Abstract
Campylobacter jejuni is a major cause of bacterial food-borne enteritis worldwide, and invasion into intestinal epithelial cells is an important virulence mechanism. Recently we reported the identification of hyperinvasive C. jejuni strains and created a number of transposon mutants of one of these strains, some of which exhibited reduced invasion into INT-407 and Caco-2 cells. In one such mutant the transposon had inserted into a homologue of cj1136, which encodes a putative galactosyltransferase according to the annotation of the C. jejuni NCTC11168 genome. In the current study, we investigated the role of cj1136 in C. jejuni virulence, lipooligosaccharide (LOS) biosynthesis, and host colonization by targeted mutagenesis and complementation of the mutation. The cj1136 mutant showed a significant reduction in invasion into human intestinal epithelial cells compared to the wild-type strain 01/51. Invasion levels were partially restored on complementing the mutation. The inactivation of cj1136 resulted in the production of truncated LOS, while biosynthesis of a full-length LOS molecule was restored in the complemented strain. The cj1136 mutant showed an increase in sensitivity to the bile salts sodium taurocholate and sodium deoxycholate and significantly increased sensitivity to polymyxin B compared to the parental strain. Importantly, the ability of the mutant to colonize 1-day-old chicks was also significantly impaired. This study confirms that a putative galactosyltransferase encoded by cj1136 is involved in LOS biosynthesis and is important for C. jejuni virulence, as disruption of this gene and the resultant truncation of LOS affect both colonization in vivo and invasiveness in vitro.
INTRODUCTION
Campylobacter jejuni is a major cause of bacterial food-borne diarrhea worldwide. Campylobacter infections vary from asymptomatic to severe enteritis characterized by inflammatory watery diarrhea. Although C. jejuni enteritis is usually self-limiting, postinfection sequelae, including neuropathies such as Guillain-Barré syndrome and Miller-Fisher syndrome, can be debilitating (4).
Despite the high prevalence, clinical consequences, and economic importance of campylobacteriosis, the mechanisms of C. jejuni pathogenesis are not fully understood. Campylobacter invasion into the epithelial mucosa appears to be an essential part of the process leading to enteritis (28). The analysis of intestinal biopsy samples from patients infected with C. jejuni (50), as well as experiments with infected primates (44, 45) and other experimentally infected model animals (3, 35, 56), together with in vitro experiments using human epithelial cells (7, 28, 29, 36), has clearly demonstrated that C. jejuni can adhere to and invade the cells of the intestinal tract. Invasion studies with intestinal epithelial cells revealed a statistically significant correlation between the ability of C. jejuni to invade cultured cells and the severity of clinical symptoms of infection (10).
Poultry is considered the major source of food-borne campylobacteriosis (5, 37). The mechanisms by which C. jejuni initiates colonization, either in humans or in the avian host, are poorly understood, but a putative correlation between the ability of C. jejuni to invade cultured Caco-2 cells and colonization of chicks was previously proposed (18). Such a correlation between invasion of cell cultures and colonization ability was also found in other investigations (57).
Like many Gram-negative pathogenic bacteria, C. jejuni has surface polysaccharides, including lipooligosaccharide (LOS). LOS consists of a highly variable outer core of nonrepeating oligosaccharides anchored to lipid A (8, 15, 38). Previously, the genes involved in LOS biosynthesis have been shown to confer variable virulence-associated phenotypes to C. jejuni. Inactivation of galE (encoding a UDP-glucose 4-epimerase) or waaF (encoding heptosyltransferase II) results in organisms expressing deeply truncated LOS molecules and with a reduced potential to invade epithelial cells in vitro (13). A recent study showed that both the C. jejuni 81-176 waaF and lgtF mutants produced truncated LOS and exhibited impaired colonization in the guts of BALB/c ByJ mice (34). Furthermore, complete deletion of the LOS biosynthesis locus in C. jejuni NCTC11168 led to attenuated growth and loss of invasion and natural transformation efficiency of the mutant (31). In contrast, mutants with mutations in cgtA, which encodes an outer core N-acetylgalactosaminyltransferase in C. jejuni 81-176, exhibited increased bacterial attachment and invasion of human cells, while in the same study mutation of neuC1, leading to the absence of sialic acid in the outer core, had no effect on invasiveness (17).
To better understand the molecular basis of invasiveness in C. jejuni, we previously identified hyperinvasive strains (11) and generated a random transposon insertion library in the hyperinvasive strain 01/51. One of the mutants identified in a screen for reduced invasion of human intestinal epithelial cells harbored a transposon insertion into a gene homologous to cj1136 of C. jejuni NCTC11168 (19). cj1136 is part of the LOS biosynthesis locus (cj1132c to cj1152c) in C. jejuni NCTC11168 and shares homology with the genes for many bacterial β-1,3-galactosyltransferases (GalTs), but the function of the encoded protein and its importance in C. jejuni pathogenesis, host colonization, and LOS biosynthesis is unknown. In the current study, a cj1136 homologue was characterized by mutagenesis, complementation, and phenotypic analysis to determine the involvement of this gene in LOS biosynthesis, virulence, and chicken colonization in C. jejuni 01/51. This cj1136 homologue in 01/51 is 100% identical at the nucleotide level to cj1136 of NCTC11168 and will therefore be referred to as cj1136 for ease of description throughout this paper.
MATERIALS AND METHODS
Bacterial culture.
C. jejuni strains (Table 1) were routinely grown on Columbia agar supplemented with sheep blood (5%, vol/vol) or Mueller-Hinton (MH) agar for 48 h at 37°C in an anaerobic work station (Don Whitley Scientific, United Kingdom) supplied with 10% CO2, 5% O2, and 85% (vol/vol) N2 or in a gas jar where a CampyGen pack (Oxoid, United Kingdom) sachet was added to generate microaerobic conditions. Escherichia coli (Table 1) was grown on Luria-Bertani agar at 37°C in an aerobic atmosphere. Culture media were also supplemented with antibiotics, including ampicillin (100 μg ml−1), kanamycin (50 μg ml−1), and chloramphenicol (20 μg ml−1), where appropriate.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| C. jejuni | ||
| 01/51 | Hyperinvasive wild-type strain 01/51 | 11 |
| 01/51 Δcj1136 | cj1136 mutation generated in strain 01/51 | This study |
| 01/51 Δcj1136::cj1136 | cj1136 mutation complemented by wild-type copy of the gene in strain 01/51 | This study |
| 81116 Δ(flaA flaB) | Double mutation in flaA and flaB genes in strain 81116 | 52 |
| E. coli TOP10F′ | F′ lacIq Tn10(Tetr) mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| Plasmids | ||
| pGEM-T Easy | Cloning vector encoding resistance to ampicillin | Promega |
| pGEM:1136::kan | pGEM-T Easy vector carrying 1.3-kb fragment of cj1136 interrupted by Kanr cassette | This study |
| pAJ20 | 2.3-kb DNA fragment flanking the intergenic region between cj0652 and cj0653c with additional BamHI restriction site in the intergenic region, cloned into pGEM-T Easy vector | This study |
| pAJ22 | Chloramphenicol resistance cassette cloned into pAJ20 | This study |
| pAJ22-1136 | Wild-type copy of cj1136 with its own promoter cloned into pAJ22 vector | This study |
| pMA24 | Source of kanamycin resistance cassette | 1 |
| pUOA18 | Source of chloramphenicol resistance cassette | 51 |
Generation of C. jejuni 01/51 Δcj1136.
