Abstract
Est1 and Ebs1 in Saccharomyces cerevisiae are paralogous proteins that arose through whole-genome duplication and that serve distinct functions in telomere maintenance and translational regulation. Here we present our functional analysis of the sole Est1/Ebs1 homologue in the related budding yeast Kluyveromyces lactis (named KlEst1). We show that similar to other Est1s, KlEst1 is required for normal telomere maintenance in vivo and full telomerase primer extension activity in vitro. KlEst1 also associates with telomerase RNA (Ter1) and an active telomerase complex in cell extracts. Both the telomere maintenance and the Ter1 association functions of KlEst1 require its N-terminal domain but not its C terminus. Analysis of clusters of point mutations revealed residues in both the N-terminal TPR subdomain and the downstream helical subdomain (DSH) that are important for telomere maintenance and Ter1 association. A UV cross-linking assay was used to establish a direct physical interaction between KlEst1 and a putative stem-loop in Ter1, which also requires both the TPR and DSH subdomains. Moreover, similar to S. cerevisiae Ebs1 (ScEbs1) (but not ScEst1), KlEst1 confers rapamycin sensitivity and may be involved in nonsense-mediated decay. Interestingly, unlike telomere regulation, this apparently separate function of KlEst1 requires its C-terminal domain. Our findings provide insights on the mechanisms and evolution of Est1/Ebs1 homologues in budding yeast and present an attractive model system for analyzing members of this multifunctional protein family.
INTRODUCTION
Telomeres are specialized nucleoprotein structures that maintain the integrity of eukaryotic chromosomal termini by protecting them from fusion and recombination and promoting their replication (for reviews, see references 12, 23 and 32). In most organisms, telomeric DNA consists of short repetitive sequences that are rich in G residues on the 3′ end-containing strand (G strand). The repeats on this strand are replenished by the telomerase ribonucleoprotein (RNP), whose essential polymerization function is mediated by two core components: a catalytic reverse transcriptase protein, telomerase reverse transcriptase (TERT), and a template-specifying RNA, telomerase RNA (TER) (for reviews, see references 2 and 30).
The telomerase RNP in the budding yeast Saccharomyces cerevisiae has been extensively characterized and contains two auxiliary protein components named Est1 and Est3. Both components are essential for telomere maintenance in vivo but dispensable for telomerase activity in vitro. A major function of Est1 is to promote the recruitment of the telomerase complex to chromosome ends (33). This function is believed to require a physical interaction between Est1 and the telomere end-binding protein Cdc13, as well as an interaction between Est1 and telomerase RNA. The domains of Est1 and its binding targets responsible for these interactions have been characterized to some extent, but not in great detail. For example, in S. cerevisiae, a bulged stem and surrounding nucleotides in telomerase RNA (Tlc1) are found to be necessary and sufficient for Est1-Tlc1 interaction, but the region of Est1 required for RNA binding is poorly understood (11, 38, 45). Besides recruitment, a second, activation function for Est1 is also evident from existing data, and recent studies suggest that this function is attributable to the ability of Est1 to promote the incorporation of Est3 into the telomerase RNP (11, 22, 31, 42, 43). In the absence of Est1, Est3 exhibits reduced association with telomerase core components in cell extracts and with telomeres in vivo. Despite these insights on Est1 functions, many questions remain with regard to the detailed biochemical properties of this protein and their physiologic relevance. For example, even though both genetic and biochemical evidence supports a direct physical interaction between Est1 and the telomere end-binding protein Cdc13, how this interaction promotes telomerase function remains incompletely understood (33, 34, 50). In addition, Est1 has been reported to possess moderate affinity and sequence specificity for telomeric G-strand DNA and to promote G-quadruplex formation, but the physiologic relevance of these activities remains unclear (10, 45, 53). Intriguingly, several mutations in Est1were recently observed to induce telomere lengthening, suggesting a previously unsuspected negative regulatory function with regard to telomere homeostasis (37). Evidently, more studies are necessary to achieve a full understanding of this critical and multifunctional protein.
Investigations of Est1 homologues have resulted in yet more questions concerning the mechanisms and evolution of this protein family. In S. cerevisiae, an EST1 homologue named EBS1 (Est1-like Bcy1 Suppressor) was shown to play a modest role in telomere maintenance but also to regulate translation and nonsense-mediated decay (NMD) (14, 24, 54). Specifically, the ebs1Δ mutant manifests higher levels of NMD-targeted RNAs and greater resistance to rapamycin, which inhibits the TOR signaling pathway that regulates cell growth and translation. In Schizosaccharomyces pombe, two Est1-like genes have been identified, only one of which is required in telomere maintenance (4). Notably, SpEst1 also mediates telomerase recruitment but does so by interacting with the S. pombe telomere protein Ccq1 rather than a Cdc13 homologue (29). In humans, three EST1-like genes (EST1A, EST1B, and EST1C) have been identified. Two of these genes, EST1A and EST1B, have been implicated in telomere regulation, though their mechanisms of action appear to be distinct from that of ScEst1 (35, 41). In particular, neither protein has been shown to mediate telomerase recruitment or telomerase RNP assembly. Interestingly, EST1A and EST1B are also known as SMG6 and SMG5 and have been implicated in NMD pathways. These proteins may regulate TERRA, which are telomere-encoded RNAs that modulate telomere lengths and chromatin structure (3, 9, 25). The involvement of diverse Est1 homologues (from yeasts to humans) in NMD suggests that this involvement may have occurred early in evolution.
Our laboratories have explored less frequently studied budding yeasts (i.e., yeasts other than S. cerevisiae) as alternative model systems for analyzing the mechanisms of telomere and telomerase proteins. We consider the effort worthwhile for two reasons. First, many S. cerevisiae telomere and telomerase proteins are difficult to express, purify, and analyze in vitro, thus limiting the possibilities for detailed mechanistic analysis. By screening homologues, we wish to identify biochemically tractable proteins and subject them to in-depth analysis. Second, we reasoned that an in vitro activity of a protein is more likely to be biologically relevant if it is conserved in evolution. By comparing the properties of orthologues to the prototypical member, we may be able to focus on features of the protein that are of greater general significance. In this study, we examined the mechanism and functions of the single Est1 homologue in the budding yeast Kluyveromyces lactis (KlEst1), which we found to be quite amenable to both biochemical and genetic analyses. We showed that KlEst1 is required for telomere maintenance in vivo and modulates the primer extension activity of telomerase in a primer-specific manner. We developed a cross-linking assay to analyze Est1-telomerase RNA interaction and characterized the regions and features of the two molecules required for direct mutual interaction. We also found that KlEst1 confers rapamycin sensitivity and thus shares the functions of ScEbs1. A careful analysis of the Est1 family of proteins in Saccharomycotina yeasts (encompassing Saccharomyces, Kluyveromyces, and Candida species) revealed the existence of two Est1/Ebs1-like proteins only in organisms that are descendants of the ancestor with a duplicated genome. Our findings support the notion that both telomere and NMD regulations are ancestral functions of Est1 and that the existence of functionally distinct Est1 and Ebs1 in S. cerevisiae is the result of whole-genome duplication (WGD) followed by subspecialization.
MATERIALS AND METHODS
Construction and growth of K. lactis strains.
The K. lactis strain 7B520 (ura3-1 his2-2 trp1) was used as the parental strain (49). A PCR-amplified K. lactis EST1 gene was cloned into the XmaI/KpnI sites of pBS SK II+ to create pEbs1. The primers used for this were ATCCCGGGtactgcttctcccaccagaga and ACGGTACCcacctcgaatgcttttacgac, with the K. lactis sequences shown in lowercase. The 1,833-bp MfeI-NheI region within the EST1 open reading frame (ORF) was then replaced with the EcoRI-XbaI HisG-URA3-HisG fragment from pBS-URA3-2 to create plasmid pΔEbs1-HUH. The K. lactis est1Δ mutant was constructed by transforming strain 7B520 with a fragment containing the disrupted EST1 gene from pΔEbs1-HUH, followed by selection on synthetic defined (SD) agar plates lacking uracil. Independent transformants were then placed on medium containing 5-fluoroorotic acid (5-FOA) to select for cells that had undergone recombination to excise the URA3 gene and one copy of HisG. Strains bearing various EST1 mutant alleles were created by transforming the est1Δ mutant with the pCXJ18-EST1-TAP series of plasmids. Strains were all passaged at 30°C in either solid or liquid yeast extract-peptone-dextrose (YPD) medium (2% peptone, 1% yeast extract, 2% dextrose).
