Abstract
Tannerella forsythia is strongly associated with chronic periodontitis, an inflammatory disease of the tooth-supporting tissues, leading to tooth loss. Fusobacterium nucleatum, an opportunistic pathogen, is thought to promote dental plaque formation by serving as a bridge bacterium between early- and late-colonizing species of the oral cavity. Previous studies have shown that F. nucleatum species synergize with T. forsythia during biofilm formation and pathogenesis. In the present study, we showed that coinfection of F. nucleatum and T. forsythia is more potent than infection with either species alone in inducing NF-κB activity and proinflammatory cytokine secretion in monocytic cells and primary murine macrophages. Moreover, in a murine model of periodontitis, mixed infection with the two species induces synergistic alveolar bone loss, characterized by bone loss which is greater than the additive alveolar bone losses induced by each species alone. Further, in comparison to the single-species infection, mixed infection caused significantly increased inflammatory cell infiltration in the gingivae and osteoclastic activity in the jaw bones. These data show that F. nucleatum subspecies and T. forsythia synergistically stimulate the host immune response and induce alveolar bone loss in a murine experimental periodontitis model.
INTRODUCTION
Periodontitis is a bacterially induced chronic inflammatory disease of the supporting tissues of teeth which leads to tooth loss (49). This disease has a strong polymicrobial etiology and results mainly from the self-damaging effects of the immune response elicited to supra- and subgingival biofilm bacteria (24). The major pathogens accompanying these biofilms are the Gram-negative anaerobes Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia—collectively known as the “red complex” consortium (37). The involvement of these bacteria in periodontitis has been demonstrated in rodent models of periodontitis, in which oral infection with either species has been shown to induce alveolar bone destruction (4, 25, 27, 34, 35). In addition, polymicrobial challenge comprising two or more species has been shown to cause a synergistic alveolar bone loss (21, 29). While the exact mechanism by which these bacterial species cooperate to induce alveolar bone loss is not completely understood, metabolic, chemical, and physical interactions are expected to play critical roles (23). Such interactions are likely to impact bacterial growth, mediate colonization through coaggregations, promote mixed biofilms, and regulate expression of microbial molecules that trigger cytokine release, inducing inflammation.
Fusobacterium nucleatum is a predominant species of subgingival biofilm in both healthy and disease states (12). Because of this bacterium's ability to coaggregate with both early and late colonizers of the human oral cavity, it is suggested to promote plaque development by acting as a bridge bacterium (6, 23, 31, 42). In this regard, studies have shown that several F. nucleatum species form a synergistic biofilm in vitro with T. forsythia (36) while F. nucleatum also strongly coaggregates with T. forsythia (36). In vivo, F. nucleatum and T. forsythia dominate the intermediate layers of subgingival biofilms (51). Additionally, F. nucleatum can modulate the inflammatory response of the host to other pathogens (13, 15, 18, 30, 46).
In the present study, we sought to determine the extent to which the alveolar bone loss and inflammation due to T. forsythia infection were impacted by the presence of F. nucleatum in a mouse model. Our data showed that mixed F. nucleatum and T. forsythia challenge causes increased secretion of inflammatory cytokines in comparison to that from single-organism challenge. Moreover, these organisms show a synergistic phenomenon with respect to the alveolar bone loss induction. They act cooperatively in mixed infections to induce alveolar bone loss more than the additive alveolar bone losses due to each species alone. This increased alveolar bone loss due to mixed infection also correlates with the increased infiltration of tissue inflammatory cells and osteoclastic activity in mice.
MATERIALS AND METHODS
Bacteria.
T. forsythia ATCC 43037 and F. nucleatum subsp. nucleatum ATCC 25586 were grown in TF broth (brain heart infusion medium containing 5 μg/ml hemin, 0.5 μg/ml menadione, 0.001% N-acetylmuramic acid, 0.1% l-cysteine, and 5% fetal bovine serum) and TS broth (tryptic soy broth containing 0.1% l-cysteine and 0.4% yeast extract), respectively, as liquid cultures or on plates containing 1.5% agar in the broth under anaerobic conditions as described previously (36).
Mice.
Specific-pathogen-free BALB/cJ mice (Jackson Laboratory, Bar Harbor, ME) were maintained in HEPA-filtered cages with autoclaved food, water, and bedding. All animal procedures were performed in accordance with the protocols approved by the University at Buffalo Institutional Animal Care and Use Committee (IACUC).
Mammalian cells, stimulation, and cytokine ELISA.
Mouse peritoneal macrophages were prepared as described previously (50), and THP1-Blue cells (InvivoGen, San Diego, CA) were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) containing 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine, 10 mM HEPES, 4.5 g/liter glucose, 100 U/ml penicillin G, 100 μg/ml streptomycin, and 0.05 mM 2-mercaptoethanol. THP1-Blue cells are derived from the human monocytic THP-1 cell line by stable transfection with a reporter plasmid expressing a secreted embryonic alkaline phosphatase (SEAP) gene under the control of NF-κB and the AP-1 inducible promoter. Mammalian cells were seeded in 48-well culture plates at a density of 105 cells per well in 250 μl growth medium. Bacteria were enumerated with the Petroff-Hausser chamber immediately prior to each experiment. For challenge with an individual species (T. forsythia or F. nucleatum), multiplicities of infection (MOIs; numbers of bacteria per mammalian cell) of 10, 100, and 250 were used. For mixed infections, both species (F. nucleatum and T. forsythia cells) were mixed in a ratio of 1:1 to obtain the required MOI of 10, 100, or 250, and mixtures were incubated at room temperature for 10 min to allow coaggregation and then added to mammalian cell cultures. Therefore, with bacteria at a 1:1 ratio, the numbers of cells of each species (F. nucleatum or T. forsythia) at a given MOI were half of their numbers in single infections with the same MOI. Coaggregations were monitored qualitatively by visual observations. At the end of the incubation period, small-sized bacterial aggregates with some flocculation and turbidity still remaining in the incubation tubes implied that coaggregation had initiated. The cell supernatants from mouse macrophages and THP1-Blue cells were challenged with Escherichia coli lipopolysaccharide (LPS) (100 ng/ml) as a positive agonist, and supernatants were collected and stored at −80°C until assayed. None of the treatments affected cell viability compared to that of the medium-only control, as judged by the trypan blue exclusion assay. The cytokines interleukin 1β (IL-1β), tumor necrosis factor alpha (TNF-α), and IL-6 were measured in triplicate by enzyme-linked immunosorbent assay (ELISA) kits from eBiosciences. The supernatants from THP1-Blue cells were assayed for SEAP by a colorimetric enzyme assay (Quanti-Blue; InvivoGen).
Western blot analysis.
A Western blot analysis for detecting NF-κB (p65) activation was performed as described previously (9) using antibodies specific to the p65 subunit or its phosphorylated form at serine 536 (Cell Signaling Technology).
Mouse infection and alveolar bone loss assessment.
