Abstract
Lactobacillus salivarius strain UCC118 is a human intestinal isolate that has been extensively studied for its potential probiotic effects in human and animal models. The objective of this study was to determine the effect of L. salivarius UCC118 on gene expression responses in the Caco-2 cell line to improve understanding of how the strain might modulate intestinal epithelial cell phenotypes. Exposure of Caco-2 cells to UCC118 led to the induction of several human genes (TNFAIP3, NFKBIA, and BIRC3) that are negative regulators of inflammatory signaling pathways. Induction of chemokines (CCL20, CXCL-1, and CXCL-2) with antimicrobial functions was also observed. Disruption of the UCC118 sortase gene srtA causes reduced bacterial adhesion to epithelial cells. Transcription of three mucin genes was reduced significantly when Caco-2 cells were stimulated with the ΔsrtA derivative of UCC118 compared to cells stimulated with the wild type, but there was no significant change in the transcription levels of the anti-inflammatory genes. UCC118 genes that were significantly upregulated upon exposure to Caco-2 cells were identified by bacterial genome microarray and consisted primarily of two groups of genes connected with purine metabolism and the operon for synthesis of the Abp118 bacteriocin. Following incubation with Caco-2 cells, the bacteriocin synthesis genes were transcribed at higher levels in the wild type than in the ΔsrtA derivative. These data indicate that L. salivarius UCC118 influences epithelial cells both through modulation of the inflammatory response and by modulation of intestinal cell mucin production. Sortase-anchored cell surface proteins of L. salivarius UCC118 have a central role in promoting the interaction between the bacterium and epithelial cells.
INTRODUCTION
The intestinal microbiota of humans is a dynamic community of several thousand phylotypes that changes in composition throughout the life span and also in some disease states (reviewed in reference 33). While the link between the microbiota and conditions such as obesity (23), colitis (15), and irritable bowel syndrome (19) is not yet understood at a precise mechanistic level, some elements in the microbiota have been shown to exert direct anti-inflammatory activity in animal models (41). Elucidating the mechanistic details of the interaction between the innate immune system and the commensal intestinal microbiota is crucial if the molecular basis for the anti-inflammatory effects of the gut microbiota is to be understood. In addition, in order to rigorously validate the efficacy of putative probiotic bacteria, it is necessary to identify biomarkers for probiotic function that may be used to demonstrate that a particular product is effective (31).
The primary site of interaction between the host immune system and the gut microbiota is the intestinal epithelium, which is one cell thick and which forms a physical barrier between the gut contents and the immune cells in the underlying lamina propria (50). The epithelium consists of several different types of differentiated epithelial cells, the most predominant of which is the enterocyte, which forms the structural epithelium in which the other cell types are located. Secreted mucins are a significant factor in restricting access of bacterial cells to the epithelial cells (21). The secreted mucin layer covers the epithelia of the stomach, the small intestine, and the colon (26). One of the main functions of the mucin layer is to limit contact between microbes present in the intestinal lumen and the epithelial cells. Sampling of the intestinal bacteria through the epithelium occurs via specialized epithelial M cells and dendritic cells (12). The main functions of enterocytes are digestion and barrier function, thus maintaining intestinal homeostasis. The tight-junction complexes between neighboring enterocytes are required to maintain this cell-cell barrier function. Increased intestinal permeability, where there are reduced tight-junction complexes, is associated with chronic intestinal inflammation under conditions such as ulcerative colitis (UC) or Crohn's disease (CD) (20), and these conditions are often characterized by the presence of higher numbers of bacteria in association with the epithelium (17, 44). It has been reported that some probiotic bacteria modulate mucin gene expression, which could enhance the barrier function of the mucus layer (6).
The ability to adhere to the intestinal epithelium is critical for probiotic function, and laboratory adhesion assays using cultured epithelial cells are a common means of assessing this putative probiotic functionality of a particular strain (13, 28, 54). Bacterial surface proteins are reported to have a role in the adhesion of probiotic bacteria to the host intestinal cells (4, 47). One class of bacterial surface proteins is those anchored by the sortase enzyme (29). A srtA deletion mutant of Lactobacillus salivarius UCC118 adhered to Caco-2 cells (47) and AGS gastric cells (36) at significantly lower levels than the wild-type strain. The sortase-dependent proteins of UCC118 have previously been studied in our laboratory (47), and they include two mucus binding proteins, LspA (lsl_0311) and LspC (lsl_1335), and a third (lsl_0152) that is encoded by a pseudogene. These gene products, in particular, could contribute to colonization of the intestinal epithelium in vivo through interaction with the epithelial mucus layer.
Recent studies have described the effect of Lactobacillus plantarum on intestinal gene transcription in vivo and reported that a number of NF-κB modulation pathways were induced (46). However, the precise nature of the response depended on the growth phase of the cells, with dead or stationary-phase bacteria being more efficient at eliciting an anti-inflammatory response. Another study compared Lactobacillus acidophilus, Lactobacillus casei, and Lactobacillus rhamnosus using a similar infusion and sampling protocol. Each individual species elicited a distinctive immune response, but the differences between the responses elicited by the same species in different subjects were greater than the variation between the responses to different strains in the same subject (45).
An alternative model uses cultured epithelial cells. For example, exposure of Caco-2 epithelial cells to L. rhamnosus induced NF-κB regulatory pathways (22) and also inhibited IκBα degradation and decreased tumor necrosis factor alpha (TNF-α)-induced interleukin 8 (IL-8) secretion (53). In the intestinal epithelial cell (IEC) line HT-29, several anti-inflammatory effects were identified, including a reduction of TNF-α-induced IL-8 secretion by Lactobacillus bulgaricus (1) and modulation of Toll-like receptor (TLR) expression, together with induction of innate immune system pathways by L. plantarum and L. rhamnosus GG (49). These and other studies indicate that while it is a relatively simplified model (compared to in vivo studies) for studying commensal-epithelium interactions, the use of cultured IECs remains an indispensable method of investigating probiotic-epithelium interaction, particularly where multiple strains are to be screened.