Plasmids and mutagenesis.
For protein expression in Escherichia coli, we cloned K. lactis EST1 fragments into pSMT3 to enable the expression of SUMO fusion proteins. PCR fragments encompassing the entire EST1 open reading frame (ORF), the N-terminal domain (NTD) (amino acids [aa] 1 to 598), the TPR subdomain (aa 1 to 303), and the downstream helical subdomain (DSH) (aa 304 to 598) were cloned between the SacI and XhoI sites of pSMT3 vector to generate pSMT3-KlEST1, pSMT3-KlEST1NTD, pSMT3-KlESTTPR, and pSMT3-KlEST1DSH, respectively. A FLAG tag was incorporated into each of the downstream primers used for PCR such that the recombinant proteins all carry a C-terminal FLAG tag. For in vivo analysis of EST1 function, we cloned different alleles of EST1 into the pCXJ18 shuttle vector, which carries the URA3 marker. The vector was first modified by introducing the TAP tag between the SalI and HindIII sites to give pCXJ18-TAP (6, 36). A PCR fragment containing the EST1 ORF and 550 bp of upstream sequence was then inserted between the KpnI and SalI sites of pCXJ18-TAP to give pCXJ18-KlEST1-TAP. Truncated EST1 genes corresponding to the NTD and the TPR domain were generated by PCR using primers containing the KpnI and SalI site and used to substitute the corresponding fragment in pCXJ18-KlEST1-TAP to give pCXJ18-KlEST1NTD-TAP and pCXJ18-KlEST1TPR-TAP, respectively. To generate pCXJ18-KlEST1DSH-TAP, two overlapping PCR fragments comprising the promoter region and the DSH subdomain were first amplified separately. The two fragments were both added to a second PCR mixture to synthesize a combined DNA fragment, which was then inserted into pCXJ18-TAP.
To analyze the roles of individual amino acids, site-specific mutagenesis was employed to create the RR (R111A and R118A), RHRQ (R111A, H117A, R118A, and Q119A), QKR (Q219A, K220A, and R223A), RR-QKR (R111A, R118A, Q219A, K220A, and R223A), KF (K522A and F529A), RR-KF (R111A, R118A, K522A, and F529A), and K467E mutants of EST1NTD in either pSMT3-KlEST1NTD or pCXJ18-KlEST1NTD-TAP.
Sequence analysis.
Est1 and Ebs1 homologues from Saccharomyces, Kluyveromyces, and Candida spp. were identified from NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi), Broad Institute (http://www.broad.mit.edu/annotation/genome/candida_group/Blast.html), and SGD (http://www.yeastgenome.org/) databases. The multiple-sequence alignment was generated using the T-COFFEE server (http://www.igs.cnrs-mrs.fr/Tcoffee/tcoffee_cgi/index.cgi) and displayed using Boxshade (http://www.ch.embnet.org/software/BOX_form.html). The phylogeny for these homologues was investigated using Phylip (http://evolution.genetics.washington.edu/phylip/getme.html) and displayed using FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
Telomere length analysis.
Chromosomal DNAs were isolated from 5 ml of saturated K. lactis culture by the smash and grab method (20), digested with EcoRI, and fractionated in 0.6 to 0.8% agarose gels. Following transfer to nylon membranes, the telomere restriction fragments were detected as previously described using an oligonucleotide probe that contains two copies of the K. lactis telomere repeat (5′-ACGGATTTGATTAGGTATGTGGTGTACGGATTTGATTAGGTATGTGGTGT-3′). The hybridization was performed at 50°C to 60°C.
Partial purification of the K. lactis telomerase complex and analysis of telomerase activity.
K. lactis telomerase fractions were generated by resolving whole-cell extracts (derived from the wild-type or an est1Δ strain) over DEAE columns as previously described for Candida albicans (21, 40). The fractions were subjected to primer extension assays using 12-nucleotide (nt) primers and a single labeled nucleotide triphosphate that is needed for incorporation at the primer +1 position. For comparison of wild-type and est1Δ telomerases, fractions containing approximately equal levels of Ter1 (as measured by reverse transcriptase [RT]-PCR) were utilized. The primer extension products were resolved on denaturing polyacrylamide gels and analyzed using a Typhoon PhosphorImager (Molecular Dynamics) and the ImageQuant software (GE Healthcare).
RNA-protein cross-linking assay.
The His6-SUMO fusion proteins were expressed and purified essentially as previously described (52). Briefly, extracts were prepared from E. coli following IPTG (isopropyl-β-d-thiogalactopyranoside) induction and the fusion proteins were purified using Ni-nitrilotriacetic acid (Ni-NTA) chromatography. The fusion proteins were then cleaved by ULP1 and the untagged polypeptides were isolated from the His6-SUMO tag by a second round of Ni-NTA affinity chromatography. The protein preparations were further purified by sedimentation over glycerol gradients (15% to 30%). 32P-labeled RNA probes for the cross-linking assay were prepared by in vitro transcription using PCR fragments containing the T7 promoter placed upstream of wild-type and mutant Ter1-EBD (the putative Est1 binding domain of K. lactis telomerase RNA) sequences. Cross-linking reaction mixtures (20 μl) contained 32P-labeled RNA probe, 12.5 ng/μl tRNA, 20 mM Tris-HCl (pH 8.0), 5 mM dithiothreitol (DTT), and 0.5 U/μl RNasin. Following incubation at 30°C for 30 min, the reaction mixtures were irradiated with 254-nm UV light on ice for 20 min and then treated with 250 ng/μl RNase A at 37°C for 30 min. The reaction mixtures were further incubated at 65°C for 5 min following the addition of SDS (4 μl of a 10% stock solution) and DTT (0.4 μl of a 1 M stock solution). The RNA-protein complexes were precipitated by adding 4 μg glycogen and 1.2 ml acetone at room temperature for 10 min, recovered by centrifugation, and then resolved by SDS-PAGE. The levels of cross-linking were analyzed using a Typhoon PhosphorImager and the ImageQuant software (GE Healthcare).
Analysis of Est1-Ter1 association in cell extracts.
K. lactis whole-cell extracts were prepared and subjected to IgG-Sepharose pulldown as previously described with slight modifications (21). Briefly, cell extracts (5 mg) were incubated with IgG-Sepharose beads (45 μl) in 1.2 ml TMG(400) buffer (10 mM Tris-HCl, pH 8.0, 1.2 mM magnesium chloride, 0.1 mM EDTA, 0.1 mM EGTA, 10% glycerol plus 400 mM sodium acetate) plus 0.05% Tween 20. The mixtures were rotated at 4°C for 2 h and the beads were washed five times in TMG(800) and twice in TMG(0) (21). The precipitated samples were then subjected to RT-PCR and Western blot analysis as previously described to quantify the levels of Ter1 and Est1, respectively (21). For RT-PCR, fragments corresponding to 201 bp of Ter1 and 385 bp of U1 (used as the control for nonspecific binding) were amplified by specific primer pairs (Ter1 forward, 5′-GCTATGACAACAATACCTTTAGAAT-3′; Ter1 reverse, 5′-GATTAGGTATGTGGTGTACGGATTTG-3′; U1 forward, 5′-GTTTGTTGACCGGGTATGAGGTTTTC-3′; U1 reverse, 5′-CTCTCACCCATAAACCTGTAAACAAG-3′). P32-labeled dCTP (1 μCi/4 nmol) was included in the reaction mixtures to allow for quantification of the amounts of PCR products by PhosphorImager analysis.
Analysis of rapamycin sensitivity and NMD transcripts.
The K. lactis strains were grown in YPD to an optical density at 600 nm (OD600) of 0.6 to 1.0. The cultures were diluted to an OD600 of 0.1, and 5-fold serial dilutions of these suspensions were spotted onto YPD plates containing 5 or 10 ng/ml rapamycin (LC laboratories). The plates were incubated at 30°C for 4 to 5 days and checked for growth. The NMD pathway was also assessed by measuring the levels of CYH2 pre-mRNA and mRNA using RT-PCR. The sequences of the primers are as follows: forward primer for mRNA, 5′-GAAAGCACAGAGGTCACGTCTCAGCCGG-3′; forward primer for pre-mRNA, 5′-CAGAACCACAGGTTTACTAACAAACAC-3′; reverse primer, 5′-CAGACTTGGAAGCAGACTTCAAGTATTC-3′. P32-labeled dCTP (1 μCi/4 nmol) was included in the reaction mixtures to allow for quantification of the amounts of PCR products by PhosphorImager analysis.
