Abstract
Monogalactosyldiacylglycerol (MGDG) in Chlamydomonas reinhardtii and other green algae contains hexadeca-4,7,10,13-tetraenoic acid (16:4) in the glycerol sn-2 position. While many genes necessary for the introduction of acyl chain double bonds have been functionally characterized, the Δ4-desaturase remained unknown. Using a phylogenetic comparison, a candidate gene encoding the MGDG-specific Δ4-desaturase from Chlamydomonas (CrΔ4FAD) was identified. CrΔ4FAD shows all characteristic features of a membrane-bound desaturase, including three histidine boxes and a transit peptide for chloroplast targeting. But it also has an N-terminal cytochrome b5 domain, distinguishing it from other known plastid desaturases. Cytochrome b5 is the primary electron donor for endoplasmic reticulum (ER) desaturases and is often fused to the desaturase domain in desaturases modifying the carboxyl end of the acyl group. Difference absorbance spectra of the recombinant cytochrome b5 domain of CrΔ4FAD showed that it is functional in vitro. Green fluorescent protein fusions of CrΔ4FAD localized to the plastid envelope in Chlamydomonas. Interestingly, overproduction of CrΔ4FAD in Chlamydomonas not only increased levels of 16:4 acyl groups in cell extracts but specifically increased the total amount of MGDG. Vice versa, the amount of MGDG was lowered in lines with reduced levels of CrΔ4FAD. These data suggest a link between MGDG molecular species composition and galactolipid abundance in the alga, as well as a specific function for this fatty acid in MGDG.
INTRODUCTION
Membranes in biological systems play a major role as barriers enclosing cells and their organelles. The lipid composition of a specific membrane is characteristic and reflects its function. The lipid fatty acid composition of the membranes of plants and algae is not static but can adapt to changes in the environment. In particular, (poly)unsaturated fatty acids are important in maintaining membrane fluidity and thus enhancing temperature and stress tolerance (see, e.g., reference 47). Unsaturated fatty acids are also precursors of signaling molecules such as jasmonate and thus are important for plant defense (15). Polyunsaturated fatty acids (PUFAs) in algae are also of practical interest, since algae are the primary producers of ω3 long-chain PUFAs, such as eicosapentaenoic acid (EPA; 20:5Δ5,8,11,14,17) or docosahexaenoic acid (DHA; 22:6Δ4,7,10,13,16,19), which are important for human nutrition (33). Efforts to produce these fatty acids in crop plants so as to increase their nutritional value are ongoing (45).
Fatty acid desaturases can be grouped into two enzyme classes. Those of the first class, soluble acyl-acyl carrier protein (ACP)-desaturases, catalyze the first step in PUFA biosynthesis (introduction of a cis-double bond between C-9 and C-10) and are found exclusively in photosynthetically active plastids. The crystal structure of these nonheme di-iron enzymes and their active site are known (11, 19).
The second and larger group of desaturases is composed of membrane-bound enzymes. They can be found in all eukaryotes and a few prokaryotes. No crystal structure is currently available, but it is proposed that the reaction mechanism is similar to that reported for the acyl-ACP-desaturases (41). Membrane-bound desaturases most likely contain four membrane-spanning domains and three histidine box motifs [HX3-4H, HX2-3HH, and (H/Q)X2-3HH], which are crucial for activity (38). In plants, membrane-bound fatty acid desaturases are present in the plastid and the endoplasmic reticulum (ER). The electron donors for plastid desaturases are typically ferredoxins, while ER enzymes use cytochrome b5 (Cytb5). Many ER-associated desaturases are directly fused to a cytochrome b5 domain. Typical “front-end” desaturases, which introduce a double bond 3 carbons closer to the carboxy terminus of the acyl chain than a preexisting double bond, predominantly contain an N-terminal cytochrome b5 (39, 40). No plastid desaturase carrying a cytochrome b5 fusion has been described previously. However, free forms of cytochrome b5 have been reported in the (outer) chloroplast envelope (17, 22), but no cytochrome b5 was detected in a recent chloroplast proteome study of Arabidopsis (14).