BALB/cJ mice (6- to 7-week-old females, 8 to 10 mice per group) were infected with bacteria (mono- or mixed infection) as previously described with the following modifications (35). Briefly, mice were first treated with kanamycin (1 mg/ml water) for 7 days ad libitum, followed by a 3-day antibiotic-free period to suppress resident oral flora. This was followed by monoinfection with live bacteria (T. forsythia or F. nucleatum) by oral gavage using a feeding needle. Monoinfections were given as 100-μl bacterial suspensions (1010 CFU/ml) in 2% carboxymethyl cellulose (CMC) three times at 48-h intervals, and this schedule was repeated the following week. Mice receiving mixed infections received 100-μl suspensions containing 1:1 ratios of T. forsythia and F. nucleatum (1010 CFU/ml total bacteria). The control group (sham-infected) mice received antibiotic pretreatment and 100 μl of 2% carboxymethyl cellulose only. Bacterial colonization was assessed by PCR analysis for the presence of T. forsythia- and F. nucleatum-specific 16S rRNA genes (36) in oral swab samples as described previously (35). The primers specific for T. forsythia were 5′-GCGTATGTAACCTGCCCGCA-3′ and 5′-TGCTTCAGTTCAGTTATACCT-3′, amplifying a 641-bp amplicon, and for F. nucleatum, the primers were 5′-AAGCGCGTCTAGGTGGTTATGT-3′ and 5′-TGTAGTTCCGCTTACCTCTCCAG-3′, amplifying a 108-bp product. A pair of ubiquitous bacterial primers, 5′-GTGCTGCAGAGAGTTTGATCATGCCTCAG-3′ and 5′-CACGGATCCTACGGGTACCTTGTTACGACTT-3′, which detect almost all bacterial 16S rRNA genes, was used as a positive control (amplicon length, 1.4 kb). Mice were sacrificed 6 weeks after first infection; serum was collected by cardiac puncture. Jaws were autoclaved and defleshed, immersed overnight in 3% hydrogen peroxide, and stained with 1% methylene blue. Horizontal bone loss was assessed morphometrically by measuring the distance between the cementoenamel junction (CEJ) and the alveolar bone crest (ABC). Measurements at 14 buccal sites per mouse (7 sites each on the left and right maxillary molars) were made under a dissecting microscope (Brook-Anco, Rochester, NY) fitted with an Aquinto imaging measurement system (a4i America, Rochester, NY). Random and blinded bone measurements were taken by two independent evaluators. Total alveolar bone loss per group was calculated by averaging total CEJ-ABC distances (14 sites per mouse) of all mice. All data were analyzed on GraphPad Prism 5 software (Graph Pad, San Diego, CA).
ELISA for bacterium-specific serum IgG titers.
T. forsythia- and F. nucleatum-specific enzyme-linked immunosorbent assays (ELISAs) were performed as described previously (35). Briefly, 96-well Immuno MaxiSorp plates (Nalgene International, Rochester, NY) were coated with formalin-fixed T. forsythia or F. nucleatum (1 × 108 cells/well). Sera were added in a 2-fold serial dilution, and T. forsythia- or F. nucleatum-specific IgG was detected using horseradish peroxidase-conjugated goat anti-mouse IgG (Bethyl Laboratories, Montgomery, TX). ELISA wells were color developed with TMB (3,3′,5,5′-tetramethylbenzidine) microwell enzyme substrate (KPL, Gaithersburg MD). After stopping the enzyme reaction with 0.1 N H2SO4, plates were read at 495 nm. The antibody titer was defined as the log2 of the highest dilution with a signal that was 0.1 optical density units above the level of the background signal.
Immunohistochemistry.
The immunohistochemical staining was performed per our previously described procedure (27). Briefly, the right and left halves of the maxillary and mandibular bones were removed, fixed, and embedded in paraffin; 4-μm sections were cut and mounted. They were deparafinized in xylene and hydrated in graded ethanol. After antigen retrieval by incubating at 90°C for 10 min with BD Retrievagen A (BD Pharmingen, San Diego, CA), specimens were sequentially incubated in (i) blocking solution containing 0.1% Triton X-100 in 0.1 M phosphate-buffered saline (PBS) for 1 h at room temperature and (ii) monoclonal rat anti-mouse CD45 (BD PharMingen) antibody diluted 1:30 in PBS containing 0.1% Triton X-100 or neutrophil marker antibody (NIMP-R14; Santa Cruz Biotechnology, Santa Cruz, CA) for 1 h at room temperature. The slides were incubated with a biotinylated secondary antibody (goat anti-rat), followed by avidin-peroxidase and color development with 3,3-diaminobenzidine (DAB) (Vector Labs, Burlingame, CA); the counter stain was hematoxylin. After each step, slides were rinsed in PBS-Tween 20 (PBST) (3 times for 10 min each).
Slides were scanned at an absolute magnification of ×400 using the Aperio Scan Scope CS system (Aperio Technologies, Vista CA). Slides (six slides per mouse) were viewed and analyzed remotely using desktop personal computers employing the virtual image viewer software (Aperio). Antibody-positive cells (stained brown) were enumerated manually in the interdental areas from the first to the third molar at randomly selected locations in each slide (6 per slide). The areas associated with these locations were then obtained from the software to calculate the average cell densities per square millimeter.
Bone histology.
Bone histology was performed per our previously described protocols (27). Briefly, mice maxillary and mandibular bones (n = 4) were fixed in 10% phosphate-buffered formalin and decalcified in 10% EDTA. The samples were then embedded in paraffin, and sections of 4 μm were prepared and stained for tartrate-resistant acid phosphatase (TRAP; Sigma). The TRAP-stained whole slides were digitally scanned immediately with a Scan Scope CS system (Aperio) to minimize color fading, and the scanned slides were viewed with image viewing software (Aperio) as described above. The right maxillary and mandibular interdental areas (average of 10 high-power fields/slide) of the crestal alveolar bone from the first molar to the third molar were used to quantify osteoclasts.
Data analysis.
Data were analyzed on GraphPad Prism software (GraphPad, San Diego, CA). Comparisons between groups were made using Student's t test or an analysis of variance, as appropriate. Statistical significance is defined as a P value of <0.05.
RESULTS
T. forsythia and F. nucleatum mixed challenge cooperatively induces NF-κB activity and phosphorylation of p65 Ser-536.
Since inflammation is mediated mainly by cytokines, we first investigated whether T. forsythia and F. nucleatum behave cooperatively to cause the induction of NF-κB, an important transcription factor for the induction of many inflammatory cytokines (3), using the reporter monocyte cell line THP1-Blue. THP1-Blue cells showed significant activation of NF-κB when challenged with F. nucleatum or T. forsythia in a dose-dependent manner (Fig. 1A). Strikingly, the NF-κB activity was significantly higher in the cells challenged with a 1:1 combination of T. forsythia and F. nucleatum at total MOIs of 10 or 100 than in the cells stimulated with either bacterium alone at a similar MOI (Fig. 1A). However, at a higher MOI of 250, no significant increase was observed with the mixed challenge compared to with the single bacterial challenge (Fig. 1A). At high bacterium cell densities, we also observed more bacterial coaggregation and sedimentation after the preincubation mixing step, which might have led to fewer bacteria being available for interacting with the mammalian cells.