Briefly, the cj1136 open reading frame, including 66 bp of upstream sequence and 90 bp of downstream sequence, was amplified by PCR from C. jejuni 01/51 using primers 1136-F and 1136-R (Table 2) and cloned into pGEM-T Easy (Promega). A BamHI restriction site was introduced into the cloned cj1136 by inverse PCR (IPCR) (55) using primers 1136-INF and 1136-INR. IPCR generated a 4.3-kb PCR product flanked by BamHI restriction sites and containing a 57-bp deletion in cj1136. The Kanr cassette was amplified from pMA24 (1) using Kan-F and Kan-R primers, BamHI digested, and ligated to BamHI-digested cj1136 IPCR product. E. coli TOP10F′ cells were transformed and transformants selected on plates containing kanamycin and ampicillin. After screening, one clone containing the Kanr cassette inserted in the same orientation as cj1136 (named pGEM:1136::kan) was used to mutate C. jejuni 01/51 by electroporation and allelic exchange as described elsewhere (53). The mutation was confirmed by PCR analysis and nucleotide sequencing (data not shown).
Table 2.
Primers used in this study
| Primer | Nucleotide sequence (5′ → 3′)a |
|---|---|
| 1136-F | GCTCGAAATCAATCATCAAATTT |
| 1136-R | CAACTCTTTGGGAAGAAATTCAA |
| 1136-INF | caaggatccGCACTATCACTCCAAGAGACTATG |
| 1136-INR | caaggatccCCATCATCATAAATACAAGCATG |
| Kan-F | ggaggatccGATAAACCCAGCGAACC |
| Kan-R | gcaggatccGACATCTAAATCTAGGTACTAAAACAATTC |
| C1136-F | caaggatccCGCAAAGTAATTTTACAAGGTTCA |
| C1136-R | caaggatcctCAACTCTTTGGGAAGAAATTCAA |
| 652-F | GAACTTACGATAGATATAGAGC |
| 653-F | TTATGATGAAATCATCATGGAGC |
| IN652-R | caaggatccCAAGATTTTTAAACCAATTGC |
| IN653-R | caaggatccAATTATAAGTTTAATACAAGG |
| CML-F | caaggatccGTCGGTATCGTATGGAGCG |
| CML-R | cacagatctCCTAAAGGGTTTTTATCAGTGCG |
BamHI (ggatcc) or BglII (agatct) restriction sites and additional nucleotides at 5′ ends of primers, where needed, are shown in lowercase.
Complementation of C. jejuni 01/51 Δcj1136.
A wild-type copy of cj1136 (under the control of its own promoter) was inserted in the 121-bp intergenic region between cj0652 and cj0653c. These are present in a tail-to-tail orientation in the chromosomes of C. jejuni NCTC11168 and 01/51. Briefly, a 2.3-kb fragment encompassing this region was amplified from C. jejuni 01/51 using 652-F and 653-F primers (Table 2) and cloned into pGEM-T Easy (Promega). A BamHI site was introduced in the intergenic region by IPCR using primers IN652-R and IN653-R and the resulting plasmid, pAJ20, confirmed by nucleotide sequencing. A chloramphenicol resistance (Cmr) gene, amplified from pUOA18 (51) using primers CML-F and CML-R (which harbor BamHI and BglII restriction sites, respectively), was digested using BamHI and BglII and ligated to BamHI-digested pAJ20 to generate pAJ22 (Table 1). cj1136 along with its presumptive promoter was amplified using primers C1136-F and C1136-R, digested with BamHI, and ligated into BamHI-digested pAJ22 to generate pAJ22-1136 (Table 1). This was electroporated into C. jejuni 01/51 Δcj1136 and the insertion of cj1136 into the intergenic region between cj0652 and cj0653c confirmed by PCR and nucleotide sequencing.
In vitro invasion and adhesion assay.
The adhesion and invasion potentials of all strains were determined using in vitro invasion and adhesion assays as described earlier (19). Briefly, Caco-2 cell monolayers were grown in two 24-well cell culture plates. The Caco-2 cell monolayer was covered with bacterial cells at a multiplicity of infection of 100 in complete cell culture medium consisting of Eagle's minimal essential medium with glutamine supplemented with 10% fetal calf serum and 1% nonessential amino acids and incubated at 37°C for 3 h in 5% (vol/vol) CO2. The monolayer was then washed 3 times with sterile phosphate-buffered saline (PBS), monolayers were lysed from one of the plates by adding 1% (vol/vol) Triton X-100, and viable bacterial cell counts were made to determine the adherent and invaded bacterial cell numbers. The monolayers of the other plate were covered with 1 ml of complete cell culture medium supplemented with 250 μg gentamicin ml−1 in each well and incubated for a further 2 h. Following incubation, the monolayers were washed three times with PBS as before and lysed with 1 ml of 1% (vol/vol) Triton X-100 in PBS, and the total number of viable bacteria per well was determined.
Assays were performed in triplicate. The number of adherent bacteria was calculated by subtracting the number of internalized bacteria from the total number counted. Invasion efficiency was expressed as the percentage of the inoculum that survived gentamicin treatment.
Extraction of C. jejuni LOS.
LOS was extracted using a modification of the phenol-water extraction procedure (32, 40). Briefly, confluent bacterial growth from agar plates was washed three times in PBS (Sigma, United Kingdom), and cells were resuspended in sterile distilled water to an optical density at 600 nm (OD600) of 1.0. One milliliter of bacterial suspension was added to an equivalent volume of phenol (preheated to 65°C) (Sigma, United Kingdom) and mixed vigorously. Samples were incubated at 65°C for 10 min, mixed at regular intervals, and, after cooling on ice, centrifuged at 12,000 × g for 3 min. The upper aqueous layer was removed and LOS precipitated by the addition of 0.1 volume of 20% sodium acetate and 5 volumes of 100% ethanol at 4°C overnight. Precipitated LOS was harvested by centrifugation (5,000 × g) for 10 min, and the pellet washed with ice-cold 70% ethanol, dried, and resuspended in 500 μl water.
Analysis of C. jejuni LOS by SDS-PAGE.
LOS extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Samples were mixed with an equivalent volume of 2× sample loading buffer (100 mM Tris-HCl [pH 8.0], 2% β-mercaptoethanol, 4% SDS, 0.02% bromophenol blue, 20% glycerol) and boiled for 10 min before being loaded onto an SDS-polyacrylamide gel consisting of a stacking gel of 4% (vol/vol) acrylamide and a separating gel of 15% (vol/vol) acrylamide. The gel was run in SDS-Tris-glycine buffer (0.025 M Tris base, 0.192 M glycine, and 0.1% SDS, pH 8.3) at a constant 200 V per gel (Bio-Rad Protean II system). LOS was detected using a previously described silver staining method (48).
MS analysis of C. jejuni LOS.
The intact LOS samples were analyzed using electrophoresis-assisted open-tubular liquid chromatography-mass spectrometry (EA-OTLC-MS) as described previously (9). Briefly, 1.0 μl of the LOS sample was injected into a capillary column, and this was followed by washing with 1.0 μl of 100% methanol. A small plug (60 nl) of 1 M ammonium acetate in deionized water was injected to elute the adsorbed intact LOS from the capillary surface. The separation was performed using 30 mM morpholine in deionized water, pH 9.0. A separation voltage of 30 kV, together with a pressure of 50,000 Pa, was applied for the EA-OTLC-MS analysis. The electrospray ionization (ESI) voltage applied on the sprayer was set at −5.2 kV. Data acquisition was performed in the range of m/z 1000 to 1600 at a scan rate of 2 s/spectrum. In the MS2 (enhanced product ion scan [EPI]) experiments, the scan speed was set to 4,000 Da/s, with Q0 trapping.
Resistance of C. jejuni to polymyxin B and bile salt detergents.
Resistance to different concentrations of polymyxin B (range, 0.5 to 10 μg ml−1) was determined by inoculating 10 μl of bacterial suspensions (at an OD550 of 0.1) onto MH agar supplemented with a specific concentration of polymyxin B. Growth was observed at 24, 48, and 72 h after incubation. All tests were carried out in triplicate. Resistance to different concentrations (range, 0 to 30 mg ml−1) of sodium deoxycholate and sodium taurocholate was determined following the protocol described elsewhere (43).
Sensitivity to Triton X-100.
