Abstract
Clustered regularly interspaced short palindromic repeats (CRISPR) confer sequence-dependent, adaptive resistance in prokaryotes against viruses and plasmids via incorporation of short sequences, called spacers, derived from foreign genetic elements. CRISPR loci are thus considered to provide records of past infections. To describe the host-parasite (i.e., cyanophages and plasmids) interactions involving the bloom-forming freshwater cyanobacterium Microcystis aeruginosa, we investigated CRISPR in four M. aeruginosa strains and in two previously sequenced genomes. The number of spacers in each locus was larger than the average among prokaryotes. All spacers were strain specific, except for a string of 11 spacers shared in two closely related strains, suggesting diversification of the loci. Using CRISPR repeat-based PCR, 24 CRISPR genotypes were identified in a natural cyanobacterial community. Among 995 unique spacers obtained, only 10 sequences showed similarity to M. aeruginosa phage Ma-LMM01. Of these, six spacers showed only silent or conservative nucleotide mutations compared to Ma-LMM01 sequences, suggesting a strategy by the cyanophage to avert CRISPR immunity dependent on nucleotide identity. These results imply that host-phage interactions can be divided into M. aeruginosa-cyanophage combinations rather than pandemics of population-wide infectious cyanophages. Spacer similarity also showed frequent exposure of M. aeruginosa to small cryptic plasmids that were observed only in a few strains. Thus, the diversification of CRISPR implies that M. aeruginosa has been challenged by diverse communities (almost entirely uncharacterized) of cyanophages and plasmids.
INTRODUCTION
Bacteria and archaea have acquired a large number of defense mechanisms against viruses, plasmids, and other mobile genetic elements (32, 37). One of the defense systems, termed clustered regularly interspaced short palindromic repeats (CRISPR), was recently identified in most archaeal genomes and nearly half of the bacterial genomes (11). The CRISPR array comprises short (21 to 48 bp) direct repeats separated by similarly sized variable unique sequences called spacers. CRISPR loci are typically accompanied by CRISPR-associated (cas) genes (19, 26, 36). Although the molecular mechanisms are far from understood, CRISPR/Cas is shown to function as a heritable, acquired defense system in some bacteria and archaea (3, 5, 10, 38). In general, the spacers are derived from partial sequences (proto-spacers) from virus and plasmid genomes that invade the cell, creating individual archives (i.e., spacers) of exposure to the noncellular parasites (3, 10, 13). Spacers are transcribed from an upstream AT-rich region (leader) as a single long precursor RNA and then processed by specific Cas proteins into small CRISPR RNAs (crRNAs) containing a single spacer (5, 6, 33). Each crRNA forms a nucleoprotein complex to guide Cas nuclease to recognize foreign DNA (5) or RNA (20) using base pairing. Through this RNA interference (RNAi)-like manner, the CRISPR/Cas system protects bacteria and archaea from viral infection and limits plasmid transfer (3, 13, 38, 39). In individual CRISPR arrays, new spacer additions are polarized to the leader end of the loci (3, 10), while simultaneously, spacer loss occurs (10), resulting in hypervariability within species. Therefore, the loci have been used for fingerprinting of pathogenic bacteria (i.e., spoligotyping [27]) and for dissecting microbial population structures (22, 49). Further, CRISPR spacers, representing direct signatures of viral infection, help to understand the coevolutionary dynamics of the host-virus community in environments (2, 21).
The unicellular cyanobacterium Microcystis aeruginosa frequently forms dense blooms in freshwater environments worldwide (50). Some strains of this species produce potent hepatotoxins called microcystins that occasionally cause death of domestic animals and humans (8). Therefore, community composition and dynamics of M. aeruginosa populations are of great concern to water quality management (50). Previous studies have shown M. aeruginosa to be genetically highly heterogeneous (44, 51); M. aeruginosa populations also undergo temporal changes in genotypic composition (4, 52, 53). Previously we isolated a phage, Ma-LMM01, infecting M. aeruginosa (57) and observed the potential qualitative impact of the cyanophage and its relatives on natural M. aeruginosa populations (54, 56). However, Ma-LMM01 has a narrow host range despite the large genetic diversity of hosts, suggesting that there is a greater diversity of host-cyanophage combinations other than Ma-LMM01 and its host strain in natural M. aeruginosa populations (57). Recently, a comparative genomic study of bacteria and archaea revealed an abundance of diverse defense systems, including a CRISPR/Cas system in the M. aeruginosa NIES843 genome (37). Together these findings imply a richness of foreign genetic elements that have coevolved with M. aeruginosa through their specific interactions that probably contribute to the large clonal diversity (4, 51, 52) and genomic plasticity (12, 28) of the species.
Here we investigated the CRISPR to understand host-parasite (i.e., cyanophages and plasmids) dynamics involving M. aeruginosa. Our aims are (i) to estimate the diversity of the CRISPR spacer repertoire within M. aeruginosa populations and (ii) to determine the potential impact of known cyanophages and plasmids on M. aeruginosa. Therefore, we determined CRISPR sequences in four M. aeruginosa strains in addition to two strains whose genomes are available in the databases and examined intraspecies variability of the loci. In addition, we assessed a natural M. aeruginosa population for diversity in the leader-end CRISPR fragments obtained using CRISPR-based PCR. We inferred previous host-parasite interactions within the M. aeruginosa populations from signatures in the CRISPR.
MATERIALS AND METHODS
M. aeruginosa strains and DNA extraction.
M. aeruginosa strains NIES87, NIES102, NIES298, and NIES1067 were obtained from the National Institute for Environmental Studies (Tsukuba, Japan). The strains used in this study represent different phylogenetic groups in this species (Table 1). In a previous report using multilocus sequence typing (MLST), M. aeruginosa strains were largely divided into five major clades (groups A to E) (44). M. aeruginosa NIES102 and NIES843 are closely related to each other and fall into a well-supported inner clade (within group A). Strains NIES298 and NIES1067 fall into other well-supported clades (group B and group D, respectively) (44). Strain NIES87 is not included in the major clades, but this strain is interesting because it carries two plasmids (46). Phylogenetic relationships of these strains are shown in Fig. S1 in the supplemental material. The strains were maintained in CB medium (29) as described previously (31). Late-exponential-phase cultures (2-ml aliquots) were mildly sonicated to remove gas vesicles and then centrifuged at 3,000 × g for 10 min (31). DNA was extracted from the cell pellets using the Nucleon Phytopure genomic DNA extraction kit (GE Healthcare, Tokyo, Japan) according to the manufacturer's instructions.
