S. tsukubaensis. Genes involved in the biosyntheses of MOM-ACP and AM-CoA are shown in red and blue, respectively. (C) Biosynthetic pathways of MOM-ACP and AM-CoA, catalyzed by FkbG-K and TcsA-D, respectively. DHCHC, which stands for (4R,5R)-4,5-dihydroxycyclohex-1-enecarboxylic acid, is the chorismate-derived starter unit.
The entire biosynthetic gene cluster of FK506 has recently been characterized from several FK506-producing streptomycete strains (20), which shows that the formation of the unusual building blocks MOM-ACP and AM-CoA requires two sets of independent, pathway-specific genes (Fig. 1B and C). The first five contiguous genes, fkbGHIJK, with an organization identical to that of the FK520 biosynthetic gene cluster, were proposed to encode MOM-ACP generation, a process that starts with the utilization of 1,3-biphosphoglycerate (1,3-BPG) as the substrate. FkbH, which has both AT and phosphatase activities, is responsible for loading glyceroyl onto FkbJ, a discrete ACP protein, to produce glyceroyl-ACP for sequential elaborate modifications by two dehydrogenases, FkbK and FkbI, and an O-methyltransferase, FkbG, to furnish the MOM-ACP unit (4, 14). In contrast, the second set of four contiguous genes, tcsABCD, identified from the FK506 biosynthetic gene cluster are unique, and more recently, their deduced products have been confirmed to form a noncanonical discrete PKS system to catalyze AM-CoA formation in coordination with the fatty acid synthase (FAS) pathway. Functioning as a priming KS, the propionyl-acylated TcsB catalyzes the condensation of the propyl moiety with malonate loaded on TcsA, an AT-ACP bidomain protein, to form an ACP-tethered β-keto-pentanoate, which may then be converted into trans-2-pentenyl-ACP by involving the KR and DH activities of FAS of the host. Further modifications of the pentenyl group, including reductive carboxylation by TcsC in a manner similar to that of crotonyl-CoA carboxylase/reductase (CCR) (17), as well as dehydrogenation by the FAD-dependent enzyme TcsD, form the ACP-tethered allylmalonate, which is likely released as AM-CoA, an unprecedented extender unit for the FK506 assembly PKS system to build the side chain as allyl (10, 20).
Recent progress in understanding the biosyntheses of MOM-ACP and AM-CoA opens the door to improve FK506 production by intracellular enrichment of the precursor supply. To achieve this, genetic manipulations have frequently been used, including phage integrase-mediated recombination. The integrase recognizes the bacterial attB site, where a conservative and reciprocal exchange (involving a cleavage and subsequent recombination process) takes place with the phage attP site of the introduced DNA to form the hybrid sequences attL and attR (6). In this study, we exploited the sequence information regarding the phage attachment sites in the FK506 producer Streptomyces tsukubaensis ZJU01, showing that two distinct site-specific recombinations, based on the ΦC31 and VWB integration systems, respectively, are operable and have no apparent effect on FK506 production. Based on this, we report the introduction of additional copies of the pathway-specific genes into S. tsukubaensis at the selected attB loci on the chromosome, aiming at improvement of FK506 production by genetic enhancement of the biosyntheses of the unusual building blocks MOM-ACP and AM-CoA.
MATERIALS AND METHODS
Bacterial strains, plasmids, and reagents.
The bacterial strains and plasmids used in this study are summarized in Table 1. The primer sequences are listed in Table 2. The biochemicals, chemicals, media, restriction enzymes, and other molecular biological reagents were purchased from standard commercial sources unless otherwise stated.
Table 1.
Bacterial strains and plasmids used and constructed in this study
| Strains/plasmids | Characteristic(s) | Source/reference |
|---|---|---|
| E. coli | ||
| DH5α | Host for general cloning | Invitrogen |
| ET12567(pUZ8002) | Donor strain for conjugation between E. coli and Streptomyces | 15 |
| BW25113 (pIJ790) | Host for Red/ET-mediated PCR-targeted mutagenesis | 11 |
| S. tsukubaensis | ||
| ZJU01 | Original FK506-producing strain | CGMCCa 51771 |
| FL101 | Derivative of ZJU01 containing pSET152 integrated by the ΦC31 integrase-mediated recombination | This study |
| FL102 | Derivative of ZJU01 containing pSOK804 integrated by the VWB integrase-mediated recombination | This study |
| FL103 | Derivative of ZJU01 containing pFL103 integrated by the ΦC31 integrase-mediated recombination with the genotype of PermE*-fkbGKJIH | This study |
| FL104 | Derivative of ZJU01 containing pFL104 integrated by the ΦC31 integrase-mediated recombination with the genotype of PermE*-tcsABCD | This study |
| FL105 | Derivative of FL101 containing pTA804 integrated by the VWB integrase-mediated recombination | This study |
| FL106 | Derivative of FL103 containing pFL106 integrated by the VWB integrase-mediated recombination with the genotype of PermE*-tcsABCD | This study |
| FL107 | Derivative of FL104 containing pFL107 integrated by the VWB integrase-mediated recombination with the genotype of PermE*-fkbGKJIH | This study |
| Plasmids | ||
| pMD18-T simple | E. coli subcloning vector | TaKaRa |
| pSET152 | E. coli-Streptomyces shuttle vector containing the aac(3)IV gene and the ΦC31 attP site and integrase gene | 3 |
| pSOK804 | E. coli-Streptomyces shuttle vector containing the aac(3)IV gene and the VWB attP site and integrase gene | 29 |
| pWHM79 | pGEM3zf derivative carrying a 0.5-kb fragment containing the ermE* promoter | 30 |
| pTA804 | pSOK804 derivative replacing the aac(3)IV gene with the bla and tsr genes | This study |
| pMD-G | pMD18-T derivative carrying fkbG without its terminator | This study |
| pMD-HIJK | pMD18-T derivative carrying fkbHIJK with its terminator | This study |
| pMOM | pMD18-T derivative carrying fkbG-fkbK-fkbJ-fkbI-fkbH | This study |
| pAM | pMD18-T derivative carrying tcsA-tcsB-tcsC-tcsD | This study |
| pFL103 | pSET152 derivative for the PermE*-controlled expression of fkbG-fkbK-fkbJ-fkbI-fkbH | This study |
| pFL104 | pSET152 derivative for the PermE*-controlled expression of tcsA-tcsB-tcsC-tcsD | This study |
| pFL106 | pTA804 derivative for the PermE*-controlled expression of tcsA-tcsB-tcsC-tcsD | This study |
| pFL107 | pTA804 derivative for the PermE*-controlled expression of fkbG-fkbK-fkbJ-fkbI-fkbH | This study |
CGMCC, China General Microbiological Culture Collection.