Homology modeling.
The KlEst1NTD sequence was submitted to the I-Tasser server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) and 1YA0.pdb (the structure file for human SMG7 N terminus) was specified as the template for modeling. The resulting model was visualized and analyzed in Pymol (http://pymol.sourceforge.net/index.html).
RESULTS
KlEst1 and other Est1/Ebs1 homologues in Saccharomycotina.
As described earlier, Est1 has been most extensively characterized in S. cerevisiae. However, many reported mechanisms of ScEst1, including its nucleic acid-binding properties and its role in enhancing telomerase activity, have been controversial. In addition, recombinant expression and purification of ScEst1 are reportedly difficult. We therefore screened other Est1/Ebs1 homologues as alternative targets of investigation. Our preliminary analysis indicated that the homologue encoded by the K. lactis genome (here referred to as KlEst1) is particularly amenable to recombinant expression and purification. Because this organism has already been utilized extensively as a model system to analyze different aspects of telomere maintenance (17, 27), we decided to engage in a detailed analysis of KlEst1.
Before proceeding with molecular genetic and biochemical investigation of KlEst1, we performed multiple-sequence alignments and phylogenetic analysis to clarify the relationship of KlEst1 to other Est1/Ebs1 homologues. As depicted in Fig. 1A, each member of the Kluyveromyces and Candida clades contains just one Est1/Ebs1-like protein. However, in the Saccharomyces clade, each genome contains both a presumptive EST1 and an EBS1 gene. Within this clade, the EST1 and EBS1 genes form well-defined monophyletic groups, suggesting a duplication of an ancestral gene prior to speciation of the Saccharomyces members. Interestingly, Saccharomyces species are known to be descendants of an ancestor that underwent whole-genome duplication (48). Indeed, syntenic analysis of available yeast genomes supports the notion that EST1 and EBS1 arose as a consequence of WGD and can be considered ohnologs (5). The Ebs1 proteins in Saccharomyces are also considerably larger than Est1 proteins, with the former ranging from 870 aa to 1,023 aa and the latter ranging from 528 aa to 767 aa. This size difference can be attributed to the larger C-terminal domain (CTD) in Ebs1 proteins; the remaining portions of the Est1 and Ebs1 proteins align well to one another and are presumably comprised of an α-helical domain (Fig. 1B). Specifically, the crystal structure of an N-terminal fragment of human EST1C (also known as SMG7), which aligns well to the N terminus of the yeast Est1/Ebs1 proteins, revealed a predominantly α-helical protein comprised of an N-terminal TPR subdomain and a downstream helical subdomain (DSH) (15). In the Kluyveromyces clade (Kluyveromyces waltii, K. lactis, Saccharomyces kluyveri, and Ashbya gossypii), the Est1/Ebs1-like proteins have extended CTDs that align well to the Saccharomyces Ebs1 proteins. Overall, the sequence and phylogenetic analyses suggest that KlEst1 may perform the functions mediated by both Est1 and Ebs1 in S. cerevisiae.
Fig 1.
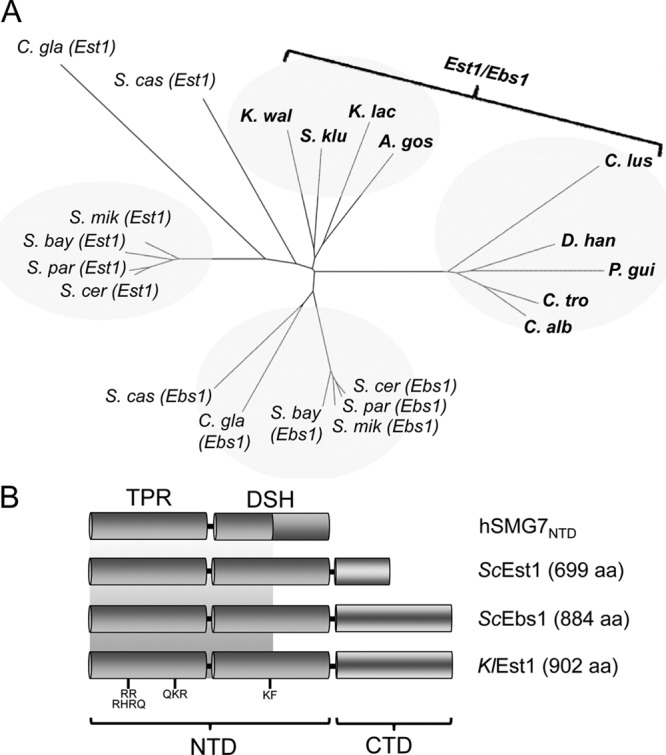
Phylogenetic analysis and domain comparison of Est1/Ebs1 homologues in budding yeast. (A) The relationships between the yeast Est1/Ebs1 homologues were analyzed using the neighbor-joining method and the results were plotted using FigTree. The abbreviations are as follows: A. gos, Ashbya gossypii; C. gla, Candida glabrata; C. alb, Candida albicans; C. lus, Candida lusitaniae; C. tro, Candida tropicalis; D. han, Debaromyces hansenii; K. lac, Kluyveromyces lactis; K. wal, Kluyveromyces waltii; P. gui, Pichia guilliermondii; S. bay, Saccharomyces bayanus; S. cas, Saccharomyces castellii; S. cer, Saccharomyces cerevisiae; S. klu, Saccharomyces kluyveri; S. mik, Saccharomyces mikata; S. par, Saccharomyces paradoxus. (B) The domain structures of the indicated Est1/Ebs1 homologues are depicted. The crystal structure of the N-terminal domain of human SMG7 (hSMG7NDT) revealed a TPR subdomain and a downstream helical subdomain (DSH). The shaded background is used to illustrate the fact that the first half but not the second half of hSMG7DSH aligns well to the yeast homologues.
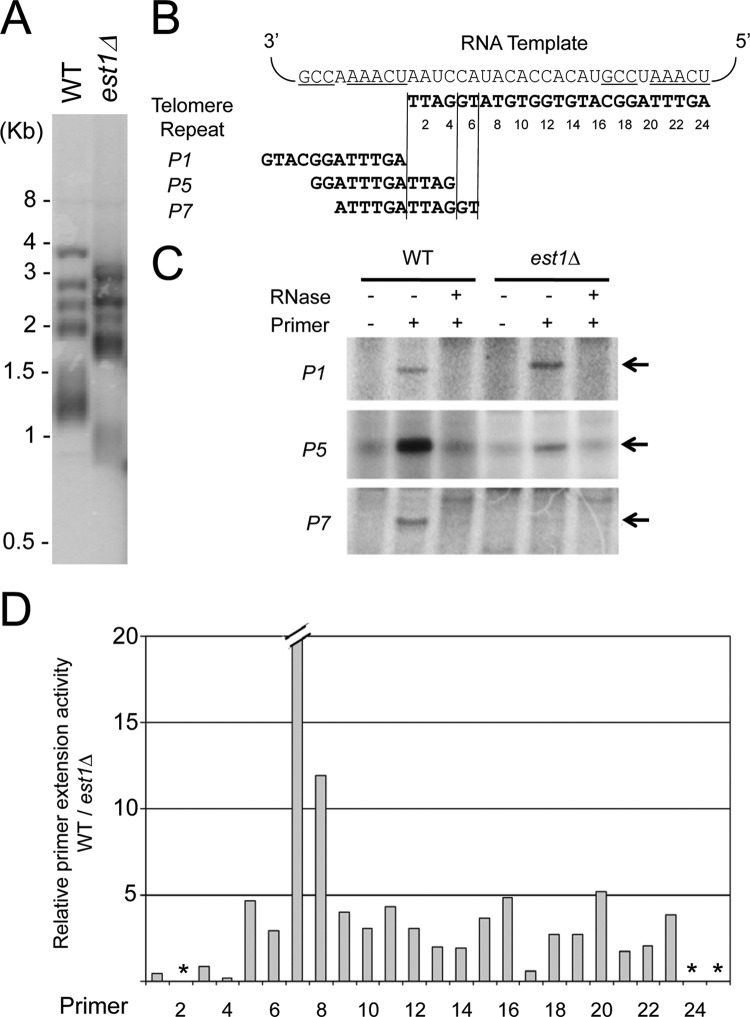
KlEst1 is required for telomere maintenance in vivo and telomerase activity in vitro.