The plastid lipid monogalactosyldiacylglycerol (MGDG) in the green alga Chlamydomonas reinhardtii occurs preferentially as a molecular species containing α-linolenic acid 18:3Δ9,12,15 in the sn-1 position and (4Z,7Z,10Z,13Z)-hexadeca-4,7,10,13-tetraenoic acid (16:4) in the sn-2 position of the glycerol backbone (10, 34). This 16:4 acyl group is found only in trace amounts in other membrane lipids. Genes necessary for the introduction of the double bonds into lipid acyl groups in Chlamydomonas have been identified and characterized (6, 16, 36), except for this MGDG-specific Δ4-desaturase.
Desaturation in the Δ4 position of acyl groups in complex lipids also occurs during the synthesis of DHA in the ER, and the respective desaturase has been characterized (see Fig. S1 in the supplemental material). Genes encoding this activity have been cloned and characterized from various algae, such as Euglena gracilis (25), Thalassiosira pseudonana (44), or species of the genus Pavlova (Rebecca) (43, 50) or Thraustochytrium (31). All the enzymes mentioned above are located in the ER and contain an N-terminal cytochrome b5 domain. Alternatively, Schizochytrium, a close relative of Thraustochytrium, is able to synthesize DHA by a desaturase-independent polyketide synthase (PKS) pathway (24). However, taking into account the fact that no PKS gene cluster has been detected in the Chlamydomonas genome (23), it is likely that the 16:4 acyl group in MGDG is synthesized exclusively by stepwise desaturation of the 16:0 acyl group. Therefore, we reasoned at the outset of this study that the candidate enzyme had to be similar to a membrane-bound desaturase and not to PKS.
A possibly related desaturase was assumed to be the ER-located Δ4-desaturase from Euglena gracilis, because it was shown to be active in the synthesis of very long chain fatty acids but could also act on shorter C16 acyl groups (25). Its activity is strongly dependent on a preexisting Δ7 double bond, as would be expected for the Chlamydomonas MGDG-specific Δ4-desaturase. Here we report the identification and characterization of the Δ4-desaturase from Chlamydomonas, which is an unusual, plastid-located cytochrome b5 fusion protein.
MATERIALS AND METHODS
Bioinformatics.
BLASTP searches (3) were conducted in the Chlamydomonas genome V4 (23). The gene model was manually corrected using RNA-Seq data from our lab (26) and GreenGenie2 (18). The model was confirmed by reverse transcription-PCR (RT-PCR), followed by TA cloning into pGEM-T Easy (Promega, Madison, WI) and sequencing.
Hydropathy plots were generated with an algorithm according to the work of von Heijne (46) using the TOPPRED program (http://mobyle.pasteur.fr/cgi-bin/portal.py?#forms::toppred). Protein alignments were generated with the Clustal algorithm (42). Phylogenetic analysis was performed by computing with the PHYLIP package (version 3.69) using the neighbor-joining method and comparison of 1,000 bootstrap replicates. TreeView was employed for visualization.
RNA isolation and cDNA preparation from Chlamydomonas.
RNA was isolated from 5-ml mid-log-phase cultures using the RNeasy Plant Mini kit (Qiagen, Valencia, CA) including the optional on-column DNase digest. A 0.5-μg portion of total RNA was reverse transcribed with the SuperScript III system (Invitrogen, Carlsbad, CA) and oligo(dT) primers at 50°C in a 20-μl reaction volume throughout all experiments. A 0.5-μl volume of cDNA was used for estimation of mRNA abundance by PCR and amplification of the full-length coding sequence.
PCR conditions.
The primers used to amplify the full-length coding sequences and the cytochrome b5 domains can be found in Table S1 in the supplemental material. All constructs mentioned above were amplified with Phusion polymerase (Finnzymes, Woburn, MA) using the GC buffer supplied (HF buffer for the cytochrome b5 gene from Arabidopsis), 2% dimethyl sulfoxide (DMSO) (Chlamydomonas genes only), and the following conditions. Initial denaturation was carried out at 98°C for 2 min, followed by 30 to 35 cycles with 10 s at 98°C, 10 s at 65°C (52°C for Arabidopsis cytochrome b5), and 20 s per kb at 72°C. The last cycle was followed by 1 min of final elongation at 72°C. For the estimation of RNA abundance, RT-PCR internal fragments of CrΔ4FAD, MGD1, DGD1, and RACK1 were amplified by using GoTaq Flexi DNA polymerase (Promega, Madison, WI) according to the manufacturer's instructions. RACK1 is a control gene with stable high expression under normal growth conditions, commonly used for normalization of Chlamydomonas mRNA (2). The required number of cycles was determined individually for each gene. The band intensity in gel images was estimated with ImageJ (1). Detailed information on primer sequences, cycle numbers, and fragment sizes can be found in Table S2 in the supplemental material.