To determine the sensitivity of C. jejuni to Triton X-100 (1%, vol/vol), overnight bacterial growth was harvested from blood agar plates and resuspended in 1 ml of distilled water to OD550 of 0.1. After a zero-hour sample was taken, Triton X-100 was added and the bacterial cell suspension incubated at room temperature for 1 h. Viable cell counts were determined by serial dilution and plating. Assays were carried out in triplicate.
Motility assays.
Motility assays were undertaken using Mueller-Hinton broth supplemented with 0.4% agar as described earlier (19).
Survival under environmental stress conditions.
Overnight bacterial growth was harvested, and a bacterial cell suspension (OD550 = 0.1) was prepared. For aeration stress, survival of strains was tested in Mueller-Hinton broth in a shaking incubator over 5 h. For survival in complete cell culture medium, survival was tested in medium in a static 5% CO2 incubator over 5 h. Survival was determined by viable cell counts.
Chick colonization.
Animal experiments were performed by licensed personnel in accordance with Home Office regulations and met with local ethics committee approval. Chick colonization was determined using a quantitative oral gavage model as previously described (54). Briefly, 4 groups of 10 specific-pathogen-free chicks, hatched from eggs obtained from Charles River SPAFAS Inc., Hanover, Germany, were housed in negative-pressure isolators and given food and water ad libitum. At 1 day old, birds were dosed by oral gavage with C. jejuni wild-type 01/51 or C. jejuni 01/51 Δcj1136 at 3.4 ± 0.9 × 104 and 3.4 ± 0.9 × 106 CFU in 100 μl of 0.1 M PBS, pH 7.2. Doses were prepared by harvesting bacteria grown overnight on blood agar plates into sterile PBS, and the actual doses were determined retrospectively by viable count. At 5 days postchallenge, the chicks were sacrificed by cervical dislocation and the colonization levels were determined by plating out dilutions of cecal contents onto blood agar plates (supplemented with kanamycin where appropriate). Individual chick colonization levels were determined as CFU per gram of cecal contents. The minimal level of detection was 100 CFU g−1 cecal contents. Statistical analysis of the data was carried out with Minitab software using the nonparametric Mann-Whitney (two-tailed) test to assess significance. P values of <0.05 were judged to be statistically significant.
Statistical analysis.
Unless otherwise stated, a paired Student t test was performed for statistical analysis. A P value of <0.05 indicated statistical significance.
RESULTS
Mutation and complementation of cj1136 confirm a role in host cell invasion.
cj1136 was initially identified as being required for optimal invasion in a screen of C. jejuni transposon mutants (19). To confirm this phenotype, we created a new targeted cj1136 mutant by insertional inactivation of the gene in strain 01/51. Phenotypic analysis confirmed that the mutant strain had a significantly reduced (P < 0.001) ability to invade Caco-2 cells compared to the wild type (Fig. 1B). Importantly, 01/51 Δcj1136 did not show a change in adhesion to Caco-2 cells compared to the wild type, confirming that the invasion-impaired phenotype was not a consequence of a reduced ability to adhere (Fig. 1A). Similarly, the cj1136 mutant did not show a growth defect in MH broth or cell culture medium or exhibit reduced motility, increased sensitivity in Triton X-100, or reduced viability under the atmospheric conditions encountered during the course of the invasion assay compared to the wild type (data not shown). To rule out further polar effects and the possibility of secondary mutations, a wild-type copy of cj1136 was introduced into the chromosome of 01/51 Δcj1136. Introduction of cj1136 at an ectopic site restored, albeit not completely, invasion levels (Fig. 1B). Taken together, these results confirm the role of Cj1136 in invasion.
Fig 1.
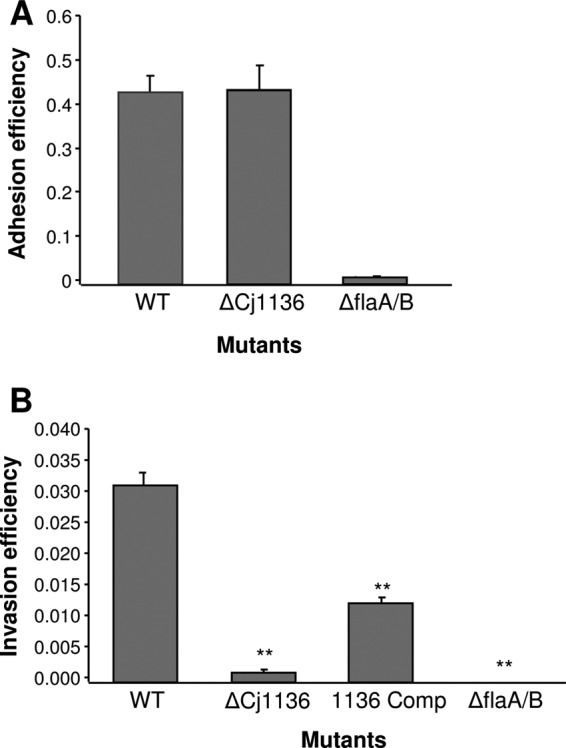
Adhesion and invasion assays using Caco-2 cells confirm the role of cj1136 in host epithelial cell invasion but not adhesion. The adhesion phenotypes of C. jejuni wild-type (WT) 01/51 and C. jejuni 01/51 Δcj1136 (ΔCj1136) (A) and the invasion phenotypes of C. jejuni WT 01/51, C. jejuni 01/51 Δcj1136, and C. jejuni 01/51 Δcj1136::cj1136 (1136 Comp) (B) were determined using Caco-2 cells. A double mutant of C. jejuni 81116 with mutations in flaA and flaB (ΔflaA/B) was used as a negative control in both assays. A paired Student t test was performed to compare the invasion levels of (i) the mutant (ΔCj1136) and negative control (ΔflaA/B) with that of the WT strain and (ii) the complemented mutant (1136 Comp) with that of the mutant (ΔCj1136). **, P < 0.001. As there was no change in adhesion between the wild type and mutant, the complemented mutant was not tested in this assay.
LOS analysis.
Having confirmed the effect on host cell invasion, we purified the LOS produced by the wild-type, Δcj1136, and Δcj1136::cj1136 strains and analyzed them by SDS-PAGE and silver staining as well as mass spectrometry.
(i) Silver staining.
Inactivation of cj1136 resulted in the generation of a truncated LOS molecule exhibiting an increased electrophoretic mobility compared to that of the LOS purified from the wild type in a silver-stained SDS-polyacrylamide gel (Fig. 2). In contrast, the LOS purified from the complemented strain was indistinguishable from that from the wild type using this analysis (Fig. 2). This confirms the putative role of Cj1136 in the biosynthesis of LOS.
Fig 2.