Fig 1.
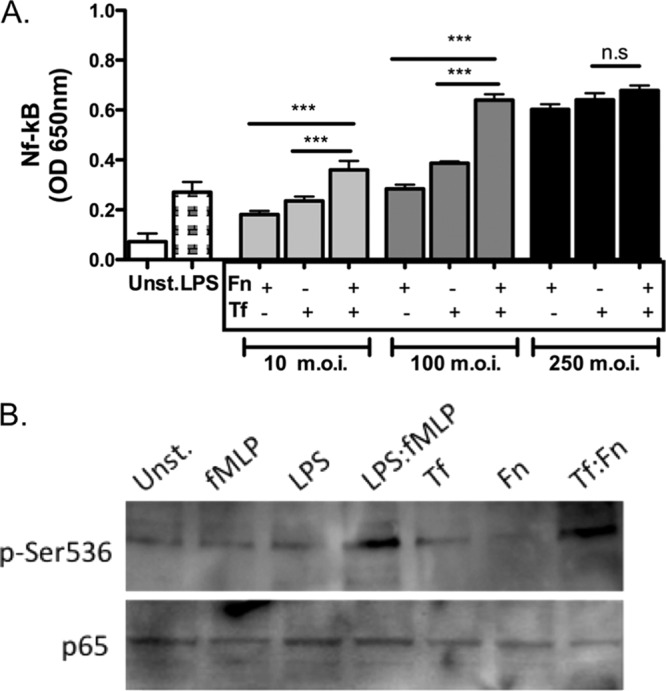
Challenge with F. nucleatum and T. forsythia mixed infection is more potent than challenge with either bacterium alone in inducing NF-κB activity. (A) NF-κB activation assay. THP1-Blue cells were stimulated in triplicate with F. nucleatum (Fn), T. forsythia (Tf), or both bacteria (Fn:Tf) at a 1:1 ratio at total-bacteria-to-host-cell MOIs of 10 and 100. E. coli LPS was used as a positive-control agonist in each case. The culture supernatants were assayed after 16 h for SEAP activity to assess NF-κB activation. Error bars indicate standard deviations. (B) Phosphorylation of Ser-536 of the NF-κB p65 subunit. THP-1 cells were stimulated with 1 nM fMLP, 10 ng/ml E. coli LPS, both fMLP (1 nM) and LPS (10 ng/ml), F. nucleatum (MOI of 10; Fn), T. forsythia (MOI of 10; Tf), or F. nucleatum and T. forsythia (1:1 at an MOI of 10; Fn:Tf) for 15 min, and total cell lysates were assessed by a Western blot using phospho-Ser-536 or p65 antibodies. Data shown in panels A and B are representative of three independent experiments with similar results; statistically significant differences between the groups are indicated by asterisks (***, P < 0.001). OD, optical density; n.s, not significant; Unst., unstimulated.
Since it has been previously shown that optimal activation of NF-κB requires phosphorylation of serine 536 of the NF-κB p65 subunit (45), we tested the effects of mixed T. forsythia and F. nucleatum challenge on Ser-536 phosphorylation to gain insight into the mechanism of NF-κB activation. Our data indicated that T. forsythia and F. nucleatum synergistically induced phosphorylation of Ser-536 (Fig. 1B). These findings demonstrate that the two bacteria synergistically induce NF-κB through phosphorylation of Ser-536 in the p65 subunit.
T. forsythia and F. nucleatum cooperatively induce cytokine release in mouse macrophages.
The release of TNF-α, IL-1β, and IL-6 cytokines in mouse primary macrophages challenged with bacteria was investigated. In agreement with the trend observed with respect to NF-κB activity in response to bacteria, T. forsythia and F. nucleatum mixed challenge at a lower MOI of 10 caused significantly increased secretion of IL-1β, TNF-α, and IL-6 compared to that in the single-species bacterial challenge in mouse macrophages. In addition, at a higher MOI, no significant differences were observed, in a trend similar to that for the NF-κB data (Fig. 2). Taken together, these results indicated that T. forsythia and F. nucleatum act synergistically to induce inflammation in host cells, i.e., their action in concert outweighs that of monoinfection at the same MOI. This synergy at lower bacterial doses might have a role during the early stages of the disease progression or might be advantageous to habitation of the mixed-species biofilm that is key to this polymicrobial disease.
Fig 2.
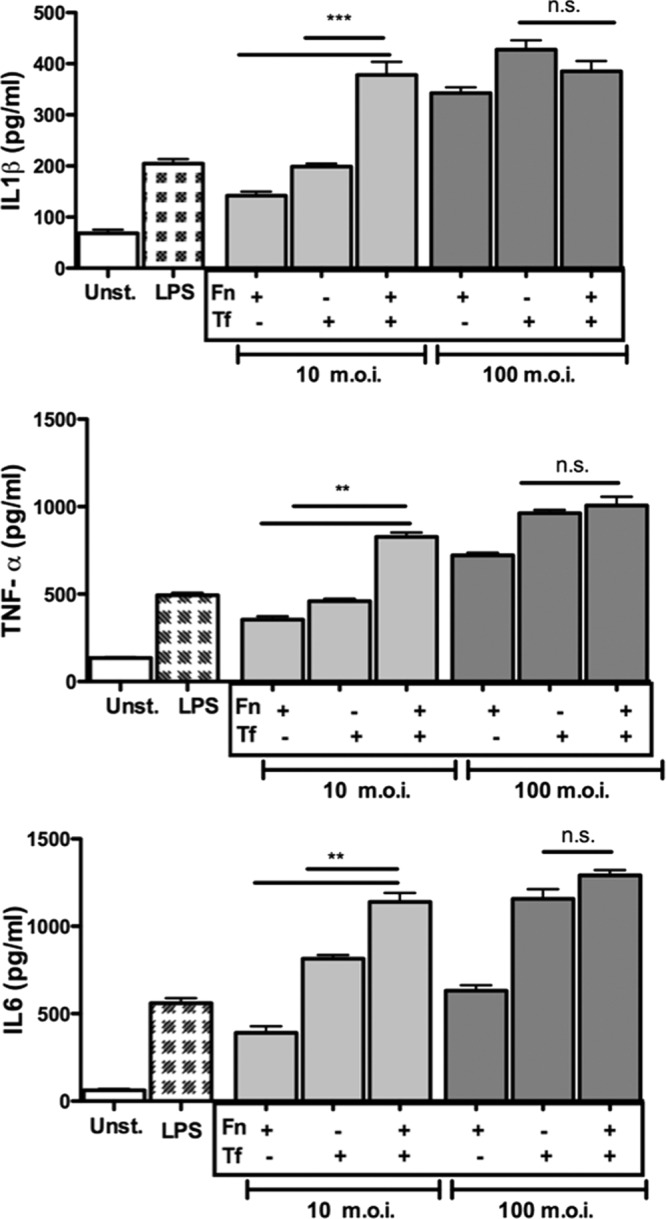
F. nucleatum and T. forsythia mixed challenge is more potent than challenge with either organism alone in inducing cytokine secretion. Mouse macrophages were stimulated in triplicate with F. nucleatum (Fn), T. forsythia (Tf), or both bacteria (Fn:Tf) at a 1:1 ratio at total-bacteria-to-host-cell MOIs of 10 and 100. E. coli LPS was used as a positive-control agonist in each case. The culture supernatants were assayed after 16 h for IL-1β, TNF-α, and IL-6. Error bars indicate standard deviations. Data shown are representative of three independent experiments with similar results; statistically significant differences between the groups are indicated by asterisks (***, P < 0.001; **, P < 0.01).
