Abstract
The Staphylococcus aureus global regulator CodY responds to nutrient availability by controlling the expression of target genes. In vitro, CodY represses the transcription of virulence genes, but it is not known if CodY also represses virulence in vivo. The dominant community-associated methicillin-resistant S. aureus (CA-MRSA) clone, USA300, is hypervirulent and has increased transcription of global regulators and virulence genes; these features are reminiscent of a strain defective in CodY. Sequence analysis revealed, however, that the codY genes of USA300 and other sequenced S. aureus isolates are not significantly different from the codY genes in strains known to have active CodY. codY was expressed in USA300, as well as in other pulsotypes assessed. Deletion of codY from a USA300 clinical isolate resulted in modestly increased expression of the global regulators agr and saeRS, as well as the gene encoding the toxin alpha-hemolysin (hla). A substantial increase (>30-fold) in expression of the lukF-PV gene, encoding part of the Panton-Valentine leukocidin (PVL), was observed in the codY mutant. All of these expression differences were reversed by complementation with a functional codY gene. Moreover, purified CodY protein bound upstream of the lukSF-PV operon, indicating that CodY directly represses expression of lukSF-PV. Deletion of codY increased the virulence of USA300 in necrotizing pneumonia and skin infection. Interestingly, deletion of lukSF-PV from the codY mutant did not attenuate virulence, indicating that the hypervirulence of the codY mutant was not explained by overexpression of PVL. These results demonstrate that CodY is active in USA300 and that CodY-mediated repression restrains the virulence of USA300.
INTRODUCTION
Asuccessful pathogen must respond to environmental stimuli by coordinately controlling expression of virulence and metabolic networks. Among microbial pathogens, Staphylococcus aureus is remarkably adapted to mammalian parasitism (15, 27). Most often, S. aureus is a commensal and colonizes without causing disease (9). However, S. aureus also can cause a wide range of infectious syndromes, ranging from uncomplicated skin and soft tissue infections to invasive diseases, such as necrotizing pneumonia (9). The microbial and host factors that determine whether colonization or symptomatic infection occurs are not known.
S. aureus regulates gene expression in response to environmental cues, such as availability of vital nutrients (31, 32). CodY, a highly conserved regulatory protein among firmicutes, has emerged in S. aureus as an important link between the nutrient supply, primarily the availability of the CodY effectors (the branched-chain amino acids [BCAAs] and GTP), and gene expression (21, 22, 30). Under nutrient-rich conditions, CodY binds DNA and regulates the expression of its target genes, in most cases acting as a negative regulator (21, 22, 30). Thus, during growth of S. aureus in rich medium, CodY target genes are repressed, but when cells reach stationary phase, BCAA and GTP concentrations become limiting, resulting in the inactivation of CodY DNA binding activity and derepression of its target genes (21, 30).
The effects of CodY on gene expression in S. aureus have been well studied in vitro. Depending on the strain studied, the S. aureus CodY regulon comprises 150 to 200 genes (21, 30). The majority of these genes encode proteins that control metabolic pathways, but CodY also represses many S. aureus virulence genes during in vitro growth (21, 22, 30). Although the mechanism is not fully understood, CodY regulates the expression of some virulence genes through direct DNA binding and regulates others indirectly. For instance, many virulence genes are controlled by RNAIII, a product of the accessory gene regulator (agr) locus, which also encodes the major quorum-sensing system in S. aureus (18). CodY represses agr transcription under nutrient-rich conditions (21, 22, 30).
CodY-mediated repression of virulence genes when nutrient supplies are plentiful would seem to be consistent with commensal growth and asymptomatic colonization. However, when key nutrients become limiting, the reduction in CodY activity leads to increased synthesis and secretion of toxins such as α-hemolysin (encoded by hla) (21, 22, 30). Theoretically, secretion of these tissue-destructive toxins would release available nutrients from host cells or allow escape of bacteria to new, nutrient-rich environments.
The transition from exponential to stationary phase in vitro is tightly regulated. This transition is controlled, at least in part, by agr. The agr locus is transcribed as two divergent transcripts, called RNAII and RNAIII (18). RNAII is the polycistronic agrBDCA transcript; agrC and agrA encode a two-component sensor and response regulator, respectively, whereas agrB and agrD encode an autoinducing peptide synthesis and secretion system that leads to activation of AgrA via phosphorylation by AgrC. RNAIII contains an mRNA for δ-hemolysin and is itself a regulatory factor controlling the expression of a large number of virulence genes (28). The sae (S. aureus accessory element) operon is also important in control of virulence; it encodes a four-component regulatory system (1, 13). SaeR and SaeS comprise a two-component signal transduction system; the functions of the upstream genes saeP and saeQ are not known. When expression of agr and sae increases, the expression of stationary-phase genes (primarily encoding surface-associated proteins) is decreased and the expression of several important virulence genes, including hla and lukSF-PV (encoding Panton-Valentine leukocidin [PVL]), is increased (4, 11, 25, 33, 36).
Therefore, the transition from exponential to postexponential growth in vitro, presumably triggered by nutrient depletion and increased bacterial population density, is advanced by agr and sae upregulation and is associated with reduced repression by CodY. The roles of agr, sae, and hla in virulence have been well characterized, whereas the role of lukSF-PV remains controversial (5–7, 10, 19, 26, 35, 37). However, although the in vitro effects of codY deletion have been documented (21, 22, 30), it is not clear if these effects correlate with in vivo virulence in experimental models of S. aureus infection.