Table 1.
M. aeruginosa strains used in this study
| Strain | Location | Year | Sequence typeb |
|---|---|---|---|
| NIES102 | Lake Kasumigaura, Japan | 1982 | 10 |
| NIES298 | Lake Kasumigaura, Japan | 1982 | 60 |
| NIES87 | Lake Kasumigaura, Japan | 1982 | 2 |
| NIES1067 | Chikatou Pond, Japan | 1982 | 27 |
| NIES843a | Lake Kasumigaura, Japan | 1997 | 18 |
| PCC7806a | Braakman Reservoir, Netherlands | 1972 | ND |
The genome sequence of this strain has been published in the NCBI/EMBL/DDBJ database.
Sequence types were determined by Tanabe et al. (44). ND, not determined.
Identification and sequencing of CRISPR arrays.
The nucleotide sequence for the CRISPR of M. aeruginosa NIES843 was obtained from the CRISPRdb database (15). To identify homologous CRISPR/Cas systems in the draft genome sequence of M. aeruginosa strain PCC7806 (12), the NIES843 CRISPR sequence was searched against 116 contig sequences deposited in the EMBL database using BLASTN (1). Homologous CRISPR arrays in another four M. aeruginosa strains (NIES87, NIES102, NIES298, and NIES1067) were amplified using a primer, MaeCRf, specific to the leader region with various reverse primers. All primer sequences used in this study are shown in Table S1 in the supplemental material. The loci were amplified from strains NIES87 and NIES102 using primer pairs MaeCRf/MaeCRrtp2 and MaeCRf/MaeCRrtp3, respectively. The PCR was performed with 25 μl containing 50 ng DNA, 0.2 μM each primer, 0.4 mM each deoxynucleoside triphosphate (dNTP), 1× LA PCR buffer, 2.5 mM MgCl2, and 1.25 U TaKaRa LA Taq polymerase (TaKaRa Bio, Otsu, Japan). The reaction conditions were as follows: 1 min of initial denaturing at 94°C, followed by 30 cycles of 94°C for 20 s and 64°C for 20 min, and a final extension at 72°C for 10 min. Because of the diversity of sequences surrounding the loci, the conventional PCR failed to amplify CRISPR from strains NIES298 and NIES1067. To obtain the NIES298 CRISPR, we searched for short genomic contigs of NIES298 (unpublished data) that matched CRISPR spacers of NIES843. Based on one of the found matches, a primer, 298CRrS6, was designed and used with the primer MaeCRf under the PCR conditions described above. For strain NIES1067, a primer, MaeCRrGT (described below and in the legend to Fig. 1A), was used with MaeCRf under the following PCR conditions: initial denaturing at 94°C for 1 min, followed by 30 cycles of 98°C for 10 s and 60°C for 20 min, and a final extension at 72°C for 10 min. PCR products were purified and then sequenced using the primer walking method. The CRISPR array was completely sequenced for strain NIES102, while those for the other three strains were partially sequenced. Therefore, amplification and sequencing were performed using successive thermal asymmetric interlaced PCR (TAIL-PCR) (34) toward the end of the arrays (primers are shown in Table S1 in the supplemental material). Reaction conditions and arbitrary primers for the TAIL-PCR were as described previously (31). A portion of the amplifications were performed using an alternative PCR with MaeCRrGT and outward primers based on CRISPR spacers in the sequenced fragments.
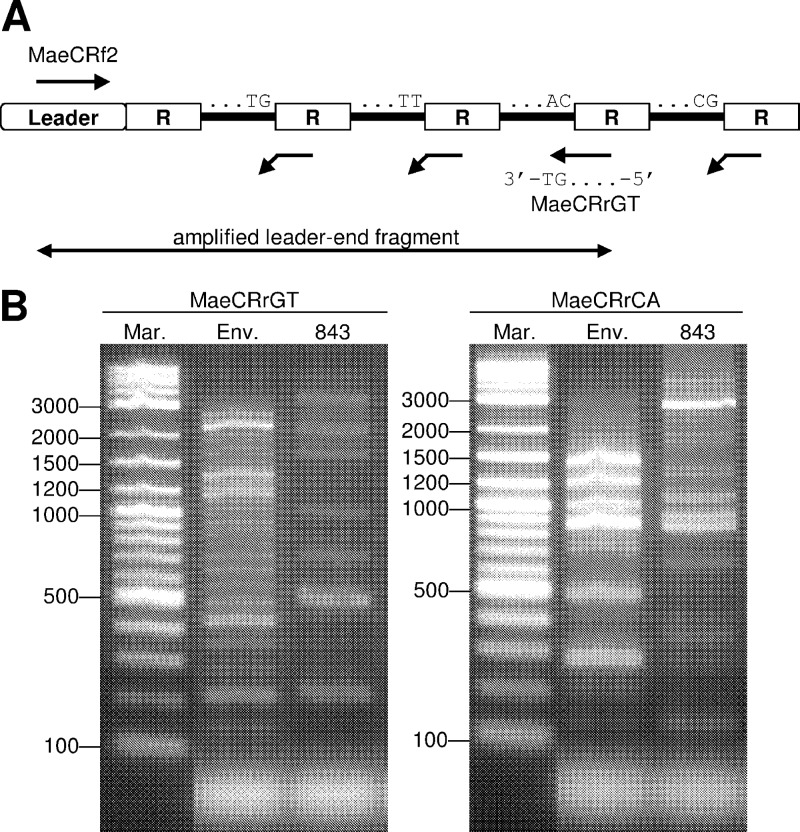
Fig 1.
Amplification of leader-end CRISPR fragments. (A) The reverse primer MaeCRrGT was designed to be complementary to the 5′ end (20 bp) of the repeat (R) but with two additional nucleotides, GT, thereby preferentially annealing to a limited number of specific spacer-repeat units. Another reverse primer, MaeCRrCA, has dinucleotide CA instead of GT. (B) PCR products from primers MaeCRrGT (left) and MaeCRrCA (right). Mar, 2-log DNA ladder; Env, DNA from Hirosawanoike Pond; 843, M. aeruginosa NIES843 genomic DNA.
