Abstract
The ability to cope with endogenous or host-generated reactive oxygen species is considered a key virulence attribute of the opportunistic pathogen Enterococcus faecalis, a leading cause of hospital-acquired infections. In this study, we used in silico and mutational analyses to identify and characterize the role of the Spx global regulator in oxidative stress tolerance and virulence in E. faecalis. While the Δspx strain grew as well as the wild-type strain under anaerobic conditions, the mutant strain exhibited impaired growth under aerobic conditions and was highly sensitive to oxidative stress agents. The spx mutant strain was also sensitive to a variety of other stressful conditions, including antibiotic stress and killing by the mouse-derived macrophage cell line J774. Using a murine model of foreign body-associated peritonitis, we demonstrated that the ability of the Δspx strain to colonize the peritoneum and disseminate in the bloodstream was significantly reduced compared to that of the parent strain. Transcriptional analysis revealed that a large number of known oxidative stress genes are under positive control by Spx. Collectively, our results show that Spx is a major stress gene regulator and is implicated in the pathophysiology of E. faecalis. The relationship of Spx to other oxidative stress regulators is also discussed.
INTRODUCTION
Enterococcus faecalis is a ubiquitous commensal bacterium that inhabits the gastrointestinal (GI) tracts of mammals and insects and is also found as a contaminant of freshwater, soils, plants, and abiotic surfaces. Despite being a harmless commensal in healthy individuals, E. faecalis is often a causative agent of opportunistic infections, especially in hospitalized patients with underlying diseases that impair the immune system, as well as in patients undergoing prolonged antibiotic treatment (7, 15, 25). In contrast to other Gram-positive pathogens, such as Streptococcus pyogenes and Staphylococcus aureus, E. faecalis expresses a relatively small number of classic virulence factors, such as secreted toxins, proteases, and immune modulators. In the case of E. faecalis infections, virulence is intimately associated with the capacity of the bacterium to withstand environmental and antibiotic stresses.
Oxidative stress is a major challenge encountered by bacteria in the human host, and the ability to survive the oxidative burst of phagocytic cells in the early stages of infections is a key virulence attribute of E. faecalis (37). In fact, many E. faecalis infections have an endogenous origin from the GI tract, and it has been proposed that this bacterium uses macrophages to translocate across intact intestinal tissue and cause systemic infections (50). Furthermore, E. faecalis must also cope with reactive oxygen species (ROS) generated by its own metabolism under aerobic conditions (13). Finally, a relatively recent study implicated the generation of endogenous ROS as the primary trigger of cell death following treatment with bactericidal antibiotics (17).
The oxidative stress responses of E. faecalis have been studied in some detail. Earlier studies revealed that a 30-min adaptation to a sublethal concentration of H2O2 conferred a >200-fold increased tolerance to a subsequent challenge with a lethal concentration of H2O2 (8). Compared to phylogenetically related streptococci and lactococci, E. faecalis is significantly more tolerant to ROS stress, a trait that can be explained partially by the larger number of ROS detoxification enzyme genes present in the genome of E. faecalis than in those of other Gram-positive cocci (3, 33, 37). For example, E. faecalis is the only pathogenic Gram-positive cocci expressing katA, a gene encoding a heme-dependent catalase (9). In addition to KatA, several other detoxification enzymes have been characterized in E. faecalis, including three peroxidases (NADH peroxidase [Npr], alkyl hydroperoxide reductase [AhpCF], and thiol peroxidase [Tpx]) (20), a manganese-containing superoxide dismutase (Sod) (34, 46), and a general stress protein homologous to the organic hydroperoxide resistance (Ohr) protein of Bacillus subtilis (38). In addition, the genome of E. faecalis encodes several other conserved enzymes that participate in detoxification, including NADH oxidase (Nox), glutathione reductase (Gor), and the thioredoxin complex (TrxA-TrxB) (3, 33).
In bacteria, the expression of genes encoding antioxidant enzymes is tightly controlled at the transcriptional level. The majority of the oxidative stress regulators sense changes in the intracellular redox environment, and as a result, they induce or derepress the transcription of genes that participate in ROS detoxification. In E. faecalis, at least two oxidative stress gene regulators, named HypR and PerR, have been characterized. HypR is a regulator of the LysR family that is homologous to the Escherichia coli OxyR regulator (42) and was shown to activate transcription of ahpCF, gor, katA, and sodA (49). An E. faecalis hypR mutant was highly sensitive to H2O2 stress and highly susceptible to killing by macrophages and showed reduced virulence in a mouse peritonitis model (47, 49). PerR, first identified in B. subtilis, is a member of the ferric uptake regulator (Fur) family that was shown to repress transcription of oxidative stress genes (24). In E. faecalis, PerR was shown to exert modest control over the transcription of oxidative stress genes (48). However, a perR mutant was significantly more resistant to H2O2 challenge and showed reduced virulence in a mouse peritonitis model (48).
In addition to HypR and PerR, the E. faecalis V583 genome encodes a few other putative oxidative stress regulators, including a second member of the Fur family (EF1585), a member of the Zur family (EF2417), and a member of the Spx/ArsC family (EF2678) (33, 37). Spx is highly conserved among low-GC Gram-positive bacteria and was first identified as a suppressor of clpP and clpX phenotypes in B. subtilis (26). Subsequent work specifically demonstrated that Spx is normally degraded by Clp proteolysis (30). While it has no DNA binding domain, the B. subtilis Spx protein functions in the disulfide stress response by interacting with the α C-terminal domain (α-CTD) of RNA polymerases to repress a variety of cellular processes, such as competence development, while activating transcription of genes involved in oxidative stress responses (27–29, 52). More recently, some attention has been given to the role of Spx in pathogenic organisms. In Streptococcus mutans, two Spx proteins, SpxA and SpxB, were shown to modulate stress tolerance and virulence by controlling the expression of several genes, including activation of several oxidative stress genes (16). Spx was also shown to affect stress tolerance and biofilm formation in S. aureus (32) and to control competence development in Streptococcus pneumoniae (44).
In this report, we used a mutagenesis approach to characterize the role of the Spx regulator of E. faecalis under conditions that are relevant for survival and colonization. The Δspx strain was highly sensitive to oxidative stress agents as well as to a variety of other stress conditions, including bactericidal antibiotics. In addition, the Δspx mutant was also highly susceptible to macrophage killing in vitro and showed attenuated virulence in a mouse model of foreign material-associated peritonitis. Finally, transcriptional analysis revealed that Spx controls a large number of genes involved in defense against oxidative stress, including several genes that are also controlled by HypR (47, 49).
MATERIALS AND METHODS
Bacterial strains.
Strains used in this study are listed in Table 1. Strains were routinely grown on Trypticase soy agar (TSA) or in brain heart infusion (BHI) medium at 37°C. E. coli strains were grown in Luria broth and used for plasmid construction and propagation. Where required for selective growth of strains, erythromycin (10 μg ml−1 for E. faecalis and 300 μg ml−1 for E. coli), spectinomycin (1,000 μg ml−1), fusidic acid (25 μg ml−1), or rifampin (200 μg ml−1) was added to the growth medium. For complementation studies, cells were grown in TY base medium (3% tryptone, 0.5% yeast extract) containing erythromycin and 1 to 2% rhamnose.
Table 1.
Strains and primers used for spx gene inactivation or complementation
| Strain or primer | Relevant characteristics or sequencea | Primer application | Source |
|---|---|---|---|
| Strains | |||
| E. faecalis strains | |||
| OG1RF | Wild-type strain; Rifr Fusr | Laboratory stock | |
| CK111 | OG1Sp upp4::P23 repA4 | Laboratory stock | |
| JAL4 | spx deletion mutant of OG1RF; Δspx Rifr Fusr | This study | |
| JAL5 | JAL4 Δspx harboring pCJK96spx; Rifr Fusr Ermr | This study | |
| JAL6 | OG1RF strain harboring pCJK96; Rifr Fusr Ermr | This study | |
| JAL7 | JAL4 strain harboring pCJK96; Rifr Fusr Ermr | This study | |
| E. coli strains | |||
| DH10B | Cloning host | Laboratory stock | |
| EC1000 | Host for cloning RepA-dependent plasmids | Laboratory stock | |
| Primers | |||
| Spx5′arm1 | CTAAGTGTTTCTAGAGATCATCTC | spx deletion | |
| Spx3′arm1 | GAAGTATAAGGATCCAACATTGTGC | spx deletion | |
| Spx5′arm2 | ACTGGAATTGGATCCAGCACAATT | spx deletion | |
| Spx3′arm2 | AGTCAAAACCCATGGCATGAAGAC | spx deletion | |
| 5′EF3spx | GGAGTGAACTAGTATGTTGACAC | Δspx complementation | |
| 3′EF2spx | GAACAAAATTCTAGATTTTTTATAAACCTGCC | Δspx complementation |
The underlined bases correspond to restriction sites included to aid in the subsequent cloning of the PCR products.