To evaluate the role of CodY in virulence, we selected a highly virulent clinical isolate from the USA300 background, the predominant background causing community-associated methicillin-resistant S. aureus (CA-MRSA) disease in the United States (9). Of note, the hypervirulent phenotype of USA300 is characterized by increased in vitro expression of agr, sae, hla, and lukSF-PV (23, 25), raising the possibility that the strain produces an inactive CodY protein. We therefore constructed a codY deletion mutant in USA300 in order to assess the effects of CodY on virulence in vitro and in clinically relevant murine models of CA-MRSA necrotizing pneumonia and skin infection. Our results demonstrate that CodY is active in the USA300 strain 923 and contributes to repression of the regulatory genes agr and sae and of the virulence genes hla and lukF-PV. Moreover, CodY proved to repress the virulence of USA300 in two animal models of disease. These data represent experimental evidence for an in vivo virulence repression role for CodY.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Table 1 lists the strains used in this study. Strains NRS123 (MW2; USA400), NRS382 (USA100), NRS383 (USA200), NRS385 (USA500), and NRS387 (USA800) were obtained from the Network on Antimicrobial Resistance in S. aureus (http://www.narsa.net). Strain LACΔpvl was a gift from Michael Otto (National Institutes of Health, Bethesda, MD). Strain 923 is a highly virulent USA300 CA-MRSA strain isolated at the University of Chicago Medical Center from a patient with an abscess (3, 23). Its virulence phenotype in rodent models of necrotizing pneumonia and skin infection has been described (23–25).
Table 1.
Strains used in this study
| Strain name | Pulsotype | Description | Reference |
|---|---|---|---|
| NRS382 | USA100 | NARSAa | |
| NRS383 | USA200 | NARSA | |
| 923 | USA300 | Clinical MRSA strain isolated from an abscess | 3, 23, 25 |
| NRS123 (MW2) | USA400 | 23, 37 | |
| NRS385 | USA500 | NARSA | |
| NRS387 | USA800 | NARSA | |
| UAMS1ΔcodY | USA200 | Isogenic codY deletion mutant in UAMS-1 | 22 |
| LACΔpvl | USA300 | Isogenic lukSF-PV mutant in LAC | 37 |
| 923ΔcodY | USA300 | Isogenic codY deletion mutant in 923 | This study |
| 923ΔcodY Δpvl | USA300 | Isogenic codY lukSF-PV deletion mutant in 923 | This study |
| 923ΔcodY + codY | USA300 | codY mutant complemented with pTL6936 | This study |
NARSA, Network on Antimicrobial Resistance in S. aureus.
Strains UAMS-1ΔcodY and LACΔpvl, previously described, were constructed by allelic exchange with replacement of the codY gene by an erythromycin resistance gene (ermC) and replacement of the lukSF-PV genes by a spectinomycin resistance gene (spc), respectively (22, 37). To generate a USA300 codY deletion mutant, the codY::ermC deletion-insertion was transduced from strain UAMS-1ΔcodY using bacteriophage ϕ11 into strain 923, using standard methods, creating 923ΔcodY. Transductants were selected on tryptic soy agar (TSA) supplemented with erythromycin (10 μg/ml). To generate a codY-PVL double-deletion mutant, the lukSF-PV::spc deletion-insertion was transduced from strain LACΔpvl into strain 923ΔcodY, creating 923ΔcodYΔpvl, with selection on TSA supplemented with spectinomycin (100 μg/ml). Deletion of codY and/or lukSF-PV was confirmed by PCR amplification and DNA sequencing, as well as by expression analysis (described below).
The plasmid pTL6936 is a derivative of the expression shuttle vector pCL15, modified by cloning the codY gene and ribosomal binding site under the control of the PSPAC promoter, as described previously (22). The plasmid was introduced by electroporation into 923ΔcodY, using S. aureus strain RN4220 as an intermediate host, creating 923ΔcodY + codY; the complemented strain was grown in the presence of chloramphenicol (10 μg/ml). For expression studies, IPTG (isopropyl-β-d-thiogalactopyranoside) was supplemented at 1 mM. Although the codY gene in pTL6936 contains one altered residue (E to K at position 67), the mutation does not affect CodY activity (22).
For all experiments, strains were subcultured from frozen skim milk stocks onto TSA plates and incubated overnight at 37°C. The following afternoon, one colony was subcultured in 5 ml tryptic soy broth (TSB) in a 50-ml Falcon tube and incubated overnight with shaking (250 rpm), at 37°C. The following morning, the overnight culture was diluted 1:100 in fresh TSB (flask/volume ratio, 7:1) and grown to the desired phase of growth (37°C; 250 rpm), as assessed by the optical density at 600 nm (OD600) and plating of serial dilutions on TSA to enumerate colonies.
In silico comparison of codY among sequenced S. aureus genomes.
The protein predicted to be encoded by codY from strain USA300_TCH1516 (accession number YP_001575080) was downloaded from NCBI (http://www.ncbi.nlm.nih.gov) (14). A BLAST (Basic Alignment Search Tool) search was performed for matches in sequenced S. aureus genomes (http://www.blast.ncbi.nlm.nih.gov).
Expression analysis by quantitative reverse transcription-PCR (qRT-PCR).
Our methods for qRT-PCR have been reported (25). Briefly, bacteria were grown in TSB to selected time points and collected by centrifugation (4,000 × g; 10 min); the cell pellets were immediately frozen at −80°C. To correct for different growth rates among isolates, all strains were grown to the same OD600. For RNA isolation and purification, the pellets were thawed on ice, resuspended in Tris-EDTA (TE), and lysed by incubation with lysostaphin (200 μg/ml) at room temperature for 10 min. Buffer RLT was added, and RNA purification was performed using the RNeasy kit with on-column DNase treatment, following the manufacturer's instructions (Qiagen). RNA quality and quantity were assessed using an Agilent 2100 Bioanalyzer at the University of Chicago Functional Genomics Facility.