Amplification of M. aeruginosa CRISPR from a natural cyanobacterial population.
M. aeruginosa CRISPR was investigated in Hirosawanoike Pond (Kyoto, Japan), which is a small (surface area, 14 ha) and shallow (mean depth, 1.5 m) reservoir. One liter of surface water was collected at a fixed point (35°02′N, 135°41′E) at noon on 13 September 2010. Cyanobacterial cells were harvested from a 50-ml aliquot of the water by mild sonication, followed by centrifugation at 1,680 × g for 10 min. DNA was extracted from the cell pellet using the xanthogenate method as described previously (45, 52). Purified DNA was suspended in 30 μl of sterilized MilliQ water.
Because conventional PCR amplification was not applicable to M. aeruginosa populations, we developed a PCR strategy based on the leader region (primer MaeCRf2) and repeat-spacer units (primer MaeCRrGT or its derivative, primer MaeCRrCA) (Fig. 1A), thereby amplifying the leader-side fragments of M. aeruginosa CRISPR irrespective of their genomic contexts. The PCR was performed with 50 μl containing 2.5 μl 1:100 dilution of the environmental DNA, 0.8 μM each primer, 0.25 mM each dNTP, 1× Ex Taq buffer, and 2 U TaKaRa Ex Taq polymerase (TaKaRa Bio). The reaction conditions were as follows: initial denaturing at 94°C for 4 min, followed by 30 cycles of 94°C for 30 s, 62°C for 1 min, and 72°C for 1 min, and a final extension at 72°C for 7 min. The PCR products were separated using electrophoresis on 2.0% (wt/vol) agarose S gels (Nippon Gene, Tokyo, Japan). The gels were stained with GelRed (Biotium, CA) and visualized using the Gel Doc XR system (Bio-Rad Laboratories, CA). To prevent bias of preferential cloning of smaller DNA fragments, small gel blocks were separately excised, and DNA was purified and cloned. Thirty-nine and 50 clones were sequenced in full for MaeCRrGT and MaeCRrCA PCR products, respectively, by the primer walking method (primers are shown in Table S1 in the supplemental material). The CRISPRtionary program (17) was used to find identical spacers in different CRISPR fragments. Next, the fragments sharing the same spacer order were manually assembled into contigs. To ensure accuracy, the leader-distal spacer was removed from the contigs, because the PCR used in this study was shown to allow one mismatch at the additional dinucleotide at the 3′ end of the reverse primers.
Bioinformatics analysis.
CRISPR repeats and spacers were identified using the CRISPRFinder (16) with manual validation. A similarity search of the unique spacer sequences was performed against the NCBI nr database using BLASTN with an E value threshold of 0.1 and the word size set at 7. The best hits for bacteriophages and plasmid sequences were investigated, and those showing ≥80% identity over the queried spacers were considered to be significant. A partial phage/plasmid sequence including the sequence match and covering the spacer length was referred to as a putative proto-spacer. Sequence logos were generated with the WebLogo (9) using 10-bp flanking sequences on both sides of the putative proto-spacers in phage Ma-LMM01 and M. aeruginosa plasmids (PMA1, pMA1, and pMA2).
Separately, spacer sequences were clustered using CD-HIT-EST web server (25), where spacers showing ≥87% identity over >60% of the shorter sequence were clustered.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the DDBJ/EMBL/GenBank database. The accession numbers are AB644436 to AB644439 for the CRISPR arrays from cultured strains and AB644412 to AB644435 for the representative clones (i.e., the longest one) of the CRISPR contigs obtained from the environmental samples. Consensus sequences for CRISPR types CT1, CT2, CT4, CT5, CT7, CT8, CT19, and CT22 to CT24 (see Results section) are provided in the supplemental material.
RESULTS
Type I-D CRISPR/Cas system in M. aeruginosa.
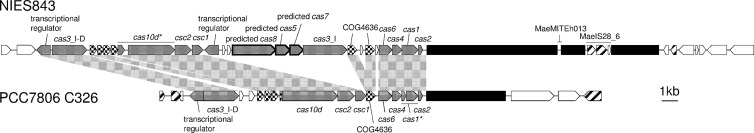
A type I-D CRISPR/Cas system homologous to that in M. aeruginosa NIES843 was identified in the genomic contig C326 of PCC7806. Makarova et al. identified “divergent rare variants” of cas8, cas7, and cas5 along with the typical type I-D cas genes in the NIES843 genome (37); however, the three cas variants and the downstream cas3 were absent in the PCC7806 draft genome (Fig. 2). The rare cas module in NIES843 may have been acquired by horizontal gene transfer (HGT), given its presence in some plasmids (e.g., Cyanothece sp. strain PCC8802 plasmid pP880201) (37). In PCC7806, cas1, presumably involved in spacer acquisition (11), was interrupted by an in-frame stop codon, suggesting that spacer uptake may no longer be active in this strain. The sequence between cas2 and the first CRISPR was highly conserved between the strains and contained AT-rich regions, including a putative promoter element (5′-TTGAAG-[17 bp]-TAYRAT-3′). Therefore, the sequence was considered to be a leader (26, 33). The CRISPR/Cas was located in different genomic contexts in the two strains (Fig. 2).
Fig 2.
Comparison of the CRISPR/Cas systems identified in M. aeruginosa NIES843 and PCC7806. Open reading frames (ORFs) and CRISPR arrays are shown as arrows and filled boxes, respectively. cas genes, toxin-antitoxin genes, and transposase genes are indicated by gray, dotted, and striped arrows, respectively. Divergent, rare cas variants (37) are enclosed with thick lines. Gene nomenclature is in accordance with the previous study (37). Pseudogenes are marked with asterisks. Gray shadows indicate conserved regions between the two genomes.
Variation in genomic position of the CRISPR arrays.