Table 2.
Primers used for gene cloning, genotype verification, and RT-PCR amplification in this study
| Name | Sequencea | Description |
|---|---|---|
| p151Redf | gtgcaatacgaatggcgaaaagccgagctcatcggtcagTTACCAATGCTTAATCAGTG | Cloning of the bla and tsr genes from pSET151 for Red/ET-mediated recombination |
| p151Redr | tcagccaatcgactggcgagcggcatcgcattcttcgcaTTATCGGTTGGCCGCGAGAT | |
| pGf | TAAGCTTCGAGGCGCCGCTGGACCGTGTGG (HindIII) | Cloning of the fkbG gene without its terminator from ZJU01 genome |
| pGr | TTCTAGACGTTCACCGCTTCCGCAACAAGG (XbaI) | |
| pHIJKf | TACTAGTGGCAGCGCCTTCGCCGACCACATC (SpeI) | Cloning of the fkbHIJK genes with their terminator from ZJU01 genome |
| pHIJKr | TTCTAGACAATGCGGGGCTGCGCGACGACG (XbaI) | |
| pABCDf | TAAGCTTCCATCCCGATCATGCCCCTCCTG (HindIII) | Cloning of the tcsABCD genes with their terminator from ZJU01 genome |
| pABCDr | TTCTAGAAGGCCCTGACGCGGGACTGACC (XbaI) | |
| ATTB1 | CGGGATCCGACCC(G/C)TTCATCATGATGGAC | Degenerate primer pair from reference 5 to identify the ΦC31 attB sites |
| ATTB2 | TGGAATTCAGGTT(G/C)ACCCA(C/G)AGCTG(G/C)AG | |
| pL-C31f | CCCACAGCTGGAGGCCGTGG | Verification of the hybrid attL site from the ΦC31-based recombination |
| pL-C31r | CAGGGCGAGCAATTCCGAGA | |
| pR-C31f | CAGAGCAGGATTCCCGTTGAG | Verification of the hybrid attR site from the ΦC31-based recombination |
| pR-C31r | CCCTTCATCATGATGGACCAG | |
| pL-VWBf | GCCTTCGTAGCTCAGGGGATAGAG | Verification of the hybrid attL site from the VWB-based recombination |
| pL-VWBr | CGCCGCCGCCTTTTCCTCAATCG | |
| p152f | GTAACGCCAGGGTTTTCCCAGTCACG | Verification of pSET152-based gene duplication at the ΦC31 attB site |
| p152r | TCCGGCTCGTATGTTGTGTGGAATTG | |
| p804f | AGCAGGCGTTCCTCGGCGTCCTCGT | Verification of pTA804-based gene duplication at the VWB attB site |
| p804r | AGCTCACTCATTAGGCACCCCAGGC | |
| pMOMf | CTCCCAGTACGGCCTGCCCACCTCC | Verification of gene duplication for the MOM-ACP biosynthetic pathway |
| pMOMr | TGCGCTCCCTGGACCTGAGAATGACG | |
| pAMf | GAAGCGCTCCCGGGGCGCGGATATC | Verification of gene duplication for the AM-CoA biosynthetic pathway |
| pAMr | CAATGGGCTGCGGCTGCTGGAGATC | |
| pRTGf | CTGGGCGGTCTGCGGGCGGAGTCG | RT-PCR amplification of a 102-bp fkbG fragment representing the fkbG mRNA level |
| pRTGr | GGTCAACCGCACCAGGAATTCCAG | |
| pRTKf | GCGGACCTCATCGGCATCGACAAC | RT-PCR amplification of a 102-bp fkbK fragment representing the fkbHIJK mRNA level |
| pRTKr | CTTGGTGAGGAGCAGGTCGCAGGG | |
| pRTAf | GGCGTCTTCGCCGGTCTGGGTGCC | RT-PCR amplification of a 106-bp tcsA fragment representing the tcsABCD mRNA level |
| pRTAr | GAAGCGGCCGGACCGGCTTCAGCC | |
| pRTDf | GTGCCGGGGGACAACGCGCTGAAC | RT-PCR amplification of a 103-bp tcsD fragment representing the tcsABCD mRNA level |
| pRTDr | GGGCGACACCCAGAGCGGATGCGG | |
| pRTPf | CAGATGCTGCACTCGCACGGTTCG | RT-PCR amplification of a 102-bp fkbP fragment as a control |
| pRTPr | GTTGAGGGCGGGGTGGTCGAGCGG |
Lowercase letters indicate the sequences for Red/ET-mediated recombination. Restriction enzyme sites are underlined.
DNA isolation, manipulation, and sequencing.
DNA isolation and manipulation in Escherichia coli and S. tsukubaensis were performed according to standard protocols (15). PCR amplifications were carried out on an Authorized Thermal TM Cycler (AG 22331; Eppendorf, Hamburg, Germany) using either Taq DNA polymerase or PrimeStar HS DNA polymerase (TaKaRa). Primer synthesis and DNA sequencing were performed at Shanghai GeneCore Biotechnology Inc.
Construction of pTA804 as a derivative of pSOK804.
According to the previously described methods (11, 12), pTA804 was constructed via Red/ET-mediated recombination by substituting bla (ampicillin resistance gene) and tsr (thiostrepton resistance gene) for aac(3)IV (apramycin resistance gene). A gene cassette containing bla and tsr was amplified from plasmid pSET151 (3) using the primer pair p151Redf/p151Redr, which was further introduced into E. coli BW25113(pIJ790) containing pSOK804 (29) by electroporation. The positive clones were selected through their ampicillin-resistant phenotype and were sequenced to confirm the fidelity of the λ Red-dependent recombination.
Construction for duplication of pathway-specific genes.
To duplicate the MOM-ACP pathway-specific genes fkbGHIJK, a 0.8-kb fragment containing fkbG without its native terminator and a 3.5-kb fragment containing fkbHIJK were amplified from the genomic DNA of ZJU01 by PCR amplification using the primer pairs pGf/pGr and pHIJKf/pHIJKr, respectively. Both PCR products were cloned into pMD18-T to yield pMD-G and pMD-HIJK. After sequencing to confirm fidelity, the 3.5-kb SpeI/XbaI fragment of pMD-HIJK was inserted into the XbaI site of pMD-G to generate pMOM, the genotype of which was validated by digestion analysis of restriction enzymes. The 4.3-kb HindIII/XbaI fragment of pMOM and a 0.5-kb EcoRI/HindIII fragment containing the constitutive ermE* promoter (PermE*) from pWHM79 (30) were then coligated into the EcoRI/XbaI site of pSET152 to generate pFL103.