T. forsythia and F. nucleatum coinfection synergistically increases alveolar bone loss.
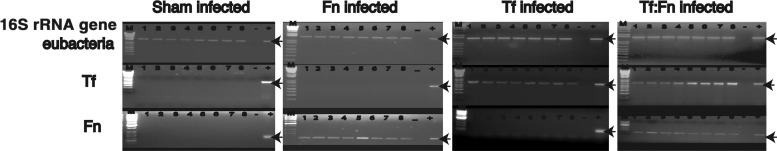
Previously, we showed that T. forsythia induces alveolar bone loss in mice (27, 35). In addition, we have shown that several F. nucleatum species and T. forsythia form synergistic mixed-species biofilms in vitro (36) (data not shown). In this case, when T. forsythia is cocultured with F. nucleatum, a robust biofilm, for which the total biomass of the biofilm is significantly greater than the additive biomasses of the biofilms formed by each bacterium alone, is formed (36). These studies, together with the data presented above, relative to monocyte and macrophage activation, indicated that mixed infection with these organisms might affect alveolar bone loss outcome in vivo. Thus, we examined the effect of mixed T. forsythia and F. nucleatum challenge in a mouse model of infection-induced alveolar bone loss. For this purpose, after suppressing the resident oral flora with antibiotic, mice were orally infected with either F. nucleatum or T. forsythia with six doses of 109 cells per dose (monoinfections). For mixed infections, mice were given six doses of 109 total cells per dose that contained a 1:1 ratio of T. forsythia and F. nucleatum (i.e., 5 × 108 cells of each bacterium). The sham-infected mice received 2% CMC alone. One week following the last infection, PCR analysis of oral swabs indicated that all mice infected with T. forsythia were positive for a T. forsythia-specific, 620-bp 16S rRNA gene product, all mice (n = 8) infected with F. nucleatum (n = 8) were positive for an F. nucleatum-specific, 108-bp 16S rRNA gene product, and all mice coinfected with F. nucleatum and T. forsythia were positive for the 620-bp and 180-bp PCR products (Fig. 3). As expected, sham-infected mice were positive only for the 1.4-kb universal eubacterium 16S rRNA gene product, and the T. forsythia- and F. nucleatum-specific 16S rRNA gene products were not detected in sham-infected mice (Fig. 3). As another confirmation of infection, T. forsythia- and F. nucleatum-specific antibody titers were evaluated in sham-infected and bacterium-infected animals. Our results showed that bacterium-specific serum IgG titers (to F. nucleatum and T. forsythia) in infected animals increased severalfold over those in sham-infected mice (Fig. 4). Mice infected with F. nucleatum or T. forsythia showed robust F. nucleatum-specific or T. forsythia-specific serum IgG titers, respectively. Cross-reactive antibody titers to T. forsythia were observed in mice infected with F. nucleatum and vice versa. Interestingly, antibody titers to T. forsythia in coinfected mice were significantly lower than those in mice infected with T. forsythia alone (Fig. 4). However, the antibody titers to F. nucleatum remained similar in both coinfected and F. nucleatum-monoinfected mice. The data suggest that F. nucleatum modulates the antibody response of the host to T. forsythia. A similar outcome was reported in a previous study in which a decreased antibody response to P. gingivalis was observed in mice previously immunized with F. nucleatum (15). The low IgG titers to T. forsythia and F. nucleatum in the sham-infected mice (Fig. 4) are presumably due to the nonspecific cross-reactivity with the antibodies to normal resident bacteria. With respect to alveolar bone loss, F. nucleatum-infected mice showed minimal alveolar bone loss (Fig. 5). This was in agreement with a previous study that showed marginal alveolar bone loss in rats infected with a clinical isolate of F. nucleatum (47). As expected (27), the alveolar bone loss (total and per site) (Fig. 5A and B) was significantly higher in T. forsythia-infected mice than in F. nucleatum-infected or sham-infected (control) mice. The net bone loss, calculated by subtracting the total alveolar bone loss of the control group (sham) from that of the infected group, was significantly higher in the mixed-infection group than in either the T. forsythia or the F. nucleatum infection group (Fig. 5C). Moreover, the net bone loss of the mixed-infection group was significantly higher than the additive net alveolar bone losses of the T. forsythia- and F. nucleatum-infected groups (Fig. 5C). The results thus demonstrate a synergistic effect of mixed infection on alveolar bone loss.
Fig 3.
PCR analysis of oral microbial samples from sham-infected, F. nucleatum-infected, T. forsythia-infected, or mixed F. nucleatum- and T. forsythia-infected mice. Agarose (1 or 2%) gels show PCR-amplified products from mouse oral microbial DNA samples with amplimers specific to16S rRNA genes for eubacteria, F. nucleatum (Fn), or T. forsythia (Tf). Lanes M, 1-kb Plus DNA ladder; lanes +, positive control using F. nucleatum or T. forsythia genomic DNA as the template; lanes −, negative control without bacterial DNA; lanes 1 to 8, oral microbial samples from individual infected mice from each group. The 1.4-kb, 640-bp, and 108-bp bands (indicated with arrows) represent eubacterial, T. forsythia, and F. nucleatum rRNA gene amplicons, respectively.
Fig 4.
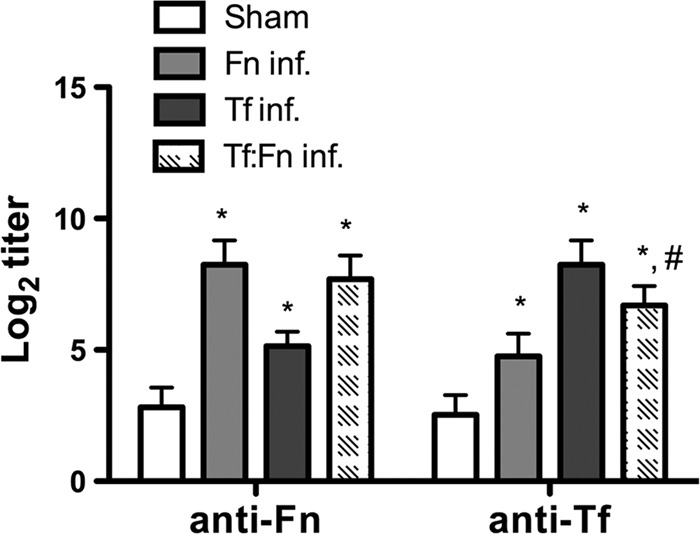
Specific and cross-reactive antibodies are elevated in mice following oral infection. Sera from mice 6 weeks after first infection (sham, F. nucleatum, T. forsythia, or coinfection) were analyzed for F. nucleatum- or T. forsythia-specific IgG (anti-Fn and anti-Tf, respectively) by ELISA. Antibody levels are presented as log2 titers. Data are the means and standard deviations for each group (n = 8), and statistical differences between the group means were determined (*, P < 0.05 versus the sham-infected group; #, P < 0.05 versus the T. forsythia-infected group).
