Abstract
Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes is an essential tool for the cultivation-independent identification of microbes within environmental and clinical samples. However, one of the major constraints of conventional FISH is the very limited number of different target organisms that can be detected simultaneously with standard epifluorescence or confocal laser scanning microscopy. Recently, this limitation has been overcome via an elegant approach termed combinatorial labeling and spectral imaging FISH (CLASI-FISH) (23). This technique, however, suffers compared to conventional FISH from an inherent loss in sensitivity and potential probe binding biases caused by the competition of two differentially labeled oligonucleotide probes for the same target site. Here we demonstrate that the application of multicolored, double-labeled oligonucleotide probes enables the simultaneous detection of up to six microbial target populations in a straightforward and robust manner with higher sensitivity and less bias. Thus, this newly developed technique should be an attractive option for all researchers interested in applying conventional FISH methods for the study of microbial communities.
INTRODUCTION
In medical and environmental microbiology, fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes is a widely used technique for direct identification and quantification of microorganisms (24). However, several factors may prevent the successful identification of certain microorganisms with conventional FISH (1, 25). Among the most frequently encountered problems are weak or undetectable probe-conferred signals emitted from microorganisms that contain target molecule numbers below the FISH detection limit (9) or that are hybridized with probes with a poor in situ accessibility of the selected probe binding site (7, 29). Improved FISH techniques for ameliorating both problems have been developed. For example, if catalyzed reporter deposition (CARD)-FISH (14, 18) is applied, a 26- to 41-fold increased sensitivity (9) can be achieved, but the protocol is more complicated and needs to be specifically adapted for some microorganisms (8, 20), and multicolor imaging options of this technique are very tedious and limited (13, 19). Furthermore, several useful approaches have been developed to tackle the poor accessibility of certain probe target sites (6, 28, 30). In this context, double labeling of oligonucleotide probes (DOPE)-FISH (20) represents a recently introduced approach that maintains the elegant simplicity of conventional FISH but offers an approximately doubled signal intensity plus an increased in situ accessibility of probe target sites without affecting the specificity of the applied probes.
Another major limitation of FISH is the fact that only a few different target organisms can be simultaneously detected by applying probes labeled with different dyes. The major reasons for this are the use of band- or long-pass filters in fluorescence image acquisition and the excitation cross talk and emission bleed-through of suitable fluorophores (23). Therefore, in practice, not more than three different dyes can be conveniently used in parallel to examine the microbial population structure within a sample. If applied together with a carefully compiled hierarchical set of three different probes, this allows, in principle, the identification of up to seven microbial populations in situ in one assay (2). However, this approach imposes strong constraints on probe design and, for example, is not applicable if phylogenetically distantly related groups of target organisms should be detected in parallel.
These very limited multiplexing options of FISH were recently overcome by the introduction of combinatorial labeling and spectral imaging FISH (CLASI-FISH) (23). Here binary combinations of oligonucleotide probe-dye constructs were used for the simultaneous detection of up to 28 target organisms, resulting in unique mixed colors that were distinguished via spectral fingerprinting. However, as the CLASI-FISH approach exploits two probes for each target organism, hybridizing to the same binding site but each labeled with a different dye, these probes compete for the same binding site. Thus, for each probe, one would expect that the signal intensity is halved and hence the sensitivity of the assay is significantly reduced. Furthermore, it has been demonstrated that the very same probes possess different binding affinities to their target sites depending on the fluorescence dye used for labeling (20). Thus, the theoretically expected binding ratio of 50:50 for two probes with the same sequence but conjugated to different fluorophores might be shifted toward the more competitive probe-dye construct, complicating the identification of target organisms.
In this study, we experimentally demonstrate the above-mentioned signal reduction effects of CLASI-FISH and present an alternative and widely applicable DOPE-FISH-based approach, which offers straightforward identification of six taxa with a single FISH experiment.