For each sample, 2 μg RNA was reverse transcribed using the High Capacity Archive cDNA kit (Applied Biosystems). qRT-PCR was performed using primers and molecular beacons (Invitrogen) for lukF-PV, luminescense upon extension (LUX) (Invitrogen) primers for hla, and Prime Time qPCR primer-probe mixtures (Integrated DNA Technologies) for codY, saeR, and RNAIII. 16S rRNA was used as an endogenous control housekeeping gene for lukF-PV and hla, and gyrB was used as an endogenous control for saeR and RNAIII. With the exception of codY, the primer and probe sequences have been described (25); the sequences of the codY primers were 5′-ATT AAT AGG CCT TCC GTA CCG CCA-3′ and 5′-GGT ACT AGG TGA ATA TGC TGC TAC AGT-3′, and that of the probe was 5′-/56-6-carboxyfluorescein (FAM)/AAA GAA GCG CGC GAT AAA GCT GCT/Iowa Black FQ (IABKFQ)/-3′. Relative quantification was calculated by the ΔΔCT method, with expression of each gene in strain 923 at 2 h (OD600 = 0.7) as the reference.
Analysis of protein abundance by Western blotting.
Bacteria were grown in TSB as described above to selected time points, at which samples were removed and cells were collected by centrifugation (4,000 × g; 10 min). The supernatants were removed and immediately frozen at −80°C. For Western blotting, the supernatants were thawed on ice, and proteins were separated by SDS-PAGE. The proteins were transferred to a nitrocellulose membrane and blocked sequentially with 3% skim milk and mouse anti-protein A antibody (1:5,000; Sigma). Membranes were washed and incubated with rabbit anti-Hla antibody (1:5,000; Sigma) or rabbit anti-LukF-PV antibody (1:500; a gift from Muzaffar Hussain). The membranes were then washed and incubated with anti-rabbit IgG conjugated to horseradish peroxidase (1:5,000; Sigma). They were washed again, and a chemiluminescent substrate was applied (Pierce). The membranes were visualized after exposure to film (Kodak). Densitometry was performed by using Molecular Imaging software (version 4.0; Kodak). The intensity of each band was compared with strain 923.
Gel mobility shift assay.
To generate a 32P-end-labeled probe, the lukSF-PV promoter region was PCR amplified from strain 923 genomic DNA using 32P-labeled primer oAR149 (5′-GTG CAG TTT GAT AAT TGT ATA TGA TG-3′) and unlabeled primer oAR150 (5′-CAG CGC CAT CAC CAA TAT TCT C-3′). The 32P-labeled primer was 5′-end labeled with radioactive [γ-32P]ATP (PerkinElmer) using T4 polynucleotide kinase (New England BioLabs) according to the manufacturer's recommendations and then purified with a QIAquick Nucleotide Removal Kit (Qiagen). Unlabeled DNA fragments were amplified using either primers oAR149 and oAR150 (for specific competition) or primers oAR172 (GGG GGG TCA CAC AAA ATA TTC) and oAR173 (GGG GGT AAT TCA TTG TCT GG) (for nonspecific competition). CodY with a six-histidine tag at the C terminus was purified as described previously (21) and mixed at various concentrations (25 to 200 nM) with the 32P-end-labeled promoter DNA (3,000 cpm; 0.1 ng per μl) in binding buffer (20 mM Tris, pH 8.0, 50 mM KCl, 2 mM MgCl2, 5% glycerol, 0.5 mM EDTA, 1 mM dithiothreitol [DTT], 0.05% Nonidet P-40, 25 μg/ml salmon sperm DNA supplemented with 10 mM [each] isoleucine, leucine, and valine and 2 mM GTP). For the competition experiment, the end-labeled DNA fragment was mixed with purified CodY-His6 at 100 nM and with increasing concentrations of either specific or nonspecific competitor DNA fragments (unlabeled). After 20 min of incubation at room temperature, the reaction mixtures were loaded on an 8% nondenaturing polyacrylamide gel containing 10 mM Tris, pH 8.0, 75 mM glycine, 0.2 mM EDTA, and 10 mM (each) isoleucine, leucine, and valine and subjected to electrophoresis for 2 h at room temperature. The electrophoresis buffer contained 35 mM HEPES-43 mM imidazole. Following electrophoresis, the gel was vacuum dried, exposed to a phosphorimager screen, and analyzed using an Applied Biosystems PhosphorImager and Image Quant software (GE Healthcare).
Mouse model of S. aureus necrotizing pneumonia.
All animal experiments were approved by the Institutional Animal Care and Use Committee at the University of Chicago. Our mouse model of S. aureus pneumonia has been described previously (25). Briefly, 6-week-old C57BL/6 mice (Jackson) were housed for 1 week prior to inoculation. On the day of inoculation, bacteria were grown to mid-exponential phase as described above, washed, and resuspended in sterile phosphate-buffered saline (PBS) to achieve a concentration of 1.5 × 108 CFU/20 μl (confirmed by plating serial dilutions). The mice were sedated with intraperitoneal ketamine and xylazine. The sedated mice were inoculated intranasally with 20 μl of the S. aureus suspension (10 μl in each nostril) and held upright for 15 s to allow full aspiration. The mice were then returned to their cages and allowed full access to food and water. The animals were observed at defined intervals and graded according to a predetermined illness severity score developed in collaboration with the Animal Resources Center at the University of Chicago (23).
