Abstract
This work reports on the identification and molecular characterization of the two-component regulatory system (2CRS) PhoRP, which controls the response to inorganic phosphate (Pi) starvation in Bifidobacterium breve UCC2003. The response regulator PhoP was shown to bind to the promoter region of pstSCAB, specifying a predicted Pi transporter system, as well as that of phoU, which encodes a putative Pi-responsive regulatory protein. This interaction is assumed to cause transcriptional modulation under conditions of Pi limitation. Our data suggest that the phoRP genes are subject to positive autoregulation and, together with pstSCAB and presumably phoU, represent the complete regulon controlled by the phoRP-encoded 2CRS in B. breve UCC2003. Determination of the minimal PhoP binding region combined with bioinformatic analysis revealed the probable recognition sequence of PhoP, designated here as the PHO box, which together with phoRP is conserved among many high-GC-content Gram-positive bacteria. The importance of the phoRP 2CRS in the response of B. breve to Pi starvation conditions was confirmed by analysis of a B. breve phoP insertion mutant which exhibited decreased growth under phosphate-limiting conditions compared to its parent strain UCC2003.
INTRODUCTION
Bifidobacteria are Gram-positive, pleomorphic, anaerobic bacteria which are considered common inhabitants of the human and animal gastrointestinal tract (53). In recent years, bifidobacteria have received increased scientific interest due to perceived beneficial or probiotic properties which they impart to their hosts (21, 32, 33, 50, 59).
Commercially exploited probiotic bacteria are typically selected for fast growth, antimicrobial activity, good adhesion properties, and utilization of prebiotic substrates (32). In addition to these properties, they must also be able to resist the adverse conditions, such as acid and bile stress, encountered during large-scale production, product storage, and passage through the oral cavity, stomach, and small intestine (4, 43, 57). All these environmental challenges have a negative impact on bifidobacterial viability and, consequently, probiotic efficacy. Therefore, successful bifidobacterial delivery, colonization, and/or survival in the gastrointestinal tract will depend on physiological responses that allow adaptation to these hazardous conditions.
Phosphorus is an essential nutrient for all cells due to its involvement in many critical biological processes, e.g., nucleic acid and (lipo)teichoic acid biosynthesis and regulation of protein activity by phosphorylation. The most commonly assimilated P source is inorganic phosphate (PO43−, or Pi). For many bacteria, it has been shown that the mechanism by which Pi is taken up from the environment will depend on the extracellular Pi concentration (15, 37). When sufficient Pi is available to the cell (in the mM range), a low-affinity transport system named Pit (for “Pi transport”) is active. In Escherichia coli, Pit is represented by a single, constitutively expressed membrane-associated protein, which transports Pi in a proton motive force-dependent manner (11). However, when Pi becomes limiting (at a concentration below 20 μM), the expression of a high-affinity ABC-type transporter, designated Pst (for “Pi-specific transport”) and consisting of four different proteins, PstSCAB, is induced (38). For many bacteria, such as E. coli, Bacillus subtilis, and Corynebacterium glutamicum, the Pi starvation response is governed by a two-component signal transduction system, such as PhoBR, PhoRP, PhoPQ, or PhoSR (14, 36, 45, 46, 63).
Many two-component regulatory systems (2CRS) are known to be responsible for gathering environmental information and translating such data into an appropriate physiological response, thus allowing adaptive responses that increase fitness and survival under adverse conditions (for reviews, see references 8, 12, 28, and 51). Typically, a 2CRS consists of a histidine protein kinase (HPK), usually a membrane-associated protein capable of sensing a specific environmental signal, and a cytoplasmic response regulator (RR), which translates the incoming signal directly or indirectly into a cellular adaptive response. A typical HPK contains an N-terminally located input or sensing domain and a C-terminal transmitter domain, which is autophosphorylated from ATP at a conserved histidine residue in response to fluctuations in chemical and/or physical conditions (sensed by the input domain). This phosphoryl group is then transferred to a conserved aspartate residue on the N-terminally positioned receiver domain of the cognate response regulator. This phosphorylation changes the biochemical properties of the RR's output domain, which can participate in DNA binding and transcriptional control, perform enzymatic activities, bind RNA, or engage in protein-protein interactions (8).
Although the number of 2CRS identified in a given bifidobacterial genome ranges from 5 to 17 (58, 25), their physiological function has remained elusive. In the current work, we report for the first time the molecular and functional characterization of a bifidobacterial 2CRS, designated here as PhoRP, from Bifidobacterium breve UCC2003, which is involved in the response to Pi starvation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. B. breve UCC2003 was grown at 37°C in MRS medium (De Man, Rogosa, and Sharpe) or RCM (reinforced clostridial medium) (Oxoid, Hampshire, England) as standing cultures supplemented with 0.05% cysteine or on RCM agar plates containing 1.5% (wt/vol) agar under anaerobic conditions in a modular atmosphere controlled system (Davidson & Hardy Ltd., Dublin, Ireland). E. coli strains were grown in LB (lysogeny broth) medium under aerobic conditions on a rotary shaker (150 rpm) at 37°C or plated on LB agar plates. Where appropriate for plasmid maintenance, media were supplemented with ampicillin (Amp) (100 μg ml−1), erythromycin (Er) (100 μg ml−1), chloramphenicol (Cm) (20 μg ml−1), kanamycin (Kn) (50 μg ml−1), or tetracycline (Tet) (10 μg ml−1) for E. coli and Cm (2 μg ml−1) or Tet (5 μg ml−1) for B. breve.
DNA techniques and transformation.
The general procedures used for DNA manipulation were essentially those described previously (42). Restriction enzymes and T4 DNA ligase were obtained from Roche Diagnostics (Bell Lane, East Sussex, United Kingdom) and used according to the manufacturer's instructions. PCRs were performed using Taq PCR master mix (Qiagen GmbH, Hilden, Germany). Synthetic oligonucleotides were synthesized by MWG Biotech AG (Ebersberg, Germany) and are listed in Table S2 in the supplemental material. PCR products were purified by using a High-Pure PCR product purification kit (Roche). Plasmid DNA was introduced into E. coli and B. breve by electrotransformation; large-scale preparation of chromosomal DNA from Bifidobacterium spp. was performed as described previously (26). Plasmid DNA was obtained from B. breve and E. coli using a QIAprep spin plasmid miniprep kit (Qiagen GmbH, Hilden, Germany). An initial lysis step was performed using 30 mg ml−1 of lysozyme for 30 min at 37°C as part of the plasmid purification protocol for B. breve.
Bioinformatics.
Sequence data were obtained from the Artemis-mediated (41) annotations of the B. breve UCC2003 genome (31). Database searches were performed using the nonredundant sequence database accessible at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov) using TBLASTN, BLASTX, and BLASTP (1). Sequence analysis was performed using the Clone Manager professional suite (Sci-Ed Central), and multiple local alignments were carried out with ClustalW software (52). The presence of conserved sequences was examined using a hidden Markov model (HMM) with the HMMER tool (http://hmmer.janelia.org/). Searches were performed for sequences up to 400 bp upstream of the deduced translational start site of every identified gene of the B. breve UCC2003 genome. A graphical representation of the identified motif was obtained using WebLogo software (3).
Construction of a phoP insertion mutant.
An internal 481-bp fragment of phoP was amplified by PCR using B. breve UCC2003 chromosomal DNA as the template and the oligonucleotide primers phoPKOf and phoPKOr (see Table S2 in the supplemental material). The generated 481-bp PCR product was cloned into pORI19, an Ori+ RepA− integration plasmid (20), using the unique PstI and XbaI restriction sites that were incorporated into the primers, and introduced into E. coli EC101 by electroporation. The expected structure of the recombinant plasmid, designated pORI19-phoP, was confirmed by restriction mapping and sequencing. The tetW gene, amplified by PCR using pAM5 plasmid DNA as the template (2) and primers tetWf and tetWr (see Table S2), thereby incorporating flanking SalI sites in the amplicon, was cloned into the SalI-cut pORI19-phoP plasmid to generate plasmid pORI19-tet-phoP. The latter plasmid was introduced into E. coli EC101 harboring pNZ-M.BbrII-M.BbrIII to facilitate methylation (30), and the resulting methylated pORI19-tet-phoP was then introduced into B. breve UCC2003 by electroporation and subsequent selection on RCA plates supplemented with Tet and Cm.