MATERIALS AND METHODS
Single- and double-labeled probes were obtained from Thermo Hybaid (Interactiva Division, Ulm, Germany). All FISH and DOPE-FISH experiments were carried out as described previously (4, 20), with a hybridization time of 4 h, followed by a 10-min washing step. For the pure-culture experiments, Escherichia coli (DSM 498) was grown in Luria-Bertani medium until stationary phase and fixed with paraformaldehyde (PFA) as described previously (4). A sampling of three individuals of the sponge Ancorina alata (Demospongiae, Astrophorida, Ancorinidae) was carried out by scuba diving at a depth of 3 m at Jones Bay (36°12′30″S, 174°14′90″E), northeastern New Zealand, in April 2010. Tissue samples were cut into small pieces of about 2 mm3, fixed in 4% PFA for 4 h at room temperature and stored in ethyl alcohol (EtOH)-phosphate-buffered saline (PBS) at −20°C. For FISH, PFA-fixed samples of A. alata were embedded in Neg-50 (Richard-Allan Scientific), a cryogenic water-soluble frozen section medium, and cut to 4-μm thin sections (Leica CM3050 S). For E. coli, the probes EUB338 (targeting the 16S rRNA of most but not all bacteria) and Gam42a (targeting the 23S rRNA of many members of the Gammaproteobacteria) were used. A. alata sections were hybridized with the probes Por1130 (doubly labeled with Fluos and Cy3, specific for the candidate phylum “Poribacteria”), GNSB-941 (doubly labeled with Cy5 and Fluos, specific for the phylum Chloroflexi), Ntspa662 (doubly labeled with Cy5 and Cy3, specific for the genus Nitrospira), Delta495a (doubly labeled with Fluos, specific for most Deltaproteobacteria and most Gemmatimonadetes), Gam42a (doubly labeled with Cy3), and Arch915 (doubly labeled with Cy5, specific for most Archaea). More information about the applied probes and their optimized hybridization conditions can be found at probeBase (12). If not stated otherwise, probes were applied in a final concentration of 5 pmol. For the sponge hybridization, an equimolar mixture of the probes was prepared, which also contained the unlabeled competitor probes for the probes Ntspa662 and Gam42a. A potential hybrid formation between deployed probes was evaluated employing the Web tool mathFISH (30), which confirmed low hybridization efficiency of all possible probe pairs. Sponge hybridizations were carried out in the presence of 35% formamide in the hybridization buffer, and the stringency of the wash buffer was adjusted accordingly (4). Probe-conferred signal intensities were quantified by analyzing at least 1,000 cells per experiment using a confocal laser scanning microscope (CLSM) (LSM 510 Meta; Zeiss, Oberkochen, Germany) and the software program daime (3). For these measurements, individual cells were detected by image segmentation via edge detection. All experiments were performed in three independent replicates, except for the competition experiment between the singly Cy3- and Fluos-labeled Eub338 probes, for which six independent replicates were analyzed. During our experiments, we noticed that conventional and DOPE-FISH probes labeled with Cy5 sometimes showed unexpected low signal intensities. This problem could be solved by reordering the probe. It is tempting to speculate that the pronounced ozone susceptibility of Cy5 contributes to this phenomenon (5).
RESULTS AND DISCUSSION
Double hybridization of E. coli with two probes binding to the same target site reduces signal intensities in a dye-dependent manner.