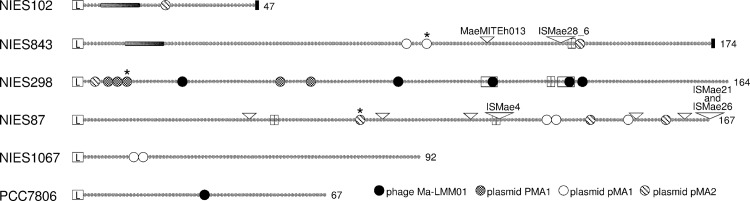
The CRISPR arrays were sequenced for another four M. aeruginosa strains (NIES87, NIES102, NIES298, and NIES1067), from cas2 to the downstream flanking sequences of the loci (Fig. 3). The flanking sequences of the loci in NIES102 and NIES298 were nearly identical to those in NIES843 and PCC7806, respectively, while the CRISPR locus of NIES1067 was located at another genomic position. We could not determine the position of the CRISPR for NIES87 because of two insertion sequences (ISs) lying at the end of the locus. CRISPR arrays in strains NIES843 and NIES87 have been subjected to transposable elements, including ISs, a miniature inverted-repeat transposable element (MITE), and other putative short sequence elements (Fig. 3). These mobile elements may contribute to the variation of the CRISPR position.
Fig 3.
CRISPR arrays in six M. aeruginosa strains. Strain-specific spacers are indicated by circles, and those spacers with significant hits to sequences of phage and plasmids are indicated by patterns as shown in the key. An asterisk indicates 100% nucleotide identity to phage/plasmid sequences. A string of 11 consecutive spacers shared between strains NIES102 and NIES843 is indicated by rectangles, with shading showing the direction of the shared spacer block. The total number of the spacers is shown at the right of each array. Degenerate terminal repeats are indicated by black bars. Spacer duplication is indicated by open boxes. Insertions of ISs, MITE, and other short sequence elements in CRISPR are indicated by triangles.
Sequence analysis of the CRISPR repeats.
Although the CRISPR location varied among these strains, the CRISPR repeat, leader, and partial cas2 sequences were nearly identical in the strains. Thus, homologous, comparable CRISPR arrays were obtained from six M. aeruginosa strains (Fig. 3). The consensus sequence of the CRISPR repeats was 37 bp long (5′-GTTCCAATTAATCTTAAACCCTATTAGGGATTGAAAC-3′) and fell into cluster 5 of the previously proposed repeat-based classification (30). Strains NIES87, NIES298, and NIES1067 had repeat variants derived from single nucleotide polymorphisms (SNPs) that were situated in the hairpin loop or external bases of the predicted stem-loop RNA structure of the repeat. In general, terminal repeats tend to be degenerate at the leader-distal end of the CRISPR loci (24). Among the M. aeruginosa strains, an identical degenerate terminal repeat was found in strains NIES102 and NIES843 (Fig. 3).
Intraspecies variability of the CRISPR spacers.
The number of spacers varied among the M. aeruginosa strains, from 47 (NIES102) to 174 (NIES843) (Fig. 3). These numbers are significantly larger relative to the average (27 repeats) among prokaryotes (14).
CRISPR spacer repertoire was compared in the context of phylogenetic relationships among the six strains (see Fig. S1 in the supplemental material). A string of 11 spacers was shared between strains NIES102 and NIES843 at the leader side of the loci, while the rest of the spacers were strain specific (Fig. 3). The other four strains, including the closest of the NIES102/NIES843 clade (strain NIES1067), had only strain-specific spacers (Fig. 3).
Diversity of the leader-end CRISPR fragments in natural cyanobacterial populations.
To investigate the CRISPR diversity in a natural population, we used the repeat-based PCR with a pond water sample. The PCR yielded multiple bands ranging from ca. 150 bp to 3,000 bp at intervals of 70 bp, which is consistent with the size of a repeat-spacer unit (Fig. 1B). Every sequence from 89 PCR clones contained a leader region and subsequent CRISPR repeats that are homologous to those of M. aeruginosa (with up to two SNPs), confirming the specificity of the PCR. Clones containing the same spacers in the same order were assembled into contigs, resulting in 24 distinct leader-end CRISPR fragments (Table 2). We designated them as CRISPR types (CTs), which probably represent distinct CRISPR genotypes. Up to 32 spacers (13 on average) were obtained for each CT (Table 2). No spacers were shared between the CRISPR types, except for three pairs of CTs sharing a portion of their spacer sets (Table 2). Separately, we determined partial CRISPR sequences from eight M. aeruginosa strains isolated from the same pond water sample. The eight strains were identified as M. aeruginosa based on specific PCR targeting partial 16S-23S rRNA gene internal transcribed spacers (55). Two strains had a spacer set identical to that of CT1, another two strains were CT4, and another one strain was CT3. Origins of the CTs obtained by the repeat-based PCR were therefore partially confirmed. The other three strains had spacer sets different from the CTs (data not shown). Considering the general trends of polarized spacer addition, our data indicate that there are no less than 24 coexisting genotypes that are different in the recent evolutionary history of the CRISPR in the M. aeruginosa population.
Table 2.
Characteristics of CTs identified in Hirosawanoike Pond
| CT | No. of clones | No. of spacersa |
No. of unique spacers with significant hitsc |
||||
|---|---|---|---|---|---|---|---|
| Total | Sharedb | Ma-LMM01 | PMA1 | pMA1 | pMA2 | ||
| CT1 | 8 | 16 | – | 4(1) | – | 1 | |
| CT2 | 15 | 13 | 10(4–13)A | – | – | – | – |
| CT3 | 1 | 12 | – | – | – | – | |
| CT4 | 10 | 27 | 1(1)B | – | – | – | – |
| CT5 | 20 | 32 | – | – | 5 | – | |
| CT6 | 1 | 5 | 1 | – | – | – | |
| CT7 | 2 | 4 | – | – | – | – | |
| CT8 | 2 | 10 | – | – | – | – | |
| CT9 | 1 | 6 | – | – | – | 1 | |
| CT10 | 1 | 6 | – | 1 | – | – | |
| CT11 | 1 | 8 | – | – | – | – | |
| CT12 | 1 | 21 | 6(1–3, 6–8)C | – | – | – | – |
| CT13 | 1 | 15 | – | – | – | – | |
| CT14 | 1 | 7 | – | – | – | – | |
| CT15 | 1 | 28 | 3 | – | – | – | |
| CT16 | 1 | 11 | – | – | – | – | |
| CT17 | 1 | 13 | 1(1)B | – | – | – | – |
| CT18 | 1 | 13 | – | – | – | – | |
| CT19 | 3 | 17 | 1 | – | – | – | |
| CT20 | 1 | 17 | – | – | – | – | |
| CT21 | 1 | 16 | 6(1–3, 4–6)C, 10(7–16)A | – | – | – | – |
| CT22 | 6 | 4 | – | – | – | – | |
| CT23 | 6 | 11 | – | – | – | – | |
| CT24 | 3 | 9 | – | – | – | – | |
Number of spacers in CT contigs.