Construction and complementation of Δspx strain.
A markerless genetic exchange system (18) was used to delete the spx (EF2678) gene in E. faecalis OG1RF. Briefly, two PCR products flanking spx were obtained with the primers listed in Table 1. The PCR amplicons were approximately 1 kb long, and following enzymatic digestion, they included the first 5 and last 16 bp of the spx gene, which were retained in the spx mutant to avoid unanticipated effects on the expression of adjacent genes. After digestion with the appropriate restriction enzymes, the two PCR products flanking the gene of interest were simultaneously cloned into pGEM5 (Promega, Madison, WI) to give plasmid pGspx. The 2-kb fragment containing the spx up- and downstream fragments was subcloned into pCJK47 (18), using E. coli EC1000 as the host strain. The resulting plasmid, pCJK-spx, was electroporated into competent E. faecalis CK111 (donor strain). E. faecalis CK111 containing pCJK-spx was conjugated with wild-type E. faecalis OG1RF, and transformants were selected on BHI agar medium containing rifampin, fusidic acid, and erythromycin. Single colonies were subjected to the PheS* negative counterselection system to isolate double-crossover integrations (18), and the spx gene deletion was confirmed by PCR and sequencing of the insertion site and flanking sequences. To complement the spx mutation in trans, the rhamnose-inducible vector pCJK96 (19), expressing full-length spx, was isolated in E. coli and subsequently electroporated into the E. faecalis Δspx strain.
Stress challenges.
To investigate the role of E. faecalis Spx in stress responses, cells of wild-type OG1RF and its isogenic spx mutant were subjected to a variety of stress challenges. To assess oxygen and heat tolerance of the spx mutant, cultures of the parent and Δspx strains grown to an optical density at 600 nm (OD600) of 0.2 were serially diluted in 0.9% saline, and 5-μl dilutions were spotted on TY plates containing 2% rhamnose and 10 μg ml−1 erythromycin. Plates were incubated for up to 72 h at 37 or 46°C under both anaerobic (in an anaerobic jar) and aerobic conditions. To evaluate the capacity of the E. faecalis strains to grow under stress conditions (shaking at 250 rpm, 1 μg ml−1 diamide, 0.5 mM H2O2, 0.01% deoxycholate, 0.02% SDS, 5% NaCl, and pH 5.5), mid-exponential-phase cultures grown in BHI were diluted 1:100 in fresh BHI medium adjusted to each specific condition, and growth was monitored using a Bioscreen C growth monitor (Oy Growth Curves AB Ltd., Helsinki, Finland) set to 37°C. The capacity of the Δspx mutant to tolerate oxidative stress was further investigated in an H2O2 killing assay. Briefly, exponentially grown cultures were incubated at 37°C in the presence of 30 mM H2O2. Aliquots were taken at 0, 15, 30, and 60 min, serially diluted, and plated on TSA. Plates were incubated at 37°C for 48 h before colonies were counted.
Antibiotic time-kill kinetics.
Cultures were grown in BHI medium to exponential phase and serially diluted in fresh BHI medium to obtain a starting inoculum of 5 × 106 to 1 × 107 CFU ml−1. Time-kill studies were initiated by adding 5 times the MIC of ampicillin (80 μg ml−1), vancomycin (40 μg ml−1), or chloramphenicol (80 μg ml−1) for the wild-type strain. Viable counts were determined at time zero and then at selected time points.
RNA analysis.
For RNA analysis, cells were grown in BHI medium to an OD600 of 0.3 (control), and an aliquot of 100 ml was transferred to a 500-ml flask and incubated in a rotary shaker set to 250 rpm at 37°C for up to 30 min. To isolate RNA from E. faecalis, cells were harvested by centrifugation at 4°C and then treated with RNA Protect reagent (Qiagen, Inc., Chatsworth, CA). Total RNA was isolated from homogenized E. faecalis cells by the hot acid-phenol method as described previously (1). RNA pellets were resuspended in nuclease-free H2O and treated with DNase I (Invitrogen, Gaithersburg, MD) at 37°C for 30 min. RNA concentrations were determined using a Nanovue Plus spectrophotometer (G&E Life Sciences, Piscataway, NJ), and 1-μl samples were run in an agarose gel to verify RNA integrity. The RNA was purified again using an RNeasy minikit (Qiagen), including a second on-column DNase treatment that was performed as recommended by the supplier. For real-time quantitative reverse transcription-PCR (qRT-PCR), gene-specific primers for known oxidative stress genes (Table 2) were designed using Beacon Designer 2.0 software (Premier Biosoft International). Reverse transcription and qRT-PCR were carried out according to protocols described elsewhere (1). Student's t test was performed to verify the significance of the real-time PCR quantifications.
Table 2.
Real-time PCR primers used in this study
| Gene | Primer sequence |
Product size (bp) | |
|---|---|---|---|
| Forward | Reverse | ||
| ahpC | AAACAGAGTAAACCTCACAA | ACACGAGTATCAACAGAAG | 151 |
| dps | TATATGCGTGGACCTGAA | ATGTTGAGAATGGAGAGC | 127 |
| gor | CCGTATATTCTGTCCCATTG | TACGGGTTTGATGTTGAAAT | 187 |
| hypR | AGAACGGCTAGACAAAGGAT | CGGCTGCAGTAATAACATTT | 158 |
| katA | AGAAGCTGTTTGGGATTTTT | GATTCCTTGATTCGTCTTGA | 190 |
| msrA | AGTTGTTTCGGGTTATAC | TATCCACTAATTGTTCTAA | 137 |
| nox | GGCGGTTATATTGGTATT | CTGTGAATGGCTTATCTAA | 115 |
| npr | CAGTAGGGGATGCTACGTTA | GTTAATCCCTGTTGAAGCAA | 194 |
| ohr | CCAGGTGTCAAGAAAGAAAA | ATGAACTTCCAATTCAGCAC | 192 |
| perR | ATTGAGGAAGTGGAAGAA | TTTACTTTGACAACTTTGAC | 139 |
| sodA | GCATTAGAACCTTACATTGAC | TTCGCCTAATTCTGGATG | 114 |
| tpx | TATTAGAATTGACAGGTGAG | TATCAGGAACCACACTAA | 135 |
| trxB | ATGGTCGTGACAGGTGTGAA | TTTTTCACGGACATCACCAA | 213 |
E. faecalis-macrophage coculture.
Cells of the murine macrophage cell line J774A.1 (American Type Culture Collection [ATCC], Manassas, VA) were grown to confluence in Dulbecco's modified Eagle's medium (DMEM) supplemented with GlutaMAX (Invitrogen, Grand Island, NY), 10% fetal bovine serum, 50 U ml−1 penicillin G, and 50 μg ml−1 streptomycin at 37°C under a 5% CO2 atmosphere. The J774A.1 cells were seeded into 24-well tissue culture plates at a density of 5 × 105 macrophages per well 24 h prior to use. Overnight cultures of E. faecalis OG1RF and the Δspx strain were washed and resuspended in an equal volume of Hanks' buffered salt solution (HBSS) and diluted to 5 × 107 CFU ml−1 in supplemented DMEM lacking antibiotics. To the macrophage monolayers, 1 ml of diluted culture was added, yielding an approximate multiplicity of infection (MOI) of 100:1. Bacterium-macrophage contact was enhanced by centrifugation at 500 × g for 5 min followed by further incubation for 25 min. Extracellular bacteria were killed by the addition of DMEM containing 300 μg ml−1 gentamicin and 50 μg ml−1 penicillin G, followed by a 1.5-h incubation period. Macrophage cells were lysed in deionized water, serially diluted, and plated on TSA for enumeration of viable internalized bacterial cells.