Cultivation and transformation of Chlamydomonas.
Chlamydomonas cells were grown in Tris-acetate-phosphate (TAP) medium (34) at 22°C in continuous light until mid-log phase (optical density at 560 nm [OD560], ≈1 to 1.5). Two different cell wall-less strains were used in this study: dw15 (courtesy of Arthur Grossman) and UVM4, which is based on strain cw15. Strain UVM4 was especially designed for efficient expression of nuclear transgenes (29). When dw15 was grown as a control, the medium had to be supplemented with 100 μM arginine. Agar plates additionally contained 0.8% Phytoblend (Caisson Laboratories Inc., North Logan, UT). For the selection of transgenic lines, plates also contained 10 μg/ml paromomycin (Sigma-Aldrich, St. Louis, MO).
Transformation of Chlamydomonas was carried out as described previously (21) with the following modifications. Thirty milliliters of a mid-log-phase culture grown in TAP medium was harvested (1,500 × g, 3 min, room temperature, swinging bucket rotor), and the cells were resuspended in 500 μl fresh TAP medium. The cells were then transferred to a glass vial containing ≈300 mg acid-washed glass beads (diameter, 0.4 to 0.5 mm). After the addition of 1 μg linearized, desalted plasmid DNA, the cells were agitated using a standard vortexer for 15 s. A 5-ml volume of TAP medium was added, and the cells were regenerated overnight in the dark under shaking (22°C). The following day, cells were harvested as described above and were resuspended in 3 ml fresh TAP medium. After careful mixing with 3 ml Top Agar (0.5% Phytoblend in TAP medium, cooled to ≤45°C), cells were spread on TAP agar containing 10 μg/ml paromomycin. The plates were kept at room temperature (≈24°C under continuous light) until colonies appeared (≈6 to 7 days). Single colonies were then transferred to a fresh TAP plate containing 10 μg/ml paromomycin and continued to grow as described above. Single colonies were used to inoculate 10 ml of TAP medium (in a 25-ml flask) and were grown as described above for biochemical and microscopic analyses.
Construction of the artificial microRNA vector.
Artificial microRNA constructs were designed as described previously (28) using the pChlamiRNA3int vector. One microgram of plasmid DNA was linearized with KpnI-HF (New England BioLabs [NEB], Ipswich, MA) and was purified (PCR purification kit; Qiagen) prior to the transformation of Chlamydomonas strain dw15. Three constructs were tested for functionality by RT-PCR analysis of transgenic Chlamydomonas lines. Only data for the most effective oligomer (5′-TCACTAAAGGATGACAACCTA-3′) are shown.
Construction of Chlamydomonas CrΔ4FAD overexpression/localization vectors.
The cDNA sequence encoding CrΔ4FAD was cloned into the NdeI site of pJR38 (29) in frame with the codon-optimized enhanced green fluorescent protein (EGFP) gene. Prior to the transformation of strain UVM4, 1 μg plasmid DNA was linearized with XbaI (NEB) and desalted (PCR purification kit; Qiagen).
Analysis of lipid composition and chlorophyll content.
Five milliliters of mid-log-phase cell cultures was harvested (1,500 × g, 3 min, room temperature, swinging bucket rotor), and lipids were extracted as described by Bligh and Dyer (4). The organic phase was transferred to a fresh glass tube and was dried under an N2 stream. The lipids were dissolved in 100 μl chloroform. Usually 10 μl was converted to their fatty acyl methyl esters (FAMEs) as described previously (35). Lipids contained in 30 μl were separated by thin-layer chromatography (TLC) (48), and MGDG and digalactosyldiacylglycerol (DGDG) were scraped and were also converted to FAMEs. FAMEs in hexane were analyzed by gas liquid chromatography (GC) on an HP6890 machine with a DB-23 column (both from Agilent Technologies, Santa Clara, CA) and the following temperature profile: 140°C for 2 min, an increase to 160°C at 25°C/min, a further increase to 250°C at 8°C/min, and holding at 250°C for 4 min, followed by a temperature decrease back to 140°C at 38°C/min. All samples were run at a constant helium flow of 1.5 ml/min and a split ratio of 1:30 (for total fatty acids) or 1:10 (for TLC-purified lipids). For the identification of FAMEs by gas chromatography-mass spectrometry (MS), the same model instrument was coupled with a model 5975B MS detector (all from Agilent Technologies) using similar running conditions.