Mouse model of S. aureus skin infection.
Our mouse model of S. aureus skin infection has been described previously (24, 25). Briefly, bacteria were grown as described above and resuspended to achieve a concentration of 1.5 × 107 CFU/50 μl. Six-week-old C57BL/6 mice were sedated with intraperitoneal ketamine and xylazine. After the backs of the mice were shaved and cleaned, each mouse was inoculated by subcutaneous injection of 50 μl of the S. aureus suspension using a 27-gauge needle. Following inoculation, the animals were returned to their cages, and skin lesions were observed daily. Photographs of the lesions were taken daily, and the area of dermonecrosis was calculated using Photoshop (Adobe), with a millimeter ruler as a reference.
Data analysis.
Relative quantitations of gene expression by qRT-PCR and skin lesion sizes in the dermonecrosis model were compared using one-way analysis of variance (ANOVA) with the Newman-Keuls posttest. Mortality rates in the pneumonia model were compared using Fisher's exact test. Differences were considered significant if the P value was <0.05. Data analysis was performed using GraphPad Prism.
RESULTS
Conservation of CodY among sequenced S. aureus isolates.
To assess the likelihood that the increased virulence of USA300 is caused by a decrease in CodY activity, we analyzed the codY coding sequence in silico in USA300 and other sequenced isolates. We found that the sequence of codY is highly conserved in more than 120 S. aureus isolates. The predicted sequence of the CodY protein obtained from strains UAMS-1 and Newman (22, 30) was identical to that in USA300 and in all the other analyzed isolates, with four exceptions. Three of the four strains with differences had a single conservative amino acid change; strain JKD6159 (multilocus sequence type [ST] 93) had I→V at position 107, strain A9635 (ST 45) had R→K at position 103, and strain A9765 (ST 8) had L→I at position 123. Given their conservative nature, these alterations seem unlikely to affect CodY activity, although the possibility has not been excluded. The fourth strain, RF122 (ST 59), had an R→G change at position 209. The last mutation is within the positioning helix of the winged helix-turn-helix motif (16) and could, in principle, affect DNA binding.
CodY expression in USA300.
Expression of codY by qRT-PCR was assessed in S. aureus isolates representing the pulsotypes USA100, USA200, USA300, USA400, USA500, and USA800 and in strain 8325-4. The codY gene was expressed at all time points assessed in strain 923 (USA300) (Fig. 1A). Among the pulsotypes, there was little difference in codY expression at an OD600 of 1.8 (approximately 3 h); expression was modestly higher only in USA800 (1.4-fold; P < 0.05) than in USA300 (Fig. 1B). At an OD600 of 6.3 (approximately 6 h), codY expression was significantly higher in USA100 (2.7-fold; P < 0.001) and lower in USA200 (2.5-fold; P < 0.01) than in USA300 (Fig. 1B). Therefore, although there were modest differences in the abundance of the codY transcript among several of the strains assessed, codY was expressed in all the strains.
Fig 1.
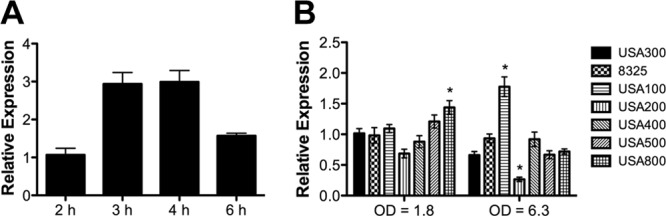
Expression of codY by qRT-PCR. (A) codY is expressed throughout the time points assessed in strain 923 (USA300). (B) codY expression among S. aureus pulsotypes. Expression is depicted relative to that of strain 923 (USA300) at 2 h (A) and strain 923 at an OD600 of 1.8 (B), using the ΔΔCT method with gyrB as the control gene. The values reported are means ± standard errors of the mean (SEM). *, P < 0.05 compared with strain 923 (USA300) at the same OD600.
Effect of a codY mutation on expression of S. aureus USA300 global regulators and virulence genes.
Strain 923ΔcodY grew more slowly than strain 923 in TSB medium (Fig. 2D). To correct for the growth differences, strain 923 was collected at the indicated time points; in each case, other strains were grown to the same OD600 and collected at the same time (3 h, OD600 = 1.8; 4 h, OD600 = 4.0; 6 h, OD600 = 7.0). The effects of the codY mutation on in vitro expression of selected S. aureus global regulatory (RNAIII and saeR) and virulence (hla and lukF-PV) genes was assessed by qRT-PCR. As expected, deletion of codY resulted in inability to detect codY mRNA and increased abundance of RNAIII (2.2-fold at 3 h of growth, P < 0.001, and 1.8-fold at 6 h, P < 0.001) (Fig. 2A). The codY deletion also resulted in increased expression of saeR (1.5-fold at 3 h, P < 0.05, and 2.1-fold at 6 h, P < 0.001) (Fig. 2A) and of hla at 4 h (2.9-fold, P < 0.001), but not at 3 or 6 h (P > 0.05) (Fig. 2B). Furthermore, the deletion of codY had a striking effect on lukF-PV expression, with a 35-fold increase at 3 h, a 16-fold increase at 4 h, and a 3.2-fold increase at 6 h of growth (P < 0.001) (Fig. 2B) compared with the wild type. Analysis of the abundances of Hla and LukF-PV proteins in culture supernatants revealed that deletion of codY resulted in a 1.8-fold increase in Hla and a 17-fold increase in LukF-PV abundance at 6 h (Fig. 2C). These differences in expression and protein abundance were reversed by complementation of codY (Fig. 2A to C).