To duplicate the AM-CoA pathway-specific genes tcsABCD, a 6.4-kb fragment containing tcsABCD was amplified by PCR amplification using the primer pair pABCDf/pABCDr with the genomic DNA of ZJU01 as the template. This fragment was cloned into pMD18-T to yield pAM. After sequencing to confirm fidelity, the 6.4-kb HindIII/XbaI fragment of pAM and a 0.5-kb EcoRI/HindIII fragment containing PermE* were coligated into the EcoRI/XbaI site of pSET152 to yield pFL104.
The pTA804 derivatives pFL106 and pFL107 were constructed similarly to pFL103 and pFL104 by inserting the corresponding DNA fragments into pTA804 instead of pSET152.
Generation of recombinant strains and confirmation of genotypes.
The pSET152 and pSOK804 derivatives were introduced into S. tsukubaensis ZJU01 by intergeneric conjugation from E. coli ET12567(pUZ8002), following the procedure described previously (15).
For pSET152-based recombination (as for FL101, FL103, and FL104), colonies with an apramycin-resistant phenotype were identified as exconjugants, the genotypes of which were further confirmed by PCR amplification-coupled sequencing using the primer pairs pL-C31f/pL-C31r and pR-C31f/pR-C31r. For pSOK804-based recombination (as for FL102), colonies resistant to apramycin were identified as exconjugants, the genotypes of which were verified by PCR-based amplification and sequencing by using the primer pair pL-VWBf/pL-VWBr. For pSET152- and pTA804-based recombination (as for FL105, FL106, and FL107), colonies resistant to both apramycin and thiostrepton were identified as exconjugants, the genotypes of which were then verified by PCR amplification and DNA sequencing by using the primer pairs pL-C31f/pL-C31r, pR-C31f/pR-C31r, and pL-VWBf/pL-VWBr.
To distinguish individual recombinant strains, different combinations of primers corresponding to the sequences of the conjugative vector (pSET152 or pTA804) and the duplicated gene cassette (fkbGHIJK or tcsABCD) were used in PCR-based amplification and sequencing to identify the specificity of each genotype (see Fig. 3A).
Fig 3.
Duplication of the MOM-ACP and AM-CoA pathway-specific genes via two actinophage (ΦC31 and VWB) integrase-mediated recombination systems for improving FK506 production. (A) Genotypes of the FK506 producer ZJU01 and its derivatives. The primer sets are labeled with their predicted fragment sizes. (B) Validation of the genotypes by PCR amplification. The primers used to amplify individual PCR products are illustrated in Fig. 2A. (C) FK506 time course titers in the fermentations of ZJU01 and the recombinant strains. The data are the average values of at least three series of three parallel tests, and the error bars represent the standard deviations.
FK506 production in S. tsukubaensis.
S. tsukubaensis ZJU01 and its derivatives were grown on agar plates (with the appropriate antibiotic[s] for recombinant strains) with alanine-starch agar (AS-1) medium consisting of 0.1% yeast extract, 0.02% l-alanine, 0.05% l-arginine, 0.5% soluble starch, 0.25% NaCl, 1.0% Na2SO4, and 2.0% agar at pH 7.5 and 30°C for sporulation. For fermentation, an agar piece of approximately 1 cm2 was inoculated into a 250-ml flask containing 25 ml of the seed medium, consisting of 1.0% (vol/vol) glycerol, 4.0% soybean meal, 1.0% soluble starch, and 0.2% CaCO3 at pH 7.0 ± 0.2, and was maintained at 28°C and 220 rpm for 24 to 28 h. Five milliliters of the seed culture was inoculated into a 500-ml flask containing 50 ml of the fresh fermentation medium [2.0% soluble starch, 1.0% glucose, 1.0% (vol/vol) soybean oil, 0.1% l-lysine, 0.17% (NH4)2SO4, 0.05% K2HPO4, 0.05% NaCl, 0.05% MgSO4 · H2O, 0.5% CaCO3, and trace elements as described previously (36), pH 7.0 ± 0.2]. The cultivations were maintained at 28°C and 220 rpm. For each strain, at least three independent isolates were subjected to parallel fermentation experiments in triplicate.
Chemical analysis of FK506 and FK520 production.
Culture samples (2 ml) were centrifuged for 10 min at 12,000 rpm. The supernatant was extracted twice with 2 ml of ethyl acetate, and the mycelium was extracted with 2 ml of acetone. The organic extracts were combined, evaporated to dryness under reduced pressure, and redissolved in 0.1 ml of acetonitrile.
High-performance liquid chromatography (HPLC) analysis was carried out on a Diamonsil C18 (2) 5μ column (250 × 4.6 mm; catalog no. 99603; Dikma Technologies) on an Agilent 1100 HPLC system (Agilent Technologies, Palo Alto, CA). An isocratic program (60% acetonitrile and 40% H2O) was carried out at a flow rate of 1 ml/min, and the temperature was maintained at 60°C. Liquid chromatography-mass spectrometry (LC-MS) analysis was carried out with the same isocratic program on an LC-MS 2010 A liquid chromatograph-mass spectrometer (Shimadzu, Japan), showing [M+Na]+ ions at m/z 826.5 and [M+NH4]+ ions at m/z 821.5, in accordance with the FK506 molecular formula C44H69NO12, and [M+Na]+ ions at m/z 814.5 and [M+NH4]+ ions at m/z 809.5, in accordance with the FK520 molecular formula C43H69NO12, respectively. The FK506 and FK520 titers were quantified using UV detection at 215 nm and were calculated with standard curves, representing the average values of at least three series of three parallel fermentation experiments.
Transcriptional analysis.
Each total-RNA sample was prepared from S. tsukubaensis mycelia after 36, 72, or 108 h of fermentation with TRIzol reagents (Invitrogen) according to the manufacturer's instructions. The DNase I-treated mRNA (5 ng) was reverse transcribed to the first-strand cDNA using the ReverTra Ace-α-kit (Toyobo). As an additional control for each primer set and RNA sample, the cDNA synthesis reaction was carried out in the absence of reverse transcriptase to verify that genomic DNA did not contaminate the RNA samples. After incubation with RNase H, the resultant cDNAs were used as templates for PCR to analyze the transcription levels of the MOM-ACP and/or AM-CoA biosynthetic genes. For each S. tsukubaensis strain, three independent isolates were subjected to RNA preparation for PCR amplification analysis in parallel.