The contents of chlorophylls a and b from Chlamydomonas cultures were determined for acetone extracts as described previously (30).
Expression of the cytochrome b5 domain in Escherichia coli.
The soluble cytochrome b5 portion (corresponding to amino acids 48 to 147) of CrΔ4FAD was cloned between the EcoRI and XhoI sites of pDsRed-plw01-His (20) and was expressed in E. coli BL21(DE3) cells (Novagen, Rockland, MA). A free form of cytochrome b5 from Arabidopsis (At1g26340) and the cytochrome b5 domain of the ER-located fatty acid Δ5-desaturase from Chlamydomonas (amino acids 1 to 99 of CrFAD13 [16]) were expressed. Thirty minutes prior to induction with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG), the heme precursor γ-aminolevulinic acid was added (final concentration, 0.5 μM) as described previously (40). After induction, cell growth was continued at 28°C for 4 h. Cells were cooled on ice, resuspended in lysis buffer (50 mM Tris-HCl, 300 mM NaCl, 10 mM imidazole [pH 7.5]), and lysed by sonication (10 cycles for 10 s, interrupted by 10 s on ice). After centrifugation at 10,000 × g, the supernatant was mixed with Ni2+-nitrilotriacetic acid agarose (Qiagen) and was incubated with shaking at 4°C for 60 min. The resin was washed twice with washing buffer (50 mM Tris-HCl, 300 mM NaCl, 20 mM imidazole [pH 7.5]) before the DsRED fusion proteins were eluted (50 mM Tris-HCl, 300 mM NaCl, 250 mM imidazole [pH 8.0]). Protein purity was confirmed by acrylamide gel electrophoresis and Coomassie staining.
Cytochrome b5 assay.
An absorbance spectrum between 400 and 600 nm was recorded for 250 μg of purified protein (air oxidized) in assay buffer (100 mM Tris-HCl, 0.5 mM EDTA [pH 8.0]) at room temperature as described previously (40). The samples were then reduced with sodium dithionite or dithiothreitol (DTT) and were immediately measured again as described above. Redox difference spectra were calculated for all treatments.
Confocal microscopy.
Confocal laser scanning microscopy images were taken at the Center of Advanced Microscopy at Michigan State University on an Olympus FluoView FV1000 confocal laser scanning microscope (Olympus, Center Valley, PA) configured on the Olympus IX81 fully automated inverted microscope platform. GFP was excited using the 488-nm argon laser line, while green fluorescence was collected using a 505- to 525-nm band-pass filter. Simultaneously, red chlorophyll autofluorescence was also collected using the 488-nm laser line for excitation and a 650-nm long-pass filter, while transmitted laser light was used to record the bright-field image. Images were recorded using the 60× UPLAPO oil objective (numerical aperture [NA], 1.42).
Nucleotide sequence accession number.
The sequence of CrΔ4FAD has been deposited in GenBank under accession number JN089704.
RESULTS
Identification of a Δ4-desaturase candidate gene.