Growth analysis under Pi starvation conditions.
Pi starvation experiments were performed in a chemically defined medium (CDM; for the composition, see Table S3 in the supplemental material), that can be modified as an essentially Pi-free chemically defined growth medium. In order to remove all Pi traces from the inoculum, cells were washed twice with Pi-free chemically defined medium (NP-CDM) before they were used to inoculate (4%) NP-CDM or NP-CDM containing various levels of Pi (final concentration of 0 mM, 1 mM, or 10 mM). Growth experiments were carried out in triplicate using a Power Wave 340 plate reader (Biotek Winooski), and obtained optical density (OD) readings were analyzed using Gen5 software (Biotek).
PhoP overexpression in B. breve.
In order to achieve PhoP overexpression, the phoP gene was amplified by PCR from chromosomal DNA of B. breve UCC2003 using the primer combination RrFoefor and RrFoerev (see Table S2 in the supplemental material). The resulting PCR product was cloned in the pTOPO system (Invitrogen, Carlsbad, CA), sequenced, removed by NcoI/XbaI restriction, and then cloned into similarly digested pNZ44, which carries a strong constitutive promoter (27). The expected structure of the resulting plasmid, pNZ44:phoP, was verified by sequence analysis prior to its transfer to B. breve UCC2003 by electroporation. Microarray-based global transcription patterns of B. breve UCC2003 and B. breve UCC2003 pNZ44:phoP were determined employing bacterial cultures that had been grown to early exponential phase in MRS.
Gene expression analysis with B. breve DNA microarrays.
In order to compare global transcription patterns of B. breve UCC2003 and the B. breve UCC2003 phoP insertion mutant under normal and phosphate starvation conditions, cells grown to early exponential phase were collected, washed twice in NP-CDM, and resuspended in the same medium containing either 20 mM Pi or no Pi. Samples were taken at 45 and 90 min after transfer. DNA microarrays containing oligonucleotide primers representing each of the 1,864 annotated genes in the genome of B. breve UCC2003 were obtained from Agilent Technologies (Palo Alto, CA). Methods for cell disruption, RNA isolation, RNA quality control, cDNA synthesis, and indirect labeling were performed as described previously (54). Labeled cDNA was hybridized using the Agilent gene expression hybridization kit (number 5188-5242) as described in the Agilent two-color microarray-based gene expression analysis v4.0 manual (publication number G4140-90050). Following hybridization, microarrays were washed as described in the manual and scanned using Agilent's DNA microarray scanner G2565A. The scans were converted to data files with Agilent's Feature Extraction software (version 9.5).
DNA microarray data were processed as previously described (9, 54). Differential expression tests were performed with the Cyber-T implementation of a variant of the t test (23). A gene was considered differentially expressed between a test condition and a control when an expression ratio of 3 or 0.33 relative to the result for the control was obtained with a corresponding P value that was equal to or less than 0.001.
Transcriptional fusions and GusA assays.
The presumed phoRP, pst, and pitA promoter regions were amplified by PCR using the primer combinations p2CSFfor/p2CSFrev, pPstfor/pPstrev and pPitfor/pPitrev, respectively (see Table S2 in the supplemental material). The PCR products generated were first cloned into pTOPO vector, then recovered by digestion with BamHI/PstI, and subsequently cloned upstream of the promoterless gusA gene present in the similarly restricted pNZ272 reporter vector (34). Ligation mixtures were independently introduced by electroporation in E. coli XL1-Blue competent cells. The resulting plasmids, once verified by restriction and sequence analysis, were then transferred to B. breve UCC2003 by electroporation.
GusA activity assays in B. breve UCC2003 or derivatives were carried out in triplicate as follows: cells were grown in CDM until an OD600 of approximately 0.45 was reached and were then harvested, washed twice in NP-CDM, resuspended in the original volume of NP-CDM, and incubated at 37°C. Samples were then taken at different time points, harvested, and resuspended in 1 ml of Pi buffer (29). After 0.5 ml of culture was mixed with 12.5 μl of 0.1% sodium dodecyl sulfate (SDS) and 25 μl of chloroform (Sigma-Aldrich, Steinhein, Germany), the suspension was incubated for 5 min at 37°C, followed by the addition of 100 μl of 4 mg/ml of 4-nitrophenyl-B-d-glucuronide (Melford, United Kingdom). Reactions were stopped by the addition of 250 μl of 1 M sodium carbonate, and samples were then centrifuged to remove debris, after which the OD420 and OD550 values were determined. GusA activity was expressed in Miller units and calculated using the following equation: (522 × OD420)/(t × v × OD600), where t is reaction time (min), v is cell volume, and OD420 and OD600 are optical densities at 420 nm and 600 nm, respectively (34). Optical measurements were carried out using a DU 530 Life Science UV/Vis spectrophotometer (Beckman Coulter, Brea, CA).
PhoP overproduction and purification.
The entire coding region of the phoP gene was amplified by PCR using chromosomal DNA of B. breve UCC2003 as a template and primer combination Bbr1683fNco1 and Bbr1683rXba1 (see Table S2 in the supplemental material). NcoI and XbaI restriction sites were incorporated at the 5′ ends of the forward and reverse primers, respectively. In addition, an in-frame His10-encoding sequence was incorporated into the forward primer to facilitate protein purification using a Ni-nitrilotriacetic acid system (Qiagen). The amplicon was digested with NcoI and XbaI and ligated into the similarly digested nisin-inducible translational fusion plasmid pNZ8048 (5). The ligation mixtures were introduced into L. lactis NZ9000 by electrotransformation, and transformants were selected based on Cm resistance (Cmr). A single Cmr transformant was shown by restriction and sequence analysis to contain the expected plasmid, which was designated pNZ8048:phoP.
Protein production and purification was carried out as previously described (29). A 400-ml portion of M17 broth supplemented with 0.5% glucose was inoculated with a 2% inoculum of L. lactis strain NZ9000 harboring pNZ8048:phoP, followed by incubation at 30°C until an OD600 of 0.5 was reached, at which point protein overexpression was induced by the addition of purified nisin (5 ng ml−1) and cultures were incubated at 30°C for 90 min. Cells were harvested by centrifugation, washed, and concentrated 40-fold in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole [pH 8.0]). Cell extracts were prepared using 106-μm glass beads and a Mini-BeadBeater-8 cell disrupter (Biospec Products, Bartlesville, OK). After homogenization, the glass beads and cell debris were sedimented by centrifugation, and the supernatant containing the cytoplasmic fraction was retained. Protein purification from the cytoplasmic fraction was performed using Ni-nitrilotriacetic acid matrices in accordance with the manufacturer's instructions (Qiagen). Elution fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), as described by Laemmli (19), on a 12.5% polyacrylamide gel. After electrophoresis, the gels were fixed and stained with Coomassie brilliant blue to identify fractions containing the purified protein. Rainbow-prestained low-molecular-weight protein markers (New England BioLabs, Hertfordshire, United Kingdom) were used to estimate the molecular weights of the purified proteins.
Phosphorylation of the response regulator protein PhoP.
Acetyl phosphate-dependent phosphorylation was achieved by incubating 2 μg of purified His-PhoP protein with 4 μCi of acetyl [γ-32P]phosphate (DuPont NEN) in 20 μl of phosphorylation buffer (50 mM Tris-HCl [pH 7.5], 100 mM KCl, 1 mM MgCl2). This mixture was incubated at 25°C, and samples were taken at regular intervals. Phosphorylation reactions were stopped with SDS loading buffer, and samples were run on a 12.5% SDS-polyacrylamide gel. After electrophoresis, gels were dried and autoradiographed in order to detect acetyl phosphate-dependent phosphotransfer to His-PhoP.
Electrophoretic mobility shift assay (EMSA).