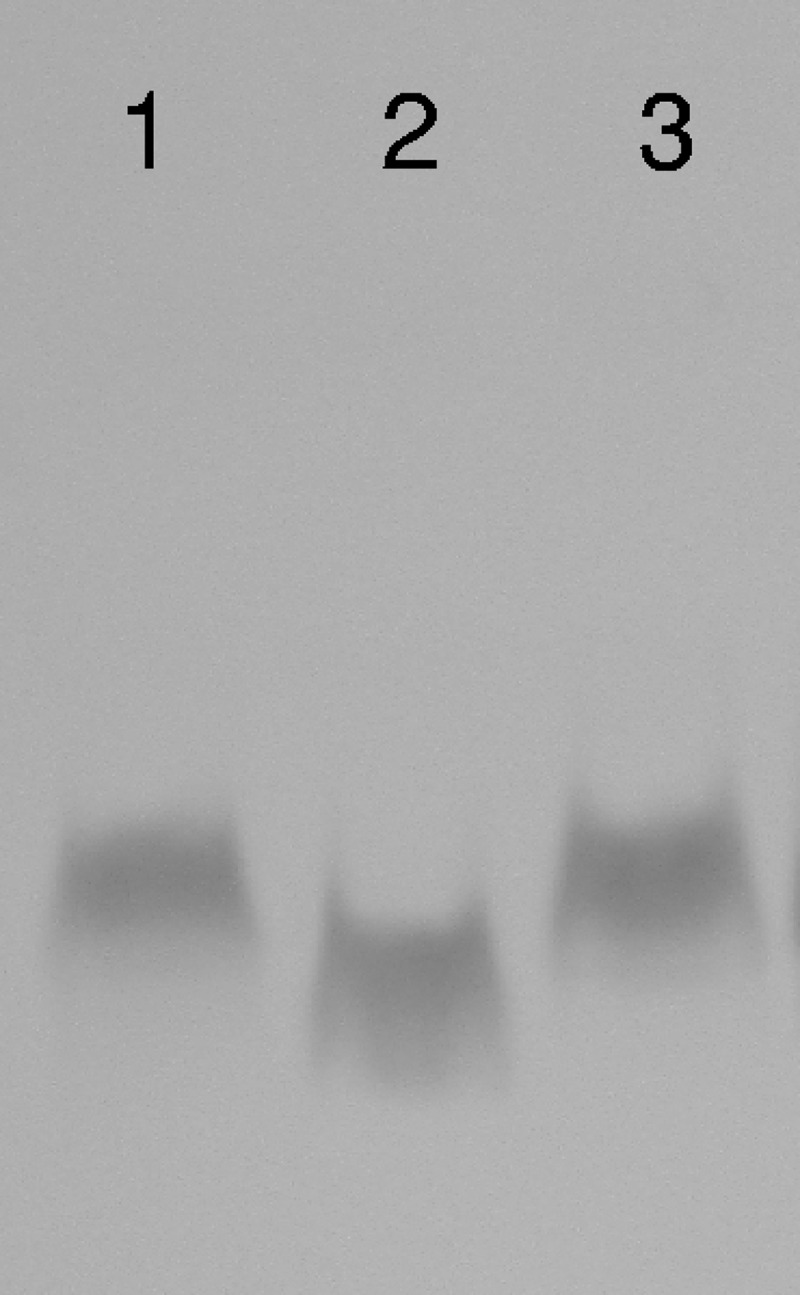
Mutation of C. jejuni 01/51 cj1136 leads to the production of truncated LOS. LOSs extracted from C. jejuni wild-type 01/51 (lane 1), C. jejuni 01/51 Δcj1136 (lane 2), and C. jejuni 01/51 Δcj1136:cj1136 (lane 3) were separated on a 15% SDS-polyacrylamide gel and silver stained. LOS from the cj1136 mutant showed an increased electrophoretic mobility compared to that extracted from the wild-type 01/51 or the complemented mutant.
(ii) MS analysis of C. jejuni LOS.
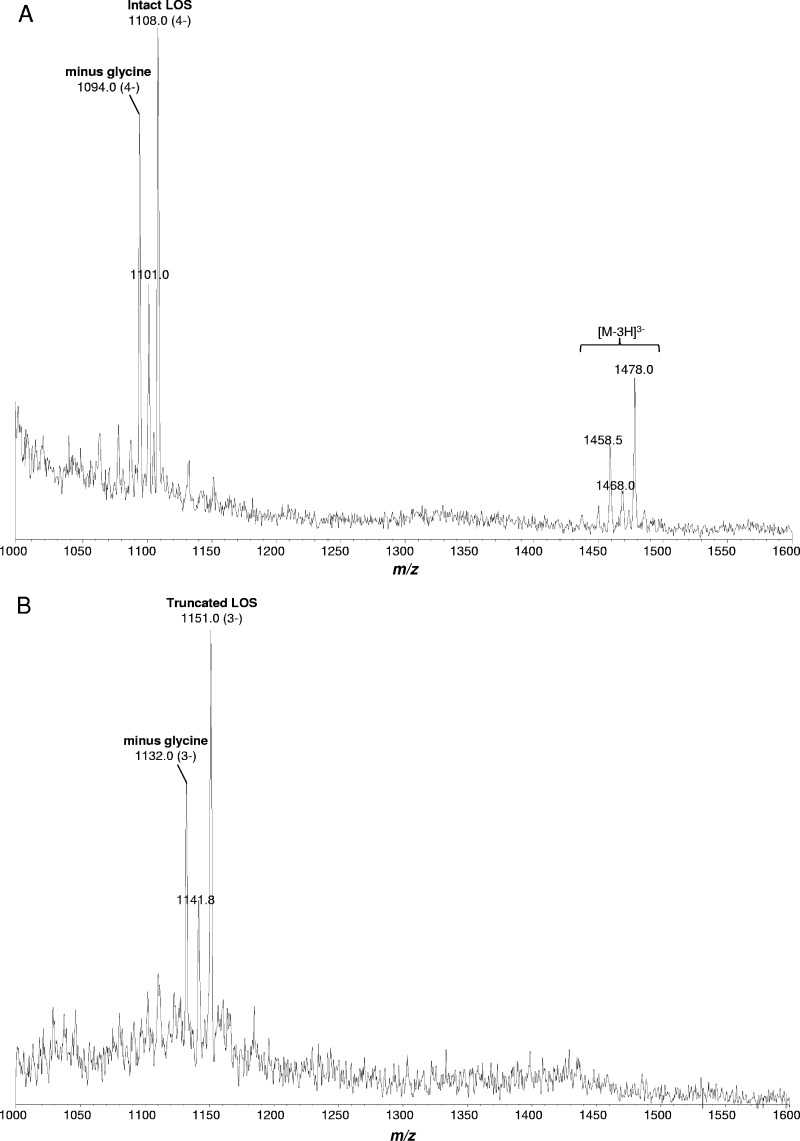
To further characterize the nature of the difference in the LOSs produced by the wild-type strain and the cj1136 mutant, LOS samples from C. jejuni wild-type 01/51, Δcj1136, and Δcj1136::cj1136 strains were analyzed by using EA-OTLC-MS. The LOS sample purified from the wild-type strain showed related triply and quadruply deprotonated ions at m/z 1478 and 1108, respectively, which correspond to a species with a mass of ∼4,436.5 Da, whereas LOS from the Δcj1136 mutant showed a major triply deprotonated ion peak at m/z 1151, which corresponds to a species with a mass of ∼3,456 Da. The MS data showed that the cj1136 mutation resulted in the generation of truncated LOS which is ∼980.5 Da smaller in mass than the wild-type LOS (Fig. 3) and confirmed the role of Cj1136 in LOS biosynthesis.
Fig 3.
Electrophoresis-assisted open-tubular liquid chromatography-mass spectrometry analysis of intact LOSs from C. jejuni wild-type 01/51 (A) and C. jejuni 01/51 Δcj1136 (B). The MS analysis of LOS samples purified from the C. jejuni 01/51 wild-type strain showed major peaks at m/z 1108 and 1478 of quadruply and triply charged ions, respectively, whereas triply charged species were observed at m/z 1151 in LOS isolated from the cj1136 mutant.