Spacers shared between CTs are indicated as x(y-z)N. x is the total number of shared spacers, y-z is the position of shared spacers (sequentially numbered from the leader-end spacer), and N denotes a pair of strains sharing the spacers.
Dashes indicate that no hit was found for the spacers. The number with 100% match is shown in parentheses.
CRISPR signatures of foreign DNA elements.
Excluding redundant spacers, we obtained a total of 995 unique spacer sequences from M. aeruginosa strains and the water sample. We determined the sequence matches comparing the 995 spacers to sequences of phages and plasmids in the NCBI nr database to show histories of host-parasite interaction. Overall, only 43 unique spacers (4%) had significant matches, of which 10 and 33 spacers matched phage and plasmid sequences, respectively (see Table S2 in the supplemental material).
The 10 spacers had 83 to 97% identity to the genome sequence of M. aeruginosa phage Ma-LMM01. Of these, four spacers were identified in the host strain NIES298 (Fig. 3). The other six spacers were identified in strain PCC7806 and CRISPR types CT6, CT15, and CT19 (Fig. 3; Table 2).
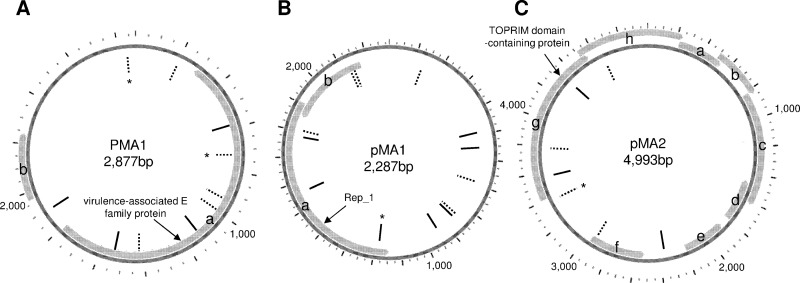
Ten, 13, and 7 unique spacers showed sequence matches to plasmids PMA1 from M. aeruginosa HUB 5-2-4 (42), pMA1, and pMA2 from NIES87 (46), respectively (Fig. 3; Table 2). Of these, four spacers showed 100% identity to sequences in the corresponding plasmids. Putative proto-spacers were evenly mapped onto each plasmid sequence (Fig. 4). The other three spacers showed moderate sequence similarity across genera to Cyanothece sp. strain PCC7424 plasmid pP742402, Streptococcus thermophilus strain LMD-9 plasmid 1, and Bacillus coagulans plasmid pMSR0 (86, 82, and 82%, respectively).
Fig 4.
Distribution of putative proto-spacers on plasmids PMA1 (A), pMA1 (B), and pMA2 (C). Each bar indicates the position of the putative proto-spacer. Solid and dashed bars indicate putative proto-spacers in clockwise and counterclockwise strands, respectively. An asterisk indicates 100% nucleotide identity. Of note, we found additional ORFs in pMA1 (ORF b) and pMA2 (ORFs a, b, e, g, and h) compared with their original description. ORF a and b of pMA1 may be generated from a longer replication gene by a frameshift. ORFs g, h, a, b, and c may be generated by frameshifts from a single hypothetical gene whose homologues are prevalent in cyanobacteria (e.g., CY0110_05002 of Cyanothece sp. strain CCY0110).
Independent acquisition of similar spacers.
Clustering analysis of the 995 spacer sequences identified 48 distinct pairs and four triads of spacers that share nearly identical sequences (see Table S3 in the supplemental material). In each pair, the similar spacers exhibited overlapping that could be merged into a contig. Furthermore, in 19 cases, the paired spacers were complementary (see Table S3). Therefore, similar spacers in each pair may be derived from the same viral (or plasmid) lineages and be acquired in separate exposure incidents. The similar spacer pairs were identified in various combinations of strains and CTs; e.g., M. aeruginosa NIES298 shared 14 similar spacers with 5 other strains and 4 CTs.
PAMs associated with the M. aeruginosa CRISPR/Cas system.
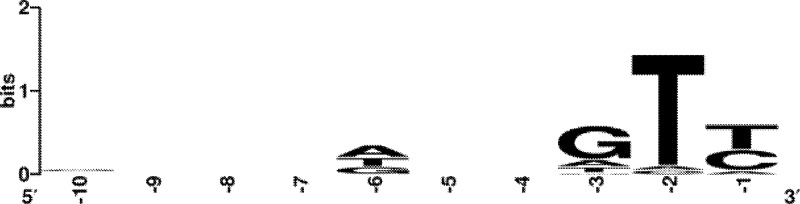
We determined if the M. aeruginosa CRISPR/Cas system associates a specific proto-spacer-associated motif (PAM), a short nucleotide motif adjacent to the proto-spacers in target sequences (40). The WebLogo analysis of the flanking sequences identified a conserved motif of GT(T/C) immediately upstream of the putative proto-spacers (Fig. 5). No particular motif was detected when searching the downstream sequences.
Fig 5.
Sequence logo of the PAM consensus. The logo was built using the WebLogo based on alignments of the flanking sequences of putative proto-spacers in Ma-LMM01, PMA1, pMA1, and pMA2. Numbers below indicate nucleotide positions where −1 is just upstream of the putative proto-spacers.
DISCUSSION
CRISPR variability and inferred host-parasite interactions.
At present, we have no experimental evidence to show whether the M. aeruginosa CRISPR/Cas functions as a defense system against parasites or, if it does, how. However, we found several spacers matching known foreign genetic elements for M. aeruginosa (e.g., Ma-LMM01) in the CRISPR loci. This strongly suggests that the M. aeruginosa CRISPR/Cas had been functional at least in the past, and thereby the spacer repertoire at each locus represents a history of previous host-parasite interactions. In our data set the spacer repertoire was unique for each M. aeruginosa strain (Fig. 3). This result should be interpreted with care because we cannot rule out the possibility of undersampling, given the high diversity of M. aeruginosa. However, considered in the context of local populations (e.g., coexisting genotypes in Hirosawanoike Pond), CRISPR variability can provides insights into the interplay between hosts and parasites (especially cyanophages) involving M. aeruginosa.