Fig 2.
Deletion of codY increased in vitro transcription and translation of selected global regulators and virulence determinants in USA300. Strain 923 is a USA300 clinical isolate, the ΔcodY strain is an isogenic codY deletion mutant of 923, the ΔcodY + codY strain is complemented with codY, and the ΔcodY Δpvl strain is an isogenic codY lukSF-PV double-deletion mutant of 923. The strains were grown to an OD600 of 1.8 (approximately 3 h), 4.0 (4 h), or 7.0 (6 h). (A) Deletion of codY abolished transcription of codY and increased expression of the global regulators RNAIII and saeR by qRT-PCR; the effects of codY deletion were reversed by complementation of codY. (B) Deletion of codY increased expression of the genes encoding α-hemolysin (hla) and Panton-Valentine leukocidin (lukF-PV) by qRT-PCR; the effects of codY deletion were reversed by complementation of codY. The values are expressed relative to strain 923 at 2 h and were quantified using the ΔΔCT method with gyrB (A) or 16S rRNA (B) as the control gene. The data are presented as means and SEM. *, P < 0.05, and ***, P < 0.001 relative to strain 923 at the same time point. (C) Deletion of codY increased the abundance of Hla and LukF-PV in culture supernatants at 6 h by Western blotting; the effects of codY deletion were reversed by complementation of codY. Deletion of lukSF-PV resulted in the absence of LukF-PV in culture supernatants. (D) Growth of strain ΔcodY was modestly slower than that of strain 923.
Since the largest effect of the codY deletion was on lukF-PV expression, we deleted lukSF-PV from the codY deletion mutant to assess the contribution of lukSF-PV overexpression to the phenotype of the codY mutant. The growth of 923ΔcodYΔpvl was indistinguishable from that of 923ΔcodY (data not shown). As expected, deletion of lukSF-PV abolished lukF-PV expression by qRT-PCR (data not shown) and resulted in undetectable LukF-PV in culture supernatant by Western blotting (Fig. 2C). The expression of codY, saeR, RNAIII, and hla by qRT-PCR and the abundance of Hla in the culture supernatant by Western blotting did not significantly differ between strains 923ΔcodY and 923ΔcodYΔpvl (data not shown).
Gel mobility shift assay of CodY binding to the lukSF-PV promoter region.
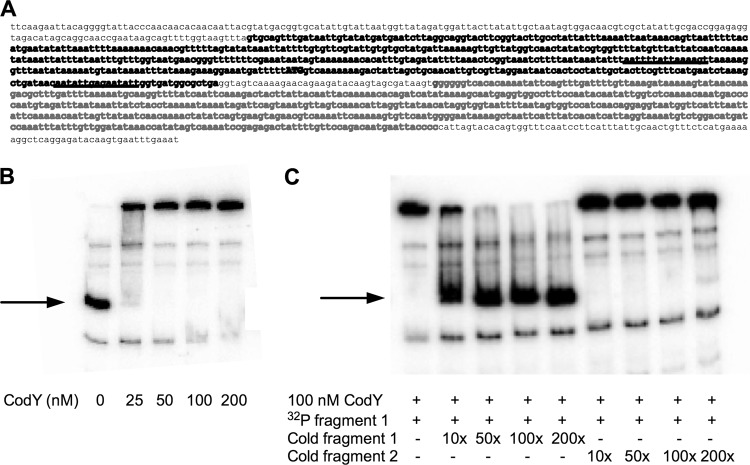
Prior work has shown that CodY regulates the expression of hla directly through binding to the hla locus and indirectly via repression of the agr locus (18, 21). The mechanism by which CodY regulates the agr locus is not known; there are no CodY binding sites in the promoter regions of the RNAIII gene or the agrBDCA operon (21). To assess the possibility that the lukSF-PV promoter region contains a site for CodY binding, we amplified the region by PCR and tested the ability of purified S. aureus CodY-His6 (21) to bind to this DNA segment in vitro. We assumed that the lukSF-PV promoter lies between the start codon of lukS-PV and the stop codon of the upstream gene, phiT (Fig. 3A). This region contains two potential CodY binding regions, each with 3 mismatches. Our results demonstrated that CodY bound to the putative lukSF-PV regulatory region, with an apparent Kd (defined as the concentration of CodY that shifts the mobility of 50% of the DNA) of <25 nM (Fig. 3B). The specificity of binding was verified by showing that the binding was not affected by the presence of a vast excess of a neighboring sequence but was completely eliminated by an excess of the unlabeled probe DNA (Fig. 3C).
Fig 3.
Gel mobility shift assay of CodY binding to the lukSF-PV promoter region. (A) Sequence of the lukSF-PV promoter region and N-terminal coding region of lukS. The sequence in boldface (fragment 1; 519 bp) includes the putative binding region of CodY containing two (underlined) putative CodY binding sites, each with 3 mismatches with respect to the consensus sequence. The letters in lightface correspond to fragment 2 (525 bp), used as a nonspecific competitor DNA. The lukS initiation codon (ATG) is shown in uppercase letters. (B) Labeled DNA fragment 1 (0.3 nM) was incubated with increasing amounts of purified S. aureus CodY-His6 in the presence of 10 mM (each) isoleucine, leucine, and valine and 2 mM GTP. The CodY concentrations used (nM) are indicated below each lane. The arrow indicates the position of the unshifted fragment 1. (C) Labeled fragment 1 DNA (0.3 nM) was incubated with purified S. aureus CodY-His6 (100 nM) and increasing amounts of either unlabeled fragment 1 (specific competitor) or fragment 2 (nonspecific competitor), as indicated below the blot. The concentration of unlabeled fragment in each reaction mixture varied from 0 to 200 times higher than the concentration of radioactive fragment 1. The arrow indicates the position of the unshifted fragment 1.