L. salivarius UCC118 was originally isolated from the terminal ileum of a healthy patient. The strain has been extensively studied as a potential probiotic organism, and there have been a number of reports of beneficial effects, including anti-infectivity (9), anti-inflammation (39), amelioration of the effects of induced rheumatoid arthritis (RA) in mice (38), and reduction of Helicobacter pylori-induced IL-8 secretion by AGS gastric cells (37). To investigate the molecular basis for altered host phenotype, we used microarrays to measure differential gene expression in both epithelial and bacterial cells following exposure of L. salivarius UCC118 to Caco-2 epithelial cells.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
L. salivarius UCC118 and its derivative strains were routinely cultured on deMan, Rogosa, and Sharpe (MRS) medium (10) at 37°C in the presence of 5% CO2.
The human colonic carcinoma cell line Caco-2 (ATCC HTB-37) was grown on Dulbecco's modified Eagle's medium (DMEM) (35) supplemented with fetal bovine serum (10%) and nonessential amino acids (1%); penicillin (100U/ml) and streptomycin (100 mg/ml) were added for routine cell culture. The cells were grown at 37°C in the presence of 5% CO2, and when the cells became confluent, the cultures were passaged by the addition of trypsin EDTA, followed by appropriate dilution in fresh DMEM.
Exposure of Caco-2 cells to L. salivarius.
For measurement of Caco-2 cell transcriptional changes, the cells were grown for 12 days in 6-well tissue culture plates to allow full differentiation to occur. The cultures were fed by removing the culture medium by aspiration at 72-h intervals and replacing it with fresh DMEM. At 12 h prior to addition of the bacteria, the medium was aspirated and replaced with antibiotic-free DMEM; 2 h prior to addition of the bacterial cells, the medium was again removed and replaced by fresh antibiotic-free DMEM. Overnight cultures of L. salivarius UCC118 and its derivatives were washed twice and resuspended in phosphate-buffered saline (PBS), and the optical density at 600 nm (OD600) of the suspension was determined. The viable-cell count of the suspension was estimated from a previously prepared standard curve that related optical density to viable-cell numbers, from which it was determined that a cell suspension with an OD600 of 1.0 contained 2 × 108 CFU/ml. Caco-2 cell cultures in 6-well plates were previously determined to contain, on average, 2 × 106 viable cells per well; the amount of bacterial suspension added was adjusted so that a multiplicity of infection (MOI) of 10:1 was achieved. The plates were incubated at 37°C for 4 h in the presence of 5% CO2, after which the growth medium was removed by aspiration and the epithelial cells were lysed by the addition of 1 ml of Trizol. The Trizol lysates were frozen at −70°C until they were required.
For the measurement of bacterial gene transcription, the Caco-2 cells were seeded in T75 tissue culture flasks and grown for 12 days to allow complete differentiation. The medium was changed twice to remove all traces of antibiotics prior to addition of the bacteria. A higher MOI of 100:1 was used to ensure that bacterial cell numbers were high enough to give sufficient yields of RNA, and the cultures were harvested following 90 min of exposure by aspirating the DMEM and replacing it with 2 ml of RNAprotect reagent (Qiagen). The tissue culture monolayer was scraped into suspension using a cell scraper and placed in a microcentrifuge tube in 1-ml aliquots. The tubes were centrifuged, excess RNAprotect was removed, and the pellets were frozen at −80°C until they were required.
RNA extraction.
RNA was extracted from all samples using Trizol reagent. For the Caco-2 cells, RNA was extracted from the TRIzol lysates by adding 0.5 ml of glass beads and 200 μl chloroform to the lysate in a screw-top microcentrifuge tube and homogenizing for 40 s in a Bead-Beater (Bio-Spec). The tubes were then centrifuged at maximum speed in a microcentrifuge tube for 10 min, after which the upper layer was removed and mixed with ethanol to obtain a final concentration of 35% (vol/vol) ethanol, and the tubes were mixed thoroughly. The ethanol-lysate mixture was applied to a purification column from a Totally RNA Isolation Kit (Agilent) and centrifuged to bind the RNA to the column. The manufacturer's protocol was followed for the remainder of the isolation. The eluted RNA was frozen at −80°C until it was required. cDNA was synthesized from the RNA using Moloney murine leukemia virus (MMLV) reverse transcriptase (Fermentas).
To extract RNA from the pelleted bacterial cells in RNAprotect, the pellets were resuspended in 250 μl Tris-EDTA (TE) buffer containing 10 mg/ml lysozyme and 25 units of mutanolysin and incubated for 45 min at 37°C. Following incubation, 750 μl of Trizol LS was added to the lysis buffer together with 0.5 ml of acid-washed glass beads. The mixture was subjected to 3 40-s cycles of mechanical disruption in a bead beater with incubation on ice between every 2 cycles of homogenization. Following homogenization, 200 μl of chloroform was added, and the mixture was shaken, followed by centrifugation at 13,200 rpm for 15 min in a microcentrifuge. The upper phase was removed, and 100% ethanol was added to a final concentration of 35%, followed by thorough mixing. The mixture was added to an RNA purification column from an Agilent/Stratagene Totally RNA isolation kit, and the manufacturer's instructions were followed as described above. cDNA was synthesized using MMLV reverse transcriptase (Fermentas).
Microarray hybridization and analysis.
Labeling of cDNA with Cy3 and Cy5 dyes for use in microarray experiments was done using a chemical labeling kit (Kreatech) following the manufacturer's instructions. The efficiency of labeling was determined using a NanoDrop spectrophotometer. Labeled cDNA was either used immediately or stored at −80°C until it was required. Slides were hybridized for 16 h at 55°C and scanned using an Agilent Microarray Scanner System (G2505B) with Agilent scan control software version 7.0 for the 44K microarray at a resolution of 5 μm. Agilent feature extraction software version 9.1 was used to process the image file from the scanner, and the extracted data were further processed using an in-house microarray transform platform that performed the following manipulations. (i) The replicate values for each gene were combined, and the mean was calculated. (ii) Outliers were identified using the Grubbs test as follows: the Z value, (|mean − x|)/SD, where x is the ratio of a spot and SD is the standard deviation, was calculated; if the Z value was greater than (n − 1)/n2, where n is the number of spots analyzed, then the spot was an outlier. (iii) The P value for each gene was calculated using a cyber t test (2). The parameters for the cyber t test were as follows: a Bayes t test was used with a beta fit of 1, a win size of 101, and a confidence level of 10. The log-transformed mean from the cyber t test was then converted to nonexponential numbers to give a fold value for up- or downregulation. Genes were selected as being significantly changed in expression if their fold change in the Cy3/Cy5 ratio was >2 and where the P value was <0.0001.