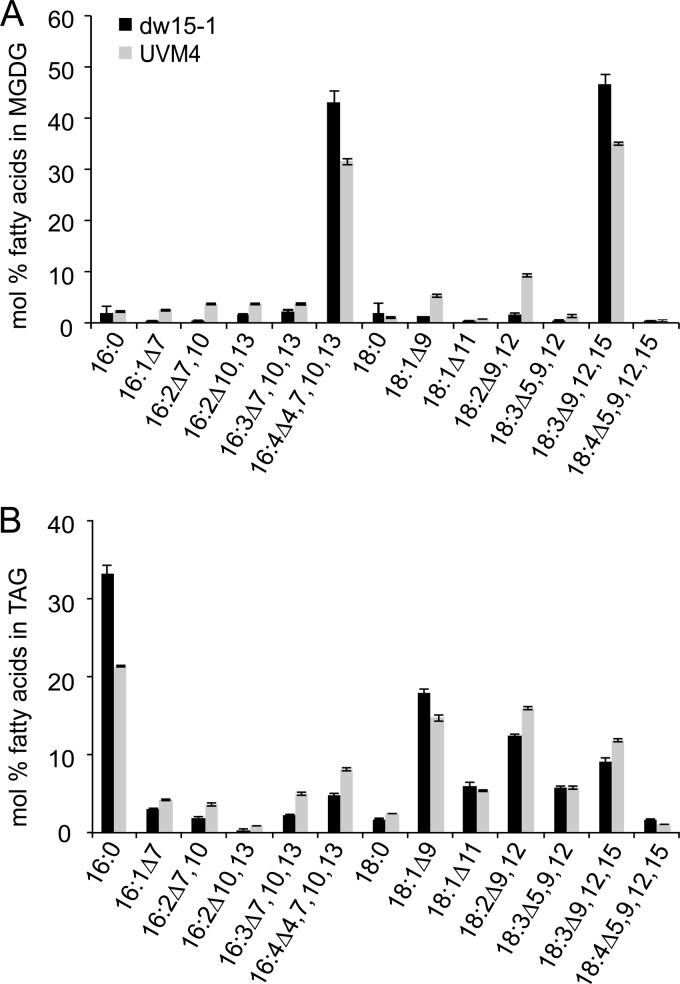
The Chlamydomonas reinhardtii strains used in this study contain 16:4 acyl groups in MGDG (Fig. 1A), as reported previously for other strains (10). Large amounts of 16:4 acyl groups were detected in MGDG, while other lipids, such as DGDG, contained only trace amounts. In addition, 16:4 acyl groups were present in triacylglycerol (TAG) in cells cultured under nitrogen deprivation (Fig. 1B), conditions that induce TAG biosynthesis in Chlamydomonas (7, 27, 49).
Fig 1.
Fatty acyl profiles of MGDG (A) and TAG (B) isolated from Chlamydomonas strains dw15 (filled bars) and UVM4 (shaded bars). Hexadecatetraenoic acid (16:4) is the major fatty acid in the sn-2 position of MGDG but was also recovered in TAG (5% in dw15; 8% in UVM4). Lipids were separated by thin-layer chromatography, converted to their fatty acid methyl esters, and quantified by GC-flame ionization detection. Fatty acids are displayed by the number of carbons, followed by a colon and the number of double bonds. The numbers following Δ indicate the positions of the double bonds in the acyl chain. Means and standard deviations of three biological replicates are shown.
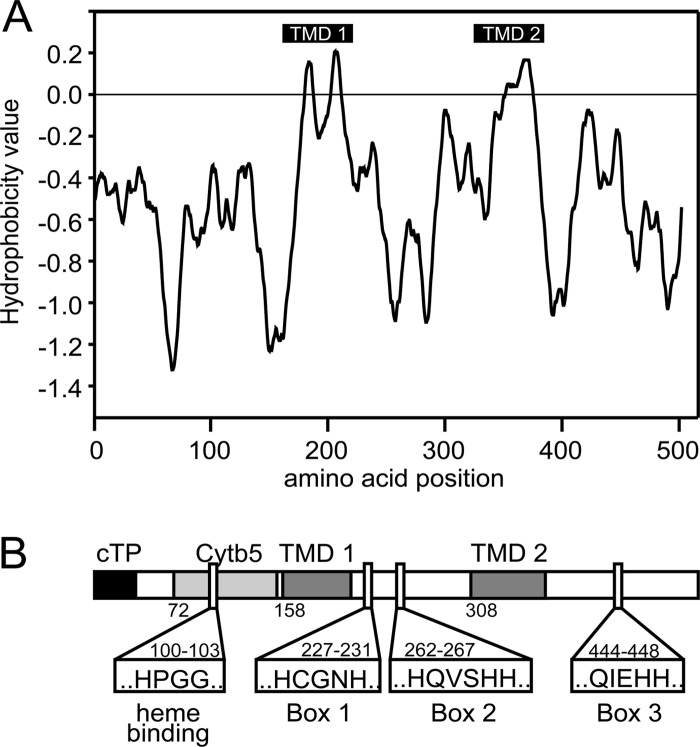
A phylogenetic comparison was used to identify the desaturase required for 16:4 acyl groups in MGDG and TAGs. Searches in the Chlamydomonas genome V4 using protein sequences for the Δ4-desaturases from Euglena gracilis (GenBank accession number AAQ19605) and Thalassiosira pseudonana (GenBank accession number AAX14506) as queries identified a candidate sequence (protein identification [ID] 35253 in the Chlamydomonas genome version 4.0 [http://genome.jgi-psf.org/Chlre4/Chlre4.home.html]). The predicted protein consisted of 1,675 amino acids (coding sequence, 5,025 bp) and contained desaturase and cytochrome b5 domains. Since the size of the coding sequence of cytochrome b5 fusion desaturases usually ranges from 1,400 to 1,600 bp, and all commonly relevant domains were encoded in the first 1,500 bp of the candidate sequence, the size of the gene model was questioned. GreenGenie2 (18) was used to correct the gene model, and the coding sequence was verified by mapping transcriptomics data (26) to the genome. The updated coding sequence was 1,551 bp (516 amino acids), while the gene was 4,812 bp and consisted of 10 exons (Fig. 2). Primers were designed based on this new model, and the PCR-amplified genomic sequence was subjected to sequencing