Deletion of codY increased the virulence of S. aureus USA300 in a necrotizing pneumonia disease model.
To test the impact of a codY mutation on pathogenesis, 6-week-old C57BL/6 mice were intranasally inoculated with 1.5 × 108 CFU of strain 923 or 923ΔcodY (15 mice/group). Fifty-three percent of the mice infected with strain 923 died within 48 h of inoculation (13% mortality at 24 h) (24). Mortality was higher among animals infected with 923ΔcodY at 24 h (80%; P < 0.001) and 48 h (93%; P = 0.02) after infection (Fig. 4A).
Fig 4.

Deletion of codY increased the virulence of USA300 in mouse models of S. aureus necrotizing pneumonia and dermonecrosis. (A) Six-week-old C57BL/6 mice were intranasally inoculated with 1.5 × 108 CFU of strain 923 (a USA300 clinical isolate), an isogenic codY deletion mutant (ΔcodY), or an isogenic codY lukSF-PV double mutant (ΔcodYΔpvl) (n = 10 to 15 mice per group). Mortality was higher among mice infected with ΔcodY or ΔcodYΔpvl than in those infected with 923 at 24 and 48 h after infection. (B) Six-week-old C57BL/6 mice were inoculated subcutaneously with 1.5 × 107 CFU of 923, ΔcodY, or ΔcodYΔpvl strains (10 to 20 mice/group). The dermonecrotic lesions were larger among mice infected with ΔcodY or ΔcodYΔpvl than among mice infected with 923. The data are presented as means ± SEM. *, P < 0.05; **, P < 0.01.
Deletion of codY increased the virulence of S. aureus USA300 in a skin infection model.
Six-week-old C57BL/6 mice were subcutaneously infected with 1.5 × 107 CFU of strain 923 or 923ΔcodY (20 mice/group). In agreement with our past studies with strain 923, lesions appeared within 24 h and peaked in size 2 to 3 days after inoculation (24). The time course was similar among animals infected with 923ΔcodY, but the lesions in animals infected with 923ΔcodY were larger (mean maximum area of dermonecrosis, 116 ± 11 mm2) than the lesions among animals infected with 923 (72 ± 10 mm2; P < 0.01) (Fig. 4B). The lesions of mice inoculated with 923ΔcodY remained larger than those of recipients of 923 throughout the course of the experiment. Other than size, there was no difference in the appearance of the lesions between the groups.
Increased expression of lukSF-PV did not explain the hypervirulent phenotype of the CodY mutant.
The strongly increased expression of lukF-PV in the codY mutant suggested a role for hyperexpression of the genes encoding PVL in its hypervirulent phenotype. To address this possibility, the virulence of strain 923ΔcodYΔpvl was assessed in the mouse models of necrotizing pneumonia and dermonecrosis. In the pneumonia model, 100% of mice infected with 923ΔcodYΔpvl died within 24 h of inoculation; these results were similar to those for recipients of 923ΔcodY (described above; P = 0.25 at 24 h; P = 1.0 at 48 h) (Fig. 4A). There were also no differences in the mean maximum areas of dermonecrosis between mice infected with 923ΔcodY and those infected with 923ΔcodYΔpvl (116 ± 11 mm2 versus 125 ± 12 mm2; P = 0.6) (Fig. 4B). These results demonstrate that increased expression of lukSF-PV was not necessary for the increased virulence of the codY mutant.
DISCUSSION
The above-mentioned results demonstrate that codY is expressed in strain 923 (USA300) and in every other pulsotype tested. There were no major differences in the predicted protein products of codY in USA300 and nearly all other relevant sequenced S. aureus clinical isolates. Furthermore, deletion of codY in strain 923 increased the in vitro expression of S. aureus global regulators and downstream virulence genes, which was reversible by complementation with codY expressed from a plasmid. Notably, the codY deletion increased virulence in mouse models of skin infection and necrotizing pneumonia. Collectively, these results demonstrate that CodY is active and represses virulence in USA300. Thus, the high virulence of USA300 compared with other clinical isolates must be due to factors other than CodY activity.
Deleting codY increased the expression of several key virulence loci, including the global regulators agr and sae and the genes encoding the secreted toxins α-hemolysin and PVL. Repression of the agr locus in USA300 confirms the role of CodY seen previously in S. aureus strains Newman, SA564, and UAMS-1 (21, 22, 30), although the magnitude of repression is different. The saeR and saeS genes are also repressed in strain UAMS-1, but not in strain Newman (21, 30). The discordant findings are likely due to the presence of a point mutation in saeS leading to its constitutive activation in Newman (34). CodY also represses expression of hla in strains UAMS-1 and SA564, but not in Newman (21, 22, 30).
Prior to this work, CodY-mediated effects on expression had been reported only in PVL-negative strains. We found that deletion of codY resulted in dramatically increased abundance of lukF-PV transcript and its encoded protein in culture supernatants. Although the expression of lukSF-PV is controlled by RNAIII and saeRS, the increased expression of lukF-PV in the codY mutant (35-fold) was much greater than the increased expression of RNAIII (2.9-fold) or saeR (2-fold). This fact, taken together with the presence of a putative CodY binding sequence (21) upstream of lukSF-PV and our demonstration that CodY binds directly in vitro to the lukSF-PV promoter region, strongly supports the case for direct regulation of lukSF-PV by CodY.