To determine if KlEST1 is involved in telomere maintenance, we constructed an est1 null strain and characterized its growth and telomere phenotypes. As predicted for a telomerase gene (26, 28), the est1 null mutant strain exhibited growth senescence and telomere attrition (Fig. 2A and data not shown; see Fig. S1A and B in the supplemental material). The growth defects of the est1Δ mutant clones were variable. In one serially passaged clone, for example, some small colonies can be seen ∼75 to 100 generations after gene deletion, followed by apparent recovery and subsequently more uniform and severe growth defects at ∼175 generations (see Fig. S1A in the supplemental material). Other clones senesced and recovered at other time points during passage (data not shown). This heterogeneous behavior is very similar to that of the K. lactis ter1Δ mutant (28). As predicted, Southern analysis of the mutant revealed progressive telomere shortening (see Fig. S1B in the supplemental material). Overall, these findings support the notion that Est1 is essential for telomerase-mediated telomere maintenance in K. lactis.
Fig 2.
Deletion of KlEST1 impairs telomere maintenance in vivo and telomerase activity in vitro. (A) Chromosomal DNAs were isolated from the wild-type (WT) strain and the est1Δ mutant (∼50 generations after strain construction) and subjected to Southern analysis using an oligonucleotide probe that corresponds to two copies of the K. lactis telomere repeats (see Materials and Methods). (B) The template region of telomerase RNA is displayed; RNA residues presumably involved in alignment are underlined (46). A series of 12-nt primers that represent different regions and permutations of the K. lactis telomere repeat unit were designated by numbers. The precise sequences of three primers (P1, P5, and P7) are shown as examples. (C) Telomerase from the wild-type and est1Δ strains were isolated by DEAE chromatography and tested by primer extension assays using the P1, P5, and P7 primers in the presence of 32P-labeled dTTP, dGTP, and dATP, respectively. In other assays, we consistently observed comigration of the major RNase-sensitive product (indicated by arrows) with the primer + 1 size standard prepared by labeling of the primer with terminal transferase. (D) The levels of primer extension products were quantified by using a PhosphorImager. The ratios of the wild-type telomerase activity to est1Δ activity on all the primers were calculated and plotted. *, the levels of primer extension products generated by telomerase on primers P2, P24, and P25 were too low to yield reliable quantifications. For primer P7, the DEAE fraction derived from the est1Δ mutant was essentially inactive, resulting in a very high WT-to-est1Δ activity ratio.
We then examined the activity of telomerase derived from the wild-type and mutant strains by primer extension assays using a series of 12-nt primers that represent different regions and permutations of the K. lactis telomere repeat unit. Each primer was designed to test the ability of telomerase to initiate extension at a defined position along the RNA template (Fig. 2B). To simplify the interpretation of results, we added just a single labeled nucleotide triphosphate to each reaction mixture (39). For most primers, we were able to detect RNase-sensitive products of the predicted size(s) (Fig. 2B and data not shown). Consistent with previous findings, we observed very little DNA synthesis on primers 24 and 25, whose 3′ ends anneal close to the 5′ template boundary of Ter1 (Fig. 2D) (16, 44). Interestingly, we observed a variable reduction of primer extension activity in the fractions derived from the est1Δ strain (Fig. 2C and D). For example, the ability of the mutant enzyme to mediate nucleotide addition was similar to that of the wild-type enzyme at position 1 within the telomere repeat but was reduced 5-fold at position 5 (Fig. 2C). The magnitude of activity reduction was especially severe (>10-fold) at positions 7 and 8. However, at most positions, the mutant enzyme was ∼2- to 5-fold less active than the wild-type enzyme. These results indicate that KlEst1 affects the activity of telomerase in a DNA substrate-dependent manner, as has been observed for Candida albicans Est1 (39). The telomere length and telomerase activity defects of the K. lactis EST1 deletion mutant argue that this gene is indeed orthologous to S. cerevisiae EST1.
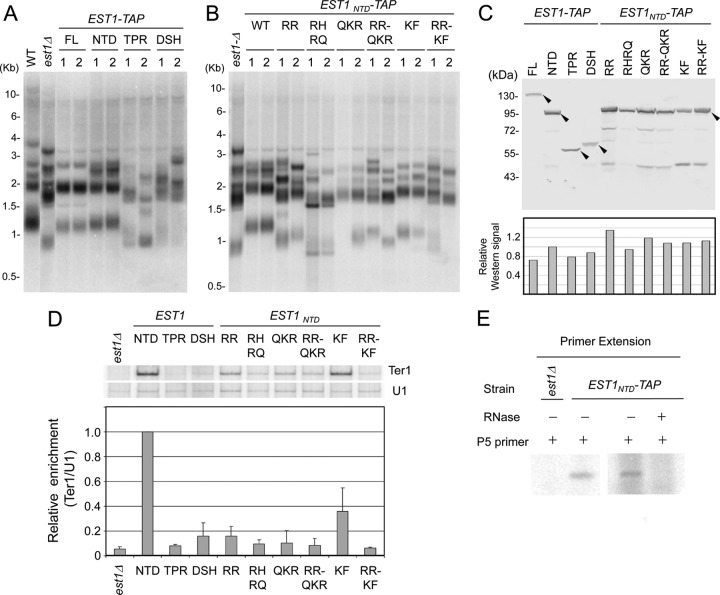
The N terminus of KlEst1 is sufficient for maintaining telomeres.
To identify the regions of Est1 required for telomere length maintenance, we introduced full-length or truncated TAP-tagged EST1 alleles into an est1Δ mutant and monitored the length of telomeres in the resulting strains by Southern blotting. The truncation variants tested include the entire N terminus of the protein (NTD, aa 1 to 598), as well as the TPR and the DSH subdomains (aa 1 to 303 and aa 304 to 598, respectively). As predicted, TAP-tagged full-length EST1 restored telomeres of the est1Δ mutant to nearly wild-type lengths (Fig. 3A). The strains reconstituted with EST1NTD contained similarly long telomeres, indicating that CTD is dispensable. In contrast, the strains reconstituted with EST1TPR or EST1DSH exhibited shorter-than-wild-type telomeres, indicating that both subdomains are required for telomere maintenance.
Fig 3.
Effects of EST1 mutations on telomere maintenance and Est1-Ter1 association. (A) Chromosomal DNAs were isolated from the wild-type, est1Δ, and two independently derived strains bearing full-length or truncated EST1 alleles (∼50 generations after the introduction of the EST1 alleles into the null strain) and subjected to Southern analysis of telomere restriction fragments. (B) Same as for panel A, except the chromosomal DNAs were isolated from strains reconstituted with EST1NTD and various point mutants of EST1NTD. (C) (Top) The TAP-tagged Est1 proteins in cell extracts were analyzed by Western blotting using antibodies directed against protein A. (Bottom) Signals were quantified, normalized to that for EST1NTD-TAP, and plotted. (D) (Top) Extracts were prepared from the est1Δ mutant as well as strains reconstituted with various EST1 alleles and subjected to the IgG-Sepharose pulldown assay. The levels of Ter1 and U1 RNA in the pulldown samples were measured by RT-PCR. P32-labeled dCTP was included in the reaction mixtures to allow identification of the PCR products by PhosphorImager scanning of gels. (Bottom) The radioactive signals for the Ter1 and U1 RT-PCR products were quantified by PhosphorImager analysis, and the ratios of Ter1 to U1 products were calculated and plotted. Data are derived from two independent experiments. (E) The IgG-Sepharose pulldown samples from the indicated strains were subjected to primer extension assays using the P5 primer and [α-32P]dGTP.