DNA fragments encompassing overlapping fragments of the pstSCAB, phoRP, and phoU promoter regions were prepared by PCR using primers described in Table S2 in the supplemental material. EMSAs were performed as described previously (55), with the following modifications: PhoP was phosphorylated by incubating 50 μM purified protein with 50 mM acetyl phosphate at 25°C for 1 h; the PCR products were generated using IRD-800-labeled fluorescent primers; and protein and probe were mixed on ice and subsequently incubated for 30 min at 37°C. Samples were loaded onto a 6% nondenaturing polyacrylamide gel prepared with 1× TAE (40 mM Tris-acetate [pH 8.0], 2 mM EDTA) and run in a 0.5×-to-2.0× gradient of TAE at 100 V for 90 min in a mini-Protean electrophoresis system (Bio-Rad Laboratories). Following electrophoresis, the presence and mobility position of the fluorescent PCR products in the gel were detected using an Odyssey instrument (LI-COR Biosciences UK Ltd., Cambridge, United Kingdom).
qRT-PCR amplifications.
Real-time quantitative RT-PCR (qRT-PCR) assays and de novo cDNA preparations were performed as previously described (35, 56). PCR primers (see Table S2 in the supplemental material) and probes were designed using the Universal ProbeLibrary Assay Design Centre (Roche). The rnpA gene (encoding the RNase P protein component) was used as a housekeeping gene with an assumed constant level of transcription to correct for variability in the initial amount of total RNA. Amplification reaction mixtures contained 1 μl of 6.7-fold-diluted cDNA, 10 μl of 2× FastStart TaqMan Probe Master (Roche), a 900 nM concentration of each primer, and 250 nM probe mix and were brought to a total volume of 20 μl by the addition of RNase-free water. All qRT-PCRs were performed in triplicate by means of a LightCycler 480 system (Roche) using 384-well plates. Thermal cycling conditions were as recommended by the manufacturer (Roche). The 2−ΔΔCT method (22) was used to calculate relative changes in gene expression determined from real-time quantitative PCR experiments. Pooled cDNA from all samples was used as a reference calibrator for analysis of differential gene expression. Results were calculated from at least two independent RNA extractions. The qRT-PCR expression data, following 2−ΔΔCT analysis, were subjected to a Mann-Whitney t test to compare all groups using GraphPad Prism 4 software (GraphPad Software). Data are reported as means and standard errors of the means. P values of <0.05 were considered significant.
Primer extension analysis.
Total RNA was isolated from B. breve UCC2003 grown to early exponential phase and incubated under phosphate starvation conditions for 45 min. RNA samples were treated with RNase-free DNase (Ambion). Primer extension was performed by annealing 1 pmol of IRD800 synthetic 18-mer oligonucleotides to 20 μg of RNA as described by Ventura et al. (60). Sequence ladders of the presumed pstS or phoR promoter regions, which were run alongside the primer extension products, were produced using the same primer as the primer extension reaction using a Thermo Sequenase primer cycle sequencing kit (Amersham). Separation was achieved on a 6.5% Li-Cor Matrix KB Plus acrylamide gel. Signal detection and image capture were performed with a Li-Cor sequencing instrument (Li-Cor Biosciences).
Microarray data accession number.
All microarray data have been deposited in the GEO database (accession number GSE34983).
RESULTS
Transcriptome analysis shows increased expression of a presumed Pi uptake system under Pi starvation conditions.
In order to investigate the genome-wide response of B. breve UCC2003 to Pi starvation, changes in gene expression were analyzed following transfer of B. breve UCC2003 to a medium without Pi (see Materials and Methods). Total RNA samples were obtained from cultures grown in a medium containing either a high level of Pi (20 mM) or no Pi for 45 min (early response) or 90 min (late response). Following cDNA synthesis and labeling, the samples were hybridized to DNA microarrays representing all identified open reading frames (ORFs) of the B. breve UCC2003 genome.
The obtained results revealed significant changes in the transcription profiles after 45 min, with a further increase of such changes following prolonged Pi starvation. From a total of 1,864 tested open reading frames, as identified on the B. breve UCC2003 genome, microarray analysis showed a significantly increased transcriptional activity for 118 genes (>3-fold; P value ≤ 0.001), while 134 genes exhibited significantly decreased levels of transcription (<3-fold; P value ≤ 0.001). Genes showing at least 3-fold upregulation in both conditions are shown in Table 1.
Table 1.
Summary of gene expression changes in B. breve UCC2003 grown under Pi limitation or subject to PhoP overexpression
| Locus tag | Annotation | Ratioa |
phoP knockout phosphate starvation |
||||
|---|---|---|---|---|---|---|---|
| Phosphate starvation |
PhoP overexpression |
phoP knockout vs wild-type phosphate starvation |
|||||
| 45 min | 90 min | 45 min | 90 min | 90 min | |||
| Bbr_0087 | Hemolysin III homolog | 3.1 | 6.8 | 0.9 | 1.8 | 2.2 | 0.8 |
| Bbr_0191 | Membrane protein | 3.5 | 8.7 | 1.0 | 0.7 | 0.5 | 1.0 |
| Bbr_0323 | atpB | 4.5 | 6.3 | 0.6 | 1.2 | 1.2 | 1.4 |
| Bbr_0330 | atpC | 3.0 | 6.1 | 0.6 | 1.7 | 1.2 | 1.7 |
| Bbr_0824 | Fic family protein | 3.1 | 8.4 | 0.9 | 0.3 | 0.5 | 0.9 |
| Bbr_0876 | Peptidase E | 3.4 | 5.1 | 0.8 | 0.7 | 0.7 | 4.7 |
| Bbr_1460 | Helicase | 3.1 | 3.6 | 1.2 | 1.7 | 1.6 | 0.8 |
| Bbr_1506 | CFA synthase | 4.1 | 12.7 | 1.0 | 0.8 | 1.2 | 1.2 |
| Bbr_1573 | phoU homolog | 1.6 | 2.3 | 1.5 | 0.5 | 0.5 | 0.9 |
| Bbr_1679 | pstB | 3.1 | 12.1 | 2.2 | 1.6 | 1.5 | 0.9 |
| Bbr_1680 | pstA | 3.9 | 12.4 | 3.2 | 1.5 | 1.8 | 0.9 |
| Bbr_1681 | pstC | 5.4 | 15.3 | 3.8 | 1.7 | 1.9 | 0.9 |
| Bbr_1682 | pstS | 9.8 | 25.9 | 9.6 | 3.4 | 4.6 | 1.0 |
| Bbr_1683 | phoP | 2.0 | 5.9 | 164.1 | 0.9 | 1.0 | 1.0 |
| Bbr_1684 | phoR | 1.7 | 3.5 | 2.9 | 0.9 | 0.8 | 1.1 |
| Bbr_1811 | Multidrug transporter | 4.8 | 11.9 | 0.8 | 1.0 | 1.3 | 1.2 |
| Bbr_0865 | Low-affinity inorganic phosphate transporter | 0.8 | 0.7 | 0.9 | 1.3 | 1.0 | 1.0 |
Calculated as average mRNA level of a given gene under phosphate starvation or PhoP overexpressing/average mRNA level of the same gene under normal growth conditions or no PhoP overexpression, respectively.