In vivo model of foreign body-associated peritonitis infection.
In vivo studies were conducted under an animal protocol approved by the University of Rochester University Committee on Animal Research. Six- to 8-week-old BALB/c mice were anesthetized using ketamine-xylazine, after which they were prepared for catheter placement by clipping of their abdominal hair followed by cleansing with a povidone-iodine solution twice and one wash with 70% ethanol. Using aseptic technique, a 1-cm-long segment of sterile silicone catheter tubing (0.024-in.-outer-diameter Sani-Tech catheter; Saint-Gobain Performance Plastics Corporation, Taunton, MA) was inserted into the peritoneum by use of an 18-gauge BD spinal needle/stylette (Becton, Dickinson and Company, Franklin Lakes, NJ). The needle wound was sealed using VetBond (3M, Saint Paul, MN). To decrease surgical site pain, animals were provided with drinking water containing acetaminophen at 1 to 2 mg ml−1 beginning the day prior to surgery and stopping the day after surgery. Four hours after catheter insertion, animals (n = 10) were inoculated with 1 × 108 CFU E. faecalis OG1RF or the Δspx strain in 1 ml phosphate-buffered saline (PBS) by intraperitoneal (i.p.) injection. Forty-eight or 96 h after injection, animals were euthanized by pentobarbital injection. The abdominal wall was dissected to expose the peritoneal lining, and the peritoneum was washed with 5 ml of cold PBS. Following washing, the peritoneum was opened using aseptic technique, and the catheter segment was removed and placed into a tube containing 1 ml PBS. After three washes in PBS, the catheter was sonicated to dislodge adherent organisms, after which the medium was diluted and plated to determine catheter organism burdens. The spleen of each animal was also removed and placed in 70% ethanol to disinfect organisms residing on the exterior of the organ, followed by rinsing once in PBS. Spleens were then homogenized in 1 ml PBS, diluted, and plated for organism counts. Alternatively, some catheters were removed and processed for scanning electron microscopy (SEM) analysis at the University of Rochester EM core laboratory. Because the data from the in vivo model were distributed nonparametrically, the Mann-Whitney-Wilcoxon test was used to compare groups. The test was performed using SigmaPlot data analysis software (Systat Software Inc., San Jose, CA). P values of ≤0.05 were considered significant.
RESULTS
Identification of E. faecalis Spx.
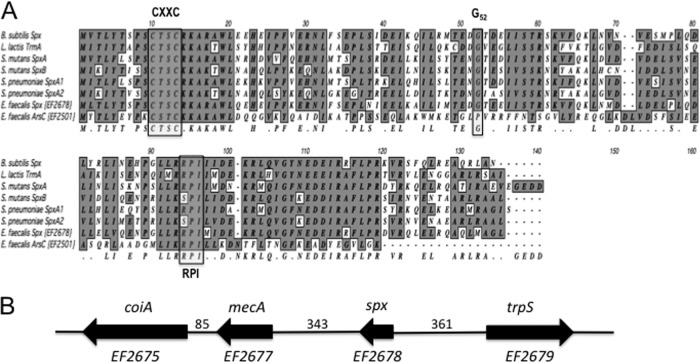
BLAST search analysis of the E. faecalis V583 genome (33) by use of the B. subtilis Spx sequence revealed two open reading frames (ORFs) encoding putative Spx/ArsC proteins (COG1393): EF2501 and EF2678. BLAST searches of the genome sequences available at the Human Oral Microbiome Database (http://www.homd.org) confirmed the presence of EF2501 and EF2678 homologues in all sequenced E. faecalis strains, including OG1RF, with the same genetic organization as that seen in V583.
Recent studies have shown that certain species of streptococci, e.g., S. mutans and S. pneumoniae (16, 44), carry two copies of the spx gene and one copy of a putative arsenate reductase gene (arsC). A phylogenetic tree of proteins containing the COG1393 domain, developed from 781 complete prokaryotic genomes, showed that these proteins can be divided into four distinct clusters: ArsC-Spx (cd03032), ArsC-ArsC (cd03034), ArsC-YffB (cd03035), and ArsC-like (cd03036) proteins (44). The previously characterized Spx proteins from B. subtilis, S. mutans, and S. pneumoniae were placed into the ArsC-Spx (cd03032) cluster (44). The proteins encoded by EF2501 and EF2678 were placed in the ArsC-like and ArsC-Spx clusters, respectively. While both the EF2501 and EF2678 proteins contain a conserved CXXC motif involved in sensing the redox state by means of disulfide bond formation (27, 31), Spx proteins possess at least two additional conserved residues/motifs: a conserved Gly52 residue responsible for the interaction of Spx with the RNA polymerase CTD (21, 31) and a C-terminal RPI motif that has been implicated in the modulation of the CXXC motif in the presence of sulfate (52). Both the EF2501 and EF2678 proteins contain the RPI motif, but the conserved Gly52 residue is found only in the EF2678 protein (Fig. 1A). Previous work with B. subtilis and S. pneumoniae demonstrated an essential role of the Gly52 residue for Spx function (44, 52), suggesting that the EF2501 protein does not interact with the α-CTD of the RNA polymerase.
Fig 1.
Identification of the Spx transcriptional regulator in E. faecalis. (A) Alignment of the amino acid sequences of known Spx proteins with those of the two putative Spx proteins in E. faecalis. Similar residues are shaded, and a consensus is shown below the sequences. Conserved motifs discussed in the text are outlined in a box. (B) Schematic diagram of the spx locus. trpS (EF2679) is predicted to code for a tryptophanyl tRNA synthetase, mecA (EF2677) codes for a putative adaptor protein that enables recognition of other proteins for proteolysis, and coiA (EF2675) codes for a transcriptional factor. Arrows indicate the direction of transcription, and numbers between genes indicate the sizes (in bp) of the intergenic regions.
In silico analysis of the EF2678 protein, henceforth designated Spx, revealed a protein of 133 amino acids with a predicted acidic pI of 5.03 and a molecular mass of 15.5 kDa. The length of the E. faecalis Spx protein is within the range of lengths for the majority of Spx proteins (around 130 to 140 amino acids). However, with the exception of S. mutans SpxA (pI of 6.06), all previously characterized Spx proteins have a basic pI (≥8.8). Nevertheless, the E. faecalis Spx protein was more closely related to B. subtilis Spx (89% similarity) than to S. mutans SpxA and SpxB (76 and 75% similarities, respectively) and S. pneumoniae SpxA1 and SpxA2 (81 and 77% similarities, respectively). Located 361 bp upstream of the spx start codon and transcribed in the opposite orientation was an ORF predicted to encode a conserved tryptophanyl tRNA synthetase (TrpS), while 343 bp downstream of the spx stop codon was a gene encoding a putative MecA protein, followed by the gene for a putative transcription factor from the CoiA superfamily, involved in competence (Fig. 1B).
The Δspx strain exhibits impaired growth in the presence of oxygen and reduced tolerance to oxidative stress agents.
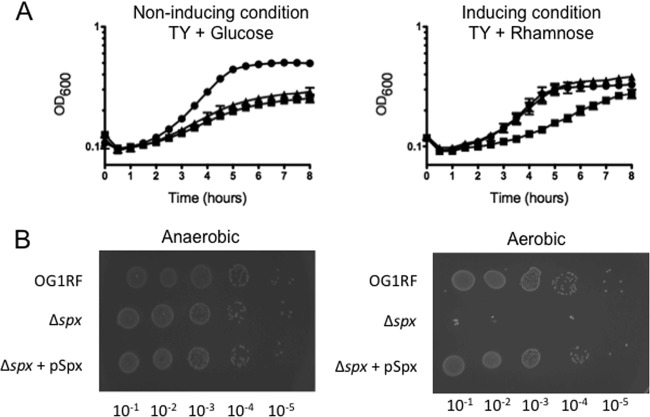
Initial attempts to obtain mutants carrying the spx deletion by use of a markerless genetic exchange system under aerobic conditions were unsuccessful. However, anaerobic incubation of the plates after the conjugation event and during counterselection yielded several isolates with the desired in-frame spx deletion, suggesting that growth of Δspx strains is severely impaired by oxygen. When it was grown aerobically in TY medium containing glucose, the Δspx strain grew more slowly than the wild-type strain, but the growth defect could be restored fully by expression of spx in trans, using the rhamnose-inducible vector pCJK96 (Fig. 2A). To confirm the oxygen sensitivity of the spx mutant, serial dilutions of the wild-type OG1RF, Δspx, and complemented Δspx strains were spotted on TY plates containing 2% rhamnose, and the plates were incubated at 37°C under aerobic and anaerobic conditions (using an anaerobic jar). The Δspx strain grew as well as the parental strain under anaerobiosis, whereas growth of the mutant strain was abolished in the presence of oxygen, a defect that was restored by complementation of the Δspx mutation (Fig. 2B).