Two previous studies identified multiple residues of S. cerevisiae Est1 required for its association with telomerase RNA and for telomere maintenance (11, 54). One study revealed clusters of charged residues within the TPR subdomain, whereas the other uncovered basic and hydrophobic residues in the DSH subdomain as being important. (The DSH subdomain was previously proposed to contain RNA recognition motifs [RRMs], but such motifs are unlikely to be present given the hEST1C structure [15].) To determine if comparable residues of KlEst1 are required for telomere maintenance, we created a series of substitution mutants based on sequence alignment (Table 1). Each cluster of mutations was introduced into the EST1NTD allele, and the resulting mutant was tested in the est1Δ background. As shown in Fig. 3B, all of the mutant strains exhibited various degrees of telomere shortening. Both the RR and QKR mutations within the TPR subdomain of KlEst1 caused significant telomere shortening, as did the KF mutations in the DSH subdomain. Several mutants that exhibited substantial telomere shortening also showed evidence of senescence upon repeated passages (e.g., the RR-KF and RR-QKR mutants) (see Fig. S2 in the supplemental material; data not shown). In general, combining different clusters of mutations resulted in a more severe defect, suggesting that these groups of residues contribute independently to telomere maintenance (Fig. 3B, compare the RR-KF mutant to the RR and KF mutants). However, it was difficult to assess quantitatively the relative severity of the telomere shortening phenotype because of clonal heterogeneity (even though the mutants were all analyzed ∼50 generations after the introduction of the mutant alleles). Notably, all mutant proteins were detected at similar levels in cell extracts, indicating that the mutations did not grossly impair protein expression or stability (Fig. 3C). Taken together, these results suggest that residues in both the TPR and DSH subdomains play a conserved function in telomere maintenance in vivo.
Table 1.
Amino acid substitutions in the EST1 mutants characterized in this and two previous studies
|
S. cerevisiae |
K. lactis |
||
|---|---|---|---|
| Allele name | Amino acid substitutions | Allele name | Amino acid substitutions |
| est1-38 | R111A, R112A, K113A, R115A | RR | R111A, R118A |
| est1-39 | R118A, K122A, K123A | RHRQ | R111A, H117A, R118A, Q119A |
| est1-41 | E222A, K223A, R226A | QKR | Q219A, K220A, R223A |
| Est1-RNP1-NC | R499G, I504A, F506A | KF | K522A, F529A |
Next, we characterized the association of Est1 mutants with telomerase RNA (Ter1) in cell extracts using an IgG-Sepharose pulldown assay. Consistent with the dispensability of CTD for telomere maintenance, we found that Est1NTD associated strongly with K. lactis Ter1 in cell extracts (Fig. 3D). Indeed, we consistently detected twice as much Ter1 in the Est1NTD pulldown sample than in the Est1FL sample (data not shown), possibly because of the higher levels of the Est1NTD protein in cell extracts (Fig. 3C). Primer extension analysis of the Est1NTD pulldown sample indicates that this protein is associated with active telomerase complex (Fig. 3E). In contrast to Est1NTD, all of the other mutants exhibited reduced association with Ter1 (3- to 16-fold reduction), suggesting that the regions deleted or the residues mutated (in the TPR and DSH subdomains) are all required for optimal Ter1 association. As in the case of telomere maintenance, combining different clusters of point mutations resulted in a more severe Ter1 association defect. For example, the RR and KF mutations separately reduced Ter1 association 6- and 3-fold, respectively, whereas the combined mutant suffered an ∼15-fold reduction in association (Fig. 3D). We conclude that the association between Est1NTD and Ter1 plays an important role in telomere maintenance in K. lactis.
The consistently detrimental effects of our point mutations on Ter1 association are not unexpected given the consequences of comparable mutations on S. cerevisiae Est1-Tlc1 interactions. Nevertheless, to minimize the possibility that our system overdiagnoses RNA-binding defects, we characterized an est1 mutant (K467E) that is analogous to the est1-60 allele of S. cerevisiae EST1 (K444E), previously shown to impair telomerase recruitment and activation but not RNA binding (33). As predicted, the K. lactis mutant manifested substantial telomere loss but normal Est1-Ter1 association in cell extracts (see Fig. S3 in the supplemental material). This result further suggests that the Cdc13-Est1 interaction previously described in S. cerevisiae is at least partially conserved in K. lactis.
KlEst1 interacts directly with the Est1 binding domain of Ter1.
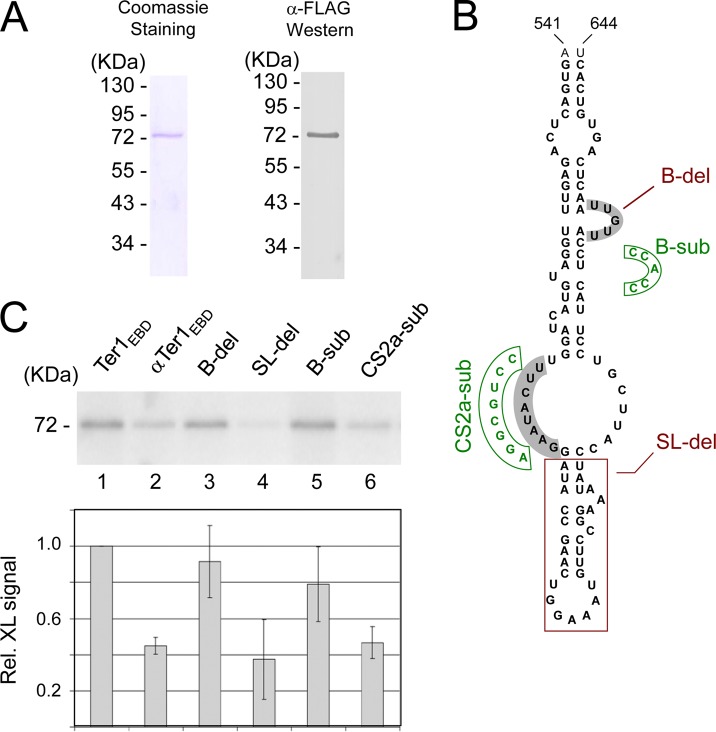
S. cerevisiae Est1 was initially reported to possess a nonspecific RNA-binding activity (45). However, further studies in vivo and in vitro implicated a specific region of Tlc1 (the S. cerevisiae telomerase RNA) in Est1 association (10, 38). In particular, a bulged stem in this region of Tlc1 appears to be a critical determinant of Est1-Tlc1 interaction. Based on potential sequence and structural similarities between this region of Tlc1 and other yeast telomerase RNAs, a conserved Est1 binding domain (EBD) has been proposed (17). The EBD, which is typically ∼100 nt long, consists of several stems and loops located 3′ to the template region of telomerase RNA. The putative EBD in K. lactis Ter1 has been shown to be required telomere maintenance in vivo (17). However, its ability to interact with Est1 has not been investigated. We therefore sought to determine biochemically if KlEst1 can interact directly with Ter1EBD. Because the CTD of KlEst1 is prone to proteolytic degradation in E. coli (data not shown) and is dispensable for telomere maintenance (Fig. 3A), we recombinantly expressed and purified Est1NTD with a C-terminal FLAG tag (Fig. 4A). The protein was first subjected to electrophoretic mobility shift assays (EMSAs) using Ter1EBD as the probe. Despite using a variety of published assay conditions, we were unable to detect complex formation between Est1 and Ter1 (data not shown) (10, 45). This observation is in agreement with an earlier study on ScEst1 but at odds with another report (10, 38). However, by employing a UV cross-linking assay, we were able to demonstrate a direct physical interaction between Est1NTD and Ter1EBD. To detect cross-linking, purified Est1NTD was incubated with uniformly labeled Ter1EBD, and the mixture was subjected to UV irradiation followed by RNase digestion, gel electrophoresis, and PhosphorImager scanning. Consistent with a covalent linkage between Est1NTD and Ter1EBD, an ∼75-kDa labeled species can be detected (Fig. 4C, lane 1). Ter1EBD was a preferred binding target because the antisense Ter1EBD (αTer1EBD) exhibited reduced cross-linking efficiency (Fig. 4C, compare lanes 1 and 2). We then tested four Ter1EBD mutants that had been shown previously to cause telomere shortening (17) and found that two of these mutants (SL-del and CS2a-sub) cross-linked to Est1 with reduced efficiency (Fig. 4B and C). The magnitude of reduction was about 60% for each mutant. A similar difference in the cross-linking efficiency of the wild-type and mutant RNAs was observed when we varied the RNA concentrations from 2 to 16 nM (see Fig. S4 in the supplemental material). The correlation between the defects of the Ter1 mutants in Est1 binding and telomere maintenance suggests that the direct RNA-binding activity of Est1 is required for telomere maintenance.
Fig 4.