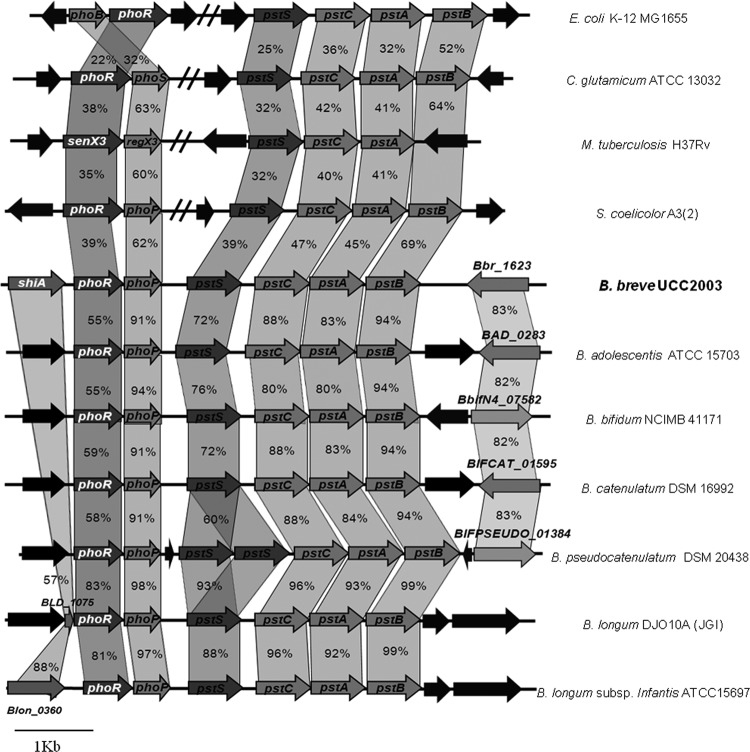
Among the various gene clusters upregulated by Pi starvation, a predicted 2CRS, represented by Bbr_1683/Bbr_1684 (referred to here as phoRP), was identified. Bbr_1684 or phoR is predicted to encode a histidine protein kinase, while the protein product of Bbr1683 (designated phoP) specifies a presumed response regulator. PhoR contains a phosphoacceptor and an ATPase domain (PFAM003709), whereas PhoP resembles a typical response regulator with a signal receiver and an effector domain (PFAM00486). The B. breve UCC2003 phoRP genes are representatives of homologous systems present in all currently available bifidobacterial genomes and in other high-GC-content Gram-positive genera, such as Streptomyces, Corynebacterium, and Mycobacterium (Fig. 1). Immediately downstream of phoRP, a cluster of four genes showed strong transcriptional upregulation under Pi starvation conditions (Table 1). This gene cluster displays significant homology to the well-characterized Pi-specific transport system of Streptomyces coelicolor and E. coli (44, 39). We refer to these four genes as the pst gene cluster, represented by pstSCAB, as illustrated in Fig. 1.
Fig 1.
Comparison of the phoRP and pstSCAB loci of B. breve UCC2003 with corresponding (putative or proven) 2CRS and Pi uptake loci from other Bifidobacterium and high-GC-content Gram-positive bacteria. Each solid arrow indicates an open reading frame (ORF). The lengths of the arrows are proportional to the length of the predicted ORF, and the gene locus name, which is indicative of its putative function, is given within the arrow. Ortholog genes are linked, and the amino acid identity of each predicted protein is indicated as a percentage relative to its equivalent protein encoded by B. breve UCC2003.
In order to identify genes whose expression is under phoRP control, a B. breve UCC2003 PhoP-overexpressing derivative was generated by cloning the phoP gene into the pNZ44 vector to generate pNZ44:phoP, which was then introduced into B. breve UCC2003 (see Materials and Methods). Microarray analysis showed that the level of phoP transcription in B. breve UCC2003 (pNZ44:phoP) was 164-fold higher than that in the B. breve control strain containing the empty pNZ44 vector (Table 1), indicating that cloning of phoP on pNZ44 leads to a significant overexpression of this gene. A number of genes showed different levels of upregulation in their expression, some of them corresponding to different cell stress proteins, although the gene showing the highest level of increased transcription was pstS. The remaining genes of the pst cluster, as well as phoR, also exhibited significant (P < 0.001) transcriptional upregulation (Table 1).
Pi-dependent control of the pst and phoRP promoters.
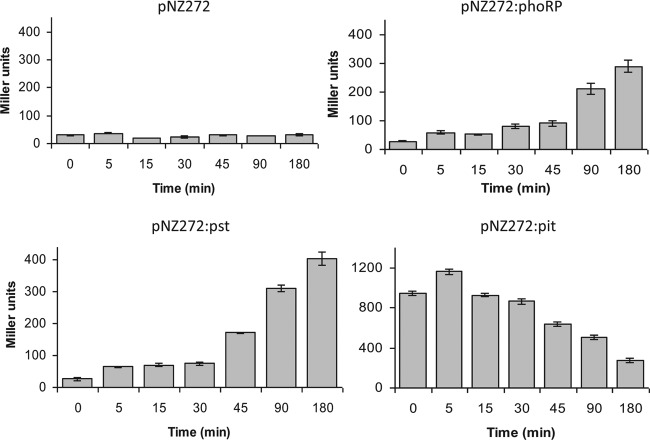
In order to further analyze Pi-dependent regulation, transcriptional fusions were constructed by cloning specific DNA fragments encompassing the prospective promoter regions upstream of phoRP, pstSCAB, and pitA, upstream of the promoterless gusA reporter gene in pNZ272 (see Materials and Methods). The pitA gene (Bbr_0865) on the B. breve UCC2003 genome specifies the presumed low-affinity inorganic phosphate transporter, which did not exhibit any appreciable increase in transcription under Pi starvation conditions or upon phoP overexpression (Table 1). B. breve UCC2003 strains harboring the gusA fusion plasmids pNZ272:phoP, pNZ272:pst, and pPNZ272:pit were used for GusA assays under Pi abundance and Pi starvation conditions. Growth under Pi-limiting conditions clearly induced the promoters present in the upstream regions of phoRP and pst, while the promoter upstream of pit appeared to be negatively affected by Pi-limiting conditions (Fig. 2; the array results were consistent with this finding, as the observed downregulation of pit was 1.2-fold at 45 min and 2.2-fold at 90 min).
Fig 2.
GusA assays testing phosphate starvation effect on phoRP, pst, and pit promoters. Enzyme activity was measured after the indicated times under phosphate starvation. Data are means from three independent experiments, and error bars show standard deviations.
Overexpression, purification, and phosphorylation of PhoP.
For functional characterization of the DNA binding activity of PhoP, this protein was expressed in L. lactis and purified as an N-terminally His-tagged version using the lactococcal nisin-controlled expression (NICE) system (5). The recombinant H10-PhoP protein was purified as a single protein of approximately 27 kDa, as determined by SDS-PAGE (see Fig. S1 in the supplemental material). Purified H10-PhoP was shown to become phosphorylated upon incubation with acetyl phosphate, a known phosphoryl donor of response regulators (24), proving that it contains a functional phosphoacceptor domain (Fig. 3).
Fig 3.
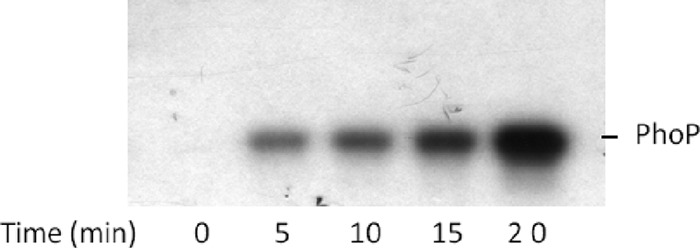
Time-dependent PhoP-His phosphorylation. Purified PhoP-His protein (2 mg) was incubated with 4 μCi of [γ-32P]acetyl phosphate, and samples were taken every 5 min and subjected to SDS-PAGE followed by autoradiography.
PhoP specifically binds the pstS, phoR, and phoU promoters.