The amplification of each sample was performed by using Taq DNA polymerase under the following conditions: for predenaturation, 2 min at 94°C, and for each cycle, 30 s at 94°C followed by 30 s at 65°C and 10 s at 72°C. A 102-bp fragment of fkbG and a 102-bp fragment of fkbK (representatives of MOM-ACP biosynthetic genes), as well as a 106-bp fragment of tcsA and a 103-bp fragment of tcsD (representatives of AM-CoA biosynthetic genes) were amplified using the primer pairs pRTGf/pRTGr, pRTKf/pRTKr, pRTAf/pRTAr, and pRTDf/pRTDr, respectively. A 102-bp fragment of the NRPS gene fkbP was amplified using the primer pair pRTPf/pRTPr and served as an internal control. The amplification of each gene was optimized by using the 72-hour sample from the original strain as the template, leading to determination of a common 30-cycle reaction that clearly shows the difference in mRNA abundance (data not shown).
RESULTS
Establishment of two actinophage (ΦC31 and VWB) integrase-mediated recombination systems in S. tsukubaensis.
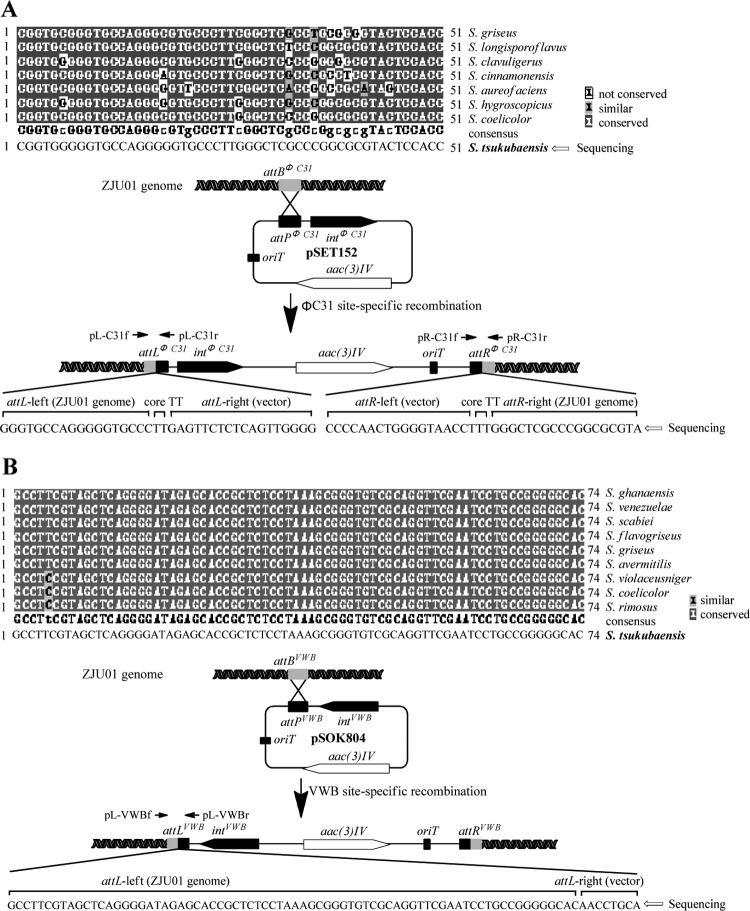
Genetic studies on the diversity of the phage ΦC31 attB sites from different Streptomyces species revealed a conservative region within an open reading frame (ORF) coding for a putative chromosome condensation protein (5). With the degenerate primer pair ATTB1/ATTB2 (5), we successfully amplified a 294-bp DNA fragment by PCR from the genomic DNA of ZJU01, which has the 51-bp conservative sequence for ΦC31 attachment (Fig. 2A). Based on this, a ΦC31-derived integrative vector, pSET152, was introduced into ZJU01 by conjugation to generate the recombinant strain FL101, with a frequency of 2 × 10−6 exconjugants per recipient spore, by using the methylation-deficient E. coli ET12567(pUZ8002) as the donor. To verify the anticipated integration, we randomly selected five apramycin-resistant exconjugants for template preparation. PCR amplification-coupled DNA sequencing clearly indicated that all of the exconjugants exhibited identical genotypes of attLΦC31-linear pSET152-attRΦC31 at the integrating locus (Fig. 2A and 3). Thus, the ΦC31-based system is applicable to site-specific recombination in S. tsukubaensis.
Fig 2.
Identification and confirmation of the ΦC31 and VWB attB sites on the chromosome of S. tsukubaensis. (A) Nucleotide sequence alignment revealed a 51-bp conserved region within the attBΦC31 locus in various streptomycete species. PCR amplification-coupled DNA sequencing confirmed the existence of the attBΦC31 locus in S. tsukubaensis and the regions surrounding the hybrid attLΦC31 and attRΦC31 after the site-specific integration of pSET152. (B) Nucleotide sequence alignment revealed a 74-bp highly conserved region widespread in various streptomycete genomes. Sequence analysis of the PCR products revealed the region surrounding the hybrid attLVWB after site-specific integration of pSOK804.
As demonstrated in S. venezuelae and S. ghanaensis, the attB site of actinophage VWB resides in a putative tRNAArg (AGG) gene (23, 33), which is different from that of ΦC31. To verify the availability of the attBVWB site on the chromosome of ZJU01, we introduced a VWB-derived integrative vector, pSOK804, into ZJU01 by conjugation, yielding the recombinant strain FL102 at a frequency of 5 × 10−5 per recipient spore. By comparing the attBVWB regions from S. venezuelae and S. ghanaensis, we identified a specific 74-bp highly similar sequence, which is widespread in various Streptomyces genomes (Fig. 2B). The 74-bp sequence was observed to appear in the conjunctive attLVWB site after integration (33), which prompted us to select the conserved attLVWB site, resulting from the hybridization of the 5′-terminal sequence of attBVWB and the 3′-terminal sequence of attPVWB of pSOK804, to validate the genotype of FL102. We performed PCR-based amplification and sequencing on five randomly selected pSOK804-based exconjugants. As expected, all five samples contained identical 74-bp sequences within the hybrid attLVWB site, supporting the genotype of FL102 as attLVWB-linear pSOK804-attRVWB, which was produced by the site-specific integration of pSOK804 (Fig. 2B and 3). Therefore, S. tsukubaensis proved to be amenable to the VWB integrase-mediated recombination system.
Effect of site-specific recombination on FK506 production in S. tsukubaensis.
The identification of two distinct attB sites on the chromosome of ZJU01 provides us a choice of the utilization of two different site-specific recombination systems in the insertion of an artificial gene construct. To apply this approach, we evaluated the effects of the ΦC31- and VWB-based integrations on S. tsukubaensis. pSET152-based integration at the attBΦC31 locus did not affect the growth characteristics, and the resulting strain, FL101 (43.4 ± 4.1 mg/liter), produced FK506 in a quantity similar to that of the original strain, ZJU01 (46.9 ± 2.5 mg/liter). In a similar manner, FL102, which bears an insertion of pSOK804 at the attBVWB locus, also showed no apparent difference in morphology and in FK506 production (40.7 ± 6.7 mg/liter) (Fig. 3C).