The human genome microarrays used for this study were Agilent whole human genome (4×44K) microarrays (catalog number G4112F). The L. salivarius microarrays were Agilent custom microarrays with 3 probes per gene and 7 replicates per probe. All microarray results are the means of a minimum of at least 3 biological replicates.
Quantitative reverse transcription (qRT)-PCR confirmation of changes in gene expression.
The nucleic acid concentration was quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific). cDNA was synthesized using 300 ng of RNA incubated with 3.2 μg of random hexamers, 0.5 μl of Transcript Reverse Transcriptase (Roche), 0.5 μl of Protector RNase inhibitor, 1 mM deoxynucleoside triphosphate (dNTP) mixture, and 4 μl of Transcriptor RT Reaction Buffer in a final volume of 20 μl. Template and random primers were incubated at 65°C for 10 min, and then the other components were added and the mixture was incubated at 55°C for 30 min. Transcript Reverse Transcriptase was inactivated by heating to 85°C for 5 min.
PCR primers and probes were designed using the Universal ProbeLibrary Assay Design Center (Roche). Primers, sequences, and probe combinations are listed in Table 1.
Table 1.
Primers used in this study
| Gene name | Forward primer 5′–3′ | Reverse primer 5′–3′ | Probe no.a |
|---|---|---|---|
| BIRC3 | GATGAAAATGCAGAGTCATCAATTA | CATGATTGCATCTTCTGAATGG | 80 |
| TNFAIP3 | TGCACACTGTGTTTCATCGAG | ACGCTGTGGGACTGACTTTC | 74 |
| CCL20 | GCTGCTTTGATGTCAGTGCT | GAAGAATACGGTCTGTGTATCCAA | 39 |
| MUC3A | CAGTCCCCCAGCCCTAAA | TATACCTTGCTAGGGACCAGGA | 55 |
| MUC12 | CCTGGAAACCTTAGCACCAG | GACAGACGCATTGTTTTCCAT | 72 |
| MUC5AC | AGCACCAGTGCCCAAGTCT | ACTCCTGGCAGTCCATGC | 43 |
| NFKBIA | GCTGATGTCAATGCTCAGGA | ACACCAGGTCAGGATTTTGC | 86 |
| CXCL1 | CATCGAAAAGATGCTGAACAGT | CTTCAGGAACAGCCACCAGT | 52 |
| CXCL-2 | CCCATGGTTAAGAAAATCATCG | CTTCAGGAACAGCCACCAAT | 69 |
| ACTB | ATTGGCAATGAGCGGTTC | TGAAGGTAGTTTCGTGGATGC | 11 |
Probe number refers to the hydrolysis probe from the Roche Universal ProbeLibrary system used with each primer pair.
The β-actin, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and TATA-binding protein (TBP) genes were used as housekeeping genes to correct for variability in the starting total RNA. Amplification reaction mixtures contained 2.5 μl of cDNA, 5 μl of 2× SensiMix II Probe Buffer (Bioline), and 5 pmol/μl (20 nM) of each primer and probe mixture and were brought up to a total of 10 μl by the addition of RNase-free water.
All reactions were performed in duplicate in 384-well plates on the LightCycler480 System (Roche). Positive and negative controls were included in each run. The thermal-cycling conditions applied were those recommended by the manufacturer (Roche). The 2−ΔΔCT method (24) was used to calculate relative changes in gene expression.
Statistical analysis.
Statistical analysis was carried out using GraphPad Prism for Windows (version 5.03; GraphPad Software, San Diego, CA). Differences between two groups were calculated using an unpaired Student's t test. Differences between three or more groups were analyzed by analysis of variance (ANOVA) with Bonferroni's post hoc test. Differences were considered significant at a P value of ≤0.05.
Microarray data accession numbers.
The data for all microarray experiments have been deposited in the EBI ArrayExpress database. The accession numbers for the array data are as follows: E-MEXP-3607 (gene expression in UCC118 following exposure to Caco-2 cells), E-MEXP-3616 (gene expression in the ΔsrtA mutant and UCC118 in response to Caco-2 cells), E-MEXP-3597 (gene expression in Caco-2 cells following exposure to UCC118), and E-MEXP-3614 (gene expression in the Caco-2 cells in response to the ΔsrtA negative mutant).
RESULTS AND DISCUSSION
Exposure to L. salivarius UCC118 alters innate immunity gene expression in Caco-2 cells.
Human genes whose expression levels changed significantly following exposure to bacterial cells are listed in Table 2. Although relatively few genes were differentially expressed, those that passed statistical cutoffs for significance all have putative roles in the regulation of the innate immune system, particularly in the downregulation of the NF-κB-mediated response through the enhanced transcription of inhibitors of NF-κB activity. In that regard, the data are resonant with those derived in vivo by other workers (46), where there was little induction of specific immune system effector genes, but rather, a number of regulatory genes were upregulated. Thus, the effect of L. salivarius on the innate immune phenotype of epithelial cells does not appear to involve the direct activation of a typical inflammatory response but rather the selective induction of an immune-regulatory response.
Table 2.