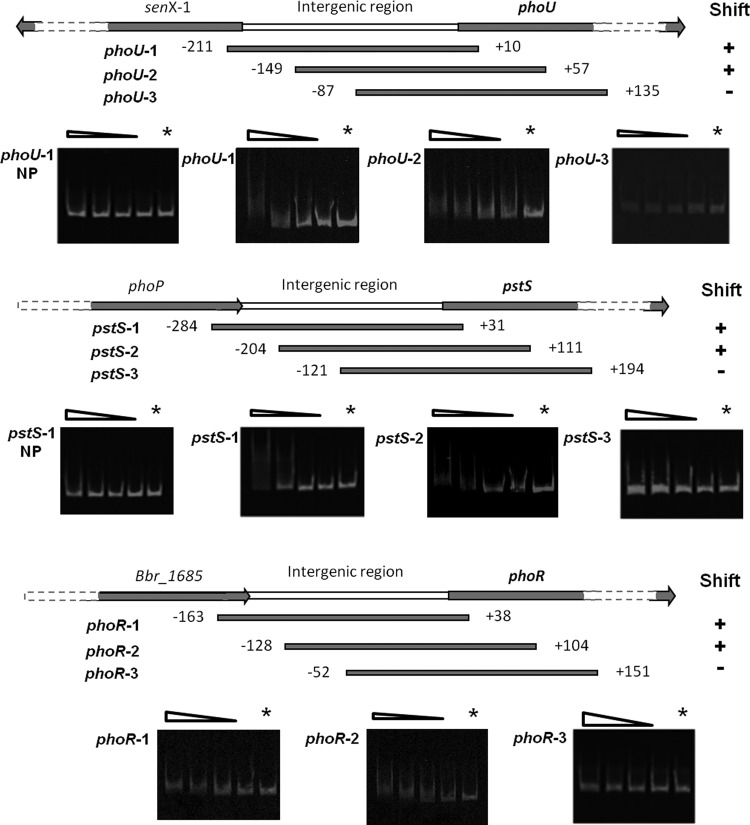
In order to obtain evidence for the expected regulatory function of phoP in the cellular response to phosphate starvation, the ability of this presumed response regulator to bind to the promoter regions of pstS, phoR, and phoU (Bbr_1573) was analyzed in vitro by EMSA. Although phoU transcription was only 2-fold upregulated upon Pi starvation (Table 1), it is known that the phoU homologue in E. coli is induced by low levels of Pi, where its gene product acts as a negative regulator of the Pho regulon (39). Three overlapping IRD800-labeled amplicons were generated for each promoter region, namely, pstS-1, pstS-2, and pstS-3, phoR-1, phoR-2, and phoR-3, and phoU-1, phoU-2, and phoU-3 (Fig. 4). EMSAs were carried out with phosphorylated or nonphosphorylated H10-PhoP protein, which revealed that phosphorylation of PhoP is essential for binding of this protein to the phoR, phoU, and pstS promoter regions (data not shown). Furthermore, these experiments revealed that phosphorylated H10-PhoP interacts with particular segments of each promoter region, indicating the presence of specific binding sequences within DNA fragments that exhibit mobility shifts (Fig. 4).
Fig 4.
PhoP binding assays to various promoter regions. Shown is a schematic representation of amplified DNA fragments used in gel mobility shift assays; the numbers correspond to the ends of the fragments relative to the presumed translation start site. Plus and minus signs indicate observed binding or no binding, respectively, of PhoP to specific sections of the phoU, pstS, or phoR promoter regions. The terms phoU-1-NP and pstS-1-NP represent binding assays with unphosphorylated PhoP, and the remainder of the bindings assays were performed using phosphorylated PhoP. PhoP was phosphorylated for 1 h at 25°C in the presence of acetyl phosphate. Each gel represents a binding assay with a particular DNA fragment as indicated in the left margin and decreasing amounts of (phosphorylated) PhoP (lanes 1 to 5 correspond to 400, 200, 100, 50, and 0 mM PhoP, respectively).
No interaction between other tested DNA fragments (representing presumed promoter regions of genes that were either up- or downregulated upon Pi starvation as determined by transcriptome analysis) and phosphorylated H10-PhoP was observed in EMSAs (data not shown).
PHO box identification and its relative distance to the transcriptional start site of the pstS and phoR promoters.
The EMSA results (Fig. 4) indicate that the PhoP binding sites in the pstS, phoR, and phoU promoter regions are localized within DNA fragments between positions −127 and +112, −66 and +51, and −121 and +105 (relative to the deduced translational start position of pstS, phoR, and phoU), respectively. Analysis of these PhoP-binding sequences within the three promoter regions revealed a commonly present direct repeat, GTTCAYY-N4-GTTCAYY, referred to here as the PHO box, shown to be highly conserved and present in the intergenic regions upstream of genes encoding proteins that are presumed to be involved in Pi uptake, Pi release, and Pi regulation in various Bifidobacterium species, like B. longum, B. adolescentis, B. dentium, and B. animalis subsp. lactis, as well as in other Gram-positive bacteria, such as Streptomyces coelicolor, Propionibacterium acnes, and Brevibacterium linens (Table 2). Interestingly, in the genome of B. adolescentis and those of various other bifidobacteria, this motif was detected upstream of a gene encoding a putative alkaline phosphatase (BAD_1548 in B. adolescentis).
Table 2.
Alignment of probable PhoP binding sites in B. breve UCC2003 with putative PhoP binding sites in high-GC-content Gram-positive species
| Organism | Locus | Putative gene name | Sequencea | Strand relative to gene |
|---|---|---|---|---|
| Bifidobacterium breve UCC2003 | Bbr_1682 | pstS | CTGTTCATCTTCTGTTTACTGAT | + |
| Bbr_1684 | phoR | AAGTTCGCCTATTGTTCATTTTT | − | |
| Bbr_1573 | phoU | AAGTTGTTCTACTGTTCATTGGG | − | |
| Bifidobacterium longum NCC2705 | BL0315 | pstS | GTGTTCATCTTCTGTTTACTGAT | + |
| BL0317 | phoR | AAGTTCGTCTATTGTTCATTTTT | − | |
| BL1658 | phoU | AAGTTGTTCTACTGTTCATTGGG | − | |
| Bifidobacterium adolescentis ATCC 15703 | BAD_0278 | pstS | CTGTTCATCTTCCGTTCACCGTT | + |
| BAD_0276 | phoR | GCGTTCACCTTCCGATAATCCTC | − | |
| BAD_1548 | phoA | GTGTTCATCTGCCGTTCTTCTGT | + | |
| Bifidobacterium dentium Bd1 | BDP_0387 | pstS | CTGTTCATCTACCGTTCACCGAT | + |
| BDP_0385 | phoR | GCGTTAACCCTCCGATAATCCAG | − | |
| Bifidobacterium animalis BB12 | BIF_01097 | pstS | ACGTTCACGCTAAATTCACCGTT | + |
| BIF_01099 | senX3 | TGGCTCACCTTCTGCGGCGTGTC | − | |
| Brevibacterium linens BL2 | BlinB_010200010959 | pstS | TCATTCATCTTCCATTCACTTAG | + |
| Arthrobacter aurescensTC1 | AAur_0195 | pstS | TCGTTCACCTTCCGTTCACCTTT | + |
| Streptomyces coelicolor A3(2) | SCO1845 | pitH | CAGTTCATCTTCCGTTCACCCTG | + |
| SCO4142 | pstS | GGGTTCACCCGGCGTTCATTTAC | + | |
| Frankia alni ACN14a | FRAAL6535 | pstS | TTGTTCATCTTTCGTTCACTCCG | + |
| Leifsonia xyli subsp. xyli CTCB07 | Lxx18420 | pstS | TCGTTAACCTTCCGTTTACCACT | + |
| Mycobacterium tuberculosis H37Rv | Rv0928 | pstS3 | AAGTTCGCCGACCGTTAACCTAG | + |
| Rv2984 | ppk | CCGTTGGTCTGCCGTTCACCCCC | − | |
| Propionibacterium acnes SK137 | HMPREF0675_3383 | pstS | GTGTTCACGTTGCGTTCACCTTG | + |
| Consensus | NNGTTCAYY-Y4-GTTCAYYNNN |
A consensus sequence for B. breve could be deduced. Conserved positions are highlighted in bold. Numbers indicate positions with respect to the translational point. R, purine; Y, pyrimidine; N, any nucleotides.
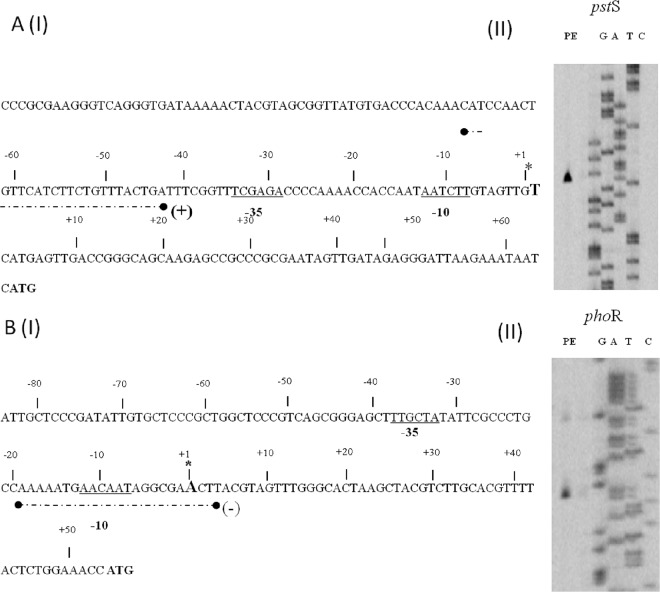
In order to investigate how the PHO box, as a binding motif for the presumed transcriptional regulator PhoP, is positioned relative to the promoters of its target genes, the transcription start sites of the pstS and phoR promoters were determined by primer extension analysis using RNA extracted from B. breve UCC2003 grown under phosphate-limiting conditions. An extension product was identified 64 nucleotides 5′ of the predicted translational start site for the pstS gene, while for phoR gene the transcription initiation site was observed 53 bp upstream of its predicted translational start site (Fig. 5). Analysis of the pstS promoter region revealed potential promoter recognition sequences resembling consensus −10 and −35 hexamers, while the PHO box was located 43 bp upstream of the transcription start site. For the phoR promoter, a potential −10 sequence could be identified, with no obvious −35 sequence present within the expected range of this −10 sequence, although a potential −35 sequence is present further upstream (Fig. 5). Interestingly, the PHO box was found to overlap the deduced −10 promoter recognition sequence.