CRISPR spacers are believed to be acquired by individuals and then selected in response to viral infection. Therefore, intrapopulation variability in the spacer repertoire implies the extent (i.e., host range of viruses) and frequency of selection events posed on the host population (21, 22, 47, 49). For example, the CRISPR sequence of Leptospirillum in acidophilic microbial biofilms showed that individuals in a nearly clonal population share spacers in the leader-distal half of the CRISPR arrays, suggesting population-wide selective sweep events (49). In contrast, we found 24 coexisting different CRISPR genotypes in a pond M. aeruginosa population, where spacers were rarely shared between the leader-end portions of the individual CRISPR types (Table 2). The lack of population-wide fixed spacers may also be suggested in another population (Lake Kasumigaura), where M. aeruginosa strains (NIES87, NIES298, and NIES102) isolated in September 1982 (Table 1) did not share a single spacer (Fig. 3). These data suggest purifying selection is unlikely to be so extensive that only strains carrying specific spacers can survive. In other words, the host-phage interaction may be subdivided into diverse “susceptible combinations,” each consisting of M. aeruginosa strains and a specific cyanophage, rather than pandemics of population-wide infectious cyanophages. Sporadic distribution of the paired similar spacers among M. aeruginosa (see Table S3 in the supplemental material) also supports the subdivided host-phage interactions.
A recent modeling study concerning CRISPR evolution showed CRISPR immunity induces allele (spacers and proto-spacers) diversification within a community of a host and a viral lineage, given that each virus carries a number of distinct, variable proto-spacers (7). Our results indicate that Ma-LMM01-matching spacers could be derived from a number of different proto-spacers, and the sequences suggest proto-spacer diversification in the phage (discussed below). Further, the data from closely related strains NIES102 and NIES843 allow inference concerning diversification of a CRISPR locus in M. aeruginosa. The strains NIES102 and NIES843, both isolated from Lake Kasumigaura, shared an identical degenerate terminal repeat, genomic location of CRISPR and a small portion of their spacer repertoires (Fig. 3). This may suggest the two strains share an ancestor at the CRISPR loci (i.e., involved in the same “susceptible combination”). However, these strains have diverged to show more than a hundred strain-specific spacers (Fig. 3), and the shared spacer block was not polarized to the leader-distal end. If the apparent rate of spacer addition is constant in the two strains over time, their spacer repertoires indicate an addition of 127 spacers in NIES843 during the 15 years after isolation of NIES102. Assuming a host growth rate of 1 per day and a bloom period of 6 months per year, the spacer addition rate is approximately one spacer per 20 generations. Although this estimate is based on many assumptions, the time scale is compatible with that of host genotype turnover (13 turnover events in 200 generations) observed in the modeling study (7). Very few studies have examined the rate of spacer addition into natural bacterial populations, and addressing this issue will help to understand host-parasite coevolutionary dynamics more clearly and deeply.
CRISPR array was significantly longer in M. aeruginosa than other organisms. A modeling study where host density is kept constant predicts that larger viral diversity leads to longer host CRISPR arrays (18). Therefore, the long CRISPR of M. aeruginosa, which grows up to form dense blooms, may reflect an adaptive response to highly diverse, quickly diversifying cyanophages. High cyanophage diversity is also compatible with a huge variety of antiviral defense systems on the genome of M. aeruginosa NIES843 (37).
Ecological impact of cyanophages and cryptic plasmids.
CRISPR signatures of M. aeruginosa provided insights into the ecological impact and dynamics of known cyanophage. Spacers matching cyanophage Ma-LMM01 were found in M. aeruginosa NIES298 and in PCC7806 from the Netherlands (Fig. 3) and three CTs from Hirosawanoike Pond (Table 2). This implies the host-virus combinations involving Ma-LMM01 and possible dispersal of its related phages. Conversely, all of the Ma-LMM01-matching spacers (10 spacers) were not completely identical to their corresponding putative proto-spacers in Ma-LMM01, suggesting high nucleotide diversity within this phage lineage. Of these, six spacers had only nucleotide mutations that are translated into silent or conservative changes in deduced amino acid sequences (Table 3). Although a single mutation in proto-spacers or PAMs basically can abolish CRISPR-mediated immunity against phages (3, 10, 13), the strict nucleotide identity required for immunity is limited to a specific seven-nucleotide region in the proto-spacers (denoted as “seed sequence”) in Escherichia coli (43). If the M. aeruginosa CRISPR system employs this “seed”-dependent immunity, Ma-LMM01 or its related phages may evade interference mediated by five of the spacers (all but CT15spc14 in Table 3), and thereby the Ma-LMM01-related phage assemblage retains the conservative nucleotide polymorphisms to circumvent host immunity.
Table 3.
Spacers showing silent or conservative mutations compared to Ma-LMM01 putative proto-spacers
| Spacer/phage genea | Nucleotide sequencec | Predicted amino acid sequencec |
|---|---|---|
| 298spc124b | AGTGGCGCGGCTACTTATCTCTACCAATTTTCTAC | SGAATYLYQFS |
| ORF40 | ........A..C..............GG....C.. | .........V. |
| 7806spc34b | ATTTGAGGGACTAAATAATGGGATCGTATTCAAT | FEGLNNGIVFN |
| ORF41 | .........CGC...C................G. | ...A......S |
| CT6spc2b | AATCCCCCCGTCAGGGATTCTCCCACGGGTTTCAAT | IPPSGILPRVS |
| ORF61 | ...A...........AG................T.. | .....V..... |
| CT15spc12b | ACTCTCCTTGCGACTATAAGTATGTGGGCAAGTCT | SPCDYKYVGKS |
| ORF25 | ............................T...... | ........... |
| CT15spc13b | TCTATCTGTTCAATACTATGCCTCTAGGAGCAGGGCAAG | YLFNTMPLGAGQ |
| ORF20 | .....................................G. | ............ |
| CT15spc14b | TTGATACAGGTGCCTTCCTAGGCTGTTATCT | DTGAFLGCY |
| ORF25 | .......G....................... | ......... |
In each row, the spacer (top) and the corresponding putative proto-spacer (bottom) are shown.
Reverse complementary sequences are shown.
Identical nucleotides and amino acids are indicated by dots. Synonymous and conservative changes are in bold and italic letters, respectively. The seed region identified in the subtype I-E CRISPR of E. coli (43) is underlined.