Fig 5.
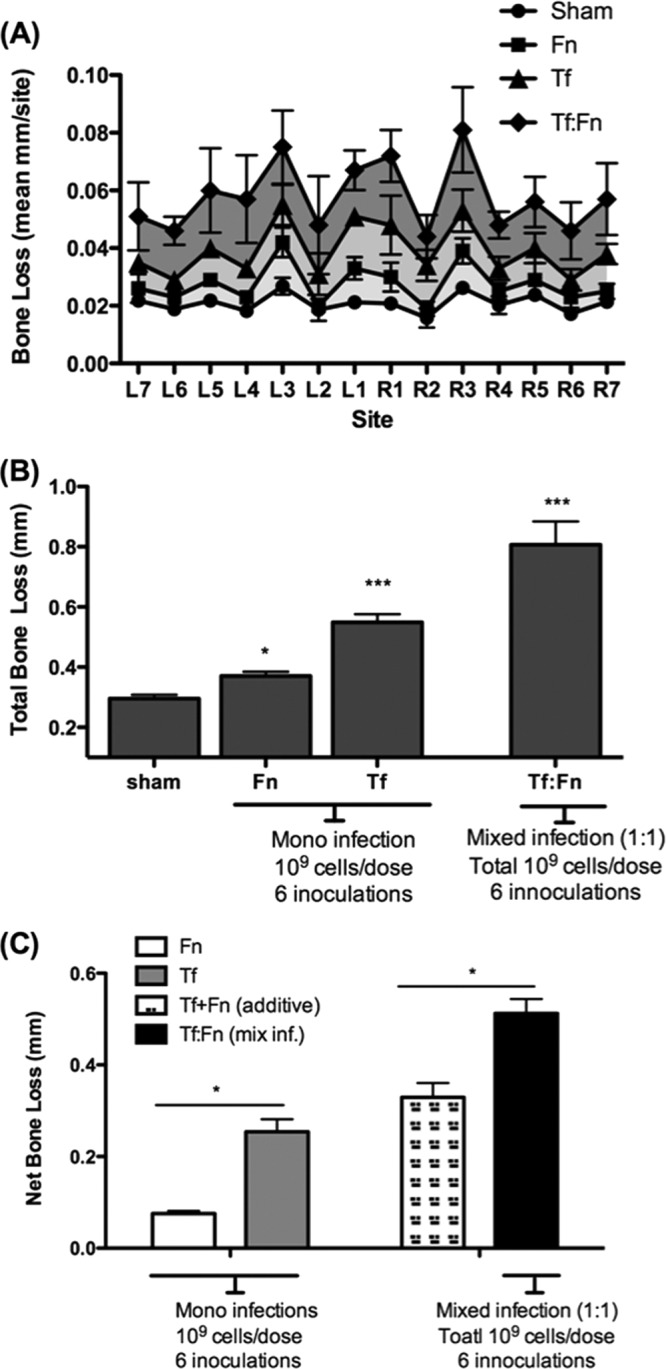
Coinfection with F. nucleatum and T. forsythia causes synergistic alveolar bone loss in mice. Mice (n = 8) were infected by oral gavage with 6 doses of F. nucleatum (109 cells/dose; Fn), T. forsythia (109 cells/dose; Tf), or a 1:1 mixture of F. nucleatum and T. forsythia (109 total cells; Tf:Fn) or sham infected. Alveolar bone destruction was assessed after 6 weeks by measuring the distance from the ABC to the CEJ at 14 maxillary buccal sites per mouse (R1 to R7, right jaw; L1 to L7, left jaw). (A) Average alveolar bone loss at 14 buccal sites. (B) Total bone loss per group, showing the mean total ABC-CEJ distance per group. (C) Net bone loss, showing the total ABC-CEJ distance of each infected group minus that of the sham-infected group. As indicated, the net bone loss due to coinfection (Fn:Tf) is significantly higher than the addition of the net bone loss values for each organism alone (Fn+Tf). Standard deviations are shown. Data were analyzed by a Mann-Whitney unpaired t test, and statistically significant differences are indicated with an asterisk (***, P < 0.001; *, P < 0.05).
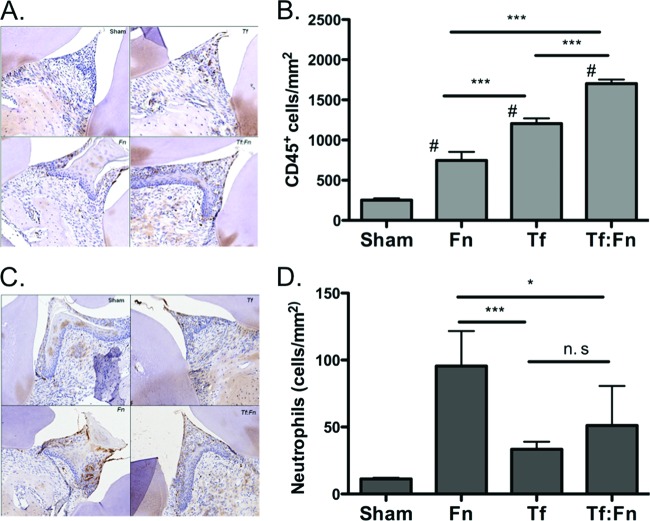
Coinfection causes increased inflammatory cell infiltration and bone osteoclastic activity.
The inflammatory response to mono- and mixed infections was determined by quantifying the number of CD45 (leukocyte common antigen)-positive cells. Neutrophils were quantified as NIMP-R14 (neutrophil-specific antibody)-positive cells. Moreover, to evaluate the number of osteoclasts in the alveolar bone, maxillae were stained for TRAP (tartrate-resistant alkaline phosphatase, a marker of osteoclasts) activity. The results indicated an increased number of chronic inflammatory cells positive for CD45 in mice challenged with T. forsythia or with F. nucleatum and T. forsythia coinfection compared to that in mice with F. nucleatum infection alone or sham infection (Fig. 6A). Following quantification of CD45-positive cells, as expected (27), T. forsythia infection resulted in increased infiltration of lymphocytic cells in gingival tissue (Fig. 6B). However, mice infected with F. nucleatum showed no significant increase (Fig. 6B). Strikingly, gingival tissues of mice coinfected with F. nucleatum and T. forsythia exhibited significantly more CD45+ lymphocytes than mice infected with T. forsythia alone (Fig. 6B). Since neutrophil infiltration is a hallmark of inflammation and neutrophils have been suggested to play protective (48) as well as destructive (20) roles during the pathogenesis of periodontal disease, we assessed neutrophil extravasation in the interdental regions as well. The results showed that in comparison to the sham-infected mice, significantly higher numbers of neutrophils were present in single- and mixed-organism-infected mice (Fig. 6C and D). Strikingly, the numbers of neutrophils decreased following the mixed infection with F. nucleatum and T. forsythia compared to those after the infection with F. nucleatum alone (Fig. 6D). This suggests that a dampening of the neutrophil response during coinfection with F. nucleatum and T. forsythia might result in reduced bacterial clearance.