The expression profile of strain 923ΔcodY suggests some possible explanations for its increased virulence. RNAIII and saeRS each promote the virulence in USA300, so one would expect increased expression of either to lead to enhanced virulence (25, 29). Hla is a major virulence determinant in mouse models of S. aureus necrotizing pneumonia and skin infection (6, 17); therefore, the increased hla expression and Hla abundance in culture supernatants observed in the codY mutant could explain the increase in virulence.
The dramatic CodY-mediated derepression of lukF-PV expression, compared with the modest effects on hla, suggested that PVL might be the key factor responsible for the hypervirulence of the codY mutant. We ruled out this possibility by demonstrating that deletion of lukSF-PV from the codY mutant did not abrogate its virulence. PVL has been reported to promote virulence in mouse and rabbit models of pneumonia and a mouse model of skin and soft tissue infection (5, 10, 19, 35). However, other groups, including ours, reported no effect of lukSF-PV deletion in USA300 isolates in mouse and rat models of pneumonia or mouse models of skin infection and bacteremia (6, 7, 26, 37). Our present results indicate that the hypervirulence of the codY mutant does not require lukSF-PV expression. It should be noted, however, that murine neutrophils are relatively resistant to the lytic effects of PVL compared with human neutrophils (20); therefore, these results do not exclude a role for PVL in virulence in humans.
Although α-hemolysin and PVL are two well-characterized virulence factors repressed by CodY, the CodY regulon includes 100 to 200 genes. Noteworthy examples are members of the phenol-soluble modulin (PSM) family (21, 30), which are important in the pathogenesis of USA300 skin infection (38). It seems likely, therefore, that the impact of a codY deletion on virulence is multifactorial, with overexpression of at least several virulence factors contributing to the hypervirulent phenotype we observed.
In support of a role for CodY in restraining virulence, deletion of rsh, a relA homolog, decreased virulence in a mouse model of hematogenous kidney infection (12). Although deletion of codY alone did not affect virulence in that model, deletion of codY from the rsh mutant restored virulence to wild-type levels. The authors proposed a model similar to that suggested for Listeria monocytogenes (2); in the absence of stringency factor (i.e., RSH, which converts GTP to [p]ppGpp), GTP accumulates, leading to hyperactivity of CodY. As a result, virulence genes repressed by CodY cannot be expressed. Although it is not immediately clear why the codY mutant in their study was not hypervirulent compared with the wild type, it is likely due to a combination of a different S. aureus background (8325 versus USA300) and a different animal model (hematogenous infection versus pneumonia and skin infection).
The reasons underlying the success (and virulence) of USA300 remain unclear. The expression profile of USA300 strains is notable for increased expression of core genomic global regulators, such as agr and sae (23, 25). In support of this notion of global upregulation, proteomic analysis of culture supernatants found significant differences in the abundances of proteins between LAC, a USA300 isolate, and MW2, its USA400 precursor (8). This led us to hypothesize that CodY was inactive in USA300 strains; however, this was not the case. Expression of codY may, however, be dysregulated in USA300; there were subtle differences in codY expression among the pulsotypes we assessed. Future work will clarify the importance of codY expression profiles among different S. aureus strains.
These results indicate that CodY deficiency is not solely responsible for the global regulatory abnormalities observed in USA300. In fact, CodY prevents USA300 from being even more virulent than it is. The expression profile of USA300 suggests that the program controlling the transition from exponential to stationary phase is altered (promoted by agr and saeRS and repressed by CodY), resulting in increased expression of global regulators and toxin genes. It is tempting to speculate that USA300 strains have fundamental alterations in quorum-sensing or nutrient response pathways that facilitate the transition from a colonizing to an invasive phenotype. Genome-wide expression arrays comparing USA300 strains with S. aureus isolates of other relevant pulsotypes would clarify whether this is the case.
In summary, CodY is active and represses virulence in USA300. Deletion of codY resulted in increased expression of selected global regulators and virulence genes and enhanced the virulence of strain 923 in mouse models of necrotizing pneumonia and skin infection.
ACKNOWLEDGMENTS
This work was funded by the Pediatric Critical Care Scientist Development Program (C.P.M.), the Grant Healthcare Foundation (to R.S.D and S.B.-V.), the National Institute of Allergy and Infectious Diseases (1K08AI076596-01A1 to C.P.M. and AI040481-08A1 to R.S.D. and S.B.-V.), and the National Institute for General Medical Sciences (GM042219 to A.L.S.).