Fig 2.
Growth characteristics of E. faecalis JAL6 (wild type + pCJK96), JAL7 (Δspx strain + pCJK96), and JAL5 (Δspx strain + pCJKSpx). (A) Growth curves for the wild-type (circles), Δspx (squares), and complemented Δspx (triangles) strains in TY containing 10 μg ml−1 erythromycin and 0.5% glucose (left) or 1% rhamnose (right) at 37°C under aerobic conditions. The data shown are averages with standard deviations for the results from three independent experiments using an automatic growth reader. (B) Growth in TY agar containing 10 μg ml−1 erythromycin and 2% rhamnose under anaerobic (left) and aerobic (right) conditions. The images shown are representative of three independent experiments. See the text for additional details.
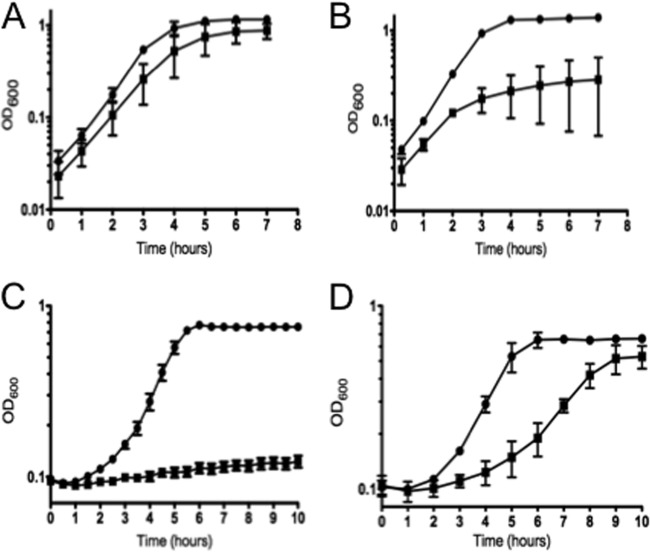
To further demonstrate the role of Spx in oxidative stress, we tested the ability of the parent and Δspx strains to grow under aeration (using a shaking incubator set to 250 rpm) or in the presence of the oxidative stress agent diamide (a thiol-specific oxidant) or H2O2. As expected, growth of the Δspx strain was severely impaired under all three conditions (Fig. 3), and these defects were alleviated in the complemented strain (see Fig. S1 in the supplemental material). We also evaluated the ability of the wild-type and Δspx strains to survive a lethal treatment with H2O2 and found that the mutant strain was significantly more sensitive to H2O2 killing that the wild-type strain (Fig. 4). Although higher concentrations of H2O2 (50 mM versus 30 mM) were used for the killing of cells grown in TY with rhamnose, the increased sensitivity to H2O2 killing of the Δspx strain was restored in the complemented strain (see Fig. S2). Collectively, these results revealed that the Δspx strain was hypersensitive to oxidative stress.
Fig 3.
Growth curves for E. faecalis OG1RF (circles) and the Δspx strain (squares) under oxidative stress conditions. Exponentially grown cultures were inoculated in BHI medium and incubated at 37°C under static conditions (A), in a shaking incubator set to 250 rpm (using a 250-ml Erlenmeyer flask filled with 100 ml of culture) (B), or under static conditions in the presence of 0.5 mM H2O2 (C) or 1 μg ml−1 diamide (D). The data shown are averages with standard deviations for the results from three independent experiments.
Fig 4.
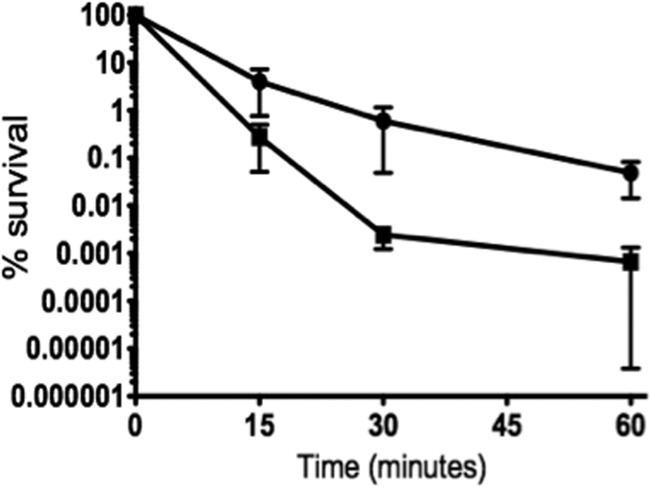
H2O2 killing of E. faecalis OG1RF (circles) and Δspx (squares) strains. Cultures grown in BHI medium to exponential growth phase were incubated at 37°C in the presence of 30 mM H2O2. Aliquots were taken at selected time points, serially diluted in PBS, and plated on TSA for CFU determination. The data shown are averages with standard deviations for the results from three independent experiments. Student's t test indicated that the differences observed at 30 and 60 min were statistically significant (P ≤ 0.05).
The Δspx strain displays impaired ability to grow under a variety of stress conditions.
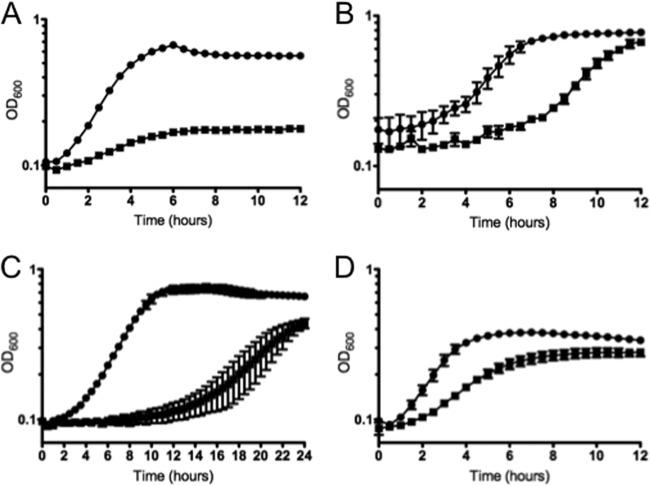
Next, we examined the ability of the strains to grow anaerobically in BHI medium at 37°C in the presence of a variety of other physiologically relevant stresses, including deoxycholate, SDS, NaCl, and pH 5.5. In all cases, the Δspx strain grew poorly compared to the parent strain (Fig. 5). Despite differences in growth rates due to the suboptimal growth conditions of TY plus rhamnose in comparison to BHI medium (compare Fig. 2A and 3B), expression of spx in trans restored the growth defect of the Δspx strain under all tested conditions (see Fig. S3 in the supplemental material). By using a plate titration assay, we also found that the Δspx strain, but not the complemented Δspx strain, displayed impaired growth at 46°C (data not shown), providing strong evidence that Spx plays a general role in stress protection.
Fig 5.
Growth of E. faecalis OG1RF (circles) and Δspx (squares) strains under different stress conditions. (A) Growth with 0.02% SDS. (B) Growth with 0.01% deoxycholate. (C) Growth with 5% NaCl. (D) Growth at pH 5.5. The data shown are averages with standard deviations for the results from at least three independent experiments.
The Δspx strain has increased susceptibility to bactericidal antibiotics.