Genes upregulated in Caco-2 cells in response to UCC118a
| Gene name | Biological function | Fold changeb | P value | qPCR FCc |
|---|---|---|---|---|
| CCL20 | Chemokine (C-C motif) ligand 20 | 38.2 | 2.82E−08 | 20 |
| CXCL1 | Chemokine (C-X-C motif) ligand 1 | 13.6 | 9.64E−07 | 47.22 |
| CXCL2 | Chemokine (C-X-C motif) ligand 2 | 6.4 | 4.29E−06 | 4.5 |
| BIRC3 | cIAP2; antiapoptosis | 6.1 | 2.27E−05 | 6.9 |
| ZC3H12A | RNase; immune regulation | 5.7 | 6.60E−05 | ND |
| TNFAIP3 | Negative regulator of NF-κB | 5.5 | 4.89E−07 | 2.5 |
| NFKBIA | Negative regulator of NF-κB | 5.4 | 1.94E−05 | 3 |
The cutoff for inclusion was a fold change of >2.0 and a P value of <9.0E−05.
Fold change, expression ratio between cells exposed to UCC118 and control cells.
PCR FC, fold change in the qPCR experiment. ND, not done.
The epithelial gene displaying the greatest expression change following L. salivarius exposure was CCL20 (also known as macrophage inflammatory protein 3A [MIP3A]), which was upregulated 38.2-fold as measured by microarray and 20-fold by quantitative PCR (qPCR). The substantial upregulation of this gene by L. salivarius UCC118 accords with previous studies in which UCC118 failed to suppress baseline CCL20 production, or pathogen-induced induction of CCL20, in HT29 epithelial cells (39). Another study indicated that L. salivarius UCC118 increased the secretion of CCL20 in AGS gastric epithelial cells in response to H. pylori (37). It is thus consistent to say, based on data from the current investigation and the two earlier studies cited, that induction of CCL20 is a reproducible response by epithelial cells to stimulation by L. salivarius UCC118. This raises the intriguing possibility that induction of CCL20, which is known to have antimicrobial properties (43, 52), is a probiotic trait of L. salivarius UCC118, in which it triggers a host antimicrobial response. Further supporting this possibility, the array data showed that the gene for the chemokine CXCL1 was also substantially upregulated (13-fold by microarray and 47-fold by qPCR), as was the gene for another chemokine, CXCL2 (6.4- and 4.5-fold in the array and qPCR data, respectively). Both of these chemokines have inherent antimicrobial properties (52) similar to those of CCL20. Furthermore, these chemokines have an important role in neutrophil attraction and have been implicated in the prevention of infection in the lung epithelium (7). Induction of these genes by L. salivarius UCC118 may therefore contribute to a generalized activation of the innate immune response in the epithelium (see below).
The TNFAIP3 gene, which was upregulated 5.5-fold according to microarray measurements following exposure to UCC118, encodes an important immune regulator that has functions in the negative regulation of NF-κB and apoptosis. Deletion of TNFAIP3 in mice leads to a plethora of immunological defects, including increased susceptibility to intestinal inflammation and rheumatoid arthritis (48). UCC118 has been reported to ameliorate the effects of colitis and collagen-induced arthritis in mice (38), so it is noteworthy that TNFAIP3 has a demonstrated role in suppressing rheumatoid arthritis and intestinal inflammation. In addition to the TNFAIP3 gene, two other NF-κB regulatory genes were upregulated, NFKBIA (5.4-fold) and BIRC3 (6.1-fold). NFKBIA is one of the most important negative regulators of NF-κB in that it interacts directly with the NF-κB transcription factor subunits, p65/p50 heterodimers, sequestering them in the cytoplasm and preventing their translocation to the nucleus. The BIRC3 (cIAP2) gene, in contrast, is an NF-κB target gene that is an inhibitor of apoptosis and whose expression in epithelial cells is likely to protect against cytokine-induced apoptosis (51). It has been demonstrated that mice defective in BIRC3 are more susceptible to dextran sodium sulfate (DSS)-induced colitis (3), so the induction of BIRC3 gene expression by L. salivarius UCC118 may be relevant to the previously reported protective effect of this strain against DSS-induced colitis in mice (38).
The ZC3H12A gene was upregulated 5.7-fold in Caco-2 cells stimulated with UCC118. The ZC3H12A gene product, MCP-1-induced protein 1 (MCPIP1), is an important immune regulator essential for the prevention of lethal autoimmune responses in mice (25). It has been clearly demonstrated that, in addition to a macrophage-associated anti-inflammatory function, ZC3H12A also has a substantial inhibitory effect on NF-κB (40). Upregulation of ZC3H12A expression by epithelial cells in response to UCC118 is potentially mechanistically relevant to the signaling involved in the demonstrated anti-inflammatory properties of UCC118 (30, 38).
The combined induction of the NF-κBIA inhibitor, together with TNFAIP3 and ZC3H12A, suggests that, although NF-κaB target genes, such as that encoding BIRC3 (and the chemokines CCL20, CXCL1, and CXCL2), are upregulated by UCC118, the net effect is to tilt the NF-κB regulatory system toward attenuating the NF-κB response. In this respect, benefits ascribed to probiotics may function similarly to amelioration of induced colitis, where a moderate NF-κB response is beneficial in inducing anti-apoptotic genes, such as BIRC3, while a more severe response would be harmful. It is becoming apparent that in terms of intestinal protection, NF-κB activity is in effect a two-edged sword in that a high level of upregulation leads to profound inflammation but a tightly regulated level of activity of NF-κB and of its target genes is critical for proper epithelial function in which the epithelial cells can mount a measured and proportionate response to commensal microorganisms (42).