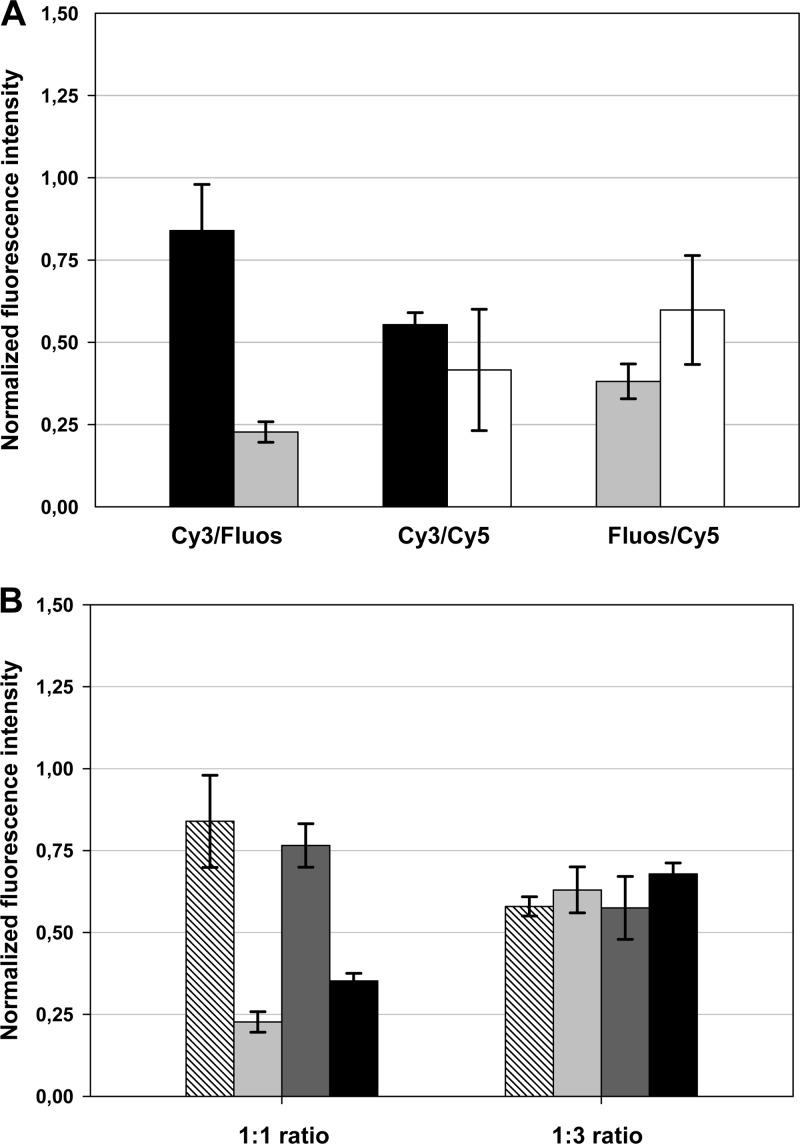
In an initial pure-culture experiment with probe EUB338 and E. coli as the target organism, the influence of double hybridization of the same rRNA target site with two probes labeled with different dyes was examined. For this purpose, we hybridized E. coli separately with the EUB338 probe monolabeled with Cy3, Cy5, and Fluos. Subsequently, we mimicked the CLASI-FISH approach (23) and used two differentially labeled derivatives of the probe in the same hybridization experiment (Cy3-Fluos, Cy3-Cy5, and Cy5-Fluos) and compared the probe-conferred signal intensities between both experiments. If Cy3- and Cy5-labeled probes were used for double hybridization, the obtained signal intensities for each dye were about 50% reduced compared to the FISH experiment without probe competition (Fig. 1A). This finding is fully consistent with previously published data demonstrating that if two differentially labeled probes are used in a competitive manner and dye-associated effects influencing the probe binding affinity are excluded, the intensity of fluorescence conferred by these probes depends linearly on their molar fraction in the probe mixture (9). As Cy3 and Cy5 share similar properties concerning their molecular weights and chemical structure, it is not surprising that no label-derived effects on the competiveness of the respective probes were detected. In contrast, the competition experiments with Cy3-Fluos-labeled and Cy5-Fluos-labeled probes revealed that cyanine-labeled probes preferentially bind to the target site (Fig. 1A). Thus, compared to the signal intensity of the Fluos-labeled EUB338 probe applied alone, the signal intensity of the Fluos-labeled EUB338 probe is reduced to below 40% when applied together with the Cy5-labeled EUB338 probe derivative and is even further reduced to about 25% when used in combination with a Cy3-labeled EUB338 probe. This effect reflects the stronger binding of Cy dye-labeled probes to rRNA that has been described previously (20) and can be reversed if the less-competitive probe is used in a higher concentration in the hybridization mixture (Fig. 1B).