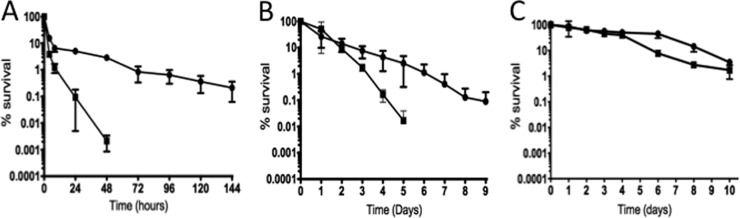
Given that the Δspx strain was highly sensitive to oxidative stress and that hydroxyl radical formation has been implicated as a common cell death mechanism induced by bactericidal antibiotics (17), we carried out antibiotic time-kill kinetics studies using two bactericidal antibiotics, namely, the cell wall inhibitors ampicillin and vancomycin. The bacteriostatic antibiotic chloramphenicol was used as a control to monitor cell viability over time. In comparison to the parent strain, the Δspx strain was killed more rapidly by both cell wall inhibitors (P ≤ 0.01) (Fig. 6A and B). As expected, treatment with chloramphenicol did not result in a statistically significant loss of cell viability compared to that of untreated cells, and both strains survived equally well over a 10-day period (Fig. 6C).
Fig 6.
Time-kill curves for logarithmic-phase cultures of the OG1RF (circles) and Δspx (squares) strains treated with 80 μg ml−1 ampicillin (A), 40 μg ml−1 vancomycin (B), or 80 μg ml−1 chloramphenicol (C). The data shown are averages with standard deviations for the results from three independent experiments.
Survival of the Δspx strain is impaired in a macrophage survival assay.
To investigate if inactivation of spx affected the ability of E. faecalis to survive within macrophages, we monitored the intracellular viability of wild-type and Δspx strains in vitro, using the mouse-derived macrophage cell line J774A.1. Despite the fact that the Δspx strain was more susceptible to antibiotics, the antibiotic treatment regimen effectively killed all wild-type and Δspx extracellular bacteria. No differences were observed in the CFU of the two strains at 30 min postinfection, indicating that the strains were internalized equally well. In coculture with J774A.1 cells, the wild-type OG1RF strain remained fully viable for up to 3 days. In contrast, the Δspx strain showed an approximately 2-log decrease (P ≤ 0.005) in viability at every 24-h period (Fig. 7).
Fig 7.
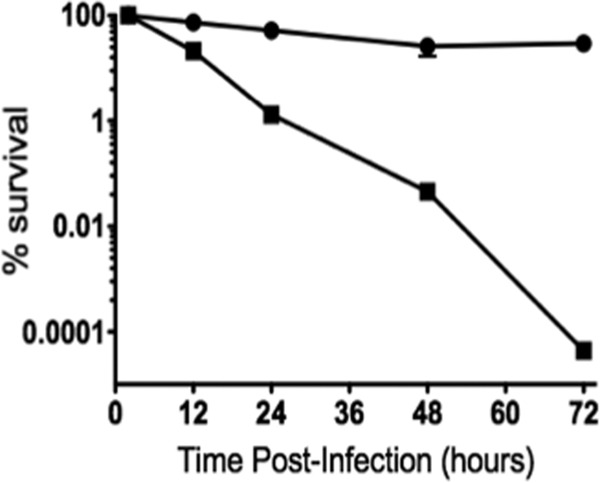
Time course of intracellular survival of the OG1RF (circles) and Δspx (squares) strains within the murine macrophage cell line J774A.1 (MOI = 100:1). The data shown are averages with standard deviations for the results from three independent experiments. Student's t test indicated that the differences observed at 24, 48, and 72 h were statistically significant (P ≤ 0.05).
Infection and dissemination of the Δspx strain are significantly impaired in a foreign body-associated peritonitis model.
To demonstrate the involvement of Spx in host infection, we adapted a mouse peritonitis infection model (40) by surgically inserting a silicone catheter into the mouse peritoneum. In a previous study, inoculation of up to 4 × 109 CFU of OG1RF into peritoneal cavities of mice resulted in no death unless the inoculum was premixed with a 50% sterile rat fecal extract (SRFE) preparation (40). Here we found that a dose of 1 × 109 CFU of OG1RF without SRFE was lethal to catheterized mice, with nearly 100% mortality within the first 24 h (data not shown). Although we cannot exclude the possibility that postsurgery recovery may also play a role in infection progression, the presence of the implant appeared to affect the pathophysiology of the infection, likely by providing the bacteria with a protected niche for colonization and survival within the host (23, 41). A dose of 1 × 108 CFU in the catheterized mice was not lethal and was used for subsequent colonization experiments.
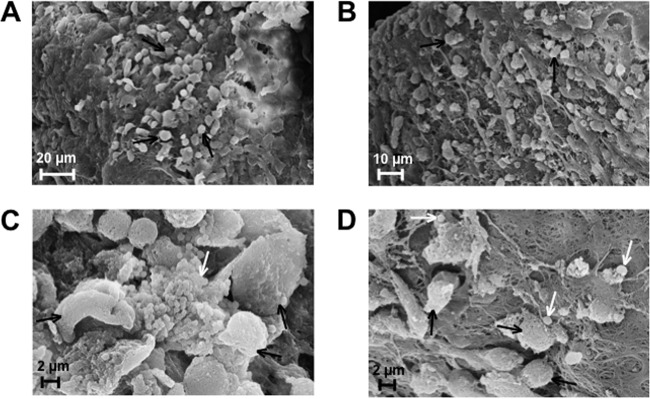
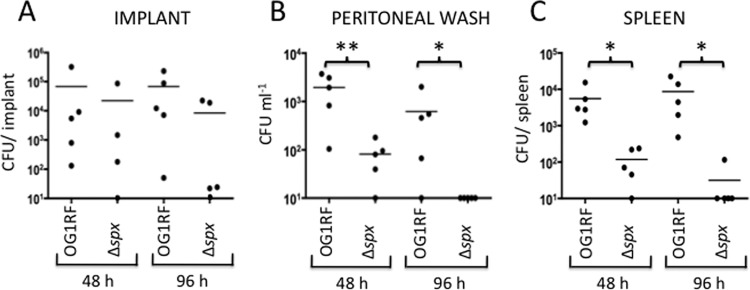
To compare the abilities of the parent and Δspx strains to establish infection, mice were inoculated with 1 × 108 CFU 4 h after catheter material insertion. To confirm the presence of bacterial biofilms on the surfaces of the implants, two independent samples were analyzed by SEM. Photomicrographs of the surfaces of catheters retrieved from the peritoneum at 48 h postinfection showed that the catheters were covered by an extracellular fibrinous matrix with embedded host cells (Fig. 8). In scattered areas, host cells appeared to be in close contact with single bacterial cells or small clusters of bacterial cells. No obvious morphological or structural differences were observed in the biofilms formed by the parent and Δspx strains. Animals were killed at 2 and 4 days postinoculation to assess bacterial loads in the implanted catheter, peritoneal cavity, and spleen (Fig. 9). The spleens were washed in ethanol after harvest to decontaminate surface-bound (peritoneal) organisms. Thus, spleen organism burdens reflect systemic dissemination of infection. The median organism burdens in the catheter samples were similar for both strains at both time points. However, the catheter of one of the five mice infected for 48 h with the Δspx strain was lost during sample processing. In contrast, the total Δspx organism burdens in the peritoneal washes and spleens (7.9 × 101 CFU ml−1 and 1.1 × 102 CFU per spleen, respectively) after 2 days were significantly lower than those of OG1RF (1.93 × 103 CFU ml−1 and 5.5 × 103 CFU per spleen, respectively; P = 0.008 for both comparisons). After 4 days of infection, this difference was even more pronounced. The number of bacteria recovered from the spleens of animals infected with OG1RF remained at a median value of 8.6 × 103 CFU ml−1; however, with one exception, no bacteria could be recovered from spleens of animals infected with the Δspx strain. Similarly, peritoneal washes of animals infected for 4 days revealed that OG1RF was still recovered at high levels (6.1 × 102 CFU ml−1), whereas the Δspx strain was no longer isolated from the peritoneal cavity. In most cases, a correlation between the CFU number found in the catheter and the CFU counts obtained in peritoneal washes or the spleen from the same animal could be observed. The exceptions were two catheters from mice infected with the Δspx strain for 96 h that showed high bacterial burdens with virtually no recovered bacteria from the peritoneal washes or spleens, suggesting that inserted foreign material provided the Δspx strain a niche to persist within the host. Note that we assessed the capacity of the Δspx strain to form biofilms in vitro by monitoring the bacterial CFU from 24-h biofilms formed on the surface of the same type of catheter used in the in vivo experiments. After several vigorous washes to remove loosely bound cells and sonication of the catheter to release tightly bound bacteria, we found no significant differences in the total CFU numbers between the wild-type and mutant strains (see Fig. S4 in the supplemental material).