L. salivarius surface proteins are involved in modulating epithelial mucin gene expression.
We compared the epithelial cell transcriptome elicited by UCC118 with that elicited by an adhesion-impaired mutant (47) in which the gene encoding sortase A, which is the only sortase present in UCC118, had been deleted. The only genes differentially transcribed were those encoding three mucins (Table 3). Surprisingly there was no difference in the expression levels of the NF-κB regulatory genes following stimulation by wild-type UCC118 or the sortase mutant. The MUC3A, MUC5AC, and MUC12 mucin genes were transcribed at lower levels following exposure to the bacterial sortase mutant (Table 3). These MUC genes encode cell surface-associated mucins (11). This was a surprising result, as upregulation of these mucin genes had not been observed following exposure to wild-type UCC118 cells. Downregulation of intestinal mucins has been reported for other gastrointestinal organisms, e.g., H. pylori (5). We can rationalize altered (reduced) mucin gene expression upon exposure to UCC118 cells lacking sortase-dependent proteins if this probiotic lactobacillus normally has both positive and negative regulatory effects on mucin gene expression. Thus, with the wild-type strain, the effect of the sortase-dependent proteins is to upregulate intestinal mucins, but this is counterbalanced by opposing effects from other cell components, analogous to how L. plantarum can have both proinflammatory and anti-inflammatory activities (46). It has been reported that probiotic bacteria increase the synthesis of cell surface mucins (6) and that modulation of mucin gene expression is dependent on the adhesion of the bacterial cells. The VSL3 preparation used for that study was a complex mixture of four Lactobacillus species, three bifidobacterial species, and Streptococcus thermophilus (6). The data presented here provide a single-strain model to further explore this phenomenon and to test the effect of lactobacillus adhesion and mucin gene induction on barrier function.
Table 3.
Genes differentially regulated in Caco-2 cells exposed to UCC118 compared to UCC118 srtAa
The cutoff for inclusion was a fold change of >2.0 and a P value of <9.0E−05.
Fold change, expression ratio between UCC118 and srtA mutant.
PCR FC, fold change in qPCR experiment.
Adhesion to epithelial cells induces L. salivarius bacteriocin gene expression.
L. salivarius genes that displayed significantly altered expression levels upon exposure to Caco-2 cells are listed in Table 4. These genes fall into two main categories, those involved in purine metabolism and those involved in the production of the bacteriocin Abp118. The expression level of purine metabolism genes is known to be affected by numerous environmental factors, and these genes are often differentially expressed in microarray experiments with lactobacilli (27). All the genes within the Abp118 bacteriocin operon were upregulated, ranging between 3.7- and 13.9-fold. Comparison of the gene expression profiles of the UCC118 wild type and srtA mutant upon exposure of both strains to Caco-2 cells (Table 5) revealed that the bacteriocin operon was differentially expressed in the wild type only. The fold difference in Abp118 gene transcription between the wild-type UCC118 and the sortase mutant (Table 5) is in the same range as the fold expression difference between UCC118 cells exposed to Caco-2 cells and nonexposed UCC118 control cells. This suggests that the sortase-mediated adhesion to Caco-2 cells is the primary trigger for increased bacteriocin gene transcription. We have not observed any significant differences between bacteriocin gene expression in the wild type and the ΔsrtA mutant when grown in MRS broth and tested by qPCR (data not shown), supporting the theory that the bacterial cell-epithelial cell interaction mediated by the sortase-anchored proteins is key for the induction of bacteriocin production. Mechanistically, this could be explained by adhesion causing a sufficiently high local concentration of induction peptide to trigger Abp118 production, since this quorum-sensing mechanism is encoded by the Abp118 operon (14).
Table 4.
Genes differentially regulated in UCC118 during coculture with Caco-2 cellsa
| Locus tagb | Gene name | Biological function | Fold changec | P value |
|---|---|---|---|---|
| LSL_0515 | purE | Phosphoribosylaminoimidazole carboxylase carboxyltransferase subunit | 6.8 | 0 |
| LSL_0516 | purK | 6.9 | 0 | |
| LSL_0517 | purB | Adenylosuccinate lyase | 2.3 | 2.23E−08 |
| LSL_0663 | purC | SAICAR synthetase | 6.5 | 6.11E−15 |
| LSL_0664 | purS | Phosphoribosylformylglycinamidine synthase | 7.1 | 2.22E−16 |
| LSL_0665 | purQ | Phosphoribosylformylglycinamidine synthase | 5.3 | 1.47E−12 |
| LSL_0666 | purL | Phosphoribosylformylglycinamidine synthase II | 5.5 | 0 |
| LSL_0667 | purF | Amidophosphoribosyltransferase | 5.6 | 0 |
| LSL_0668 | purM | Phosphoribosylformylglycinamidine cycloligase | 6.7 | 0 |
| LSL_0669 | purN | Phosphoribosylglycinamide formyltransferase | 6.6 | 0 |
| LSL_0670 | purH | Bifunctional purine biosynthesis protein PurH | 7.2 | 0 |
| LSL_0671 | NAd | Hypothetical membrane-spanning protein | 6.6 | 0 |
| LSL_1908 | NA | Hypothetical protein | 2.0 | 0.007 |
| LSL_1909 | abpD | Abp118 bacteriocin export accessory protein | 2.4 | 0.011 |
| LSL_1910 | abpT | Abp118 bacteriocin export accessory protein | 3.1 | 0.002 |
| LSL_1911 | NA | Hypothetical membrane-spanning protein | 3.7 | 6.08E−06 |
| LSL_1912 | abpR | AbpR response regulator | 3.7 | 2.44E−07 |
| LSL_1913 | abpK | Sensory transduction histidine kinase | 2.2 | 0.003 |
| LSL_1914 | abpIP | Abp118 bacteriocin induction peptide | 4.4 | 1.33E−06 |
| LSL_1915 | abpIM | Abp118 bacteriocin immunity protein | 3.7 | 5.16E−08 |
| LSL_1916 | abp118b | Abp118 bacteriocin beta peptide | 6.5 | 1.12E−08 |
| LSL_1917 | Abp118a | Abp118 bacteriocin alpha peptide | 11.4 | 4.60E−10 |
| LSL_1918 | NA | Bacteriocin-like prepeptide | 12.9 | 1.27E−08 |
| LSL_1919 | NA | Hypothetical membrane-associated protein | 1.6 | 1.03E−08 |
| LSL_1920 | NA | Bacteriocin-like prepeptide | 13.9 | 4.10E−08 |
| LSL_1921 | NA | Nonfunctional salivaricin precursor | 11.1 | 8.53E−08 |
| LSL_1922 | NA | Hypothetical membrane-spanning protein | 3.2 | 0.009 |
The cutoff for inclusion was a fold change of >2.0 and a P value of <0.01.