Fig 1.
FISH signal intensity of E. coli cells hybridized with two probes binding to the same rRNA target region is dependent on dye combinations and probe concentrations. (A) E. coli cells were simultaneously hybridized with two EUB338 probes labeled with different dyes, and the signal intensity was recorded for each dye. In the next step, for each fluorophore in each combination (Fluos-Cy3-, Cy3-Cy5-, and Fluos-Cy5-labeled probe combinations were used), the signal intensity was normalized to the signal intensity obtained for E. coli after hybridization with only one probe labeled with the respective dye. Black, light-gray, and white bars display results for the Cy3-, Fluos-, and Cy5-labeled probe EUB338, respectively, in the competition experiments. (B) Increase of the concentration of the less-competitive probe derivative in the hybridization mixture (Fluos-labeled probe to Cy3-labeled probe, 3:1) leads to signal recovery. Hatched and light-gray bars display results for the Cy3- and Fluos-labeled probe EUB338, respectively. Dark-gray and black bars display results for the Cy3- and Fluos-labeled probe Gam42a, respectively.
In summary, the CLASI-FISH approach leads, in the best case, to a loss of probe-conferred signal intensities of 50% for both dyes of the binary combination, if the dyes possess similar properties. Thus, the detection limit of the CLASI-FISH technique for microbes with low cellular ribosome content is significantly reduced. However, this problem could be solved if future CLASI-FISH applications were to be based on probes labeled with two identical dyes (23). The observed shift in the probe binding ratio, as a consequence of a difference in competitiveness of certain probe-dye constructs, is more problematic. In particular, cells with low ribosome content might not display the intended color mix but instead might appear single colored and thus would not be correctly identified. It did not escape our attention that neither Cy dyes nor Fluos have been used in the original CLASI-FISH approach (23). However, the chemical structures and properties of the applied dyes in that study were highly diverse, and thus it would be necessary to test all binary dye combinations in well-controlled experiments analogous to those described above in order to exclude dye combination-conferred biases or to select suitable dye combinations.
Multicolor DOPE-FISH.
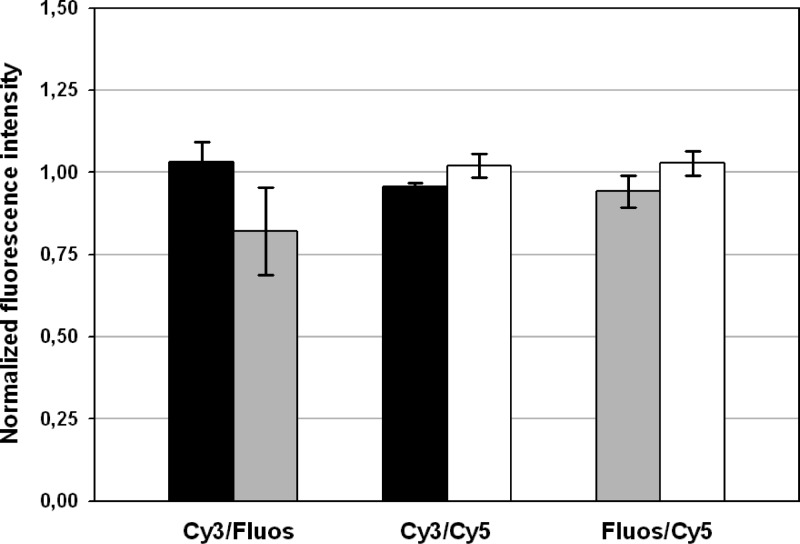
Due to the above-described limitations of multicolor FISH based on double hybridizations with binary mixtures of differently labeled probes, we evaluated whether double labeling of the same oligonucleotide probe with two different dyes could be a suitable alternative. For this purpose, a set of experiments with probe EUB338 and E. coli, analogous to those described above, were performed. Theoretically, one would expect that probe-conferred signal intensities obtained after hybridization with a probe labeled with two different dyes would be comparable to those measured in the control experiment with the respective monolabeled probe. Indeed, the Cy5-Fluos- and Cy5-Cy3-double-labeled probe signal intensities for each dye were comparable to those obtained from hybridization with the Fluos- and Cy5-single-labeled probe derivatives, indicating that quenching effects can be neglected for these dye combinations (Fig. 2). This is not surprising, since the emission maxima of Fluos (521 nm) and Cy3 (570 nm) are well separated from the absorption maximum of Cy5 (649 nm). Concerning the Fluos-Cy3 combination, the situation is different with the emission maximum of Fluos being quite close to the absorption maximum of Cy3 (550 nm). As a consequence, the photons emitted by the Fluos dye are partially quenched by Cy3 in double-labeled probes, leading to an approximately 20% reduction in Fluos signal intensity (Fig. 2), which is, however, much less pronounced than in the competition experiment of Fluos-labeled probes with Cy3-labeled probes (Fig. 1).