KlEst1 interacts directly with the putative Est1 binding site of telomerase RNA. (A) Est1NTD was expressed in E. coli and purified as described in Materials and Methods. The purified protein was analyzed by SDS-PAGE, followed by Coomassie staining (left) or Western blotting using antibodies against the FLAG tag fused to the C terminus of the recombinant protein (right). (B) The secondary structure of Ter1EBD as predicted by RNAfold is displayed. The mutations tested as shown in panel C, including B-del, SL-del, B-sub, and CS2a-sub, are indicated. (C) Purified Est1NTD (12.5 nM) was incubated with 32P-labeled Ter1EBD, antisense Ter1EBD (α-Ter1EBD), or mutants of Ter1EBD (16 nM) and subjected to UV irradiation and RNase digestion. The products were analyzed by SDS-PAGE and PhosphorImager scanning. Representative assays are shown at the top and the summary data from three independent experiments are shown at the bottom.
We have demonstrated that mutations in the TPR and DSH subdomains of Est1 caused defects in Ter1 association in cell extracts (Fig. 3C). To investigate whether the defects can be accounted for by deficiencies in the direct binding activity of Est1, we purified the Est1NTD RR and RHRQ mutants and subjected them to the UV cross-linking assay using wild-type Ter1EBD, antisense Ter1EBD, and the CS2a-sub mutant as the probes. As shown in Fig. 5A, compared to the wild-type protein, the cross-linking efficiencies of both mutant proteins to Ter1EBD were reduced about 3-fold (lanes 1, 4, and 7). The mutant proteins nevertheless prefer Ter1 EBD to the antisense control and the CS2a-sub mutant by the same magnitude as the wild-type protein (compare lanes 4 to 6 and lanes 7 to 9 to lanes 1 to 3). Hence, these residues within the TPR domain appear to contribute to the affinity but not the sequence specificity of telomerase RNA binding; if these residues contribute to specificity, then one would expect the mutant proteins to exhibit reduced preference for wild-type RNA.
Fig 5.
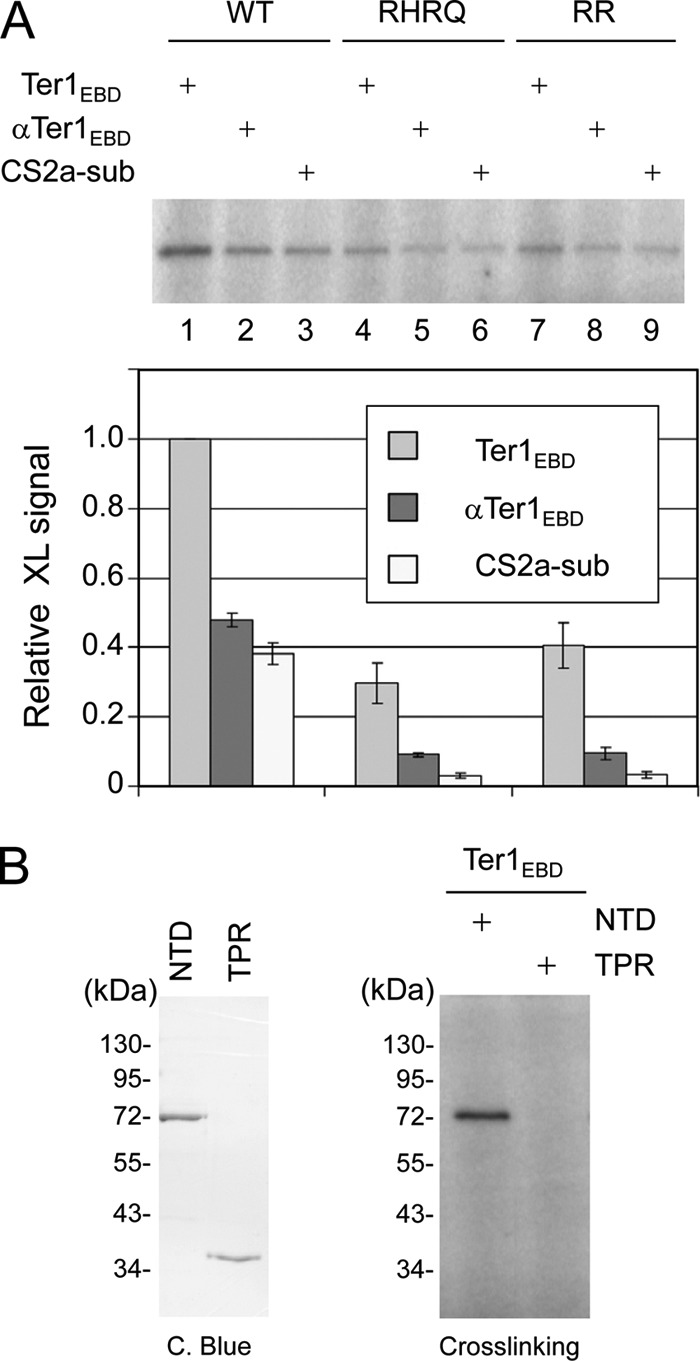
Analysis of Ter1EBD binding by KlEst1 mutants. (A) (Top) Purified wild-type Est1NTD and the RR and RHRQ mutants were subjected to UV irradiation in the presence of the indicated RNAs. The products were subjected to RNase digestion and analyzed by SDS-PAGE and PhosphorImager scanning. (Bottom) The relative signals from three independent experiments were calculated and plotted. (B) Equal molar amounts of purified Est1NTD and Est1TPR were analyzed by SDS-PAGE and Coomassie staining (left) and then subjected to the UV cross-linking assay with 32P-labeled Ter1EBD.
We next attempted to determine if the TPR or DSH subdomain alone can be cross-linked to Ter1EBD. However, we were only able to purify adequate quantities of KlEst1TPR (also with a C-terminal FLAG tag) for the analysis, KlEst1DSH being largely insoluble when expressed in E. coli. As shown in Fig. 5B, whereas KlEst1NTD exhibited robust cross-linking to Ter1EBD, the same level of KlEst1TPR failed to generate any significant amount of labeled products. Our data thus suggest that the TPR subdomain alone interacts with Ter1 with greatly reduced affinity, if at all. The cross-linking analyses of the substitution mutants and the TPR subdomain both support the notion that the KlEst1-Ter1 association in cell extracts is due at least in part to a direct physical interaction between these two molecules.
ScEst1 has been shown to bind telomere G-strand repeats with moderate affinity (10, 45). We therefore tested purified KlEst1NTD for binding to K. lactis telomere repeats using both EMSA and photo-cross-linking. Despite using a variety of target DNA and assay conditions, we were unable to detect significant DNA binding by KlEst1NTD, suggesting that this activity is not a conserved property of Est1 orthologues (data not shown).
KlEst1 mediates the functions of ScEbs1.
As described earlier, ScEbs1 but not ScEst1 has been shown to render yeast cells rapamycin sensitive, possibly through a role in the NMD pathway (14, 24). We therefore analyzed the rapamycin sensitivity of our KlEst1 mutants. Consistent with a role for KlEst1 in NMD, the est1Δ mutant reconstituted with an empty vector exhibited greater resistance to rapamycin than that reconstituted with full-length KlEst1-TAP (Fig. 6A). Interestingly, deleting the CTD abolished almost completely the sensitivity of the reconstituted strain. In addition, neither the TPR nor the DSH subdomain alone was capable of conferring rapamycin sensitivity to the reconstituted strain. We conclude that even though the CTD of KlEst1 is dispensable for telomere maintenance, it is crucial for the role of this protein in the rapamycin-responsive pathway(s). The contribution of KlEst1 to NMD is further supported by measurements of the levels of CYH2 pre-mRNA, a putative NMD target (24). Consistent with an NMD defect, the amount of the CYH2 pre-mRNA in the est1Δ mutant was approximately three times that in the wild-type strain (Fig. 6B).
Fig 6.