Locus tag, UCC118 genome annotation locus tag.
Fold change, expression ratio between UCC118 cells exposed to Caco-2 cells and unexposed UCC118.
NA, not applicable.
Table 5.
Genes differentially expressed in UCC118 cells exposed to Caco-2 cells relative to ΔsrtA mutant cells exposed to Caco-2 cellsa
| Locus tagb | Gene name | Biological function | Fold changec | P value |
|---|---|---|---|---|
| LSL_1908 | NAd | Hypothetical protein | 1.9 | 0.002 |
| LSL_1909 | abpD | Abp118 bacteriocin export accessory protein | 6.1 | 1.85E−10 |
| LSL_1910 | abpT | Abp118 Bacteriocin export accessory protein | 7.4 | 9.39E−12 |
| LSL_1911 | NA | Hypothetical membrane-spanning protein | 4.7 | 6.08E−06 |
| LSL_1912 | abpR | AbpR response regulator | 4.8 | 1.05E−09 |
| LSL_1913 | abpK | Sensory transduction histidine kinase | 3.7 | 2.33E−09 |
| LSL_1914 | abpIP | Abp118 bacteriocin induction peptide | 4.2 | 1.52E-07 |
| LSL_1915 | abpIM | Abp118 bacteriocin immunity protein | 4.8 | 8.8E−14 |
| LSL_1916 | abp118b | Abp118 bacteriocin beta peptide | 10.8 | 6.11E−15 |
| LSL_1917 | abp118a | Abp118 bacteriocin alpha peptide | 11.7 | 2.18E−09 |
| LSL_1918 | NA | Bacteriocin-like prepeptide | 13.3 | 4.35E−10 |
| LSL_1919 | NA | Hypothetical membrane-associated protein. | 2.0 | 4.9E−11 |
| LSL_1920 | NA | Bacteriocin-like prepeptide | 17.1 | 1.1E−10 |
| LSL_1921 | NA | Nonfunctional salivaricin precursor | 13.6 | 4.09E−10 |
| LSL_1922 | NA | Hypothetical membrane-spanning protein | 7.3 | 1.45E−13 |
The cutoff for inclusion was a fold change of >2.0 and a P value of <0.01.
Locus tag, UCC118 genome annotation locus tag.
Fold change, expression ratio (linear) between UCC118 cells exposed to Caco-2 cells and ΔsrtA mutants exposed to Caco-2 cells.
NA, not applicable.
Localized induction of bacteriocin production by adhesion to mucosal surfaces could be relevant for probiotic effects (reviewed in reference 34), including playing a role in the anti-infective activity shown by UCC118 in models of enteric pathogenesis (9). There is also evidence that L. salivarius strains isolated from different intestinal sources produce different variants of the bacteriocin salivaricin from the same genetic locus, suggesting that bacteriocin production in L. salivarius has evolved in a host-specific manner (32). In addition to the anti-infective effect, it is possible that enhanced bacteriocin production may aid colonization of the bacteriocin producer. This has been demonstrated for a number of bacteria, including Escherichia coli, where colicin producers were more persistent than nonproducers (16), and Streptococcus mutans, where a mutant with enhanced mutacin production was more successful at colonizing the oral tract than the wild-type strain (18). UCC118 is strongly adherent to epithelial cells (47) and is known to be able to persist in the gut for a substantial length of time after ingestion (8). The induction of bacteriocin gene expression reported here is likely to enhance the colonization potential of L. salivarius through inhibition of competing organisms and may well be a factor in the persistence of UCC118 in the gastrointestinal tract (GIT). Bacteriocins may also function as signaling molecules that could directly influence the host immune system; for example, bacteriocin genes have been identified in L. plantarum among a group of genes that are implicated in influencing epithelial gene transcription (27).
Conclusions.
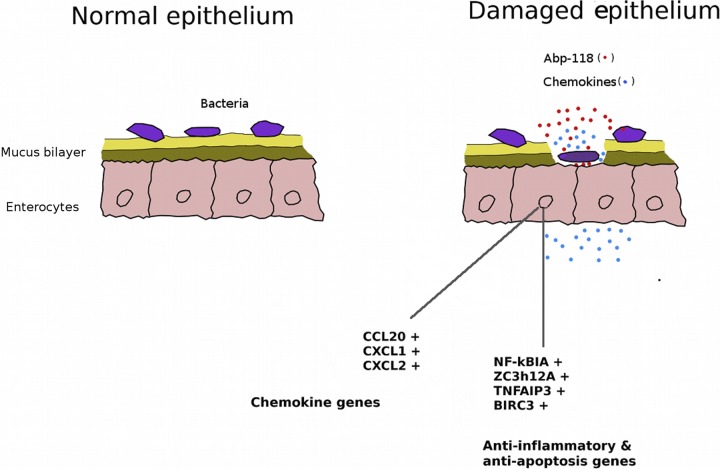
The data accrued in this study have been integrated into a model (Fig. 1) illustrating the relevant proposed mechanisms. It could be argued that L. salivarius cells in the intestine might be restricted from contacting the epithelium, so the transcriptional response observed in cell culture systems would be limited. However, any disruption to the epithelium, including sloughing off mucus by digesta, would allow intestinal bacteria, including lactobacilli, to contact epithelial cells in greater numbers, which would allow induction of the responses detailed here. Enhanced mucus production would be accompanied by sortase-mediated lactobacillus adhesion through the action of the mucus binding proteins LspA and LspC, bacteriocin production, and localized competitive exclusion of bacteriocin-sensitive pathogens (Fig. 1). It is conceivable that commensal bacteria, such as UCC118, with proven colonization and persistence, effectively act as “sentinels,” conditioning the epithelium for possible exposure to pathogens or other inflammatory stimuli. Through their presence in substantial numbers in the mucus layer, they are able to trigger protective immune responses in epithelia following disruption of this layer or the underlying epithelium.
Fig 1.