Fig 2.
Signal intensities of E. coli cells hybridized with DOPE-FISH probes labeled with two different dyes. For each fluorophore in each combination (Fluos-Cy3-, Cy3-Cy5-, and Fluos-Cy5-double-labeled probes were applied), the signal intensity was normalized to the signal intensity obtained for E. coli by applying the respective single-labeled probe derivative. Black, light-gray, and white bars display results for Cy3, Fluos, and Cy5, respectively. It should be noted that the observed quenching of the Fluos signal of probes carrying Cy3 on the other end does not occur if cells are hybridized simultaneously with a Cy3-monolabeled and a Fluos-monolabeled probe binding to different target sites (data not shown).
In summary, dual-colored DOPE probes are an easy-to-apply alternative for multicolor FISH experiments, which lead to signal intensities between 75 and 104% for each dye compared to the use of conventional monolabeled FISH probes and thus outperform the use of two differentially labeled probes competing for the same target site.
Multicolor DOPE-FISH for the simultaneous visualization of six phylogenetically distinct sponge symbiont populations without spectral unmixing.
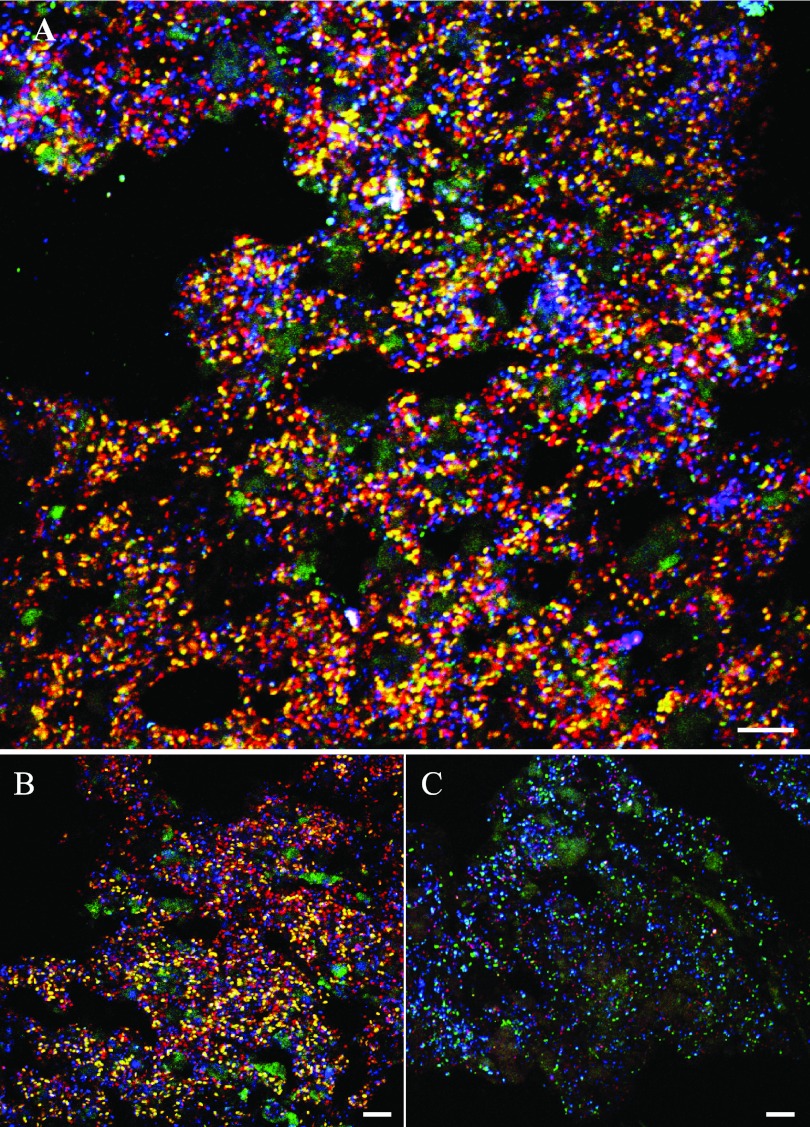
In a proof-of-principle experiment, the multicolor DOPE-FISH approach was applied to visualize six different microbial symbiont populations in a marine sponge. These animals are well known to harbor dense and diverse microbial communities with substantial ecological and biotechnological importance (21). Sponge-associated microbes include bacteria and archaea that possess different degrees of host specificity (22) and can comprise up to 40% of the sponge tissue volume. At the time of this report, members of more than 25 different bacterial phyla and 3 archaeal phyla (11, 17, 26, 27) had been detected in sponges. However, due to the rather low ribosome content of certain microbes thriving in sponges (10) and the considerable high background fluorescence often observed, sponge tissue represents a challenging material for FISH analyses of inhabiting microbes. Consequently, any loss in sensitivity of the FISH assay