Fig 5.
Schematic representation of the B. breve UCC2003 ptsS [A(I)] and phoR [B(I)] promoter regions. Underlined sequences indicate the −10 and −35 hexamers as deduced from the primer extension results [A(II) and B(II)]; the transcriptional start sites (TSS) are indicated by asterisks; broken black lines underneath the respective sequence indicate the PhoP binding sequence (PHO box).
Disruption of the phoP gene in B. breve UCC2003.
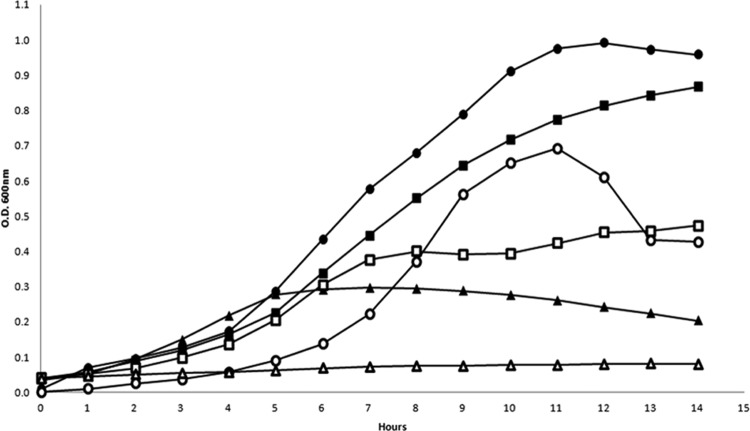
In order to test if disruption of phoP in UCC2003 would result in the loss of this strain's capacity to appropriately respond to Pi limitation, a phoP insertion mutant was generated, designated B. breve UCC2003∷phoP (see Materials and Methods). To verify the expected phenotype of the B. breve UCC2003∷phoP mutant strain, both wild-type and mutant strains were analyzed for their ability to grow in NP-CDM alone or NP-CDM to which Pi had been added at 1 mM and 10 mM, the latter representing the approximate Pi concentration present in complex medium. Growth experiments were carried out in triplicate, and results are presented in Fig. 6. As expected, both UCC2003 and UCC2003∷phoP exhibited little or no growth in the absence of Pi in the medium (NP-CDM). B. breve UCC2003 was able to grow in CDM with various Pi concentrations, although its growth capacity was directly proportional to the concentration of Pi, reaching a maximum OD600 of 0.99 when grown in CDM containing 10 mM Pi. The growth ability of the UCC2003∷phoP strain was markedly lower than that of wild-type UCC2003 in CDM with 1 mM Pi, while the observed growth pattern of these two strains was similar in CDM containing 10 mM Pi, at least for the first 9 h. These results demonstrate that phoRP plays an important role in the adaptive response of B. breve UCC2003 to phosphate limitation. In order to determine the effects of the phoP insertional mutation, transcriptional profiling was performed for this mutant using DNA microarray technology (see Materials and Methods). The obtained results revealed a total absence of induction of any of the genes of the pstSCAB cluster, consistent with the observed reduced growth of this mutant when subjected to Pi starvation (Table 3). qRT-PCR was performed in order to confirm these differences in gene expression; primers were designed for pstS, as this gene displayed the most significant transcription difference of this cluster. Results indeed confirmed increased transcription in the wild-type strain, while in the phoP mutant strain there was effectively no change in pstS transcription, although this gene seems to exhibit a basal transcription level (data not shown).
Fig 6.
Growth rates of B. breve UCC2003 and UCC2003∷phoP in CDM with different Pi concentrations: ●, 10 mM Pi, wild type; ■, 1 mM Pi, wild type; ▲, no Pi, wild type; ○, 10 mM Pi, mutant; □, 1 mM Pi, mutant; △, no Pi, mutant.
Table 3.
Summary of differential transcription behavior of phoRP and pstSCAB in B. breve UCC2003 and UCC2003∷phoP grown under Pi limitation
| ORF | Annotation | Ratioa |
|||
|---|---|---|---|---|---|
| UCC2003 |
UCC2003∷phoP |
||||
| 45 min | 90 min | 45 min | 90 min | ||
| Bbr_1679 | pstB | 5.3 | 8 | 0.6 | 0.7 |
| Bbr_1680 | pstA | 6.2 | 8.8 | 0.6 | 0.5 |
| Bbr_1681 | pstC | 7.8 | 12.3 | 0.6 | 0.5 |
| Bbr_1682 | pstS | 14.4 | 20 | 0.3 | 0.2 |
| Bbr_1683 | phoP | 3.3 | 4.1 | ND | ND |
| Bbr_1684 | phoR | 2.5 | 2.8 | ND | ND |
Calculated as average mRNA level of a given gene under phosphate starvation conditions/average mRNA level of the same gene under non-starvation conditions. The relative mRNA levels are averages from three independent experiments. ND, not determined.
DISCUSSION
Two-component regulatory systems are employed extensively in nature by microorganisms to modify their cellular physiology in response to alterations in environmental conditions. In this work we describe the first characterization of a bifidobacterial 2CRS and provide evidence that the B. breve UCC2003 PhoRP proteins constitute an autoregulatory, Pi-responsive 2CRS, which regulates the expression of the Pi starvation response operon pstSCAB. The PhoRP system shows high homology to corresponding 2CSs in other members of the genus Bifidobacterium, while it also appears to be conserved among many other members of the Actinobacteridae.
The histidine protein kinase PhoR contains a single predicted transmembrane domain (TMD) in its N-terminal domain that allows it to be anchored to the membrane while also enabling it to sense the presence of extracellular Pi. PhoR contains a classical phosphoacceptor domain with a conserved histidine residue, as well as an ATPase motif (7). Although HPKs commonly have two TMDs, HPKs with a single TMD have been reported, e.g., for Streptomyces lividans (48). We show in this report that heterologously expressed H10-PhoP is readily and stably phosphorylated using acetyl phosphate as a phosphoryl donor, while it also exhibits in vitro DNA binding ability. Generally, response regulators exhibit DNA binding activity when phosphorylated, although specific DNA binding activity of unphosphorylated RRs has been described previously (40).
Microarray analysis data obtained from B. breve cells subjected to Pi limitation revealed a significant change in global gene transcription, where transcription of around 10% of the total B. breve gene content was affected. Similar results have previously been obtained for other bacteria, such as B. subtilis (13) and C. glutamicum (16). The transcriptional fusion results as well as microarray experiments convincingly show that transcription of phoRP and the pst operon is induced under conditions of Pi starvation. In addition, among the Pi starvation-induced genes, we identified some genes that correspond to characterized stress proteins like GroEL, ClpB, DnaK, different ABC transporters, while expression of such stress response proteins is even higher in the phoP insertion mutant strain (data not shown). It is also remarkable that Pi starvation reduces the transcription of genes encoding ribosomal proteins. This extensive alteration in the observed transcription profile can be explained by significant growth rate delay as a result of Pi scarcity, which is consistent with our finding that under Pi starvation conditions, B. breve UCC2003∷phoP showed clearly altered growth behavior compared with the wild-type strain.