Footnotes
Published ahead of print 23 April 2012
REFERENCES
- 1. Adhikari RP, Novick RP. 2008. Regulatory organization of the staphylococcal sae locus. Microbiology 154:949–959 [DOI] [PubMed] [Google Scholar]
- 2. Bennett HJ, et al. 2007. Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol. Microbiol. 63:1453–1467 [DOI] [PubMed] [Google Scholar]
- 3. Boyle-Vavra S, Yin S, Daum RS. 2006. The VraS/VraR two-component regulatory system required for oxacillin resistance in community-acquired methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 262:163–171 [DOI] [PubMed] [Google Scholar]
- 4. Bronner S, Stoessel P, Gravet A, Monteil H, Prevost G. 2000. Variable expressions of Staphylococcus aureus bicomponent leucotoxins semiquantified by competitive reverse transcription-PCR. Appl. Environ. Microbiol. 66:3931–3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brown EL, et al. 2009. The Panton-Valentine leukocidin vaccine protects mice against lung and skin infections caused by Staphylococcus aureus USA300. Clin. Microbiol. Infect. 15:156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bubeck Wardenburg J, Bae T, Otto M, Deleo FR, Schneewind O. 2007. Poring over pores: alpha-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat. Med. 13:1405–1406 [DOI] [PubMed] [Google Scholar]
- 7. Bubeck Wardenburg J, Palazzolo-Ballance AM, Otto M, Schneewind O, DeLeo FR. 2008. Panton-Valentine leukocidin is not a virulence determinant in murine models of community-associated methicillin-resistant Staphylococcus aureus disease. J. Infect. Dis. 198:1166–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burlak C, et al. 2007. Global analysis of community-associated methicillin-resistant Staphylococcus aureus exoproteins reveals molecules produced in vitro and during infection. Cell Microbiol. 9:1172–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. David MZ, Daum RS. 2010. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 23:616–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Diep BA, et al. 2010. Polymorphonuclear leukocytes mediate Staphylococcus aureus Panton-Valentine leukocidin-induced lung inflammation and injury. Proc. Natl. Acad. Sci. U. S. A. 107:5587–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dunman PM, et al. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341–7353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Geiger T, et al. 2010. Role of the (p)ppGpp synthase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus. Infect. Immun. 78:1873–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Giraudo AT, Calzolari A, Cataldi AA, Bogni C, Nagel R. 1999. The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol. Lett. 177:15–22 [DOI] [PubMed] [Google Scholar]
- 14. Highlander SK, et al. 2007. Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol. 7:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Iwatsuki K, Yamasaki O, Morizane S, Oono T. 2006. Staphylococcal cutaneous infections: invasion, evasion and aggression. J. Dermatol. Sci. 42:203–214 [DOI] [PubMed] [Google Scholar]
- 16. Joseph P, Ratnayake-Lecamwasam M, Sonenshein AL. 2005. A region of Bacillus subtilis CodY protein required for interaction with DNA. J. Bacteriol. 187:4127–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kennedy AD, et al. 2010. Targeting of alpha-hemolysin by active or passive immunization decreases severity of USA300 skin infection in a mouse model. J. Infect. Dis. 202:1050–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kornblum J, Kreisworth B, Projan SJ, Ross H, Novick RP. 1990. Agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus, p. 373–402 In Novick R. (ed), Molecular biology of the staphylococci. VCH Publishers, New York, NY [Google Scholar]
- 19. Labandeira-Rey M, et al. 2007. Staphylococcus aureus Panton-Valentine leukocidin causes necrotizing pneumonia. Science 315:1130–1133 [DOI] [PubMed] [Google Scholar]
- 20. Loffler B, et al. 2010. Staphylococcus aureus Panton-Valentine leukocidin is a very potent cytotoxic factor for human neutrophils. PLoS Pathog. 6:e1000715 doi:10.1371/journal.ppat.1000715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Majerczyk CD, et al. 2010. Direct targets of CodY in Staphylococcus aureus. J. Bacteriol. 192:2861–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Majerczyk CD, et al. 2008. Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 190:2257–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Montgomery CP, et al. 2008. Comparison of virulence in community-associated methicillin-resistant Staphylococcus aureus pulsotypes USA300 and USA400 in a rat model of pneumonia. J. Infect. Dis. 198:561–570 [DOI] [PubMed] [Google Scholar]
- 24. Montgomery CP, Boyle-Vavra S, Daum RS. 2009. The arginine catabolic mobile element is not associated with enhanced virulence in experimental invasive disease caused by the community-associated methicillin-resistant Staphylococcus aureus USA300 genetic background. Infect. Immun. 77:2650–2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Montgomery CP, Boyle-Vavra S, Daum RS. 2010. Importance of the global regulators agr and saeRS in the pathogenesis of CA-MRSA USA300 infection. PLoS One 5:e15177 doi:10.1371/journal.pone.0015177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Montgomery CP, Daum RS. 2009. Transcription of inflammatory genes in the lung after infection with community-associated methicillin-resistant Staphylococcus aureus: a role for Panton-Valentine leukocidin? Infect. Immun. 77:2159–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Novick R. 2000. Gram Positive pathogens. American Society for Microbiology, Washington, DC [Google Scholar]
- 28. Novick RP, et al. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967–3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nygaard TK, et al. 2010. SaeR binds a consensus sequence within virulence gene promoters to advance USA300 pathogenesis. J. Infect. Dis. 201:241–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pohl K, et al. 2009. CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J. Bacteriol. 191:2953–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Regassa LB, Betley MJ. 1992. Alkaline pH decreases expression of the accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 174:5095–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Regassa LB, Couch JL, Betley MJ. 1991. Steady-state staphylococcal enterotoxin type C mRNA is affected by a product of the accessory gene regulator (agr) and by glucose. Infect. Immun. 59:955–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rogasch K, et al. 2006. Influence of the two-component system saeRS on global gene expression in two different Staphylococcus aureus strains. J. Bacteriol. 188:7742–7758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schafer D, et al. 2009. A point mutation in the sensor histidine kinase SaeS of Staphylococcus aureus strain Newman alters the response to biocide exposure. J. Bacteriol. 191:7306–7314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tseng CW, et al. 2009. Staphylococcus aureus Panton-Valentine leukocidin contributes to inflammation and muscle tissue injury. PLoS One 4:e6387 doi:10.1371/journal.pone.0006387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vandenesch F, Kornblum J, Novick RP. 1991. A temporal signal, independent of agr, is required for hla but not spa transcription in Staphylococcus aureus. J. Bacteriol. 173:6313–6320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Voyich JM, et al. 2006. Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J. Infect. Dis. 194:1761–1770 [DOI] [PubMed] [Google Scholar]
- 38. Wang R, et al. 2007. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 13:1510–1514 [DOI] [PubMed] [Google Scholar]