CRISPR signatures of M. aeruginosa also showed repeated exposures to plasmids PMA1, pMA1, and pMA2. Uniform distribution of putative proto-spacers throughout these plasmids (Fig. 4) suggests that the spacers were acquired from these plasmids themselves, rather than other plasmids having homologous genes (e.g., conserved replication genes). Given the proposed mechanisms of interference, a plasmid cannot become established in host cells that carry a spacer completely matching a partial sequence of the plasmid (13, 39). In accordance, M. aeruginosa NIES843 possessing a spacer with 100% identity to plasmid pMA1 (Fig. 3) carries no plasmid (28). Unstable presence of the small plasmids reported in some M. aeruginosa strains (42) may be attributed to such CRISPR-mediated exclusion. However, strain NIES87 retains pMA2 (46), despite possessing a spacer with 100% identity to the plasmid (Fig. 3). This apparent conflict may have resulted from defective CRISPR interference, given the proliferation of IS elements around the CRISPR array in NIES87 (Fig. 3). The small plasmids carry very few genes, and thus their role in ecology and evolution of M. aeruginosa remains unclear. Interestingly, similar small cryptic plasmids were found to dominate in marine Synechococcus metagenomes and were hypothesized to facilitate HGT and, in some cases, phage resistance (35, 41). When considering the suggested spread of the small plasmids among M. aeruginosa populations, they may facilitate host genetic diversity via HGT or chromosomal integration.
Functional characteristics of M. aeruginosa CRISPR.
M. aeruginosa and several other cyanobacteria possess the recently identified subtype I-D CRISPR/Cas system, which is a hybrid of the type I system and type III executive Cas module (36). Our data provide some information on the functional characteristics of the M. aeruginosa CRISPR/Cas system. Uniform distribution of the putative proto-spacers without bias toward either strand of the plasmids (Fig. 4) suggests that the M. aeruginosa CRISPR/Cas recognizes DNA rather than RNA. Combined with the presence of the upstream PAM (Fig. 5), this is in accordance with the type I information processing (i.e., spacer acquisition) system (36).
Conclusions.
M. aeruginosa shows a high degree of CRISPR heterogeneity within populations. Thus, we infer that the host-phage community can be subdivided into a number of different “susceptible combinations” of M. aeruginosa and cyanophages. In each combination, the CRISPR spacers and cyanophages may quickly diversify through coevolution. This intricate interaction is expected to shape complex host-phage communities. The spacer sequences imply what kinds of phages are involved in each “susceptible combination” with a strain of M. aeruginosa (Fig. 3; see also Table S3 in the supplemental material). Nevertheless, a significant fraction of the spacer sets in M. aeruginosa are of unknown origin, largely because of the underrepresentation of the diversity of bacteriophages and plasmids in the current sequence data. Thus, M. aeruginosa may be challenged by diverse, almost entirely uncharacterized communities of cyanophages and plasmids. Some of the unknown spacers would be attributed to previously observed (but not isolated) M. aeruginosa cyanophages (23, 48), and a metagenomic survey will provide links between the CRISPR signatures and uncultured phage community.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a Grant-in-Aid for Science Research (20310045). S.K. is a research fellow supported by JSPS for Young Scientists (224469).
Footnotes
Published ahead of print 25 May 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2.Andersson AF, Banfield JF. 2008. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320:1047–1050 [DOI] [PubMed] [Google Scholar]
- 3.Barrangou R, et al. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712 [DOI] [PubMed] [Google Scholar]
- 4.Bozarth CS, Schwartz AD, Shepardson JW, Colwell FS, Dreher TW. 2010. Population turnover in a Microcystis bloom results in predominantly nontoxigenic variants late in the season. Appl. Environ. Microbiol. 76:5207–5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brouns SJ, et al. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carte J, Pfister NT, Compton MM, Terns RM, Terns MP. 2010. Binding and cleavage of CRISPR RNA by Cas6. RNA 16:2181–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Childs LM, Held NL, Young MJ, Whitaker RJ, Weitz JS. 2012. Multiscale model of CRISPR-induced coevolutionary dynamics: diversification at the interface of Lamarck and Darwin. Evolution doi:10.1111/j.1558–5646.2012.01595.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Codd GA, Lindsay J, Young FM, Morrison LF, Metcalf JS. 2005. Harmful cyanobacteria, p 1–23 In Huisman J, Matthijs HCP, Visser PM. (ed), Harmful cyanobacteria. Springer, Dordrecht, Netherlands [Google Scholar]
- 9.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deveau H, et al. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190:1390–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deveau H, Garneau JE, Moineau S. 2010. CRISPR/Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol. 64:475–493 [DOI] [PubMed] [Google Scholar]
- 12.Frangeul L, et al. 2008. Highly plastic genome of Microcystis aeruginosa PCC 7806, a ubiquitous toxic freshwater cyanobacterium. BMC Genomics 9:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garneau JE, et al. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468:67–71 [DOI] [PubMed] [Google Scholar]
- 14.Godde JS, Bickerton A. 2006. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J. Mol. Evol. 62:718–729 [DOI] [PubMed] [Google Scholar]
- 15.Grissa I, Vergnaud G, Pourcel C. 2007. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8:172 doi:10.1186/1471-2105-8-172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grissa I, Vergnaud G, Pourcel C. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35:W52–W57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grissa I, Vergnaud G, Pourcel C. 2008. CRISPRcompar: a website to compare clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 36:W145–W148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haerter JO, Trusina A, Sneppen K. 2011. Targeted bacterial immunity buffers phage diversity. J. Virol. 85:10554–10560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haft DH, Selengut J, Mongodin EF, Nelson KE. 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 1:e60 doi:10.1371/journal.pcbi.0010060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hale CR, et al. 2009. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139:945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Held NL, Whitaker RJ. 2009. Viral biogeography revealed by signatures in Sulfolobus islandicus genomes. Environ. Microbiol. 11:457–466 [DOI] [PubMed] [Google Scholar]