In many bacteria whose Pi starvation response has been studied in detail, e.g., E. coli (62), B. subtilis (13) or Corynebacterium (16), PhoRP homologues activate additional operons related to the acquisition and metabolism of alternative P sources. Compared to PhoRP-dependent genes found in C. glutamicum, a homologous glpQ gene (glycerophosphoryldiester phosphodiesterase) is present in the B. breve genome, while the ugpAEBC (ABC-type sn-glycerol 3-phosphate uptake system) is absent. Homologous genes coding for PhoC (putative cell wall-associated phosphatase), UshA (UDP sugar hydrolase and 5′ nucleotidase activity), and NucH (extracellular nuclease) appear to be absent from the B. breve UCC2003 genome. In contrast to its equivalent in C. glutamicum (46), the pitA promoter in B. breve does not appear to be regulated by PhoRP. Transcriptional fusions of the pitA promoter indicated that this gene is highly expressed when cells are actively growing. The observed drop in transcription following transfer to Pi-limiting medium is possibly caused by an arrest of metabolic processes, as is evident from the cessation of growth. Presumably, the PitA system is used for Pi uptake under normal physiological conditions, while the more energy-demanding Pst system is utilized during conditions of Pi starvation. In contrast, expression of the phoU gene in E. coli is induced in response to low levels of environmental phosphate, where PhoU acts as a negative regulator of the Pho regulon, although it is unclear exactly how PhoU accomplishes this. Recently it was postulated that in E. coli, PhoU controls the activity of the PstSCAB2 transporter, as well as its abundance within the cell, through the PhoBR two-component signaling pathway (39). A phoU homolog, designated Bbr_1573, is present on the B. breve UCC2003 genome, although microarray analysis of data obtained from B. breve UCC2003 subjected to Pi starvation, PhoP overexpression, or phoP mutation indicated that phoU transcription is only moderately affected (and always below the selected cutoff value). However, PhoP was shown to specifically bind the presumed phoU promoter region, thus indicating that phoU is part of the PhoRP regulon.
No information is available concerning regulatory sequences involved in Pi starvation response in Bifidobacterium species, although PHO boxes have been described for other bacteria as two tandem direct repeats of 11 bp in which the first five nucleotides of the repeat are highly conserved, while the following six positions are much less or not at all conserved (2, 6, 46, 49, 63, 64). A similar situation was found in the phoRP, pst, and phoU promoters in B. breve UCC2003, as well as in upstream regions of genes related to Pi metabolism from other Bifidobacterium species and various high-GC-content Gram-positive bacteria, which allows us to propose a consensus PHO motif in high-GC-content Gram-positive bacteria: GTTCAYY-Y4-GTTCAYY. Gel retardation experiments have demonstrated PhoP's ability to bind to its own promoter as well as the pst promoter; however, the position of these PHO boxes relative to the transcriptional start site of the operons is different: the PHO box in the pst promoter region is located 43 bp upstream of the transcription start site, while in the phoRP promoter, it overlaps the deduced −10 sequence. Differences in the position of PHO boxes in promoters that are subject to PhoP regulation have been reported for several species. For example, the E. coli phoBR operon is transcriptionally autoregulated via the direct binding of PhoB to a single PHO box which is 10 bp upstream of the putative −10 region. The E. coli Pho regulon consists of about 40 genes, and PhoB-binding sites can be found in two or more copies within some promoter regions (17, 18). The presence of multiple PHO boxes within a single promoter region has also been found: the Vibrio cholerae phoBR operon is autoregulated by a novel and complex mechanism that involves PhoB binding to three Pho boxes, at 35 (box 1), 60 (box 2), and 80 (box 3) base pairs upstream of the putative phoB translation start site. These boxes are located in the sense (box 1) or antisense (boxes 2 and 3) strands, and PhoB interacts with them in a sequential and cooperative manner and with different affinities (6).
B. breve UCC2003∷phoP was shown to exhibit reduced growth rates under Pi-limiting conditions compared to the wild-type strain, and this result is consistent with our genetic data and substantiates the notion of a direct correlation between phoRP and the response to Pi starvation in B. breve UCC2003.
Contradictory studies about limiting Pi concentrations for bacteria in the gut have been reported (61, 63), and an explanation for this may be the multiplicity of different microhabitats and metabolic niches associated with the gut. We have found that the phoRP genes are highly conserved among available bifidobacterial genomes, whose single known habitat is the gastrointestinal tract. Nevertheless, only a small number of genes appear to be directly under PhoRP control, perhaps reflecting a rather degenerated coping strategy for, at least as far as these gut commensals are concerned, a seemingly rare environmental condition.
Supplementary Material
ACKNOWLEDGMENTS
The Alimentary Pharmabiotic Centre is a research center funded by Science Foundation Ireland (SFI), through the Irish Government's National Development Plan (grant no. s02/CE/B124 and 07/CE/B1368). Pablo Alvarez-Martin was supported by a postdoctoral fellowship from the Spanish Ministry of Science and Innovation (MICINN), Matilde Fernández was supported by a postdoctoral fellowship from the Spanish Ministry of Education and Culture (MEC), Madrid, Spain, while Aldert Zomer acknowledges financial support through an EMBARK fellowship from the Irish Research Council for Science Engineering and Technology.
Footnotes
Published ahead of print 25 May 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alvarez-Martin P, O'Connell-Motherway M, van Sinderen D, Mayo B. 2007. Functional analysis of the pBC1 replicon from Bifidobacterium catenulatum L48. Appl. Microbiol. Biotechnol. 76:1395–1402 [DOI] [PubMed] [Google Scholar]
- 3. Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. De Dea Lindner J, et al. 2007. Exploiting Bifidobacterium genomes: the molecular basis of stress response. Int. J. Food Microbiol. 120:13–24 [DOI] [PubMed] [Google Scholar]
- 5. de Ruyter PG, Kuipers OP, de Vos WM. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Diniz MM, et al. 2011. Fine-tuning control of phoBR expression in Vibrio cholerae by binding of PhoB to multiple Pho boxes. J. Bacteriol. 193:6929–6938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dutta R, Inouye M. 2000. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 25:24–28 [DOI] [PubMed] [Google Scholar]
- 8. Gao R, Mack TR, Stock AM. 2007. Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem. Sci. 32:225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. García de la Nava GJ, van Hijum SAFT, Trelles O. 2003. PreP: gene expression data pre-processing. Bioinformatics 19:2328–2329 [DOI] [PubMed] [Google Scholar]
- 10. Reference deleted.