has to be avoided. For the multicolor DOPE-FISH application, we decided to employ the marine sponge A. alata due to its high microbial abundance and diversity revealed in previous studies by transmission electron microscopy (TEM), 16S rRNA (gene) clone library analyses, and amplicon pyrosequencing (10, 17). Using six specific DOPE-FISH probes uniquely labeled with different dyes, six phylogenetically distinct microbial symbiont populations—members of the candidate phylum “Poribacteria,” the phylum Chloroflexi, the genus Nitrospira, the Deltaproteobacteria, the Gammaproteobacteria, and the Archaea—could be simultaneously detected in a single hybridization event (Fig. 3A). Archaea, Gammaproteobacteria, and Poribacteria clearly dominated the microbial biomass (Fig. 3A and B), whereas the Deltaproteobacteria, Chloroflexi, and Nitrospira were less abundant (Fig. 3A and C). Generally, this observation is in good accordance with previous findings: Archaea, Poribacteria, and Gammaproteobacteria are often found as numerically dominant players in microbial sponge communities (10, 15–17). Furthermore, Kamke and coworkers reported low abundances of Nitrospira- and Deltaproteobacteria-related clones in DNA- and RNA-based clone libraries generated from A. alata (10).
Fig 3.
Multicolor DOPE-FISH analyses of microbial sponge symbionts. (A) FISH of tissue from the marine sponge A. alata with rRNA-targeted probes specific for Poribacteria (Por1130, yellow), Nitrospira (Ntspa662, pink), Chloroflexi (GNSB941, cyan), Deltaproteobacteria (Delta495a, green; larger green areas are sponge autofluorescence), Gammaproteobacteria (Gam42a, red), and domain Archaea (Arch915, blue). (B) Poribacteria (Por1130, yellow), Gammaproteobacteria (Gam42a, red), and domain Archaea (Arch915, blue). Autofluorescence of sponge tissue is displayed in green. (C) Nitrospira (Ntspa662, magenta), Chloroflexi (GNSB941, cyan), and Deltaproteobacteria (Delta495a, green; larger green areas are sponge autofluorescence). For all panels, CLSM images were recorded separately for each color channel and subsequently superimposed to obtain the final composite image. Bars, 10 μm.