Duplication of the fkbGHIJK cassette encoding MOM-ACP formation via the ΦC31-based recombination system.
FkbG and FkbH to FkbK are convergently transcribed on the S. tsukubaensis chromosome (Fig. 1B). To simplify plasmid construction, fkbG without its native terminator was amplified and placed upstream of fkbHIJK, and the resultant fragment was inserted into pSET152 under the control of the constitutive ermE* promoter (PermE*) to give pFL103. pFL103 was introduced into ZJU01 by conjugation to yield the recombinant strain FL103, the genotype of which was verified by PCR amplification-coupled sequencing as attLΦC31-linear pFL103-attRΦC31 (Fig. 3A and B). The highest FK506 titer of FL103 was observed after 6 days of fermentation, similar to that of the wild-type strain ZJU01 (Fig. 3C). FL103 produced FK506 with a maximal titer of 61.3 ± 2.3 mg/liter under the specified culture conditions, showing an approximate 30% improvement in comparison with ZJU01 (46.9 ± 2.5 mg/liter). The data indicated that the duplication of the fkbGHIJK cassette has a positive effect on FK506 production.
Duplication of the tcsABCD cassette encoding AM-CoA formation via the ΦC31-based recombination system.
pFL104, a pSET152-derived plasmid containing the AM-CoA pathway-specific genes tcsABCD under the control of PermE*, was introduced into ZJU01 to yield the recombinant strain FL104, the genotype of which was confirmed as attLΦC31-linear pFL104-attRΦC31 (Fig. 3A and B). Similarly, FK506 production by FL104 reached the maximum after 6 days of fermentation (Fig. 3C), with a yield of 95.7 ± 4.6 mg/liter, approximately 100% above that of ZJU01. The duplication of the tcsABCD cassette significantly improved the FK506 fermentation yield, suggesting that AM-CoA formation in cells can be one of the main rate-limiting factors for FK506 biosynthesis in S. tsukubaensis.
Coduplication of the tcsABCD and fkbGHIJK cassettes via both the ΦC31- and VWB-based recombination systems.
Inspired by the success of FL103 (duplicating fkbGHIJK alone) and FL104 (duplicating tcsABCD alone) in FK506 production at an apparently improved level, we investigated whether the enhancement of both the MOM-ACP and AM-CoA pathways would result in a greater degree of production improvement. We thus developed a strategy in which both the ΦC31- and VWB-based systems could be utilized simultaneously. Since S. tsukubaensis is sensitive to apramycin and thiostrepton, we constructed a new VWB-based shuttle plasmid as a complement of pSET152. By replacing the original apramycin resistance gene on pSOK804 with the ampicillin and thiostrepton resistance genes, we generated a new conjugative vector, pTA804. We obtained FL105, a derivative of FL101, by inserting linear pTA804 at the attBVWB locus by conjugation and thiostrepton resistance selection. This recombinant strain, with an expected genotype of attLΦC31-linear pSET152-attRΦC31-attLVWB-linear pTA804-attRVWB, shown in Fig. 3A, produced FK506 (40.1 ± 4.5 mg/liter) at a level similar to that of the original strain, ZJU01 (Fig. 3C). The finding supported the practicability of the strategy of using both the ΦC31- and VWB-based systems for recombination, given that pTA804 is compatible with pSET152 in S. tsukubaensis.
To increase the expression of both MOM-ACP- and AM-CoA-forming genes, we inserted the tcsABCD cassette into pTA804 to yield pFL106, which was introduced into FL103 by conjugation to generate the recombinant strain FL106 (containing the fkbGHIJK and tcsABCD cassettes together). The genotype of this strain was confirmed as attLΦC31-linear pFL103-attRΦC31-attLVWB-linear pFL106-attRVWB (Fig. 3A and B). After 6 days of fermentation, FK506 titers (105.9 ± 7.3 mg/liter) increased approximately 125% over that of ZJU01 (Fig. 3C). Alternatively, we constructed pFL107, a pTA804 derivative carrying the fkbGHIJK cassette. pFL107 was introduced into FL104 to produce a recombinant strain, FL107, whose genotype was then verified by PCR amplification and DNA sequencing as attLΦC31-linear pFL104-attRΦC31-attLVWB-linear pFL107-attRVWB (Fig. 3A and B). FL107 showed an ability to produce FK506 similar to that of FL106 (Fig. 3C), with a maximal FK506 yield (107.9 ± 2.8 mg/liter) in fermentation on day 6. These results indicated that FK506 production can be further improved by coenhancing the biosyntheses of MOM-ACP and AM-CoA without the positional effect by using the two integrase-based recombination systems in S. tsukubaensis.
Transcriptional assay of the fkbGHIJK and tcsABCD cassettes by RT-PCR amplification.
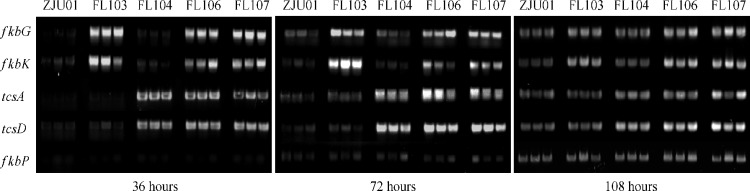
To correlate the overexpression of the MOM-ACP- and AM-CoA-coding genes with the duplication of the corresponding gene cassettes, we performed transcriptional analysis by semiquantitative reverse transcription (RT)-PCR amplification, with the transcription level of the NRPS gene fkbP as an internal control. Because the AM-CoA pathway-specific genes tcsABCD are organized in a contiguous form without a terminator, they can be transcribed into one polycistronic mRNA. We chose a 106-bp fragment within the first gene, tcsA, and a 103-bp fragment within the final gene, tcsD, to represent the transcription level of the AM-CoA biosynthetic genes. After reconstruction of the fkbGHIJK cassette, fkbG and fkbHIJK at the insertion locus were supposed to be transcribed into the same polycistronic mRNA. We chose here a 102-bp fragment within the first gene, fkbG, and a 102-bp fragment within the last gene, fkbK, to illustrate the transcription level of the MOM-ACP biosynthetic genes.
Indeed, RT-PCR analyses at the different time points in fermentation clearly indicated that the transcription levels of the genes coding for the MOM-ACP and AM-CoA biosynthetic pathways had apparently increased in FL103 and FL104, respectively (Fig. 4). This strongly supported the idea that the enhancement of pathway-specific gene transcription leads to the improvement in FK506 production. Compared to the original strain, ZJU01, the two recombinants, FL106 and FL107, with a higher yield of FK506 production, also exhibited the increased transcription levels of both the tcsABCD and fkbGHIJK cassettes in all parallel assays (Fig. 4).