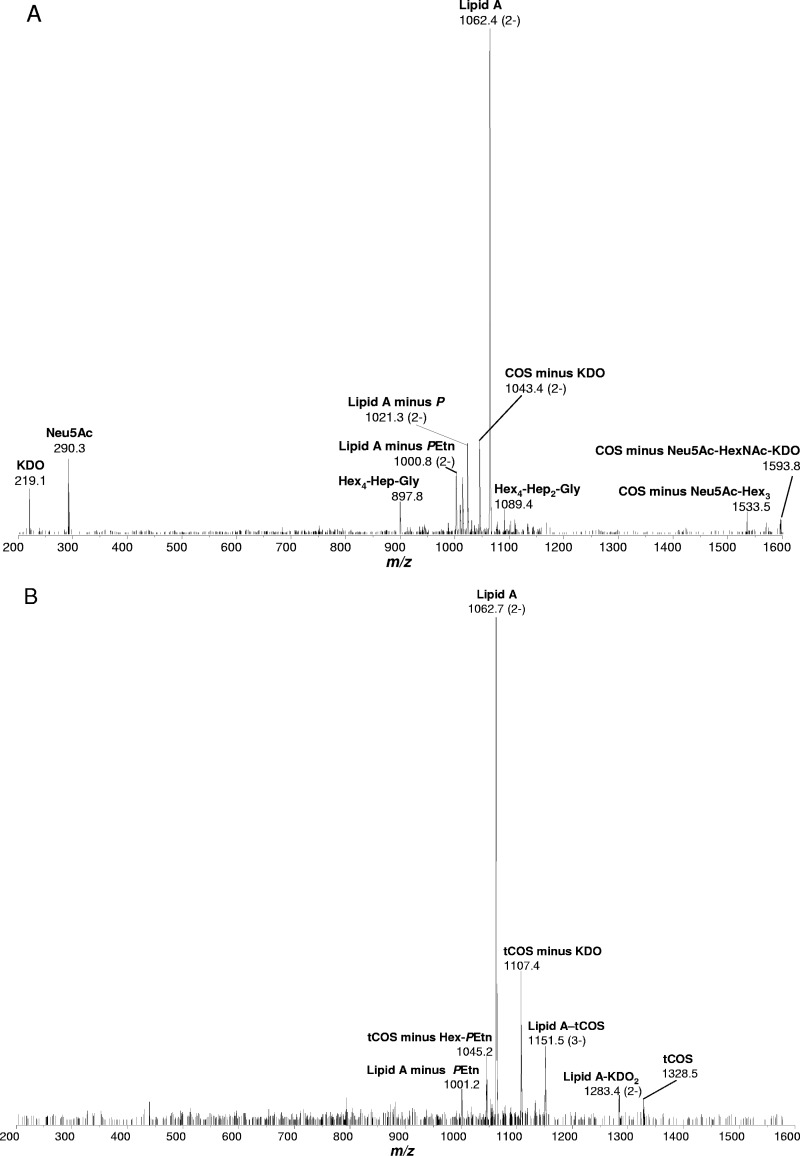
To further characterize the composition of LOS, tandem MS/MS was performed on the intact LOS from C. jejuni wild-type 01/51 (m/z 1108) and 01/51 Δcj1136 (m/z 1151). The LOS samples from both the wild type and the cj1136 mutant showed a major peak at m/z 1062, the mass corresponding to the lipid A component containing a diamino-dideoxyhexose–amino-deoxyhexose (160.2 + 161.2 Da), two palmitate chains (2 × 238.4 Da), four 3-hyroxymyristate chains (4 × 226.4 Da), two phosphoethanolamine (PEtn) residues (2 × 123.05 Da), two phosphate (P) groups (2 × 79.98 Da), and a water molecule (18.0 Da), summing up to a mass of ∼2,127.8 Da. The remaining mass of the core oligosaccharide in wild-type LOS is consistent with glycoforms containing two 2-keto-3-deoxyoctulosonic acid (KDO) residues (2 × 220.2 Da), one glycine (57.05 Da), one PEtn (123.05 Da), two heptose (Hep) residues (2 × 192.17 Da), five hexose (Hex) residues (5 × 162.14 Da), one N-acetylhexosamine (HexNAc) residue (203.19 Da), and one N-acetylneuraminic acid (Neu5Ac) residue (291.26 Da), summing up to a mass of ∼2,309.6 Da. Thus, the mass of LOS from wild-type strain 01/51 corresponds to the composition Neu5Ac1-HexNAc1-Hex5-Gly1-Hep2-PEtn1-KDO2-lipid A. The loss of a mass of ∼980.5 Da in LOS generated by the cj1136 mutant of strain 01/51 corresponds to three Hex residues (3 × 162.14 Da), one HexNAc residue (203.19 Da), and one Neu5Ac residue (291.26 Da). The total mass of truncated LOS produced in C. jejuni 01/51 Δcj1136 corresponds to the composition Hex2-Gly1-Hep2-PEtn1-KDO2-lipid A. The LOS sample purified from the complemented mutant C. jejuni 01/51 Δcj1136::cj1136 showed the same molecular mass as that isolated from the wild-type 01/51strain (not shown). The MS/MS data also showed the loss of monosaccharide hexose (m/z 219) and monosialic acid (Neu5Ac) (m/z 290) in the mutant (Fig. 4), which indicates the LOS truncation in the outer core.
Fig 4.
MS/MS spectra from the intact LOSs from C. jejuni wild-type 01/51 (A) and C. jejuni 01/51 Δcj1136 (B). The intact LOS of C. jejuni wild-type 01/51 ions m/z 1108 and truncated LOS of cj1136 mutant ions m/z 1151 were further analyzed by MS/MS. The major cluster of molecular ions at m/z 1062.4 corresponds to lipid A in the LOSs from both wild-type 01/51 and its cj1136 mutant. Neu5Ac, sialic acid; KDO, 2-keto 3-deoxy-octulosonic acid, P, phosphate; PEtn, phosphoethanolamine; COS, core oligosaccharide; tCOS, truncated core oligosaccharide; Hex, hexose; Hep, heptose; Gly, glycine. The composition of COS corresponds to Neu5Ac1-HexNAc1-Hex5-Gly1-Hep2-PEtn1-KDO2, and that of tCOS corresponds to Hex2-Gly1-Hep2-PEtn1-KDO2.
Resistance to polymyxin B and bile salts.
LOS forms a major part of the C. jejuni outer envelope and protects the bacteria from different antimicrobial peptides (34). We tested the sensitivities of our strains to polymyxin B to determine if production of truncated LOS by the mutant affected the sensitivity of the strain. The cj1136 mutant showed a significant (6-fold) decrease in resistance to polymyxin B compared to the wild type (Table 3). The cj1136 mutant showed growth in medium containing only 1.0 μg polymyxin B ml−1, whereas the wild-type strain 01/51 was able to grow at a concentration of 6 μg polymyxin B ml−1. Complementation of the cj1136 mutation with a wild-type copy of the gene restored resistance to wild-type levels. Furthermore, the cj1136 mutant also showed a decrease in resistance to the bile salts sodium deoxycholate and sodium taurocholate compared to strain 01/51 and the complemented mutant (Table 3).
Table 3.
Sensitivities of C. jejuni 01/51, 01/51 Δcj1136, and the complemented mutant (01/51 Δcj1136::cj1136) to polymyxin B and bile salts
| Compound | Maximum concn (μg ml−1) of compound at which growth was observed |
||
|---|---|---|---|
| Wild-type 01/51 | 01/51 Δcj1136 | 01/51 Δcj1136::cj1136 | |
| Polymyxin B | 6.0 | 1.0 | 6.0 |
| Sodium deoxycholate | 28 | 20 | 28 |
| Sodium taurocholate | 30 | 22 | 30 |
C. jejuni 01/51 Δcj1136 is impaired in its ability to colonize the chick gut.
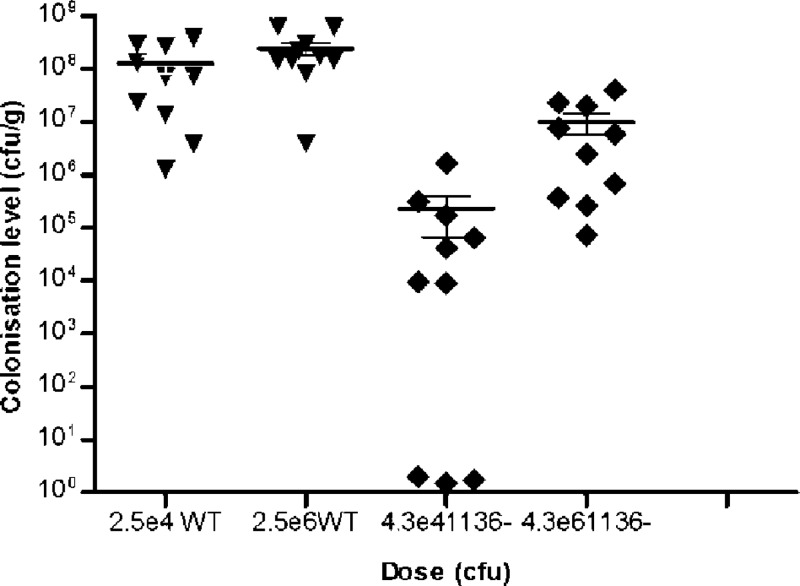
The in vivo significance of the truncated LOS brought about by the disruption of cj1136 was tested using an in vivo model of chick colonization (54). Two groups of birds were dosed orally with the wild-type strain (one group with 104 CFU and one group with 106 CFU) and two with the cj1136 mutant at doses similar to those for the wild type. The wild-type C. jejuni strain colonized 100% of the birds, and the median levels of colonization was 7.2 × 107 and 1.59 × 108 CFU g−1 cecal content for the low and high doses, respectively. The colonization ability was impaired in the cj1136 mutant, which showed a significantly (P < 0.001) lower level of chick colonization than the wild type (Fig. 5). Thirty percent of the birds (n = 3/10) challenged with the lower dose of the cj1136 mutant were not detectably colonized. The median colonization levels reached in the birds challenged with either dose of the mutant were significantly below those observed in birds gavaged with similar doses of the wild-type strain (Fig. 5), suggesting that an intact LOS is required for optimal chick colonization by C. jejuni.