Fig 8.
Scanning electron micrographs of silicone tubes implanted in the peritoneal cavities of mice infected with E. faecalis OG1RF (A and C) and the Δspx strain (B and D) for 48 h. (A and B) Low-magnification images showing catheters coated in fibrin-like matrix-containing host cells (black arrows). (C and D) High-magnification images showing bacteria (white arrows) and host cells (black arrows). Panel C shows a cluster of OG1RF cells surrounded by host cells, and panel D shows single Δspx cells also in contact with host cells. Panels A and B are representative images of implants retrieved at 48 h postinfection. Panels C and D are used to illustrate that enterococcal cells recovered from implants can be found at the outer surface of biofilms.
Fig 9.
Colonization and dissemination of E. faecalis OG1RF and Δspx strains in a foreign body-associated murine peritonitis model at 48 h (n = 5) and 96 h (n = 5) postinfection. (A) Bacterial burdens from silicone implants retrieved from the peritoneum at 48 and 96 h postinfection. (B) Bacterial burdens from peritoneal washes performed at 48 and 96 h postinfection. (C) Bacterial burdens from homogenized spleens collected at 48 and 96 h postinfection. Statistical analyses were performed using the nonparametric Mann-Whitney-Wilcoxon test. Comparisons with P values of ≤0.05 were considered significant. *, P ≤ 0.05; **, P ≤ 0.01.
Expression of several genes involved in the oxidative stress response is altered in the Δspx mutant.
The expression of a subset of known oxidative stress genes was analyzed by qRT-PCR. Experiments were carried out with RNAs isolated from cells grown at 37°C to mid-logarithmic phase under static conditions (control) and from cells that were transferred to a 37°C rotary shaker (set to 250 rpm) for 15 or 30 min (aerobic conditions). Compared to the parent strain grown under static conditions, the genes encoding the Dps DNA binding protein (dps), glutathione peroxidase (gor), methionine sulfoxide reductase A (msrA), NADH oxidase (nox), NADH peroxidase (npr), organic hydroperoxide resistance protein (ohr), superoxide dismutase (sodA), and thiolperoxidase (tpx) were downregulated in the Δspx strain (2-fold cutoff; P ≤ 0.05) (Table 3). Upon a shift to a more aerobic environment, expression of the genes encoding the heme-dependent catalase (katA) and thioredoxin reductase (trxB) was also downregulated in the mutant, whereas the initial downregulation of dps, gor, and msrA transcription was no longer observed. Among the downregulated genes, the most significant differences were observed in the expression levels of nox, ohr, and sodA (ranging from approximately 1 log to 2 1/2 log; P ≤ 0.005). These results reveal that a large number of known oxidative stress genes are under positive control by Spx. Interestingly, the ahpC (alkyl hydroperoxide reductase) and dps genes were upregulated in the Δspx strain during aerobic incubation (2-fold cutoff; P ≤ 0.05). In addition to genes responsible for detoxification and protection, we also assessed the expression levels of hypR and perR, encoding two previously characterized oxidative stress regulators of E. faecalis (47–49). No significant differences in the expression levels of hypR were observed under all conditions tested. However, compared to that in the wild type, the expression of perR was induced in the Δspx strain during aerobic growth, with a 2.6-fold induction after 15 min of shaking (not statistically significant) and a 6.5-fold induction after 30 min of shaking (P ≤ 0.05). Notably, ahpC, dps, and perR itself are known targets of the PerR repressor (11), suggesting that the increased transcription of these genes in the Δspx strain under aerobic conditions may be linked to PerR regulation (see Discussion).
Table 3.
Fold change in expression of selected genes involved in oxidative stress in the Δspx strain relative to the parent strain (OG1RF)a
| Gene | Function | Fold change in expression in Δspx strain relative to OG1RFb |
||
|---|---|---|---|---|
| Static conditions | 15 min of shaking | 30 min of shaking | ||
| ahpC | Alkyl peroxide reductase | ND | 71* | 10.1* |
| dps | DNA binding protein | 0.27* | 7.63 | 3.86* |
| gor | Glutathione reductase | 0.173** | ND | ND |
| hypR | Transcriptional regulator | ND | ND | ND |
| katA | Heme-dependent catalase | ND | 0.246* | 0.218* |
| msrA | Methionine-S-sulfoxide reductase | 0.27* | ND | ND |
| nox | H2O-forming NADH oxidase | 0.0055** | 0.13** | 0.037** |
| npr | NADH peroxidase | 0.124** | 0.351 | 0.312* |
| ohr | Organic hydroperoxide resistance protein | 0.028** | 0.016** | 0.130** |
| perR | Transcriptional regulator | ND | 2.65 | 6.5* |
| sodA | Superoxide dismutase | 0.042** | 0.006** | 0.084** |
| tpx | Thiol peroxidase | 0.26* | 0.34* | 0.491 |
| trxB | Thioredoxin reductase | ND | ND | 0.460* |
Twofold cutoff (P ≤ 0.05).
*, P ≤ 0.05; **, P ≤ 0.005; ND, no difference in expression levels detected.
DISCUSSION
ROS production by host phagocytes is a major line of defense against bacterial infections. In addition to dealing with the host-generated oxidative burst, E. faecalis is a notorious producer of extracellular H2O2 when it is grown in an oxygenated environment (12). Thus, the ability to cope with endogenously or exogenously generated ROS is considered a major factor in the virulence of E. faecalis. In this study, we identified and characterized the phenotypes associated with the inactivation of the spx gene.
Despite its long-established association with ClpXP proteolysis and with oxidative stress responses, only more recently has the characterization of Spx in Gram-positive pathogens received some attention. In Listeria monocytogenes, the gene encoding Spx was upregulated during intracellular growth, suggesting that Spx may be important for intracellular survival (4). In S. aureus, which like E. faecalis encodes only one apparent Spx homologue, inactivation of spx affected general stress tolerance and enhanced biofilm formation (32). Studies with Lactococcus lactis, S. mutans, and S. pneumoniae revealed that some species harbor two copies of the Spx protein (16, 44, 45). In S. mutans, SpxA and SpxB were shown to modulate stress tolerance through activation of oxidative stress responses and to participate in the pathogenic process as assessed by two in vivo virulence models (16). In S. pneumoniae, a double inactivation of both Spx proteins (named SpxA1 and SpxA2) was lethal, but single-gene inactivation suggested that SpxA1 negatively regulates competence development (44). In this study, we showed that Spx participates in E. faecalis stress responses and virulence. As observed with Δspx mutants of B. subtilis, S. aureus, and S. mutans, the E. faecalis Δspx mutant was highly sensitive to oxidative stresses induced by aeration, H2O2, or the thiol stressor diamide. In addition to oxidative stress, the E. faecalis Δspx strain was also highly susceptible to multiple stresses and showed impaired growth or survival at acidic pH, high temperatures, and high salt concentrations and in the presence of detergents and bactericidal antibiotics.
Using E. coli and S. aureus as model organisms, Kohanski and colleagues observed that bactericidal antibiotics such as β-lactams, aminoglycosides, and fluoroquinolones trigger overproduction of hydroxyl radicals in both species (17). Addition of the iron chelator 2,2′-dipyridyl or the hydroxyl radical scavenger thiourea conferred significant protection against killing by these drugs, leading the authors to postulate that the generation of ROS by “cidal” antibiotics induces a common cell death response in bacteria (17). The relationship of oxidative stress and antimicrobial-mediated cell killing was recently investigated in E. faecalis (2). By using a series of mutant strains lacking known ROS scavengers or detoxifying enzymes, the authors suggested that accumulation of the superoxide anion (O2−), but not hydroxyl radicals, was associated with drug-induced killing in E. faecalis (2). We further confirmed the link between oxidative stress responses and antibiotic killing, as the Δspx strain was more susceptible to bactericidal drugs than the wild-type OG1RF strain. Interestingly, two previous reports have also implicated Spx in bacterial tolerance against cell wall inhibitors. A microarray study revealed that expression of the B. subtilis spx gene and of several genes regulated by Spx was induced by challenge with different inhibitors of cell wall biosynthesis (39). In addition, a more recent report proposed that the activation of oxidative stress responses via Spx could also contribute to glycopeptide resistance in S. aureus (36).