Fig 4.
Semiquantitative RT-PCR analysis of the transcript levels of the fkbGHIJK and tcsABCD cassettes compared with that of fkbP. For each strain, the experiments were carried out on three independent isolates after 36, 72, and 108 h of fermentation. A 102-bp fkbG fragment and a 102-bp fkbK fragment were amplified to represent the fkbGHIJK transcript level, while a 106-bp tcsA fragment and a 103-bp tcsD fragment were amplified to represent the tcsABCD transcript level; a 102-bp fkbP fragment served as an internal control.
Effect on FK520 production of duplication of the MOM-ACP and AM-CoA cassettes.
S. tsukubaensis also has the ability to produce a minor product, FK520 (ascomycin), which differs from FK506 in structure only by the C-21 substitution of an ethyl group instead of an allyl group (Fig. 1A). This difference is dependent on the corresponding AT domain of the PKS system in substrate utilization, which selects ethylmalonyl (EM)-CoA as the extender unit for producing FK520 or AM-CoA for producing FK506 (20). To evaluate the relevance of FK520 with FK506 in production during the engineering process, the recombinant strains FL103, FL104, FL106, and FL107, as well as the original strain, ZJU01, were fermented and then subjected to HPLC quantification (Fig. 5). In FL103, the yield of FK520 (9.3 ± 0.6 mg/liter versus 7.4 ± 0.7 mg/liter for ZJU01) increased along with FK506, showing a ratio of FK520 to FK506 of 1:6.6 (a similar ratio of 1:6.3 was found in ZJU01). This is consistent with the prediction that the duplication of fkbGHIJK enhances the formation of MOM-ACP, the extender unit shared by the biosyntheses of FK520 and FK506. In FL104, the yield of FK520 (11.1 ± 0.9 mg/liter) increased; however, the growth rate was lower than that in FL103, giving a ratio of FK520 to FK506 of 1:8.6. Thus, the duplication of tcsABCD may mainly enhance the supply of AM-CoA, the extender unit used only in FK506 formation, providing a great potential to optimize its purity and production at the fermentation stage. In this case, the increase of FK520 production could be due to the promiscuity of the CCR protein TcsC in activity, which might accept crotonyl-CoA as the substrate to produce EM-CoA via reductive carboxylation for FK520 biosynthesis, as proposed previously (17). In line with the above, the yields of FK520 in FL106 (13.6 ± 1.0 mg/liter) and in FL107 (13.8 ± 0.7 mg/liter) also increased, showing a comprehensive effect of duplicating both fkbGHIJK and tcsABCD to give a similar ratio of FK520 to FK506 of 1:7.8.
Fig 5.

HPLC analysis of FK506 and FK520 from day 6 fermentation cultures of the S. tsukubaensis strains ZJU01, FL103, FL104, FL106, and FL107. AU, arbitrary units.
Further improvement of FK506 production by optimizing the glucose concentration.
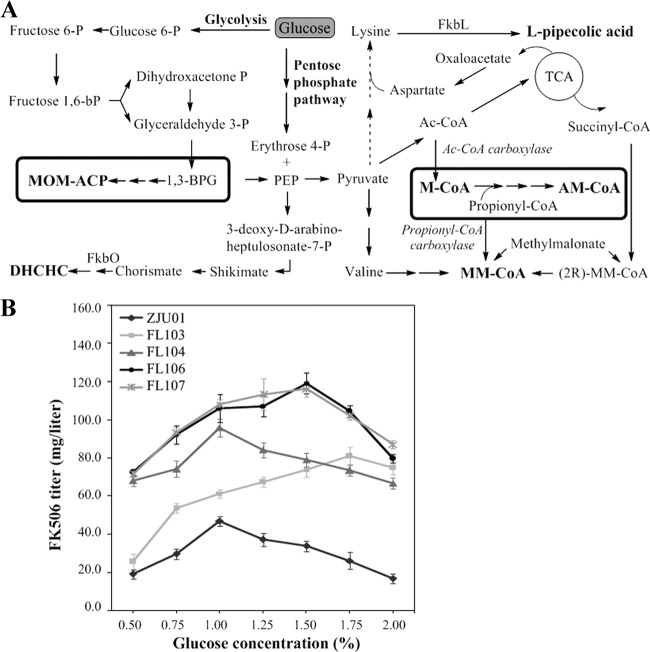
In regard to FK506 production, the fermentation culture of S. tsukubaensis in this study contained soybean oil and l-lysine, which provide the precursors for FK506 biosynthesis through cellular metabolism, including M-CoA, MM-CoA, and l-pipecolic acid (7, 16, 32, 36). Glucose, one of the most commonly used carbon sources for fermentative production, also serves as a direct source to biosynthesize the precursors of various secondary metabolites. However, when the culture is grown in a high concentration of glucose (or other easily assimilated carbohydrates, such as xylose and sucrose), the production of these metabolites is usually repressed (1, 25). Although the mechanism has yet to be understood, it was proposed that this repression effect may be related to glucose transport and phosphorylation (25). An overview of glucose metabolism relevant to FK506 biosynthesis is illustrated in Fig. 6A.
Fig 6.
Glucose-dependent profile of FK506 production by the S. tsukubaensis strains ZJU01, FL103, FL104, FL106, and FL107. (A) Overview of glucose metabolism in relation to FK506 biosynthesis. The boxed areas represent the MOM-ACP and AM-CoA pathways. (B) FK506 titers of ZJU01 and its recombinant strains FL103, FL104, FL106, and FL107 on day 6 with respect to different glucose concentrations. The data are the average values calculated from three independent isolates of parallel tests in triplicate, and the error bars represent the standard deviations.
Because MOM-ACP biosynthesis utilizes 1,3-BPG, which is a direct product of glycolysis (Fig. 6A), as its substrate (Fig. 1C), we reasoned that the overexpression of MOM-ACP biosynthetic enzymes would accelerate the depletion of intracellular 1,3-BPG. This depletion could consequently increase the glycolytic flux and render the cells more resistant to high concentrations of glucose. To test this hypothesis, strains were fermented in cultures with glucose concentrations that varied from 0.5% to 2.0%. FK506 production by ZJU01 and FL104 (the strain overexpressing AM-CoA biosynthetic genes alone) exhibited similar trends in response to the increased glucose concentration, showing maximal titers at 1.0% glucose and a linear decrease afterward (Fig. 6B). Production in ZJU01 decreased to about 35% of the maximum level in the culture that contained 2.0% glucose. Interestingly, for the strains overexpressing MOM-ACP biosynthetic genes, either alone or together with the AM-CoA genes, the FK506 production profiles were quite different (Fig. 6B). FK506 production by FL103 (the strain overexpressing MOM-ACP genes alone) increased almost linearly before the glucose concentration reached 1.75% (80.9 ± 4.7 mg/liter), while FL106 (119.1 ± 5.4 mg/liter) and FL107 (116.2 ± 4.1 mg/liter) (the strains overexpressing both MOM-ACP and AM-CoA genes) showed maximum FK506 production at 1.5% glucose. Consequently, FK506 production by FL106 and FL107 increased approximately 10% when the glucose concentration of the culture changed from 1.0% to 1.5%.