Fig 5.
Mutation of cj1136 results in a reduced ability to colonize chicks. The C. jejuni wild-type (WT) 01/51 and its cj1136 mutant derivative (1136−) were tested in an in vivo model of chick colonization. A low dose and a high dose of each strain were administered to 10 birds in each group. The number of bacteria that had colonized the birds was determined in cecal contents at 5 days after infection. The C. jejuni 01/51 wild-type strain colonized to ∼108 CFU g−1 irrespective of dose; however, both low and high doses of the mutant in cj1136 showed significantly lower levels of chick colonization compared to cognate doses of the wild-type strain, confirming that cj1136 plays a role in chick colonization by C. jejuni. A nonparametric Mann-Whitney (two-tailed) test was used to assess statistical significance of colonization levels of the mutant compared with the wild type at each dose (P < 0.001).
DISCUSSION
Previously, a study conducted in our laboratory using random transposon mutagenesis identified a number of genes involved in invasiveness in C. jejuni (19). A C. jejuni 01/51 homologue of NCTC11168 cj1136 was found to be interrupted by transposon insertion in a mutant with a significantly reduced invasion phenotype in INT-407 and Caco-2 cells (19). Here we characterized the cj1136 gene in detail by targeted mutagenesis, complementation, and phenotypic analysis.
In this study, mutation of cj1136 resulted in reduced invasion in Caco-2 cells compared to the C. jejuni 01/51 wild-type strain (without a change in adhesion), confirming our original observations using the transposon mutant. Partial restoration of the invasion phenotype was observed in the complemented mutant; however, this was not to the level of the wild-type strain, which is most likely due to the introduction of the wild-type gene into a nonnative position in the genome. Even though some promoter sequence was included upstream of the cj1136 gene in the complemented mutant, the ectopic copy of cj1136 may no longer be regulated or expressed in the exact same manner as the wild-type gene. This may not have led to a detectable difference in phenotype when the bacteria were grown on rich Mueller-Hinton agar (for LOS analysis and sensitivity testing) but may have done so in the invasion assays, when the medium utilized was Eagle's minimal essential medium.
Both adherence and invasion are multifactorial and motility-dependent processes (53), yet adherence is not always followed by invasion. No correlation between adhesion and invasion was established in our study, and elsewhere it has been reported that some adherent strains were also deficient in invasion into Caco-2 cells (18). LOS structures are generally important for invasion in many pathogenic bacteria (22, 30, 31, 41, 47). A previous study proposed a link between the presence of cgtB, encoding a putative galactosyltransferase in C. jejuni, and a higher level of invasion into human intestinal epithelial cells and chick colonization potential of the isolates (33). Inactivation of galE in C. jejuni resulted in the production of truncated LOS and reduced invasion into INT-407 cells (13), and mutation of waaC also reduced the invasion potential of C. jejuni 81-176 (24). In contrast, a site-specific insertional mutation of the cgtA gene in C. jejuni strain 81-176, which encodes an N-acetylgalactosaminyltransferase, led to a significant (more than 2-fold) increase in invasion of INT-407 cells compared to that of the wild type (17). These studies suggest that a minimal LOS core structure is required for optimal invasion and that mutations that cause truncation of this minimal structure (mutations in waaC, waaF, galE, and, as now shown, cj1136) lead to impaired invasion. Mutations in the outer core (e.g., cgtA) are more likely to have strain-specific effects, modulating invasion in some but not all strains.
Lipopolysaccharide (LPS) and LOS are important for host colonization in many Gram-negative pathogens. In Salmonella species, a reduction in the ability to colonize chickens correlated with changes in the LPS profile (6, 49). A recent study has shown the importance of LOS for host colonization in C. jejuni using a mouse competition model (34). We have studied colonization in the chicken, which is the natural host and main source of human C. jejuni infection. C. jejuni 01/51 Δcj1136 showed a significant reduction, indicating that cj1136 and an intact LOS are crucial for chick colonization by this organism.
cj1136 is part of the LOS biosynthesis locus in C. jejuni NCTC11168 (39). Based on nucleotide sequence homology to bacterial galactosyltransferase genes, cj1136 was predicted to transfer β-1,3-galactose to heptose II in LOS biosynthesis (15, 26). We determined the compositions of LOS samples purified from the C. jejuni 01/51 wild-type strain as well as from the cj1136 mutant and the complemented mutant by mass spectrometry, and the mutant showed the loss of 1 Neu5Ac, 1 HexNAc, and 3 Hex residues from its LOS outer core. These data suggest that CJ1136 is a hexose transferase; however, further structural and linkage analysis of LOS will help determine the exact hexose residue transferred and linkage formed. Nevertheless, these data provide the first experimental confirmation that cj1136 is involved in LOS biosynthesis in C. jejuni and that inactivation of this gene results in a truncated LOS.
The structure of LOS varies between C. jejuni strains, and this variability can result from difference in gene content, phase variation due to homopolymeric tracts, amino acid substitution, or mutation in glycosyltransferase-encoding genes. cj1136 is not phase variable (15). Interestingly, in this study we found that the LOS of C. jejuni 01/51 has a glycine modification. The glycine modifications of LOSs from other C. jejuni strains have previously been reported (9); however, the role of LOS modification with glycine in C. jejuni pathogenesis is as yet unknown.
LOS plays an important role in maintaining the integrity of the outer membrane. It also provides protection to the organism from antimicrobial peptides and bile salt detergents. Polymyxin B is an antimicrobial peptide that has binding affinity to negatively charged structures, including LPS/LOS. The cj1136 mutant showed a 6-fold reduction in resistance to polymyxin B compared to wild-type strain 01/51. This finding is consistent with other previous studies that also reported a higher sensitivity to polymyxin B in mutants with truncated LOS (20, 31, 34), confirming that an intact LOS is important in the protection of the organism from antimicrobial peptides.
Bile salts, secreted in bile in the animal intestine, also have potential bactericidal activity. They can kill bacterial organisms by destroying the lipid bilayer outer membrane. The cj1136 mutant showed greater sensitivity to the bile salt detergents sodium deoxycholate and sodium taurocholate, which is in line with other studies (23, 31, 46). The increased sensitivity observed in the cj1136 mutant may be explained by a recent hypothesis that the negatively charged cell surface molecules normally buried by LOS are exposed to allow binding and thus disruption of the cell membrane by polymyxin B and bile salt detergents (2). However, the product of cj1136 or LOS may also have secondary roles involved in efflux pumps and/or electrostatic interactions of bacterial surface molecules.