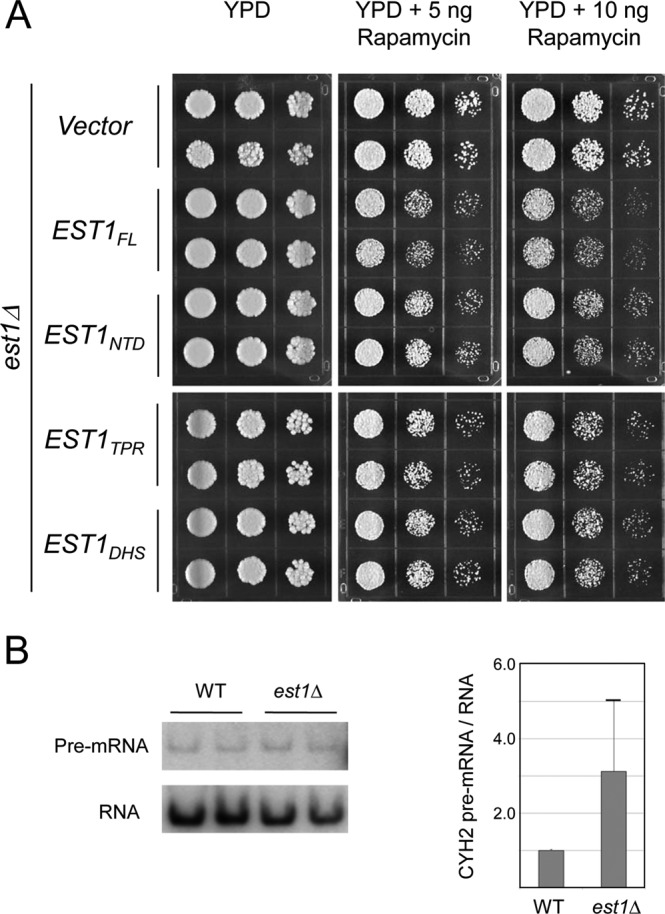
Est1 confers rapamycin sensitivity to the cells. (A) The est1Δ null strain reconstituted with wild-type and mutant EST1s was spotted on YPD plates without rapamycin and grown at 30°C for 2 days or with the indicated concentrations of rapamycin and grown for 4 days. (B) The levels of CYH2 pre-mRNA and RNA in the wild-type and est1Δ strains were measured by RT-PCR. A representative set of assays is displayed (left). The results from two experiments were quantified and plotted (right).
DISCUSSION
Our analysis of KlEst1 has provided insights on the evolution and mechanisms of Est1 family members, including the demonstration of (i) a direct physical interaction between Est1 and telomerase RNA, (ii) a conserved role for the TPR and DSH domain in this interaction, and (iii) a widespread function of Est1 family members in non-sense-mediated decay. The implications of these findings are discussed below.
Activity conservation between ScEst1 and KlEst1.
The initial investigation of ScEst1 suggests that the protein possesses a high-affinity but nonspecific RNA-binding activity, as well as a G-strand DNA-binding activity (45). Subsequent studies using pulldown of RNA by Sepharose-bound Est1 and RNA EMSA indicate at least some preference of ScEst1 for its presumed target site in Tlc1 (S. cerevisiae telomerase RNA) (10, 38). The authenticity of the DNA-binding activity is less clear. No mutations that selectively abolish the DNA-binding activity are available to assess the physiologic relevance of this activity. In addition, one study suggests that excess RNA can inhibit the formation of an Est1-DNA complex, hinting at a single nucleic acid binding site on this protein (10). Our analysis of KlEst1 clearly establishes RNA binding as a conserved and functionally important property of Est1 homologues. Conversely, our failure to detect a DNA-binding activity in KlEst1 casts some doubts on the significance of this activity.
The RNA-binding activity of KlEst1.
Our data argue that the direct binding of KlEst1NTD to Ter1EBD is a key function of this protein. But surprisingly, this interaction appears to be relatively weak and non-sequence specific. In contrast to a previous report on ScEst1, we were unable to detect a KlEst1NTD-Ter1EBD EMSA complex using a variety of assay conditions. Moreover, the cross-linking efficiency of Est1NTD to the wild-type EBD was only 2- to 3-fold higher than a variety of inactive control and mutant RNAs. At first glance, such a weak and nonspecific interaction would appear inadequate to underpin the role of KlEst1 in telomere maintenance. However, contacts between KlEst1 and other telomerase components may provide the additional specificity necessary for the assembly of the holoenzyme. There is compelling support, for example, for an interaction between Est1 and Est3 (another telomerase subunit) in both S. cerevisiae and C. albicans (22, 43). Another potential reason for the low sequence specificity of KlEst1 may be related to its role in NMD, which could involve its binding to other RNA targets with distinct sequences and structures.
As noted before, the structure of KlEst1NTD is likely to be largely alpha-helical and to contain a TPR-like subdomain (Fig. 1B). Because TPR domains are known to mediate diverse cellular functions through interaction with target peptides using its concave surface, the potential involvement of this structural module in RNA binding is unexpected (1, 8). To gain insights on the mechanisms of RNA binding, we generated a homology model of KlEst1NTD and mapped the positions of functionally important residues (see Fig. S5 in the supplemental material). Notably, while the three clusters of residues implicated in RNA binding are scattered in different surface areas, they are all located away from the putative peptide-binding, concave groove. This observation suggests that Est1 could use distinct, nonoverlapping surfaces for binding peptide and RNA targets. One cluster of residues required for RNA binding is located in a loop protruding from the edge of the TPR (RR), another is on the convex surface (QKR), and a third one is near the end of the DSH domain (KF). Assuming that all three clusters contact RNA directly, the rather large spatial separations between them imply a large RNA target site, which is consistent with the proposed Ter1EBD.
While this work was under review, Webb and Zakian reported an unbiased search for mutations that reduce S. pombe Est1-Ter1 interaction (47). Notably, they also identified residues in the TPR domain as being required for the interaction. Indeed, one of the amino acids implicated in fission yeast Ter1 binding (R194) aligns to K223 of ScEst1 and K220 of KlEst1, which we and others have shown to be involved in the corresponding interactions in budding yeast (reference 11 and this work) (see Fig. S6 in the supplemental material). This striking mechanistic conservation in turn suggests that Ter1 binding was an ancestral function of yeast Est1.
In addition to binding RNA, budding yeast Est1 is likely to interact with multiple protein targets, including Cdc13, Est3, and other less-well-characterized factors (37, 43, 50, 51). Data from mutational analysis suggest that both the TPR and DSH domains are involved in these interactions as well (33, 34, 37, 51). How the protein and RNA interaction functions of Est1 are coordinated is clearly worthy of further investigation.
Evolution of Est1 homologues.
As described earlier, ScEst1 and ScEbs1 are evidently ohnologs that emerged as a consequence of whole-genome duplication and perform distinct functions in telomere maintenance and NMD in Saccharomyces cerevisiae. An obvious question, then, is whether their functional divergence represents a case of subfunctionalization or neofunctionalization (7, 18). In other words, did the ancestral protein perform both functions, which were then delegated to different descendants? Or did one of the descendants evolve new activities? Our finding that KlEst1 participates in both telomere maintenance and NMD regulation supports the notion of subfunctionalization. Specifically, our results suggest that Est1 and Ebs1 together exemplify the duplication, degeneration, and complementation mechanisms of subfunctionalization, which also apply to other telomere-related proteins such as Sir2/Hst1 (13, 19). Indeed, the observation that human Est1 homologues have roles in both pathways argues that these functions are inherent to the ancient Est1 that existed prior to the divergence of yeasts and mammals. The fixation of EST1 paralogs in evolutionarily distant lineages suggests that the multiplicity of functions performed by the ancestral gene may be difficult to optimize in a single polypeptide.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yehudi Tzfati for plasmids containing various Ter1 mutants and for comments on the manuscript.
This work was supported by NIH GM-062631 and GM062631-08S1 (N.F.L.) and NIH GM 61645 and ACS RSF-02-253-01-GMC (M.J.M.).