DISCUSSION
As a common approach for improving polyketide production, the genetic enhancement of the precursor supply has proven to be effective and promising for optimizing the strains for natural product biosynthesis. The intracellular pools of M-CoA and MM-CoA, which are the most widespread extenders and usually have low concentrations in cells (24), are frequently enriched by engineering to improve the production of desired polyketides. For example, the overexpression of acetyl (Ac)-CoA carboxylase, which diverts Ac-CoA away from the tricarboxylic acid (TCA) cycle and directs it toward M-CoA, resulted in a significant increase in actinorhodin production (27). The overexpression of the MM-CoA mutase pathway led to a 50% increase in erythromycin production by Saccharopolyspora erythraea (26), as well as a 50% increase in FK506 production by S. clavuligerus CKD1119 (19). The overexpression of propionyl-CoA carboxylase combined with the supplementation of the culture with propionate caused an 80% increase in rapamycin production by S. hygroscopicus ATCC 29253 (13).
M-CoA and MM-CoA metabolism is known to provide universal substrates independently of the polyketide biosynthetic pathways in microorganisms; however, FK506 biosynthesis requires two unusual pathway-specific building blocks, MOM-ACP and AM-CoA, raising the question of whether their formation in cells can be a bottleneck to production improvement. Here, the overexpression of a minimal set of genes that are responsible for the biosynthesis of either MOM-ACP or AM-CoA enhances the production of FK506 in the recombinant strains. The data suggest that the intracellular formation of the unusual PKS extender units is likely to be the rate-limiting factor and can be rationally engineered to improve FK506 production. It is noteworthy that only one molecule of AM-CoA is incorporated into the FK506 scaffold, yet the production in FL104 upon duplicating the AM-CoA biosynthetic genes increased by approximately 100% over that in ZJU01. It is likely that for the original strain, AM-CoA biosynthesis has a lower turnover number than PKS and thus is unable to provide sufficient precursors for FK506 biosynthesis.
Current successes in generating high-producing strains via site-specific recombination mainly rely on the application of the actinophage ΦC31 system (13, 19, 26, 27, 34). However, with this system, our ability to evaluate the comprehensive effect of the genetic enhancement of both the MOM-ACP and AM-CoA supplies was limited. The characterization of new site-specific recombination systems provided us with an exciting possibility that diverse genetic tools could be used at the same time for the stable expression of recombinant constructs in candidate strains. Considering that most actinomycetes are sensitive to apramycin and thiostrepton, the newly constructed vector pTA804 (VWB based) carrying ampicillin and thiostrepton resistance genes may serve as a useful tool that complements the plasmid pSET152 (ΦC31 based) for genetic manipulation in actinomycetes. The strategy of utilizing two actinophage (ΦC31 and VWB)-based recombination systems provides a convenient platform for a rapid evaluation of the gene dose effect in S. tsukubaensis, which facilitates the simultaneous overexpression of the MOM-ACP and AM-CoA pathways. The two systems can operate independently and exhibit a comprehensive effect, as exemplified by those in FL106 and FL107, to enhance the transcriptional levels of fkbGHIJK and tcsABCD to further improve the production of FK506.
The strategy described here could also be applicable and useful for other secondary metabolites that contain pathway-specific building blocks, as exemplified by ansatomycin, midecamycin, tautomycin, and oxazolomycin, the biosyntheses of which share a route to build the methoxy group at the β-carbon of the growing polyketide chain via the incorporation of MOM-ACP as the extender unit by PKSs (4, 14). To put it into practice, we developed a method using two distinct site-specific recombination systems in the FK506 producer S. tsukubaensis. These studies broaden the repertoire of actinophage-based vectors and allow the engineering of Actinomyces in a multiple-site-specific recombination manner.
ACKNOWLEDGMENTS
This work was supported in part by grants from the National Natural Science Foundation (20921091 and 21176214) and the National Basic Research Program (2010CB833200 and 2012CB721100) of China and the Chinese Academy of Sciences (KJCX2-YW-201).