Schematic summary of the proposed protective mechanism of L. salivarius UCC118 on the intestinal epithelium. In the normal (undamaged) epithelium, the commensal bacteria are prevented from reaching the epithelial cells by the mucus bilayer. In a damaged epithelium, the integrity of the mucus layer is lost, and UCC118 can interact directly with the epithelial cells, with resulting induction of genes for chemokines and anti-inflammatory regulators in the epithelial cells and induction of the genes for Abp118 bacteriocin in the L. salivarius UCC118 cells.
ACKNOWLEDGMENTS
This research was supported by Science Foundation Ireland through a Centre for Science, Engineering and Technology award to the Alimentary Pharmabiotic Centre, UCC (SFI CSET grant 07/CE/B1368).
We are grateful to Alimentary Health Ltd., Ireland, for providing L. salivarius UCC118. We thank Heleen de Weerd for assistance with the microarray data analysis platform and Aldert Zomer and Ian Jeffery for guidance in microarray analysis.
Footnotes
Published ahead of print 18 May 2012
REFERENCES
- 1. Bai A-P, Ouyang Q, Zhang W, Wang C-H, Li S-F. 2004. Probiotics inhibit TNF-alpha-induced interleukin-8 secretion of HT29 cells. World J. Gastroenterol. 10:455–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baldi P, Long AD. 2001. A Bayesian framework for the analysis of microarray expression data: regularized T-test and statistical inferences of gene changes. Bioinformatics 17:509–519 [DOI] [PubMed] [Google Scholar]
- 3. Bertrand MJM, et al. 2009. Cellular inhibitors of apoptosis cIAP1 and cIAP2 are required for innate immunity signaling by the pattern recognition receptors NOD1 and NOD2. Immunity 30:789–801 [DOI] [PubMed] [Google Scholar]
- 4. Buck BL, Altermann E, Svingerud T, Klaenhammer TR. 2005. Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 71:8344–8351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Byrd JC, Yunker CK, Xu QS, Sternberg LR, Bresalier RS. 2000. Inhibition of gastric mucin synthesis by Helicobacter pylori. Gastroenterology 118:1072–1079 [DOI] [PubMed] [Google Scholar]
- 6. Caballero-Franco C, Keller K, De Simone C, Chadee K. 2007. The VSL#3 probiotic formula induces mucin gene expression and secretion in colonic epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 292:G315–G322 [DOI] [PubMed] [Google Scholar]
- 7. Cai S, Batra S, Lira SA, Kolls JK, Jeyaseelan S. 2010. CXCL1 regulates pulmonary host defense to Klebsiella infection via CXCL2, CXCL5, NF-κB, and MAPKs. J. Immunol. 185:6214–6225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Collins JK, et al. 2002. A randomised controlled trial of a probiotic Lactobacillus strain in healthy adults: assessment of its delivery, transit and influence on microbial flora and enteric immunity. Microb. Ecol. Health Dis. 14:81–89 [Google Scholar]
- 9. Corr SC, et al. 2007. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl. Acad. Sci. U. S. A. 104:7617–7621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. De Man JC, Rogosa M, Sharpe ME. 1960. A medium for the cultivation of lactobacilli. J. Appl. Microbiol. 23:130–135 [Google Scholar]
- 11. Dharmani P, Srivastava V, Kissoon-Singh V, Chadee K. 2009. Role of intestinal mucins in innate host defense mechanisms against pathogens. J. Innate Immun. 1:123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Didierlaurent A, Sirard J, Kraehenbuhl J, Neutra MR. 2002. How the gut senses its content. Cell. Microbiol. 4:61–72 [DOI] [PubMed] [Google Scholar]
- 13. Ewaschuk JB, et al. 2008. Secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function. Am. J. Physiol. Gastrointest. Liver Physiol. 295:G1025–G1034 [DOI] [PubMed] [Google Scholar]
- 14. Flynn S, et al. 2002. Characterization of the genetic locus responsible for the production of ABP-118, a novel bacteriocin produced by the probiotic bacterium Lactobacillus salivarius subsp. salivarius UCC118. Microbiology 148:973–984 [DOI] [PubMed] [Google Scholar]
- 15. Frank DN, St Alamand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci., U. S. A. 104:13780–13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gillor O, Giladi I, Riley MA. 2009. Persistence of colicinogenic Escherichia coli in the mouse gastrointestinal tract. BMC Microbiol. 9:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hansson GC. 2012. Role of mucus layers in gut infection and inflammation. Curr. Opin. Microbiol. 15:57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hillman JD, Dzuback AL, Andrews SW. 1987. Colonization of the human oral cavity by a Streptococcus mutans mutant producing increased bacteriocin. J. Dent. Res. 66:1092–1094 [DOI] [PubMed] [Google Scholar]
- 19. Jeffery IB, et al. 2011. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut. doi:10.1136/gutjnl-2011-301501 [DOI] [PubMed] [Google Scholar]
- 20. Kiesslich R, et al. 2011. Local barrier dysfunction identified by confocal laser endomicroscopy predicts relapse in inflammatory bowel disease. Gut. doi:10.1136/gutjnl-2011-300695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim YS, Ho SB. 2010. Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr. Gastroenterol. Rep. 12:319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kumar A, et al. 2007. Commensal bacteria modulate cullin-dependent signaling via generation of reactive oxygen species. EMBO J. 26:4457–4466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444:1022–1023 [DOI] [PubMed] [Google Scholar]
- 24. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 25. Matsushita K, et al. 2009. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature 458:1185–1190 [DOI] [PubMed] [Google Scholar]
- 26. McGuckin MA, Lindén SK, Sutton P, Florin TH. 2011. Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol. 9:265–278 [DOI] [PubMed] [Google Scholar]