As a consequence of differences in the cellular ribosome content of the different target populations, the optimal adjustment of the CLSM settings for image recording was quite challenging. The rather low probe-conferred signal intensity of, for instance, the Nitrospira-like target cells (detected by a Cy3-Cy5-double-labeled probe), required a high detection sensitivity for the Cy3 and Cy5 channels. This again led to overexposure for three other distinct target populations detected by probes carrying a Cy3 or Cy5 dye as well (Poribacteria, Cy3-Fluos; Gammaproteobacteria, Cy3; Archaea, Cy5) but displaying a higher ribosome content. Regardless of which approach is chosen (multicolor DOPE-FISH or CLASI-FISH), such conditions, with target populations differing in their cellular ribosome content, require careful consideration regarding the design of the applied probe-dye constructs, but any loss in sensitivity (as a result of two probes competing for the same binding site) or skew in the dye-conferred signal ratio (as a result of different competitiveness of certain probe-dye constructs) will further aggravate the simultaneous detection of different target populations under such difficult conditions.
In summary, we were able to demonstrate that multicolor DOPE-FISH is a straightforward stand-alone approach for the simultaneous detection of up to six microbial populations in a single FISH experiment. If combined with spectral unmixing, as impressively applied in a recent CLASI-FISH study (23), the use of oligonucleotide probes labeled with different binary combinations of dyes will almost certainly further enhance the potential of multicolor FISH for the analyses of samples with elevated levels of background fluorescence and/or for microbes with a low cellular ribosome content.
ACKNOWLEDGMENTS
We thank Mike Taylor for providing samples of the sponge Ancorina alata and Holger Daims for help with image analysis.
F.B. was funded by the graduate school “Symbiotic Interactions” of the University of Vienna and partially by a grant from the Austrian Science Fund (FWF) (Y277-B03). K.S. and A.V. were supported by the German Federal State of Hessen as part of the LOEWE program. K.S. was also partially supported by a grant from the Austrian Science Fund (P20775).
Footnotes
Published ahead of print 11 May 2012
REFERENCES
- 1. Amann R, Fuchs BM. 2008. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 6:339–348 [DOI] [PubMed] [Google Scholar]
- 2. Amann R, Snaidr J, Wagner M, Ludwig W, Schleifer KH. 1996. In situ visualization of high genetic diversity in a natural microbial community. J. Bacteriol. 178:3496–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Daims H, Lücker S, Wagner M. 2006. daime, a novel image analysis program for microbial ecology and biofilm research. Environ. Microbiol. 8:200–213 [DOI] [PubMed] [Google Scholar]
- 4. Daims H, Stoecker K, Wagner M. 2005. Fluorescence in situ hybridization for the detection of prokaryotes, p 213–239. In Osborn AM, Smith CJ. (ed), Advanced methods in molecular microbial ecology. Bio-Garland, Abingdon, United Kingdom [Google Scholar]
- 5. Fare TL, et al. 2003. Effects of atmospheric ozone on microarray data quality. Anal. Chem. 75:4672–4675 [DOI] [PubMed] [Google Scholar]
- 6. Fuchs BM, Glockner FO, Wulf J, Amann R. 2000. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 66:3603–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fuchs BM, et al. 1998. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 64:4973–4982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Furukawa K, Hoshino T, Tsuneda S, Inamori Y. 2006. Comprehensive analysis of cell wall-permeabilizing conditions for highly sensitive fluorescence in situ hybridization. Microb. Environ. 21:227–234 [Google Scholar]
- 9. Hoshino T, Yilmaz LS, Noguera DR, Daims H, Wagner M. 2008. Quantification of target molecules needed to detect microorganisms by fluorescence in situ hybridization (FISH) and catalyzed reporter deposition-FISH. Appl. Environ. Microbiol. 74:5068–5077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kamke J, Taylor MW, Schmitt S. 2010. Activity profiles for marine sponge-associated bacteria obtained by 16S rRNA vs 16S rRNA gene comparisons. ISME J. 4:498–508 [DOI] [PubMed] [Google Scholar]