Footnotes
Published ahead of print 11 May 2012
REFERENCES
- 1. Abbanat D, Maiese W, Greenstein M. 1999. Biosynthesis of the pyrroindomycins by Streptomyces rugosporus LL-42D005; characterization of nutrient requirements. J. Antibiot. 52:117–126 [DOI] [PubMed] [Google Scholar]
- 2. Andexer JN, et al. 2011. Biosynthesis of the immunosuppressants FK506, FK520, and rapamycin involves a previously undescribed family of enzymes acting on chorismate. Proc. Natl. Acad. Sci. U. S. A. 108:4776–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bierman M, et al. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49 [DOI] [PubMed] [Google Scholar]
- 4. Carroll BJ, et al. 2002. Identification of a set of genes involved in the formation of the substrate for the incorporation of the unusual “glycolate” chain extension unit in ansamitocin biosynthesis. J. Am. Chem. Soc. 124:4176–4177 [DOI] [PubMed] [Google Scholar]
- 5. Combes P, Till R, Bee S, Smith MCM. 2002. The streptomyces genome contains multiple pseudo-attB sites for the (phi)C31-encoded site-specific recombination system. J. Bacteriol. 184:5746–5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Craig NL. 1988. The mechanism of conservative site-specific recombination. Annu. Rev. Genet. 22:77–105 [DOI] [PubMed] [Google Scholar]
- 7. Gatto GJ, Boyne MT, Kelleher NL, Walsh CT. 2006. Biosynthesis of pipecolic acid by RapL, a lysine cyclodeaminase encoded in the rapamycin gene cluster. J. Am. Chem. Soc. 128:3838–3847 [DOI] [PubMed] [Google Scholar]
- 8. Gatto GJ, McLoughlin SM, Kelleher NL, Walsh CT. 2005. Elucidating the substrate specificity and condensation domain activity of FkbP, the FK520 pipecolate-incorporating enzyme. Biochemistry 44:5993–6002 [DOI] [PubMed] [Google Scholar]
- 9. Gold BG. 2000. Neuroimmunophilin ligands: evaluation of their therapeutic potential for the treatment of neurological disorders. Expert Opin. Investig. Drugs. 9:2331–2342 [DOI] [PubMed] [Google Scholar]
- 10. Goranovic D, et al. 2010. Origin of the allyl group in FK506 biosynthesis. J. Biol. Chem. 285:14292–14300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gust B, Challis GL, Fowler K, Kieser T, Chater KF. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gust B, et al. 2004. Lambda red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv. Appl. Microbiol. 54:107–128 [DOI] [PubMed] [Google Scholar]
- 13. Jung WS, et al. 2011. A combined approach of classical mutagenesis and rational metabolic engineering improves rapamycin biosynthesis and provides insights into methylmalonyl-CoA precursor supply pathway in Streptomyces hygroscopicus ATCC 29253. Appl. Microbiol. Biotechnol. 91:1389–1397 [DOI] [PubMed] [Google Scholar]
- 14. Kato Y, et al. 2002. Functional expression of genes involved in the biosynthesis of the novel polyketide chain extension unit, methoxymalonyl-acyl carrier protein, and engineered biosynthesis of 2-desmethyl-2-methoxy-6-deoxyerythronolide B. J. Am. Chem. Soc. 124:5268–5269 [DOI] [PubMed] [Google Scholar]
- 15. Kieser T, Bibb MJ, Buttner MJ, Chater KF, Howood DA. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kindom [Google Scholar]
- 16. Kim HS, Park YI. 2007. Lipase activity and tacrolimus production in Streptomyces clavuligerus CKD 1119 mutant strains. J. Microbiol. Biotechnol. 17:1638–1644 [PubMed] [Google Scholar]
- 17. Kosec G, et al. 2012. Novel chemobiosynthetic approach for exclusive production of FK506. Metab. Eng. 14:39–46 [DOI] [PubMed] [Google Scholar]
- 18. Liu J, et al. 1991. Calcineurin is a common target of cyclophilin-cyclosporine-A and FKBP-FK506 complexes. Cell 66:807–815 [DOI] [PubMed] [Google Scholar]
- 19. Mo S, Ban YH, Park JW, Yoo YJ, Yoon YJ. 2009. Enhanced FK506 production in Streptomyces clavuligerus CKD1119 by engineering the supply of methylmalonyl-CoA precursor. J. Ind. Microbiol. Biotechnol. 36:1473–1482 [DOI] [PubMed] [Google Scholar]
- 20. Mo S, et al. 2011. Biosynthesis of the allylmalonyl-CoA extender unit for the FK506 polyketide synthase proceeds through a dedicated polyketide synthase and facilitates the mutasynthesis of analogues. J. Am. Chem. Soc. 133:976–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Motamedi H, Cai SJ, Shafiee A, Elliston KO. 1997. Structural organization of a multifunctional polyketide synthase involved in the biosynthesis of the macrolide immunosuppressant FK506. Eur. J. Biochem. 244:74–80 [DOI] [PubMed] [Google Scholar]
- 22. Motamedi H, Shafiee A. 1998. The biosynthetic gene cluster for the macrolactone ring of the immunosuppressant FK506. Eur. J. Biochem. 256:528–534 [DOI] [PubMed] [Google Scholar]
- 23. Ostash B, Makitrinskyy R, Walker S, Fedorenko V. 2009. Identification and characterization of Streptomyces ghanaensis ATCC14672 integration sites for three actinophage-based plasmids. Plasmid 61:171–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Park JW, Jung WS, Park SR, Park BC, Yoon YJ. 2007. Analysis of intracellular short organic acid-coenzyme A esters from actinomycetes using liquid chromatography-electrospray ionization-mass spectrometry. J. Mass Spectrom. 42:1136–1147 [DOI] [PubMed] [Google Scholar]
- 25. Ramos I, et al. 2004. Glucose kinase alone cannot be responsible for carbon source regulation in Streptomyces peucetius var. caesius. Res. Microbiol. 155:267–274 [DOI] [PubMed] [Google Scholar]
- 26. Reeves AR, et al. 2007. Engineering of the methylmalonyl-CoA metabolite node of Saccharopolyspora erythraea for increased erythromycin production. Metab. Eng. 9:293–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ryu YG, Butler MJ, Chater KF, Lee KJ. 2006. Engineering of primary carbohydrate metabolism for increased production of actinorhodin in Streptomyces coelicolor. Appl. Environ. Microbiol. 72:7132–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schwecke T, et al. 1995. The biosynthetic gene-cluster for the polyketide immunosuppressant rapamycin. Proc. Natl. Acad. Sci. U. S. A. 92:7839–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sekurova ON, et al. 2004. In vivo analysis of the regulatory genes in the nystatin biosynthetic gene cluster of Streptomyces noursei ATCC 11455 reveals their differential control over antibiotic biosynthesis. J. Bacteriol. 186:1345–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shen B, Hutchinson CR. 1996. Deciphering the mechanism for the assembly of aromatic polyketides by a bacterial polyketide synthase. Proc. Natl. Acad. Sci. U. S. A. 93:6600–6604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sierra-Paredes G, Sierra-Marcuno G. 2008. Ascomycin and FK506: pharmacology and therapeutic potential as anticonvulsants and neuroprotectants. CNS Neurosci. Ther. 14:36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Singh BP, Behera BK. 2009. Regulation of tacrolimus production by altering primary source of carbons and amino acids. Lett. Appl. Microbiol. 49:254–259 [DOI] [PubMed] [Google Scholar]
- 33. Van Mellaert L, Mei L, Lammertyn E, Schacht S, Anne J. 1998. Site-specific integration of bacteriophage VWB genome into Streptomyces venezuelae and construction of a VWB-based integrative vector. Microbiology 144:3351–3358 [DOI] [PubMed] [Google Scholar]
- 34. Wu JQ, et al. 2011. Toward improvement of erythromycin A production in an industrial Saccharopolyspora erythraea strain via facilitation of genetic manipulation with an artificial attB site for specific recombination. Appl. Environ. Microbiol. 77:7508–7516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu K, Chung L, Revill WP, Katz L, Reeves CD. 2000. The FK520 gene cluster of Streptomyces hygroscopicus var. ascomyceticus (ATCC 14891) contains genes for biosynthesis of unusual polyketide extender units. Gene 251:81–90 [DOI] [PubMed] [Google Scholar]
- 36. Yoon YJ, Choi CY. 1997. Nutrient effects on FK-506, a new immunosuppressant, production by Streptomyces sp. in a defined medium. J. Ferment. Bioeng. 83:599–603 [Google Scholar]