- 27. Meijerink M, et al. 2010. Identification of genetic loci in Lactobacillus plantarum that modulate the immune response of dendritic cells using comparative genome hybridization. PLoS One 5:e10632 doi:10.1371/journal.pone.0010632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moussavi M, Adams MC. 2010. An in vitro study on bacterial growth interactions and intestinal epithelial cell adhesion characteristics of probiotic combinations. Curr. Microbiol. 60:327–335 [DOI] [PubMed] [Google Scholar]
- 29. Neville B, O'Toole P. 2010. Probiotic properties of Lactobacillus salivarius and closely related Lactobacillus species. Future Microbiol. 5:759–774 [DOI] [PubMed] [Google Scholar]
- 30. O'Mahony L, et al. 2001. Probiotic impact on microbial flora, inflammation and tumour development in IL-10 knockout mice. Aliment. Pharmacol. Ther. 15:1219–1225 [DOI] [PubMed] [Google Scholar]
- 31. Oozeer R, et al. 2010. Gut health: predictive biomarkers for preventive medicine and development of functional foods. Br. J. Nutr. 103:1539–1544 [DOI] [PubMed] [Google Scholar]
- 32. O'Shea EF, et al. 2011. Production of multiple bacteriocins from a single locus by gastrointestinal strains of Lactobacillus salivarius. J. Bacteriol. 193:6973–6982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. O'Toole PW, Claesson MJ. 2010. Gut microbiota: changes throughout the lifespan from infancy to elderly. Int. Dairy J. 20:281–291 [Google Scholar]
- 34. O'Toole PW, Cooney JC. 2008. Probiotic bacteria influence the composition and function of the intestinal microbiota. Interdiscip. Perspect. Infect. Dis. 2008:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pinto M, et al. 1983. Enterocyte-like differentiation and polarization of the human colon carcinoma cell line Caco-2 in culture. Biol. Cell 47:323–330 [Google Scholar]
- 36. Ryan KA, Daly P, Li Y, Hooton C, O'Toole PW. 2008. Strain-specific inhibition of Helicobacter pylori by Lactobacillus salivarius and other lactobacilli. J. Antimicrob. Chemother. 61:831–834 [DOI] [PubMed] [Google Scholar]
- 37. Ryan KA, O'Hara AM, van Pijkeren J-P, Douillard FP, O'Toole PW. 2009. Lactobacillus salivarius modulates cytokine induction and virulence factor gene expression in Helicobacter pylori. J. Med. Microbiol. 58:996–1005 [DOI] [PubMed] [Google Scholar]
- 38. Sheil B, et al. 2004. Is the mucosal route of administration essential for probiotic function? Subcutaneous administration is associated with attenuation of murine colitis and arthritis. Gut 53:694–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sibartie S, et al. 2009. Modulation of pathogen-induced CCL20 secretion from HT-29 human intestinal epithelial cells by commensal bacteria. BMC Immunol. 10:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Skalniak L, et al. 2009. Regulatory feedback loop between NF-kappaB and MCP-1-induced protein 1 RNase. FEBS J. 276:5892–5905 [DOI] [PubMed] [Google Scholar]
- 41. Sokol H, et al. 2008. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U. S. A. 105:16731–16736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Spehlmann ME, Eckmann L. 2009. Nuclear factor-kappa B in intestinal protection and destruction. Curr. Opin. Gastroenterol. 25:92–99 [DOI] [PubMed] [Google Scholar]
- 43. Starner TD, Barker CK, Jia HP, Kang Y, McCray PB., Jr 2003. CCL20 is an inducible product of human airway epithelia with innate immune properties. Am. J. Respir. Cell Mol. Biol. 29:627–633 [DOI] [PubMed] [Google Scholar]
- 44. Swidsinski A, et al. 2002. Mucosal flora in inflammatory bowel disease. Gastroenterology 122:44–54 [DOI] [PubMed] [Google Scholar]
- 45. van Baarlen P, et al. 2011. Human mucosal in vivo transcriptome responses to three lactobacilli indicate how probiotics may modulate human cellular pathways. Proc. Natl. Acad. Sci. U. S. A. 108:4562–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van Baarlen P, et al. 2009. Differential NF-κB pathways induction by Lactobacillus plantarum in the duodenum of healthy humans correlating with immune tolerance. Proc. Natl. Acad. Sci. U. S. A. 106:2371–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. van Pijkeren J-P, et al. 2006. Comparative and functional analysis of sortase-dependent proteins in the predicted secretome of Lactobacillus salivarius UCC118. Appl. Environ. Microbiol. 72:4143–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vereecke L, Beyaert R, van Loo G. 2009. The ubiquitin-editing enzyme A20 (TNFAIP3) is a central regulator of immunopathology. Trends Immunol. 30:383–391 [DOI] [PubMed] [Google Scholar]
- 49. Vizoso Pinto MG, Rodriguez Gomez M, Seifert S, Watzl B, Holzapfel WH, Franz CM. 2009. Lactobacilli stimulate the innate immune response and modulate the TLR expression of HT29 intestinal epithelial cells in vitro. Int. J. Food Microbiol. 133:86–93 [DOI] [PubMed] [Google Scholar]
- 50. Wells JM, Rossi O, Meijerink M, van Baarlen P. 2011. Epithelial crosstalk at the microbiota-mucosal interface. Proc. Natl. Acad. Sci. U. S. A. 108:4607–4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yan F, et al. 2011. Colon-specific delivery of a probiotic-derived soluble protein ameliorates intestinal inflammation in mice through an EGFR-dependent mechanism. J. Clin. Invest. 121:2242–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang D, et al. 2003. Many chemokines including CCL20/MIP-3α display antimicrobial activity. J. Leukoc. Biol. 74:448–455 [DOI] [PubMed] [Google Scholar]
- 53. Zhang L, Li N, Caicedo R, Neu J. 2005. Alive and dead Lactobacillus rhamnosus GG decrease tumor necrosis factor-alpha-induced interleukin-8 production in Caco-2 cells. J. Nutr. 135:1752–1756 [DOI] [PubMed] [Google Scholar]
- 54. Zoumpopoulou G, Tsakalidou E, Dewulf J, Pot B, Grangette C. 2009. Differential crosstalk between epithelial cells, dendritic cells and bacteria in a co-culture model. Int. J. Food Microbiol. 131:40–51 [DOI] [PubMed] [Google Scholar]