Fig 6.
Coinfection causes increased lymphocytic infiltration. Sections were prepared from tissues 3 weeks following infection. Sham, sham-infected mice; Fn, F. nucleatum infection; Tf, T. forsythia infection; Tf:Fn, T. forsythia and F. nucleatum infection. Sections were analyzed for CD45-positive cells (A) or neutrophils (C) by immunostaining (brown cells) and counterstained with hematoxylin and eosin (H&E). All images were analyzed by Aperio image software. Magnification, ×400. Inflammatory cells were quantified as numbers of CD45+ cells (B) or neutrophils (D) per mm2. Error bars indicate standard deviations (4 mice/group). Statistically significant differences between groups are indicated (***, P < 0.001 versus sham-infected mice; *, P < 0.01 versus sham-infected mice; n.s, not significant versus Fn:Tf mixed infection).
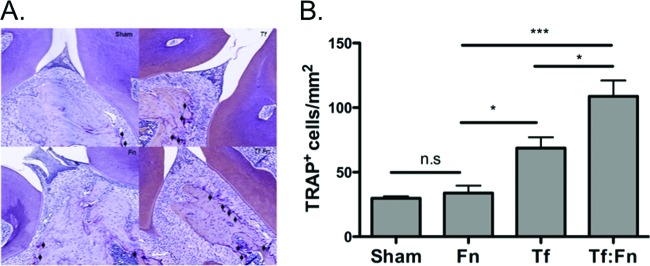
With regard to the osteoclastic activity, as expected (27), increased osteoclastic activity was observed in T. forsythia-infected mice compared to that in sham-infected mice. Interestingly, a significant increase in the number of TRAP+ cells was observed in coinfected mice compared to that in monoinfected mice (Fig. 7A and B). Therefore, the results suggested that coinfection with F. nucleatum and T. forsythia is more potent than infection with either organism alone in inducing inflammation and enhancing the bone osteoclastic activity associated with alveolar bone loss.
Fig 7.
Coinfection causes increased osteoclastic activity. (A) Representative histological sections showing TRAP+ cells (indicated with arrows) from sham-, F. nucleatum-, T. forsythia-, or F. nucleatum- and T. forsythia-infected mice. (B) Average numbers of TRAP+ cells in 10 high-power magnification fields per slide (4 mice/group). Statistically significant differences between infected and sham-infected groups are indicated (***, P < 0.001; *, P < 0.05; n. s, not significant).
DISCUSSION
T. forsythia is implicated in the pathogenesis of various forms of periodontal diseases (41). We previously showed that T. forsythia induces cytokine release from monocytic and epithelial cells (28) and forms synergistic biofilm with F. nucleatum species in a contact-dependent manner (36). In vivo, T. forsythia induces alveolar bone loss in mice (27). On the other hand, F. nucleatum is present in healthy individuals but is considered an opportunistic pathogen in polymicrobial infections that is known to contribute to various conditions, such as bacterial vaginosis (10), acute appendicitis (39), and anaerobic bacteremia, (8) and to be a risk factor for preterm birth (5, 16). In the oral cavity, F. nucleatum is believed to contribute to the maintenance of healthy mucosal surfaces by increasing the expression of antimicrobial peptides and inhibitors that block neutrophil proteases, minimizing tissue damage (46). F. nucleatum is also a bridging bacterium which facilitates plaque formation (51). The influence of F. nucleatum in mixed infections during pathogenesis has been evaluated previously in animal models. For example, F. nucleatum in combination with P. gingivalis causes increased alveolar bone loss compared to that induced by either bacterium alone (30). However, F. nucleatum did not further affect the alveolar bone loss due to polymicrobial infection comprising P. gingivalis, T. denticola, and T. forsythia (21). As for mixed infection involving F. nucleatum and T. forsythia, a previous study demonstrated the influence of F. nucleatum on T. forsythia virulence (40). This study showed synergistic enhancement of abscess formation in rabbits by mixed infection. The present study was undertaken to evaluate the effect of F. nucleatum on the ability of T. forsythia to induce alveolar bone loss in a mouse model to more closely mimic the pathogenesis associated with periodontitis. Our results demonstrated a significant increase in NF-κB activity in monocytic cells as well as secretion of the proinflammatory cytokines IL-1β, TNF-α, and IL-6 in mouse macrophages in response to mixed-species infection compared to that following individual-species challenge. F. nucleatum and T. forsythia in a 1:1 proportion at low multiplicities of infection induced significantly greater inflammatory responses than either species alone at similar multiplicities of infection. At higher multiplicities, the differences in the inflammatory response between mixed- and single-species challenges were not evident. It is possible that receptor saturation and/or tolerance occurring due to increasing bacterial loads is a cause. Our data are in agreement with previous studies that reported increased cytokine release by epithelial cells in response to mixed infection of T. forsythia and F. nucleatum (22) and by dendritic cells in response to mixed infection of F. nucleatum and P. gingivalis (18) in comparison to that in response to monoinfections. In vivo, T. forsythia and F. nucleatum induced alveolar bone loss in a synergistic manner (greater than the additive responses by each bacterium). Strikingly, F. nucleatum coinfection led to a dampening of the antibody response to T. forsythia. Although we do not know the reason for this phenomenon, a previous study also observed a dampened antibody response to P. gingivalis following exposure to F. nucleatum (15). It is also likely that F. nucleatum suppresses the expression or the availability of antigenically dominant T. forsythia antigens, a result which causes a reduced bacterium-specific antibody response. Strikingly, some degree of cross-reactive antibodies to T. forsythia was observed in F. nucleatum-monoinfected mice and vice versa. The reason for this observation is not fully known except that this might represent cross-reactivity of bacterial antigens to antibodies elicited to conserved microbial components/antigens of these species and the resident mouse flora. Our in vivo data showed increased inflammatory cell (CD45+) and neutrophil infiltration in the gingival tissues of mice challenged with mixed-species infection compared to that in mice with monospecies infections. This increased inflammation also led to a corresponding increase in osteoclastic activity (TRAP+). This study demonstrates a synergy between F. nucleatum and T. forsythia with regard to virulence in a periodontitis model. As mixed-infection partners, they induce alveolar bone loss that is significantly greater than the additive bone losses induced by each species alone.