- 22.Held NL, Herrera A, Cadillo-Quiroz H, Whitaker RJ. 2010. CRISPR associated diversity within a population of Sulfolobus islandicus. PLoS One 5:e12988 doi:10.1371/journal.pone.0012988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Honjo M, et al. 2006. Diversity of virus-like agents killing Microcystis aeruginosa in a hyper-eutrophic pond. J. Plankton Res. 28:407–412 [Google Scholar]
- 24.Horvath P, et al. 2008. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 190:1401–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y, Niu B, Gao Y, Fu L, Li W. 2010. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26:680–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jansen R, Embden JD, Gaastra W, Schouls LM. 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 43:1565–1575 [DOI] [PubMed] [Google Scholar]
- 27.Kamerbeek J, et al. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaneko T, et al. 2007. Complete genomic structure of the bloom-forming toxic cyanobacterium Microcystis aeruginosa NIES-843. DNA Res. 14:247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasai F, Kawachi M, Erata M, Watanabe MM. 2004. NIES collection list of strains, 7th ed The National Institute for Environmental Studies, Tsukuba, Japan [Google Scholar]
- 30.Kunin V, Sorek R, Hugenholtz P. 2007. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 8:R61 doi:10.1186/gb-2007-8-4-r61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuno S, Yoshida T, Kamikawa R, Hosoda N, Sako Y. 2010. The distribution of a phage-related insertion sequence element in the cyanobacterium, Microcystis aeruginosa. Microbes Environ. 25:295–301 [DOI] [PubMed] [Google Scholar]
- 32.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8:317–327 [DOI] [PubMed] [Google Scholar]
- 33.Lillestøl RK, Redder P, Garrett RA, Brügger K. 2006. A putative viral defence mechanism in archaeal cells. Archaea 2:59–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu YG, Whittier RF. 1995. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25:674–681 [DOI] [PubMed] [Google Scholar]
- 35.Ma Y, Paulsen IT, Palenik B. 2012. Analysis of two marine metagenomes reveals the diversity of plasmids in oceanic environments. Environ. Microbiol. 14:453–466 [DOI] [PubMed] [Google Scholar]
- 36.Makarova KS, et al. 2011. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9:467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makarova KS, Wolf YI, Snir S, Koonin EV. 2011. Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J. Bacteriol. 193:6039–6056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manica A, Zebec Z, Teichmann D, Schleper C. 2011. In vivo activity of CRISPR-mediated virus defence in a hyperthermophilic archaeon. Mol. Microbiol. 80:481–491 [DOI] [PubMed] [Google Scholar]
- 39.Marraffini LA, Sontheimer EJ. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mojica FJM, Díez-Villaseñor C, García-Martínez J, Almendros C. 2009. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155:733–740 [DOI] [PubMed] [Google Scholar]
- 41.Palenik B, Ren Q, Tai V, Paulsen IT. 2009. Coastal Synechococcus metagenome reveals major roles for horizontal gene transfer and plasmids in population diversity. Environ. Microbiol. 11:349–359 [DOI] [PubMed] [Google Scholar]
- 42.Schwabe W, Weihe A, Börner T, Henning M, Kohl J-G. 1988. Plasmids in toxic and nontoxic strains of the cyanobacterium Microcystis aeruginosa. Curr. Microbiol. 17:133–137 [Google Scholar]
- 43.Semenova E, et al. 2011. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. U. S. A. 108:10098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanabe Y, Kasai F, Watanabe M. 2007. Multilocus sequence typing (MLST) reveals high genetic diversity and clonal population structure of the toxic cyanobacterium Microcystis aeruginosa. Microbiology 153:3695–3703 [DOI] [PubMed] [Google Scholar]
- 45.Tillett D, Neilan BA. 2000. Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J. Phycol. 36:251–258 [Google Scholar]
- 46.Tominaga H, Kawagishi S, Ashida H, Sawa Y, Ochiai H. 1995. Structure and replication of cryptic plasmids, pMA1 and pMA2, from a unicellular cyanobacterium, Microcystis aeruginosa. Biosci. Biotechnol. Biochem. 59:1217–1220 [DOI] [PubMed] [Google Scholar]
- 47.Touchon M, et al. 2011. CRISPR distribution within the Escherichia coli species is not suggestive of immunity-associated diversifying selection. J. Bacteriol. 193:2460–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tucker S, Pollard P. 2005. Identification of cyanophage Ma-LBP and infection of the cyanobacterium Microcystis aeruginosa from an Australian subtropical lake by the virus. Appl. Environ. Microbiol. 71:629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tyson GW, Banfield JF. 2008. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ. Microbiol. 10:200–207 [DOI] [PubMed] [Google Scholar]
- 50.Visser PM, Ibelings BW, Mur LR, Walsby AE. 2005. The ecophysiology of the harmful cyanobacterium Microcystis, p 109–142 In Huisman J, Matthijs HCP, Visser PM. (ed), Harmful cyanobacteria. Springer, Dordrecht, Netherlands [Google Scholar]
- 51.Wilson AE, et al. 2005. Genetic variation of the bloom-forming cyanobacterium Microcystis aeruginosa within and among lakes: implications for harmful algal blooms. Appl. Environ. Microbiol. 71:6126–6133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoshida M, Yoshida T, Takashima Y, Kondo R, Hiroishi S. 2005. Genetic diversity of the toxic cyanobacterium Microcystis in Lake Mikata. Environ. Toxicol. 20:229–234 [DOI] [PubMed] [Google Scholar]
- 53.Yoshida M, Yoshida T, Takashima Y, Hosoda N, Hiroishi S. 2007. Dynamics of microcystin-producing and non-microcystin-producing Microcystis populations is correlated with nitrate concentration in a Japanese lake. FEMS Microbiol. Lett. 266:49–53 [DOI] [PubMed] [Google Scholar]
- 54.Yoshida M, et al. 2008. Ecological dynamics of the toxic bloom-forming cyanobacterium Microcystis aeruginosa and its cyanophages in freshwater. Appl. Environ. Microbiol. 74:3269–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshida M, et al. 2008. Intra-specific phenotypic and genotypic variation in toxic cyanobacterial Microcystis strains. J. Appl. Microbiol. 105:407–415 [DOI] [PubMed] [Google Scholar]
- 56.Yoshida M, Yoshida T, Yoshida-Takashima Y, Kashima A, Hiroishi S. 2010. Real-time PCR detection of host-mediated cyanophage gene transcripts during infection of a natural Microcystis aeruginosa population. Microbes Environ. 25:211–215 [DOI] [PubMed] [Google Scholar]
- 57.Yoshida T, et al. 2006. Isolation and characterization of a cyanophage infecting the toxic cyanobacterium Microcystis aeruginosa. Appl. Environ. Microbiol. 72:1239–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.