- 11. Lee OO, et al. 2011. Pyrosequencing reveals highly diverse and species-specific microbial communities in sponges from the Red Sea. ISME J. 5:650–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Loy A, Maixner F, Wagner M, Horn M. 2007. probeBase—an online resource for rRNA-targeted oligonucleotide probes: new features 2007. Nucleic Acids Res. 35:D800–D804 doi:10.1093/nar/gkl856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pernthaler A, Pernthaler J. 2007. Fluorescence in situ hybridization for the identification of environmental microbes. Methods Mol. Biol. 353:153–164 [DOI] [PubMed] [Google Scholar]
- 14. Pernthaler A, Pernthaler J, Amann R. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Radax R, Hoffmann F, Rapp HT, Leininger S, Schleper C. 2012. Ammonia-oxidizing archaea as main drivers of nitrification in cold-water sponges. Environ. Microbiol. doi:10.1111/j.1462–2920.2011.02661.x. [DOI] [PubMed]
- 16. Radax R, et al. 2012. Metatranscriptomics of the marine sponge Geodia barretti: tackling phylogeny and function of its microbial community. Environ. Microbiol. doi:10.1111/j.1462–2920.2012.02714.x. [DOI] [PubMed]
- 17. Schmitt S, et al. 2012. Assessing the complex sponge microbiota: core, variable and species-specific bacterial communities in marine sponges. ISME J. 6:564–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schönhuber W, Fuchs B, Juretschko S, Amann R. 1997. Improved sensitivity of whole-cell hybridization by the combination of horseradish peroxidase-labeled oligonucleotides and tyramide signal amplification. Appl. Environ. Microbiol. 63:3268–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Speel EJM, Ramaekers FCS, Hopman AHN. 1997. Sensitive multicolor fluorescence in situ hybridization using catalyzed reporter deposition (CARD) amplification. J. Histochem. Cytochem. 45:1439–1446 [DOI] [PubMed] [Google Scholar]
- 20. Stoecker K, Dorninger C, Daims H, Wagner M. 2010. Double labeling of oligonucleotide probes for fluorescence in situ hybridization (DOPE-FISH) improves signal intensity and increases rRNA accessibility. Appl. Environ. Microbiol. 76:922–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Taylor MW, Radax R, Steger D, Wagner M. 2007. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71:295–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taylor MW, Schupp PJ, Dahllof I, Kjelleberg S, Steinberg PD. 2004. Host specificity in marine sponge-associated bacteria, and potential implications for marine microbial diversity. Environ. Microbiol. 6:121–130 [DOI] [PubMed] [Google Scholar]
- 23. Valm AM, et al. 2011. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc. Natl. Acad. Sci. U. S. A. 108:4152–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wagner M, Haider S. 2012. New trends in fluorescence in situ hybridization for identification and functional analyses of microbes. Curr. Opin. Biotechnol. 23:96–102 [DOI] [PubMed] [Google Scholar]
- 25. Wagner M, Horn M, Daims H. 2003. Fluorescence in situ hybridisation for the identification and characterisation of prokaryotes. Curr. Opin. Microbiol. 6:302–309 [DOI] [PubMed] [Google Scholar]
- 26. Webster NS, Taylor MW. 2012. Marine sponges and their microbial symbionts: love and other relationships. Environ. Microbiol. 14:335–346 [DOI] [PubMed] [Google Scholar]
- 27. Webster NS, et al. 2010. Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ. Microbiol. 12:2070–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yilmaz LS, Noguera DR. 2004. Mechanistic approach to the problem of hybridization efficiency in fluorescent in situ hybridization. Appl. Environ. Microbiol. 70:7126–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yilmaz LS, Ökten HE, Noguera DR. 2006. Making all parts of the 16S rRNA of Escherichia coli accessible in situ to single DNA oligonucleotides. Appl. Environ. Microbiol. 72:733–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yilmaz LS, Parnerkar S, Noguera DR. 2011. mathFISH, a web tool that uses thermodynamics-based mathematical models for In silico evaluation of oligonucleotide probes for fluorescence in situ hybridization. Appl. Environ. Microbiol. 77:1118–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]