Mechanistically, it remains to be determined how F. nucleatum and T. forsythia mixed challenge modulates innate and adaptive responses to induce synergistic alveolar bone loss. In this regard, polymicrobial challenge might engage multiple pattern recognition receptors (PRRs) through multiple pathogen-associated molecular patterns (PAMPs). Such PRR-PAMP interactions might induce a variety of transducing signals, each resulting in a cellular response that could be either cooperative or competing, ultimately influencing the overall host response. It has been demonstrated that bacterial products such as LPS, muramyl peptide, and bacterial-derived formylated peptides (fMLP) can synergistically induce proinflammatory cytokine responses. These synergistic effects have been shown to be mediated via the regulation of posttranscriptional turnover and translation of cytokine mRNA (14, 43) as well as the NF-κB signaling pathways (9). Moreover, the cytokine responses may possibly be influenced by the action of bacterial sialidases on Toll-like receptors (TLRs) (1, 2); T. forsythia NanH sialidase (32, 38) might play a role in this regard. Regarding NF-κB activation, NF-κB is present in the cytoplasm as an inactive heterodimer of an IκB family inhibitor subunit, a 50-kDa DNA-binding subunit (p50), and a 65-kDa transactivation subunit (RelA/p65) (17). Receptor-ligand engagement causes kinase-dependent phosphorylation and ubiquitin-mediated degradation of the IκB inhibitor, releasing the active p50/p65 heterodimer to translocate to the nucleus and initiate transcription (45). It has been shown that optimal transactivation of gene expression requires phosphorylation of the Ser-536 of p65 (45). In addition, mixed challenge with bacterial LPS and fMLP synergistically activates NF-κB by phosphorylating both Ser-536 and Ser-276 of p65 (9). Another mechanism by which polymicrobial species might synergize during pathogenesis is through enhancing the invasion ability of partner species in a mixed infection (19, 22), which, in turn, may allow increased stimulation of intracellular receptors, such as NOD1 and TLR9. During synergistic alveolar bone loss due to mixed challenge in our study, increased T. forsythia load due to F. nucleatum may be a contributing factor, since F. nucleatum may have a growth-promoting effect on T. forsythia. This notion is supported by the fact that F. nucleatum has the ability to generate reducing conditions (11) and provide the exogenous amino sugar MurNAc for the growth of T. forsythia (7, 44). T. forsythia lacks key enzymes needed for the de novo production of this essential amino sugar needed for peptidoglycan synthesis. Still other nutritional relationships may exist; for example, the genome of F. nucleatum strain 25586 contains a complete sialic acid metabolic system (38) and may utilize free sialic acid released by the action of the T. forsythia NanH sialidase enzyme on salivary or epithelial glycoproteins (32). In addition, NanH may act to expose subterminal galactose residues on human glycoproteins to which F. nucleatum is well known to bind via a galactose-binding lectin (26, 33). Whether F. nucleatum presence results in increased growth and colonization of T. forsythia in the oral cavity or vice versa is difficult to assess due to the lack of suitable methods to quantify bacteria from the mouse oral cavity. While PCR analyses for the presence of 16S rRNA genes in mouse oral swabs are routinely used to assess colonization, it is not feasible to obtain samples from the subgingival sites of mice to truly estimate bacterial counts relevant for the study.
In conclusion, our data strongly indicate that mixed T. forsythia-F. nucleatum challenge induces a more robust inflammatory response, which, in turn, is responsible for synergistic effects on alveolar bone loss.
ACKNOWLEDGMENTS
This study was supported by USPHS research grants DE014749 and DE019424 from the National Institute of Dental and Craniofacial Research. G.P.S. is supported by Dunhill Medical Trust grant no. R185/0211.
Footnotes
Published ahead of print 30 April 2012
REFERENCES
- 1. Amith SR, et al. 2010. Neu1 desialylation of sialyl alpha-2,3-linked beta-galactosyl residues of TOLL-like receptor 4 is essential for receptor activation and cellular signaling. Cell Signal. 22:314–324 [DOI] [PubMed] [Google Scholar]
- 2. Amith SR, et al. 2009. Dependence of pathogen molecule-induced toll-like receptor activation and cell function on Neu1 sialidase. Glycoconj. J. 26:1197–1212 [DOI] [PubMed] [Google Scholar]
- 3. Baeuerle PA, Henkel T. 1994. Function and activation of NF-κB in the immune system. Annu. Rev. Immunol. 12:141–179 [DOI] [PubMed] [Google Scholar]
- 4. Baker PJ, Evans RT, Roopenian DC. 1994. Oral infection with Porphyromonas gingivalis and induced alveolar bone loss in immunocompetent and severe combined immunodeficient mice. Arch. Oral Biol. 39:1035–1040 [DOI] [PubMed] [Google Scholar]
- 5. Barak S, Oettinger-Barak O, Machtei EE, Sprecher H, Ohel G. 2007. Evidence of periopathogenic microorganisms in placentas of women with preeclampsia. J. Periodontol. 78:670–676 [DOI] [PubMed] [Google Scholar]
- 6. Bolstad AI, Jensen HB, Bakken V. 1996. Taxonomy, biology, and periodontal aspects of Fusobacterium nucleatum. Clin. Microbiol. Rev. 9:55–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Braham PH, Moncla BJ. 1992. Rapid presumptive identification and further characterization of Bacteroides forsythus. J. Clin. Microbiol. 30:649–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brook I. 2010. The role of anaerobic bacteria in bacteremia. Anaerobe 16:183–189 [DOI] [PubMed] [Google Scholar]
- 9. Chen LY, et al. 2009. Synergistic induction of inflammation by bacterial products lipopolysaccharide and fMLP: an important microbial pathogenic mechanism. J. Immunol. 182:2518–2524 [DOI] [PubMed] [Google Scholar]
- 10. Citron DM. 2002. Update on the taxonomy and clinical aspects of the genus Fusobacterium. Clin. Infect. Dis. 35:S22–S27 [DOI] [PubMed] [Google Scholar]
- 11. Diaz PI, Zilm PS, Rogers AH. 2002. Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbon-dioxide-depleted environments. Microbiology 148:467–472 [DOI] [PubMed] [Google Scholar]
- 12. Dzink JL, Socransky SS, Haffajee AD. 1988. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J. Clin. Periodontol. 15:316–323 [DOI] [PubMed] [Google Scholar]
- 13. Feuille F, Ebersole JL, Kesavalu L, Stepfen MJ, Holt SC. 1996. Mixed infection with Porphyromonas gingivalis and Fusobacterium nucleatum in a murine lesion model: potential synergistic effects on virulence. Infect. Immun. 64:2094–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gao JJ, Xue Q, Papasian CJ, Morrison DC. 2001. Bacterial DNA and lipopolysaccharide induce synergistic production of TNF-alpha through a post-transcriptional mechanism. J. Immunol. 166:6855–6860 [DOI] [PubMed] [Google Scholar]
- 15. Gemmell E, Bird PS, Carter CL, Drysdale KE, Seymour GJ. 2002. Effect of Fusobacterium nucleatum on the T and B cell responses to Porphyromonas gingivalis in a mouse model. Clin. Exp. Immunol. 128:238–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Han YW, et al. 2004. Fusobacterium nucleatum induces premature and term stillbirths in pregnant mice: implication of oral bacteria in preterm birth. Infect. Immun. 72:2272–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hatada EN, Krappmann D, Scheidereit C. 2000. NF-κB and the innate immune response. Curr. Opinion. immunol. 12:52–58 [DOI] [PubMed] [Google Scholar]