While the results that we provide here indicate that the main targets of Spx regulatory control are genes involved in oxidative stress responses, we also showed that a lack of spx impairs tolerance to other stresses, including low pH, detergents, salt, and antibiotics. Spx homologues in L. lactis have been shown to be critical in responding to cell envelope stress (45) as well as in general stress tolerance (10). In S. mutans, inactivation of the spxA and spxB genes significantly impaired oxidative stress survival but also affected survival under acidic conditions (16). Finally, the S. aureus homologue of Spx has an impact on biofilm formation (32). These observations may not be entirely surprising, as different stresses can have similar effects on cellular processes and cross talk between different stress response systems has been well documented (22, 51).
Enterococcal infections are associated with the presence of foreign medical devices, including urethral catheterization, peritoneal dialysis, and prosthetic heart valves. In this study, we modified a mouse peritonitis model (40) by surgically introducing a silicone catheter material into the peritoneal cavity. Previous studies showed that the presence of foreign medical devices affects the ability of the host to eradicate bacterial infections (41). These devices are thought to provide a protective environment for bacteria to form biofilms, which are intrinsically more resistant to host immune defenses (6, 41) and recalcitrant to antimicrobial therapies (23, 41). Moreover, the presence of catheters has been associated with histological and immunological changes in the mucosa that exacerbate inflammatory responses while facilitating bacterial colonization and persistence (5). Our results confirmed that the presence of the foreign device facilitated infection, as the intraperitoneal lethal dose for catheterized mice was at least 1 log lower than the lethal dose found in studies with noncatheterized mice (14, 40). Compared to the parent strain, the ability of the Δspx strain to colonize the peritoneal cavity and cause disseminated infection was significantly reduced (Fig. 9). Note that the oxygen tension in the peritoneal cavity is relatively low, ranging from 40 to 50 mm Hg (35), indicating that the reduced ability of the Δspx strain to survive in the peritoneal fluid or to disseminate into the bloodstream may not be associated directly with environmental oxygen. Given the presence of large numbers of resident macrophages in the peritoneal cavity and the impaired capacity of the Δspx strain to survive within macrophages in vitro (Fig. 7), the finding that the Δspx strain could still be isolated from catheters after being eliminated from the peritoneal fluid suggests that the implant indeed facilitated bacterial persistence, likely by offering a protected niche which was less accessible to immune cells. In fact, while we recovered an average of 104 to 105 bacterial CFU per implant (1 cm long by 0.024-in. diameter), our SEM analysis suggested that the majority of viable bacteria were within the inner layers of the biofilm. In addition, in vitro biofilm quantifications suggested that there were no significant differences in the abilities of the wild-type and Δspx strains to form biofilms on the catheter surface. Altogether, these results support the significance of Spx regulation in E. faecalis virulence and demonstrate the usefulness of a foreign body-associated murine peritonitis model for investigating E. faecalis virulence.
Two other oxidative stress transcriptional regulators with DNA binding activity, PerR and HypR, have been characterized previously in E. faecalis. Previous qRT-PCR analysis revealed that HypR and, to a lesser extent, PerR control a subset of genes involved in oxidative stress responses (47–49). HypR was shown to be a major regulator of oxidative stress genes by activating transcription of the ahpC/ahpF, gor, katA, nox, npr, ohr, sodA, and trxB genes during H2O2 stress (47). On the other hand, the scope of PerR regulation was relatively modest, though ahpC/ahpF, dps, nox, and npr were under apparent negative control by PerR (48). Here we demonstrated that several oxidative stress genes regulated by HypR were also under Spx control, including dps, gor, katA, msrA, nox, npr, ohr, sodA, and trxB (Table 3). Although HypR was shown to exert positive control only under conditions of oxidative stress (30 min with 2.4 mM H2O2) (47), our data reveal that Spx is a major regulator of oxidative stress genes even under nonstressful conditions (e.g., static growth). Among the genes regulated by Spx, expression of nox, ohr, and sodA was dramatically reduced in the absence of Spx, regardless of the growth condition tested. Interestingly, ahpC and dps were upregulated in the Δspx strain under aeration. Given that Spx is known to function as a positive regulator of genes involved in oxidative stress and that the ahpCF operon and dps were shown to be under positive control of Spx in S. mutans (16), the upregulation of dps and ahpC in the Δspx strain under any given condition was unexpected. However, a similar pattern of expression was observed for the perR gene, e.g., perR transcription was induced in the Δspx strain during aeration. Because PerR is known to function as a repressor of ahpC, dps, and perR itself (24, 48), it is quite possible that the upregulation of ahpC, dps, and perR in the Δspx strain during aerobic growth was not directly associated with Spx control. In B. subtilis, it was demonstrated that oxidation of two histidine residues (H37 and H91) in PerR disturbs the formation of the protein-DNA complex, thereby resulting in activation of the PerR regulon (24, 43). It is tempting to speculate that in the absence of Spx, PerR is more susceptible to oxidation due to increased accumulation of ROS that ultimately lead to alleviation of PerR-repressed genes. Because HypR and Spx regulate genes repressed by PerR, we concluded that regulation of oxidative stress pathways in E. faecalis is under the control of at least three transcriptional regulators.
In summary, in silico and mutational analyses were used here to identify and characterize a novel virulence factor of E. faecalis. Specifically, we showed that the Δspx strain was hypersensitive to a variety of medically relevant stress conditions and was highly sensitive to macrophage killing in vitro. We also demonstrated that the ability of the mutant strain to colonize the peritoneum and disseminate in the bloodstream was significantly reduced compared to that of the parent strain. By comparing our qRT-PCR results with previously published studies (47–49), we demonstrated that Spx, HypR, and (to some extent) PerR are responsible for controlling the expression of the major oxidative stress genes. Studies to elucidate the interactions and regulatory hierarchy of HypR, PerR, and Spx in the control of oxidative stress genes are under way.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by NIH-NIDCR award DE019783 to J.A.L. A.O.G. was supported by NIDCR grant T32DE007202, and J.E.M. was supported by NIGMS grant R25 GM064133.
We thank Gary Dunny and Christopher Kristich for plasmid pCJK96.