Mutants with mutations in putative glycosyltransferase genes waaF, lgtF, and galT in C. jejuni 81-176 have been reported to have reduced invasion potential compared to the wild-type strain (22, 24), and this reduction in invasiveness was found to be due to increased sensitivity to Triton X-100 used for lysis of the cultured cell monolayer. We tested the survival of the cj1136 mutant in Triton X-100 and found the mutation did not alter sensitivity, confirming that the reduction in invasion levels exhibited by the mutant was not due to killing of the mutant by Triton X-100. Furthermore, the reduction in invasion exhibited by the mutant was not due to a change in growth, motility, or survivability under the environmental conditions encountered during the invasion assays.
A number of other genes as well as cj1136 encode putative galactosyltransferases, including galT in C. jejuni 81-176 (14), and eight genes encode putative galactosyltransferases (wlaH, wlaG, cj1136, cj1138, wlaN [cj1139c], cj1434c, cj1438c, and cj1440c) in C. jejuni NCTC11168 (39). Sequencing of C. jejuni RM1221 resulted in the annotation of two further putative galactosyltransferase genes (cje1278 and cje1280) with no similarity to genes of NCTC11168 (12). Apart from cj1136, those genes encoding putative galactosyltransferases in C. jejuni 01/51 are not known, but if others do exist, they were unable to compensate for the cj1136 mutation in this strain.
Capsular polysaccharide of C. jejuni has previously been shown to play a role in C. jejuni pathogenesis and host colonization (21) and is believed to be the major serodeterminant in the Penner or heat-stable (HS) serotyping scheme (27). Furthermore, in addition to LOS biosynthesis, a putative galactosyltransferase encoded by cj1136 might also be involved in the biosynthesis of other carbohydrate structures, including capsular polysaccharides. Heptosyltransferase I encoded by waaC in C. jejuni 81-176 was found to be involved in biosynthesis of both the LOS and capsular carbohydrates (23), and an epimerase encoded by gne was found to be important for the three major carbohydrates' biosynthetic pathways (LOS, capsule, and N-linked protein glycosylation) (16). Similarly, a single phosphatase enzyme, GmhB, is involved in both LOS and capsule biosynthetic pathways (25). Interestingly, the serotype of the C. jejuni 01/51 wild-type strain is HS4 but the cj1136 mutant's serotype is HS50, which raises the question as to whether the capsule structure is also altered in the cj1136 mutant strain or whether the LOS is the major serodeterminant in strain 01/51. HS4 and HS50 are known to be cross-reacting serotypes, and it has been suggested previously that LOS does play a role in their serospecificity (42), so it is probable that a change in the LOS structure does bring about a change in reactivity with the serotyping antisera; nevertheless, the capsule region of C. jejuni 01/51 is currently being investigated. It is possible that cj1136 may have a more direct role in virulence and host colonization. However, at this stage it appears that the observed reduction in virulence and host colonization in the cj1136 mutant of C. jejuni 01/51 is due to the production of truncated LOS.
This study, for the first time, confirmed the role of cj1136 in LOS biosynthesis, the invasion of human intestinal epithelial cells in vitro, and the in vivo colonization of the chicken gut. It also emphasizes the importance of LOS as a fundamental structure of C. jejuni that affects many different phenotypes of this major human pathogen.
ACKNOWLEDGMENTS
This work was supported by the Nottingham Trent University RAE fund.
We thank Judith Richardson at the Laboratory of Gastrointestinal Pathogens, HPA Colindale, for the serotyping data.
Footnotes
Published ahead of print 16 April 2012
REFERENCES
- 1. Abuoun M. 2007. The heterogeneity of the putative Campylobacter virulence factor: cytolethal distending toxin (CDT). Ph.D. thesis University of Warwick, Warwick, United Kingdom [Google Scholar]
- 2. Amini S, Goodarzi H, Tavazoie S. 2009. Genetic dissection of an exogenously induced biofilm in laboratory and clinical isolates of E. coli. PLoS Pathog. 5:e1000432 doi:10.1371/journal.ppat.1000432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Babakhani FK, Joens LA. 1993. Primary swine intestinal cells as a model for studying Campylobacter jejuni invasiveness. Infect. Immun. 61:2723–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blaser MJ, Engberg J. 2008. Clinical aspects of C. jejuni and C. coli infections, p 99–121 In Nachamkin I, Szymanski CM, Blaser MJ. (ed), Campylobacter. ASM Press, Washington, DC [Google Scholar]
- 5. Butzler JP. 2004. Campylobacter, from obscurity to celebrity. Clin. Microbiol. Infect. 10:868–876 [DOI] [PubMed] [Google Scholar]
- 6. Craven SE, et al. 1993. Characterization of Salmonella california and S. typhimurium strains with reduced ability to colonize the intestinal tract of broiler chicks. Avian Dis. 37:339–348 [PubMed] [Google Scholar]
- 7. De Melo MA, Gabbiani G, Pechere JC. 1989. Cellular events and intracellular survival of Campylobacter jejuni during infection of HEp-2 cells. Infect. Immun. 57:2214–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dorrell N, et al. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dzieciatkowska M, et al. 2007. Mass spectrometric analysis of intact lipooligosaccharide: direct evidence for O-acetylated sialic acids and discovery of O-linked glycine expressed by Campylobacter jejuni. Biochemistry 46:14704–14714 [DOI] [PubMed] [Google Scholar]
- 10. Everest PH, et al. 1992. Differentiated Caco-2 cells as a model for enteric invasion by Campylobacter jejuni and C. coli. J. Med. Microbiol. 37:319–325 [DOI] [PubMed] [Google Scholar]
- 11. Fearnley C, et al. 2008. Identification of hyperinvasive Campylobacter jejuni strains isolated from poultry and human clinical sources. J. Med. Microbiol. 57:570–580 [DOI] [PubMed] [Google Scholar]
- 12. Fouts D, et al. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 3:e15 doi:10.1371/journal.pbio.0030015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fry BN, et al. 2000. The galE gene of Campylobacter jejuni is involved in lipopolysaccharide synthesis and virulence. Infect. Immun. 68:2594–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gilbert M, et al. 2002. The genetic bases for the variation in the lipo-oligosaccharide of the mucosal pathogen, Campylobacter jejuni. Biosynthesis of sialylated ganglioside mimics in the core oligosaccharide. J. Biol. Chem. 277:327–337 [DOI] [PubMed] [Google Scholar]
- 15. Gilbert M, Parker CT, Moran AP. 2008. Campylobacter jejuni lipooligosaccharides: structure and biosynthesis, p 483–504 In Nachamkin I, Szymanski CM, Blaser MJ. (ed), Campylobacter. ASM Press, Washington, DC [Google Scholar]
- 16. Guerry P, Szymanski CM. 2008. Campylobacter sugars sticking out. Trends Microbiol. 16:428–435 [DOI] [PubMed] [Google Scholar]
- 17. Guerry P, et al. 2002. Phase variation of Campylobacter jejuni 81-176 lipooligosaccharide affects ganglioside mimicry and invasiveness in vitro. Infect. Immun. 70:787–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hanel I, Muller J, Muller W, Schulze F. 2004. Correlation between invasion of Caco-2 eukaryotic cells and colonization ability in the chick gut in Campylobacter jejuni. Vet. Microbiol. 101:75–82 [DOI] [PubMed] [Google Scholar]
- 19. Javed MA, et al. 2010. Transposon mutagenesis in a hyper-invasive clinical isolate of Campylobacter jejuni reveals a number of genes with potential roles in invasion. Microbiology 156:1134–1143 [DOI] [PubMed] [Google Scholar]