- 11. Harris RM, Webb DC, Howitt SM, Cox GB. 2001. Characterization of PitA and PitB from Escherichia coli. J. Bacteriol. 183:5008–5014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hoch JA. 2000. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3:165–170 [DOI] [PubMed] [Google Scholar]
- 13. Hoi LT, et al. 2006. The phosphate-starvation response of Bacillus licheniformis. Proteomics 6:3582–3601 [DOI] [PubMed] [Google Scholar]
- 14. Hsieh YJ, Wanner BL. 2010. Global regulation by the seven-component Pi signaling system. Curr. Opin. Microbiol. 13:198–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hulett FM. 1996. The signal-transduction network for Pho regulation in Bacillus subtilis. Mol. Microbiol. 19:933–939 [DOI] [PubMed] [Google Scholar]
- 16. Ishige T, Krause M, Bott M, Wendisch VF, Sahm H. 2003. The phosphate starvation stimulon of Corynebacterium glutamicum determined by DNA microarray analyses. J. Bacteriol. 185:4519–4529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kasahara M, Makino K, Amemura M, Nakata A, Shinagawa H. 1991. Dual regulation of the ugp operon by phosphate and carbon starvation at two interspaced promoters. J. Bacteriol. 173:549–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kimura S, Makino K, Shinagawa H, Amemura M, Nakata A. 1989. Regulation of the phosphate regulon of Escherichia coli: characterization of the promoter of the pstS gene. Mol. Gen. Genet. 215:374–380 [DOI] [PubMed] [Google Scholar]
- 19. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 20. Law J, et al. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177:7011–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leahy SC, Higgins DG, Fitzgerald GF, van Sinderen D. 2005. Getting better with bifidobacteria. J. Appl. Microbiol. 98:1303–1315 [DOI] [PubMed] [Google Scholar]
- 22. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 23. Long AD, et al. 2001. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J. Biol. Chem. 276:19937–19944 [DOI] [PubMed] [Google Scholar]
- 24. Lukat GS, McCleary WR, Stock AM, Stock JB. 1992. Phosphorylation of bacterial response regulator proteins by low-molecular weight phosphodonors. Proc. Natl. Acad. Sci. U. S. A. 89:718–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. MacConaill LE, Butler D, O'Connell-Motherway M, Fitzgerald GF, van Sinderen D. 2003. Identification of two-component regulatory systems in Bifidobacterium infantis by functional complementation and degenerate PCR approaches. Appl. Environ. Microbiol. 69:4219–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mazé A, O'Connell-Motherway M, Fitzgerald GF, Deutscher J, van Sinderen D. 2007. Identification and characterization of a fructose phosphotransferase system in Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 73:545–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McGrath S, Fitzgerald GF, van Sinderen D. 2001. Improvement and optimization of two engineered phage resistance mechanisms in Lactococcus lactis. Appl. Environ. Microbiol. 67:608–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mitrophanov AY, Groisman EA. 2008. Signal integration in bacterial two-component regulatory systems. Genes Dev. 22:2601–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. O'Connell Motherway M, et al. 2008. Characterization of ApuB, an extracellular type II amylopullulanase from Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 74:6271–6279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. O'Connell-Motherway M, O'Driscoll J, Fitzgerald GF, van Sinderen D. 2009. Overcoming the restriction barrier to plasmid transformation and targeted mutagenesis in Bifidobacterium breve UCC2003. Microb. Biotechnol. 2:321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. O'Connell-Motherway M, et al. 2011. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc. Natl. Acad. Sci. U. S. A. 108:11217–11222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ouwehand AC, Salminen S, Isolauri E. 2002. Probiotics: an overview of beneficial effects. Antonie Van Leeuwenhoek 82:279–289 [PubMed] [Google Scholar]
- 33. Picard C, et al. 2005. Review article: bifidobacteria as probiotic agents—physiological effects and clinical benefits. Aliment. Pharmacol. Ther. 22:495–512 [DOI] [PubMed] [Google Scholar]
- 34. Platteeuw C, Simons G, de Vos WM. 1994. Use of the Escherichia coli beta-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl. Environ. Microbiol. 60:587–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pokusaeva K, et al. 2010. Ribose utilization by the human commensal Bifidobacterium breve UCC2003. Microb. Biotechnol. 3:311–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Prágai Z, et al. 2004. Transcriptional regulation of the phoPR operon in Bacillus subtilis. J. Bacteriol. 186:1182–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qi Y, Kobayashi Y, Hulett FM. 1997. The pst operon of Bacillus subtilis has a phosphate-regulated promoter and is involved in phosphate transport but not in regulation of the Pho regulon. J. Bacteriol. 179:2534–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rao NN, Torriani A. 1990. Molecular aspects of phosphate transport in Escherichia coli. Mol. Microbiol. 4:1083–1090 [DOI] [PubMed] [Google Scholar]
- 39. Rice CD, Pollard JE, Lewis ZT, McCleary WR. 2009. Employment of a promoter-swapping technique shows that PhoU modulates the activity of the PstSCAB2 ABC transporter in Escherichia coli. Appl. Environ. Microbiol. 75:573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Risøen PA, Brurberg MB, Eijsink VG, Nes IF. 2000. Functional analysis of promoters involved in quorum sensing-based regulation of bacteriocin production in Lactobacillus. Mol. Microbiol. 37:619–628 [DOI] [PubMed] [Google Scholar]
- 41. Rutherford K, et al. 2000. Artemis: sequence visualization and annotation. Bioinformatics 10:944–945 [DOI] [PubMed] [Google Scholar]
- 42. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 43. Sánchez B, et al. 2007. Low-pH adaptation and the acid tolerance response of Bifidobacterium longum biotype longum. Appl. Environ. Microbiol. 73:6450–6459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Santos-Beneit F, Rodríguez-GarcíA A, Franco-Domínguez E, Martín JF. 2008. Phosphate-dependent regulation of the low- and high-affinity transport systems in the model actinomycete Streptomyces coelicolor. Microbiology 154:2356–2370 [DOI] [PubMed] [Google Scholar]
- 45. Santos-Beneit F, Rodríguez-GarcíA A, Sola-Landa A, Martín JF. 2009. Cross-talk between two global regulators in Streptomyces: PhoP and AfsR interact in the control of afsS, pstS and phoRP transcription. Mol. Microbiol. 72:53–68 [DOI] [PubMed] [Google Scholar]
- 46. Schaaf S, Bott M. 2007. Target genes and DNA-binding sites of the response regulator PhoR from Corynebacterium glutamicum. J. Bacteriol. 189:5002–5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reference deleted.
- 48. Sola-Landa A, Moura RS, Martín JF. 2003. The two-component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc. Natl. Acad. Sci. U. S. A. 100:6133–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sola-Landa A, Rodríguez-GarcíA A, Franco-Domínguez E, Martín JF. 2005. Binding of PhoP to promoters of phosphate-regulated genes in Streptomyces coelicolor: identification of PHO boxes. Mol. Microbiol. 56:1373–1385 [DOI] [PubMed] [Google Scholar]
- 50. Stanton C, Ross RP, Fitzgerald GF, van Sinderen D. 2005. Fermented functional foods based on probiotics and their biogenic metabolites. Curr. Opin. Biotechnol. 16:198–203 [DOI] [PubMed] [Google Scholar]
- 51. Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183–215 [DOI] [PubMed] [Google Scholar]
- 52. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids. Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Turroni F, Ribbera A, Foroni E, van Sinderen D, Ventura M. 2008. Human gut microbiota and bifidobacteria: from composition to functionality. Antonie Van Leeuwenhoek 94:35–50 [DOI] [PubMed] [Google Scholar]
- 54. van Hijum SAFT, et al. 2005. A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics 6:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. van Sinderen D, et al. 1995. comK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis. Mol. Microbiol. 15:455–462 [DOI] [PubMed] [Google Scholar]
- 56. Ventura M, Canchaya C, Meylan V, Klaenhammer TR, Zink R. 2003. Analysis, characterization, and loci of the tuf genes in Lactobacillus and Bifidobacterium species and their direct application for species identification. Appl. Environ. Microbiol. 69:6908–6922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ventura M, et al. 2005. The ClgR protein regulates transcription of the clpP operon in Bifidobacterium breve UCC 2003. J. Bacteriol. 187:8411–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ventura M, et al. 2006. How high G+C Gram-positive bacteria and in particular bifidobacteria cope with heat stress: protein players and regulators. FEMS Microbiol. Rev. 30:734–759 [DOI] [PubMed] [Google Scholar]
- 59. Ventura M, et al. 2009. The Bifidobacterium dentium Bd1 genome sequence reflects its genetic adaptation to the human oral cavity. PLoS Genet. 5:e1000785 doi:10.1371/journal.pgen.1000785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ventura M, et al. 2009. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat. Rev. Microbiol. 7:61–71 [DOI] [PubMed] [Google Scholar]
- 61. von Krüger WM, Humphreys S, Ketley JM. 1999. A role for the PhoBR regulatory system homologue in the Vibrio cholerae phosphate-limitation response and intestinal colonization. Microbiology 145:2463–2475 [DOI] [PubMed] [Google Scholar]
- 62. Wanner BL. 1996. Phosphorus assimilation and control of the phosphate regulon, p 1357–1381 In Neidhardt FC, Curtiss R., III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE. (ed), Escherichia coli and Salmonella. ASM Press, Washington, DC [Google Scholar]
- 63. Wosten MMSM, et al. 2006. The Campylobacter jejuni PhoS/PhosR operon represents a non-classical phosphate-sensitive two-component system. Mol. Microbiol. 62:278–291 [DOI] [PubMed] [Google Scholar]
- 64. Yuan ZC, Zaheer R, Morton R, Finan TM. 2006. Genome prediction of PhoB regulated promoters in Sinorhizobium meliloti and twelve proteobacteria. Nucleic Acids Res. 34:2686–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.