Footnotes
Published ahead of print 16 April 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Abranches J, Candella MM, Wen ZT, Baker HV, Burne RA. 2006. Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans. J. Bacteriol. 188:3748–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bizzini A, Zhao C, Auffray Y, Hartke A. 2009. The Enterococcus faecalis superoxide dismutase is essential for its tolerance to vancomycin and penicillin. J. Antimicrob. Chemother. 64:1196–1202 [DOI] [PubMed] [Google Scholar]
- 3. Bourgogne A, et al. 2008. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 9:R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chatterjee SS, et al. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74:1323–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Delnay KM, Stonehill WH, Goldman H, Jukkola AF, Dmochowski RR. 1999. Bladder histological changes associated with chronic indwelling urinary catheter. J. Urol. 161:1106–1108 [PubMed] [Google Scholar]
- 6. del Pozo JL, Patel R. 2007. The challenge of treating biofilm-associated bacterial infections. Clin. Pharmacol. Ther. 82:204–209 [DOI] [PubMed] [Google Scholar]
- 7. Fisher K, Phillips C. 2009. The ecology, epidemiology and virulence of Enterococcus. Microbiology 155:1749–1757 [DOI] [PubMed] [Google Scholar]
- 8. Flahaut SJ, Laplace M, Frere J, Auffray Y. 1998. The oxidative stress response in Enterococcus faecalis: relationship between H2O2 tolerance and H2O2 stress proteins. Lett. Appl. Microbiol. 26:259–264 [DOI] [PubMed] [Google Scholar]
- 9. Frankenberg L, Brugna M, Hederstedt L. 2002. Enterococcus faecalis heme-dependent catalase. J. Bacteriol. 184:6351–6356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frees D, Varmanen P, Ingmer H. 2001. Inactivation of a gene that is highly conserved in Gram-positive bacteria stimulates degradation of non-native proteins and concomitantly increases stress tolerance in Lactococcus lactis. Mol. Microbiol. 41:93–103 [DOI] [PubMed] [Google Scholar]
- 11. Fuangthong M, Herbig AF, Bsat N, Helmann JD. 2002. Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J. Bacteriol. 184:3276–3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huycke MM, Abrams V, Moore DR. 2002. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis 23:529–536 [DOI] [PubMed] [Google Scholar]
- 13. Huycke MM, et al. 2001. Extracellular superoxide production by Enterococcus faecalis requires demethylmenaquinone and is attenuated by functional terminal quinol oxidases. Mol. Microbiol. 42:729–740 [DOI] [PubMed] [Google Scholar]
- 14. Ike Y, Hashimoto H, Clewell DB. 1984. Hemolysin of Streptococcus faecalis subspecies zymogenes contributes to virulence in mice. Infect. Immun. 45:528–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jett BD, Huycke MM, Gilmore MS. 1994. Virulence of enterococci. Clin. Microbiol. Rev. 7:462–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kajfasz JK, et al. 2010. Two Spx proteins modulate stress tolerance, survival, and virulence in Streptococcus mutans. J. Bacteriol. 192:2546–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810 [DOI] [PubMed] [Google Scholar]
- 18. Kristich CJ, Chandler JR, Dunny GM. 2007. Development of a host-genotype-independent counterselectable marker and a high-frequency conjugative delivery system and their use in genetic analysis of Enterococcus faecalis. Plasmid 57:131–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kristich CJ, Wells CL, Dunny GM. 2007. A eukaryotic-type Ser/Thr kinase in Enterococcus faecalis mediates antimicrobial resistance and intestinal persistence. Proc. Natl. Acad. Sci. U. S. A. 104:3508–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. La Carbona S, et al. 2007. Comparative study of the physiological roles of three peroxidases (NADH peroxidase, alkyl hydroperoxide reductase and thiol peroxidase) in oxidative stress response, survival inside macrophages and virulence of Enterococcus faecalis. Mol. Microbiol. 66:1148–1163 [DOI] [PubMed] [Google Scholar]
- 21. Lamour V, Westblade LF, Campbell EA, Darst SA. 2009. Crystal structure of the in vivo-assembled Bacillus subtilis Spx/RNA polymerase alpha subunit C-terminal domain complex. J. Struct. Biol. 168:352–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lemos JA, Abranches J, Burne RA. 2005. Responses of cariogenic streptococci to environmental stresses. Curr. Issues Mol. Biol. 7:95–107 [PubMed] [Google Scholar]
- 23. Lewis K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45:999–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mongkolsuk S, Helmann JD. 2002. Regulation of inducible peroxide stress responses. Mol. Microbiol. 45:9–15 [DOI] [PubMed] [Google Scholar]
- 25. Murray BE. 1990. The life and times of the enterococcus. Clin. Microbiol. Rev. 3:46–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakano MM, Hajarizadeh F, Zhu Y, Zuber P. 2001. Loss-of-function mutations in yjbD result in ClpX- and ClpP-independent competence development of Bacillus subtilis. Mol. Microbiol. 42:383–394 [DOI] [PubMed] [Google Scholar]
- 27. Nakano S, Erwin KN, Ralle M, Zuber P. 2005. Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx. Mol. Microbiol. 55:498–510 [DOI] [PubMed] [Google Scholar]
- 28. Nakano S, Kuster-Schock E, Grossman AD, Zuber P. 2003. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 100:13603–13608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nakano S, Nakano MM, Zhang Y, Leelakriangsak M, Zuber P. 2003. A regulatory protein that interferes with activator-stimulated transcription in bacteria. Proc. Natl. Acad. Sci. U. S. A. 100:4233–4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakano S, Zheng G, Nakano MM, Zuber P. 2002. Multiple pathways of Spx (YjbD) proteolysis in Bacillus subtilis. J. Bacteriol. 184:3664–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Newberry KJ, Nakano S, Zuber P, Brennan RG. 2005. Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the alpha C-terminal domain of RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 102:15839–15844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pamp SJ, Frees D, Engelmann S, Hecker M, Ingmer H. 2006. Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J. Bacteriol. 188:4861–4870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Paulsen IT, et al. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074 [DOI] [PubMed] [Google Scholar]
- 34. Poyart C, Quesnes G, Trieu-Cuot P. 2000. Sequencing the gene encoding manganese-dependent superoxide dismutase for rapid species identification of enterococci. J. Clin. Microbiol. 38:415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Renvall S, Niinikoski J. 1980. Kinetics of oxygen in peritoneal cavity. Effects of chemical peritonitis and intraperitoneally administered colloids in rats. J. Surg. Res. 28:132–139 [DOI] [PubMed] [Google Scholar]
- 36. Renzoni A, et al. 2011. Whole genome sequencing and complete genetic analysis reveals novel pathways to glycopeptide resistance in Staphylococcus aureus. PLoS One 6:e21577 doi:10.1371/journal.pone.0021577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Riboulet E, et al. 2007. Relationships between oxidative stress response and virulence in Enterococcus faecalis. J. Mol. Microbiol. Biotechnol. 13:140–146 [DOI] [PubMed] [Google Scholar]
- 38. Rince A, Giard JC, Pichereau V, Flahaut S, Auffray Y. 2001. Identification and characterization of gsp65, an organic hydroperoxide resistance (ohr) gene encoding a general stress protein in Enterococcus faecalis. J. Bacteriol. 183:1482–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rukmana A, Morimoto T, Takahashi H, Giyanto, Ogasawara N. 2009. Assessment of transcriptional responses of Bacillus subtilis cells to the antibiotic enduracidin, which interferes with cell wall synthesis, using a high-density tiling chip. Genes Genet. Syst. 84:253–267 [DOI] [PubMed] [Google Scholar]
- 40. Singh KV, Qin X, Weinstock GM, Murray BE. 1998. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J. Infect. Dis. 178:1416–1420 [DOI] [PubMed] [Google Scholar]
- 41. Stewart PS, Costerton JW. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135–138 [DOI] [PubMed] [Google Scholar]
- 42. Storz G, Imlay JA. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188–194 [DOI] [PubMed] [Google Scholar]
- 43. Traore DA, et al. 2009. Structural and functional characterization of 2-oxo-histidine in oxidized PerR protein. Nat. Chem. Biol. 5:53–59 [DOI] [PubMed] [Google Scholar]
- 44. Turlan C, Prudhomme M, Fichant G, Martin B, Gutierrez C. 2009. SpxA1, a novel transcriptional regulator involved in X-state (competence) development in Streptococcus pneumoniae. Mol. Microbiol. 73:492–506 [DOI] [PubMed] [Google Scholar]
- 45. Veiga P, et al. 2007. SpxB regulates O-acetylation-dependent resistance of Lactococcus lactis peptidoglycan to hydrolysis. J. Biol. Chem. 282:19342–19354 [DOI] [PubMed] [Google Scholar]
- 46. Verneuil N, et al. 2006. Implication of (Mn)superoxide dismutase of Enterococcus faecalis in oxidative stress responses and survival inside macrophages. Microbiology 152:2579–2589 [DOI] [PubMed] [Google Scholar]
- 47. Verneuil N, et al. 2005. Implication of hypR in the virulence and oxidative stress response of Enterococcus faecalis. FEMS Microbiol. Lett. 252:137–141 [DOI] [PubMed] [Google Scholar]
- 48. Verneuil N, et al. 2005. Contribution of a PerR-like regulator to the oxidative-stress response and virulence of Enterococcus faecalis. Microbiology 151:3997–4004 [DOI] [PubMed] [Google Scholar]
- 49. Verneuil N, et al. 2004. Effects of the Enterococcus faecalis hypR gene encoding a new transcriptional regulator on oxidative stress response and intracellular survival within macrophages. Infect. Immun. 72:4424–4431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wells CL, Jechorek RP, Erlandsen SL. 1990. Evidence for the translocation of Enterococcus faecalis across the mouse intestinal tract. J. Infect. Dis. 162:82–90 [DOI] [PubMed] [Google Scholar]
- 51. Wick LM, Egli T. 2004. Molecular components of physiological stress responses in Escherichia coli. Adv. Biochem. Eng. Biotechnol. 89:1–45 [DOI] [PubMed] [Google Scholar]
- 52. Zuber P. 2004. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J. Bacteriol. 186:1911–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.