- 20. Jeon B, Muraoka W, Scupham A, Zhang Q. 2009. Roles of lipooligosaccharide and capsular polysaccharide in antimicrobial resistance and natural transformation of Campylobacter jejuni. J. Antimicrob. Chemother. 63:462–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jones MA, et al. 2004. Adaptation of Campylobacter jejuni NCTC11168 to high-level colonization of the avian gastrointestinal tract. Infect. Immun. 72:3769–3776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kanipes MI, Holder LC, Corcoran AT, Moran AP, Guerry P. 2004. A deep-rough mutant of Campylobacter jejuni 81-176 is noninvasive for intestinal epithelial cells. Infect. Immun. 72:2452–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kanipes MI, Papp-Szabo E, Guerry P, Monteiro MA. 2006. Mutation of waaC, encoding heptosyltransferase I in Campylobacter jejuni 81-176, affects the structure of both lipooligosaccharide and capsular carbohydrate. J. Bacteriol. 188:3273–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kanipes MI, et al. 2008. Genetic analysis of lipooligosaccharide core biosynthesis in Campylobacter jejuni 81-176. J. Bacteriol. 190:1568–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karlyshev AV, et al. 2005. Analysis of Campylobacter jejuni capsular loci reveals multiple mechanisms for the generation of structural diversity and the ability to form complex heptoses. Mol. Microbiol. 55:90–103 [DOI] [PubMed] [Google Scholar]
- 26. Karlyshev AV, Ketley JM, Wren BW. 2005. The Campylobacter jejuni glycome. FEMS Microbiol. Rev. 29:377–390 [DOI] [PubMed] [Google Scholar]
- 27. Karlyshev AV, Linton D, Gregson NA, Lastovica AJ, Wren BW. 2000. Genetic and biochemical evidence of a Campylobacter jejuni capsular polysaccharide that accounts for Penner serotype specificity. Mol. Microbiol. 35:529–541 [DOI] [PubMed] [Google Scholar]
- 28. Ketley JM. 1997. Pathogenesis of enteric infection by Campylobacter. Microbiology 143:5–12 [DOI] [PubMed] [Google Scholar]
- 29. Konkel ME, Joens LA. 1989. Adhesion to and invasion of HEp-2 cells by Campylobacter spp. Infect. Immun. 57:2984–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lambotin M, et al. 2005. Invasion of endothelial cells by Neisseria meningitidis requires cortactin recruitment by a phosphoinositide-3-kinase/Rac1 signalling pathway triggered by the lipo-oligosaccharide. J. Cell Sci. 118:3805–3816 [DOI] [PubMed] [Google Scholar]
- 31. Marsden GL, Li J, Everest PH, Lawson AJ, Ketley JM. 2009. Creation of a large deletion mutant in Campylobacter jejuni reveals the lipooligosaccharide gene cluster is not required for viability. J. Bacteriol. 191:2392–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McNally A, La Ragione RM, Best A, Manning G, Newell DG. 2007. An aflagellate mutant Yersinia enterocolitica biotype 1A strain displays altered invasion of epithelial cells, persistence in macrophages, and cytokine secretion profiles in vitro. Microbiology 153:1339–1349 [DOI] [PubMed] [Google Scholar]
- 33. Muller J, Meyer B, Hanel I, Hotzel H. 2007. Comparison of lipooligosaccharide biosynthesis genes of Campylobacter jejuni strains with varying abilities to colonize the chicken gut and to invade Caco-2 cells. J. Med. Microbiol. 56:1589–1594 [DOI] [PubMed] [Google Scholar]
- 34. Naito M, et al. 2010. Effects of sequential Campylobacter jejuni 81-176 lipooligosaccharide core truncations on biofilm formation, stress survival, and pathogenesis. J. Bacteriol. 192:2182–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Newell DG, Pearson A. 1984. The invasion of epithelial cell lines and the intestinal epithelium of infant mice by Campylobacter jejuni/coli. J. Diarrhoeal Dis. Res. 2:19–26 [PubMed] [Google Scholar]
- 36. Oelschlaeger TA, Guerry P, Kopecko DJ. 1993. Unusual microtuble-dependent endocytosis mechanisms triggered by Campylobacter jejuni and Citrobacter freundii. Proc. Natl. Acad. Sci. U. S. A. 90:6884–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Park SF. 2002. The physiology of Campylobacter species and its relevance to their role as foodborne pathogens. Int. J. Food Microbiol. 74:177–188 [DOI] [PubMed] [Google Scholar]
- 38. Parker CT, Gilbert M, Yuki N, Endtz HP, Mandrell RE. 2008. Characterization of lipooligosaccharide-biosynthetic loci of Campylobacter jejuni reveals new lipooligosaccharide classes: evidence of mosaic organizations. J. Bacteriol. 190:5681–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Parkhill J, et al. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668 [DOI] [PubMed] [Google Scholar]
- 40. Prendergast MM, Kosunen TU, Moran AP. 2001. Development of an immunoassay for rapid detection of ganglioside GM(1) mimicry in Campylobacter jejuni strains. J. Clin. Microbiol. 39:1494–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Preston A, Mandrell RE, Gibson BW, Apicella MA. 1996. The lipooligosaccharides of pathogenic gram-negative bacteria. Crit. Rev. Microbiol. 22:139–180 [DOI] [PubMed] [Google Scholar]
- 42. Preston MA, Penner JL. 1989. Characterization of cross-reacting serotypes of Campylobacter jejuni. Can. J. Microbiol. 35:265–273 [DOI] [PubMed] [Google Scholar]
- 43. Raphael BH, et al. 2005. The Campylobacter jejuni response regulator, CbrR, modulates sodium deoxycholate resistance and chicken colonization. J. Bacteriol. 187:3662–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Russell RG, Blaser MJ, Sarmiento JI, Fox J. 1989. Experimental Campylobacter jejuni infection in Macaca nemestrina. Infect. Immun. 57:1438–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Russell RG, O'Donnoghue M, Blake DC, Jr, Zulty J, DeTolla LJ. 1993. Early colonic damage and invasion of Campylobacter jejuni in experimentally challenged infant Macaca mulatta. J. Infect. Dis. 168:210–215 [DOI] [PubMed] [Google Scholar]
- 46. Steeghs L, et al. 2001. Outer membrane composition of a lipopolysaccharide-deficient Neisseria meningitidis mutant. EMBO J. 20:6937–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Swords WE, et al. 2000. Non-typeable Haemophilus influenzae adhere to and invade human bronchial epithelial cells via an interaction of lipooligosaccharide with the PAF receptor. Mol. Microbiol. 37:13–27 [DOI] [PubMed] [Google Scholar]
- 48. Tsai C-M, Frasch CE. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115–119 [DOI] [PubMed] [Google Scholar]
- 49. Turner AK, Lovell MA, Hulme SD, Zhang-Barber L, Barrow PA. 1998. Identification of Salmonella Typhimurium genes required for colonization of the chicken alimentary tract and for virulence in newly hatched chicks. Infect. Immun. 66:2099–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van Spreeuwel JP, et al. 1985. Campylobacter colitis: histological immunohistochemical and ultrastructural findings. Gut 26:945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang Y, Taylor DE. 1990. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene 94:23–28 [DOI] [PubMed] [Google Scholar]
- 52. Wassenaar TM, Bleumink-Pluym NM, Newell DG, Nuijten PJ, van der Zeijst BA. 1994. Differential flagellin expression in a flaA flaB+ mutant of Campylobacter jejuni. Infect. Immun. 62:3901–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wassenaar TM, Bleumink-Pluym NM, van der Zeijst BA. 1991. Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. EMBO J. 10:2055–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wassenaar TM, van der Zeijst BA, Ayling R, Newell DG. 1993. Colonization of chicks by motility mutants of Campylobacter jejuni demonstrates the importance of flagellin A expression. J. Gen. Microbiol. 139:1171–1175 [DOI] [PubMed] [Google Scholar]
- 55. Wren BW, Henderson J, Ketley JM. 1994. A PCR-based strategy for the rapid construction of defined bacterial deletion mutants. Biotechniques 16:994–996 [PubMed] [Google Scholar]
- 56. Yao R, Burr DH, Guerry P. 1997. CheY-mediated modulation of Campylobacter jejuni virulence. Mol. Microbiol. 23:1021–1031 [DOI] [PubMed] [Google Scholar]
- 57. Ziprin RL, et al. 2001. Role of Campylobacter jejuni potential virulence genes in cecal colonization. Avian Dis. 45:549–557 [PubMed] [Google Scholar]