Footnotes
Published ahead of print 27 April 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Allan RK, Ratajczak T. 2011. Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones 16:353–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Autexier C, Lue NF. 2006. The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 75:493–517 [DOI] [PubMed] [Google Scholar]
- 3. Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. 2007. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318:798–801 [DOI] [PubMed] [Google Scholar]
- 4. Beernink H, Miller K, Deshpande A, Bucher P, Cooper J. 2003. Telomere maintenance in fission yeast requires an est1 ortholog. Curr. Biol. 13:575–580 [DOI] [PubMed] [Google Scholar]
- 5. Byrne KP, Wolfe KH. 2005. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 15:1456–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen XJ. 1996. Low- and high-copy-number shuttle vectors for replication in the budding yeast Kluyveromyces lactis. Gene 172:131–136 [DOI] [PubMed] [Google Scholar]
- 7. Conant GC, Wolfe KH. 2008. Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9:938–950 [DOI] [PubMed] [Google Scholar]
- 8. D'Andrea L, Regan L. 2003. TPR proteins: the versatile helix. Trends Biochem. Sci. 28:655–662 [DOI] [PubMed] [Google Scholar]
- 9. Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. 2009. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell 35:403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. DeZwaan DC, Freeman BC. 2009. The conserved Est1 protein stimulates telomerase DNA extension activity. Proc. Natl. Acad. Sci. U. S. A. 106:17337–17342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Evans S, Lundblad V. 2002. The Est1 subunit of Saccharomyces cerevisiae telomerase makes multiple contributions to telomere length maintenance. Genetics 162:1101–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ferreira M, Miller K, Cooper J. 2004. Indecent exposure: when telomeres become uncapped. Mol. Cell 13:7–18 [DOI] [PubMed] [Google Scholar]
- 13. Force A, et al. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151:1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ford AS, Guan Q, Neeno-Eckwall E, Culbertson MR. 2006. Ebs1p, a negative regulator of gene expression controlled by the Upf proteins in the yeast Saccharomyces cerevisiae. Eukaryot. Cell 5:301–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fukuhara N, et al. 2005. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol. Cell 17:537–547 [DOI] [PubMed] [Google Scholar]
- 16. Fulton TB, Blackburn EH. 1998. Identification of Kluyveromyces lactis telomerase: discontinuous synthesis along the 30-nucleotide-long templating domain. Mol. Cell. Biol. 18:4961–4970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gunisova S, et al. 2009. Identification and comparative analysis of telomerase RNAs from Candida species reveal conservation of functional elements. RNA 15:546–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hahn MW. 2009. Distinguishing among evolutionary models for the maintenance of gene duplicates. J. Hered. 100:605–617 [DOI] [PubMed] [Google Scholar]
- 19. Hickman MA, Froyd CA, Rusche LN. 2011. Reinventing heterochromatin in budding yeasts: Sir2 and the origin recognition complex take center stage. Eukaryot. Cell 10:1183–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hoffman C, Winston F. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267–272 [DOI] [PubMed] [Google Scholar]
- 21. Hsu M, Lue NF. 2011. Analysis of yeast telomerase by primer extension assays. Methods Mol. Biol. 735:97–106 [DOI] [PubMed] [Google Scholar]
- 22. Hsu M, Yu EY, Singh SM, Lue NF. 2007. Mutual dependence of Candida albicans Est1p and Est3p in telomerase assembly and activation. Eukaryot. Cell 6:1330–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hug N, Lingner J. 2006. Telomere length homeostasis. Chromosoma 115:413–425 [DOI] [PubMed] [Google Scholar]
- 24. Luke B, et al. 2007. Saccharomyces cerevisiae Ebs1p is a putative ortholog of human Smg7 and promotes nonsense-mediated mRNA decay. Nucleic Acids Res. 35:7688–7697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luke B, et al. 2008. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell 32:465–477 [DOI] [PubMed] [Google Scholar]
- 26. Lundblad V, Szostak JW. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57:633–643 [DOI] [PubMed] [Google Scholar]
- 27. McEachern M, Haber J. 2006. Telomerase-independent telomere maintenance in yeast, p 199–224 In de Lange T, Lundblad V, Blackburn E. (ed), Telomeres, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 28. McEachern MJ, Blackburn EH. 1996. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 10:1822–1834 [DOI] [PubMed] [Google Scholar]
- 29. Moser BA, Chang YT, Kosti J, Nakamura TM. 2011. Tel1(ATM) and Rad3(ATR) kinases promote Ccq1-Est1 interaction to maintain telomeres in fission yeast. Nat. Struct. Mol. Biol. 18:1408–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Osterhage JL, Friedman KL. 2009. Chromosome end maintenance by telomerase. J. Biol. Chem. 284:16061–16065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Osterhage JL, Talley JM, Friedman KL. 2006. Proteasome-dependent degradation of Est1p regulates the cell cycle-restricted assembly of telomerase in Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 13:720–728 [DOI] [PubMed] [Google Scholar]
- 32. Palm W, de Lange T. 2008. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42:301–334 [DOI] [PubMed] [Google Scholar]
- 33. Pennock E, Buckley K, Lundblad V. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104:387–396 [DOI] [PubMed] [Google Scholar]
- 34. Qi H, Zakian VA. 2000. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 14:1777–1788 [PMC free article] [PubMed] [Google Scholar]
- 35. Redon S, Reichenbach P, Lingner J. 2007. Protein RNA and protein protein interactions mediate association of human EST1A/SMG6 with telomerase. Nucleic Acids Res. 35:7011–7022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rigaut G, et al. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030–1032 [DOI] [PubMed] [Google Scholar]
- 37. Sealey DC, Kostic AD, Lebel C, Pryde F, Harrington L. 2011. The TPR-containing domain within Est1 homologs exhibits species-specific roles in telomerase interaction and telomere length homeostasis. BMC Mol. Biol. 12:45 doi:10.1186/1471-2199-12-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Seto AG, Livengood AJ, Tzfati Y, Blackburn EH, Cech TR. 2002. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev. 16:2800–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Singh S, Lue N. 2003. Ever shorter Telomere 1 (EST1)-dependent reverse transcription by Candida telomerase in vitro: evidence in support of an activating function. Proc. Natl. Acad. Sci. U. S. A. 100:5718–5723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Singh S, Steinberg-Neifach O, Mian I, Lue N. 2002. Analysis of telomerase in Candida albicans: potential role in telomere end protection. Eukaryot. Cell 1:967–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Snow B, et al. 2003. Functional conservation of the telomerase protein est1p in humans. Curr. Biol. 13:698–704 [DOI] [PubMed] [Google Scholar]
- 42. Taggart A, Teng S, Zakian V. 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297:1023–1026 [DOI] [PubMed] [Google Scholar]
- 43. Tuzon CT, Wu Y, Chan A, Zakian VA. 2011. The Saccharomyces cerevisiae telomerase subunit Est3 binds telomeres in a cell cycle- and Est1-dependent manner and interacts directly with Est1 in vitro. PLoS Genet. 7:e1002060 doi:10.1371/journal.pgen.1002060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Underwood DH, Zinzen RP, McEachern MJ. 2004. Template requirements for telomerase translocation in Kluyveromyces lactis. Mol. Cell. Biol. 24:912–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Virta-Pearlman V, Morris DK, Lundblad V. 1996. Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev. 10:3094–3104 [DOI] [PubMed] [Google Scholar]
- 46. Wang ZR, Guo L, Chen L, McEachern MJ. 2009. Evidence for an additional base-pairing element between the telomeric repeat and the telomerase RNA template in Kluyveromyces lactis and other yeasts. Mol. Cell. Biol. 29:5389–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Webb CJ, Zakian VA. 2012. Schizosaccharomyces pombe Ccq1 and TER1 bind the 14-3-3-like domain of Est1, which promotes and stabilizes telomerase-telomere association. Genes Dev. 26:82–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wolfe KH, Shields DC. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387:708–713 [DOI] [PubMed] [Google Scholar]
- 49. Wray LV, Jr, Witte MM, Dickson RC, Riley MI. 1987. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol. Cell. Biol. 7:1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu Y, Zakian VA. 2011. The telomeric Cdc13 protein interacts directly with the telomerase subunit Est1 to bring it to telomeric DNA ends in vitro. Proc. Natl. Acad. Sci. U. S. A. 108:20304–20310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yu EY, et al. 2007. Regulation of telomere structure and functions by subunits of the INO80 chromatin remodeling complex. Mol. Cell. Biol. 27:5639–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yu EY, Yen WF, Steinberg-Neifach O, Lue NF. 2010. Rap1 in Candida albicans: an unusual structural organization and a critical function in suppressing telomere recombination. Mol. Cell. Biol. 30:1254–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang ML, et al. 2010. Yeast telomerase subunit Est1p has guanine quadruplex-promoting activity that is required for telomere elongation. Nat. Struct. Mol. Biol. 17:202–209 [DOI] [PubMed] [Google Scholar]
- 54. Zhou J, Hidaka K, Futcher B. 2000. The Est1 subunit of yeast telomerase binds the Tlc1 telomerase RNA. Mol. Cell. Biol. 20:1947–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.