- 18. Huang CB, Altimova Y, Strange S, Ebersole JL. 2011. Polybacterial challenge effects on cytokine/chemokine production by macrophages and dendritic cells. Inflamm. Res. 60:119–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Inagaki S, Onishi S, Kuramitsu HK, Sharma A. 2006. Porphyromonas gingivalis vesicles enhance attachment, and the leucine-rich repeat BspA protein is required for invasion of epithelial cells by “Tannerella forsythia.” Infect. Immun. 74:5023–5028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kantarci A, Oyaizu K, Van Dyke TE. 2003. Neutrophil-mediated tissue injury in periodontal disease pathogenesis: findings from localized aggressive periodontitis. J. Periodontol. 74:66–75 [DOI] [PubMed] [Google Scholar]
- 21. Kesavalu L, et al. 2007. Rat model of polymicrobial infection, immunity, and alveolar bone resorption in periodontal disease. Infect. Immun. 75:1704–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kirschbaum M, Schultze-Mosgau S, Pfister W, Eick S. 2010. Mixture of periodontopathogenic bacteria influences interaction with KB cells. Anaerobe 16:461–468 [DOI] [PubMed] [Google Scholar]
- 23. Kolenbrander PE. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413–437 [DOI] [PubMed] [Google Scholar]
- 24. Kumar PS, et al. 2006. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J. Clin. Microbiol. 44:3665–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee SF, Andrian E, Rowland E, Marquez IC. 2009. Immune response and alveolar bone resorption in a mouse model of Treponema denticola infection. Infect. Immun. 77:694–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mangan DF, Novak MJ, Vora SA, Mourad J, Kriger PS. 1989. Lectinlike interactions of Fusobacterium nucleatum with human neutrophils. Infect. Immun. 57:3601–3611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Myneni SR, et al. 2011. TLR2 signaling and Th2 responses drive Tannerella forsythia-induced periodontal bone loss. J. Immunol. 187:501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Onishi S, et al. 2008. Toll-like receptor 2-mediated interleukin-8 expression in gingival epithelial cells by the Tannerella forsythia leucine-rich repeat protein BspA. Infect. Immun. 76:198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Orth RK, O'Brien-Simpson NM, Dashper SG, Reynolds EC. 2011. Synergistic virulence of Porphyromonas gingivalis and Treponema denticola in a murine periodontitis model. Mol. Oral Microbiol. 26:229–240 [DOI] [PubMed] [Google Scholar]
- 30. Polak D, et al. 2009. Mouse model of experimental periodontitis induced by Porphyromonas gingivalis/Fusobacterium nucleatum infection: bone loss and host response. J. Clin. Periodontol. 36:406–410 [DOI] [PubMed] [Google Scholar]
- 31. Rickard AH, Gilbert P, High NJ, Kolenbrander PE, Handley PS. 2003. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 11:94–100 [DOI] [PubMed] [Google Scholar]
- 32. Roy S, Honma K, Douglas I, Sharma A, Stafford G. 2011. Role of sialidase in glycoprotein utilisation by Tannerella forsythia. Microbiology 157:3195–3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shaniztki B, Hurwitz D, Smorodinsky N, Ganeshkumar N, Weiss EI. 1997. Identification of a Fusobacterium nucleatum PK1594 galactose-binding adhesin which mediates coaggregation with periopathogenic bacteria and hemagglutination. Infect. Immun. 65:5231–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sharma A. 2010. Virulence mechanisms of Tannerella forsythia. Periodontol. 2000 54:106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sharma A, et al. 2005. Tannerella forsythia-induced alveolar bone loss in mice involves leucine-rich-repeat BspA protein. J. Dent. Res. 84:462–467 [DOI] [PubMed] [Google Scholar]
- 36. Sharma A, Inagaki S, Sigurdson W, Kuramitsu HK. 2005. Synergy between Tannerella forsythia and Fusobacterium nucleatum in biofilm formation. Oral. Microbiol. Immunol. 20:39–42 [DOI] [PubMed] [Google Scholar]
- 37. Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134–144 [DOI] [PubMed] [Google Scholar]
- 38. Stafford G, Roy S, Honma K, Sharma A. 2012. Sialic acid, periodontal pathogens and Tannerella forsythia: stick around and enjoy the feast! Mol. Oral Microbiol. 27:11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Swidsinski A, et al. 2011. Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut 60:34–40 [DOI] [PubMed] [Google Scholar]
- 40. Takemoto T, Kurihara H, Dahlen G. 1997. Characterization of Bacteroides forsythus isolates. J. Clin. Microbiol. 35:1378–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tanner AC, Izard J. 2006. Tannerella forsythia, a periodontal pathogen entering the genomic era. Periodontol. 2000 42:88–113 [DOI] [PubMed] [Google Scholar]
- 42. Weiss EI, et al. 2000. Attachment of Fusobacterium nucleatum PK1594 to mammalian cells and its coaggregation with periodontopathogenic bacteria are mediated by the same galactose-binding adhesin. Oral Microbiol. Immunol. 15:371–377 [DOI] [PubMed] [Google Scholar]
- 43. Wolfert MA, Murray TF, Boons GJ, Moore JN. 2002. The origin of the synergistic effect of muramyl dipeptide with endotoxin and peptidoglycan. J. Biol. Chem. 277:39179–39186 [DOI] [PubMed] [Google Scholar]
- 44. Wyss C. 1989. Dependence of proliferation of Bacteroides forsythus on exogenous N-acetylmuramic acid. Infect. Immun. 57:1757–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang F, Tang E, Guan K, Wang CY. 2003. IKK beta plays an essential role in the phosphorylation of RelA/p65 on serine 536 induced by lipopolysaccharide. J. Immunol. 170:5630–5635 [DOI] [PubMed] [Google Scholar]
- 46. Yin L, Dale BA. 2007. Activation of protective responses in oral epithelial cells by Fusobacterium nucleatum and human beta-defensin-2. J. Med. Microbiol. 56:976–987 [DOI] [PubMed] [Google Scholar]
- 47. Yoshida-Minami I, et al. 1997. Alveolar bone loss in rats infected with a strain of Prevotella intermedia and Fusobacterium nucleatum isolated from a child with prepubertal periodontitis. J. Periodontol. 68:12–17 [DOI] [PubMed] [Google Scholar]
- 48. Yu JJ, et al. 2007. An essential role for IL-17 in preventing pathogen-initiated bone destruction: recruitment of neutrophils to inflamed bone requires IL-17 receptor-dependent signals. Blood 109:3794–3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zambon JJ, et al. 1994. Epidemiology of subgingival bacterial pathogens in periodontal diseases, p 3–12 In Genco RJ, Hamada S, Lehrer JR, McGhee JR, Mergenhagen S. (ed), Molecular pathogenesis of periodontal diseases. ASM Press, Washington, DC [Google Scholar]
- 50. Zheng X, Goncalves R, Mosser DM. 2008. The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. Chapter 14:Unit 14.1. doi:10.1002/0471142735.im1401s83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zijnge V, et al. 2010. Oral biofilm architecture on natural teeth. PLoS One 5:e9321 doi:10.1371/journal.pone.0009321 [DOI] [PMC free article] [PubMed] [Google Scholar]