for confirmation. The corrected coding sequence has been deposited in GenBank under accession number JN089704. No information about 5′ and 3′ untranslated regions was obtained. Figure 2 shows the hydrophobicity plot and a schematic structural overview of the likely desaturase, tentatively designated CrΔ4FAD. A complete protein alignment of CrΔ4FAD with known Δ4-fatty acid desaturases can be found in Fig. S2 in the supplemental material. Figure S3 in the supplemental material shows a regioselectivity dendrogram of desaturase-like sequences from organisms of different classes, including CrΔ4FAD (Fig. S3A) and a schematic structural comparison of the major desaturase classes (Fig. S3B).
Fig 2.
Protein topology prediction for CrΔ4FAD. The hydropathy plot (A) was generated using the TOPPRED program (http://mobyle.pasteur.fr/cgi-bin/portal.py?#forms::toppred) and is based on the algorithm of von Heijne (46). (B) Schematic overview of CrΔ4FAD topology. cTP, chloroplast transit peptide; Cytb5, cytochrome b5 domain; TMD, membrane-spanning domains, each composed of two transmembrane helices. The three histidine boxes and the heme binding motif are indicated.
Distinguishing this enzyme from other described desaturases, the CrΔ4FAD sequence contained both a putative chloroplast transit peptide and an N-terminal cytochrome b5 domain, suggesting a chloroplast location.
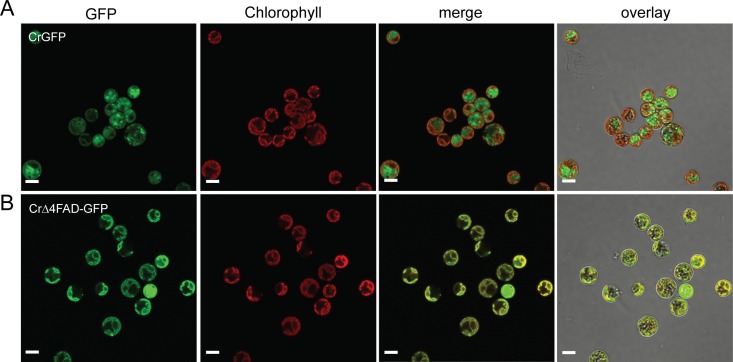
CrΔ4FAD-GFP is targeted to the chloroplast in Chlamydomonas.
The presence of a likely chloroplast transit peptide suggested that this protein is located in the plastid. To determine whether CrΔ4FAD is indeed imported into the plastid, we expressed a green fluorescent protein (GFP)-tagged version in Chlamydomonas and analyzed the transgenic lines by confocal laser scanning microscopy. Chlamydomonas cells producing GFP alone showed a strong fluorescence signal in the cytosol, whereas the GFP signal overlapped with the chlorophyll autofluorescence signal in cells expressing CrΔ4FAD-GFP (Fig. 3A and B). Due to the high-fluorescence background in Chlamydomonas, we were not able to determine whether the protein is located in the envelope or thylakoid membranes.
Fig 3.
CrΔ4FAD-GFP localizes to the chloroplast in Chlamydomonas. Free GFP is localized in the cytosol of Chlamydomonas strain UVM4 (A), in contrast to CrΔ4FAD-GFP, which is found in the plastid (B). Bars, 5 μm.
Modulation of CrΔ4FAD expression in Chlamydomonas alters the MGDG content.
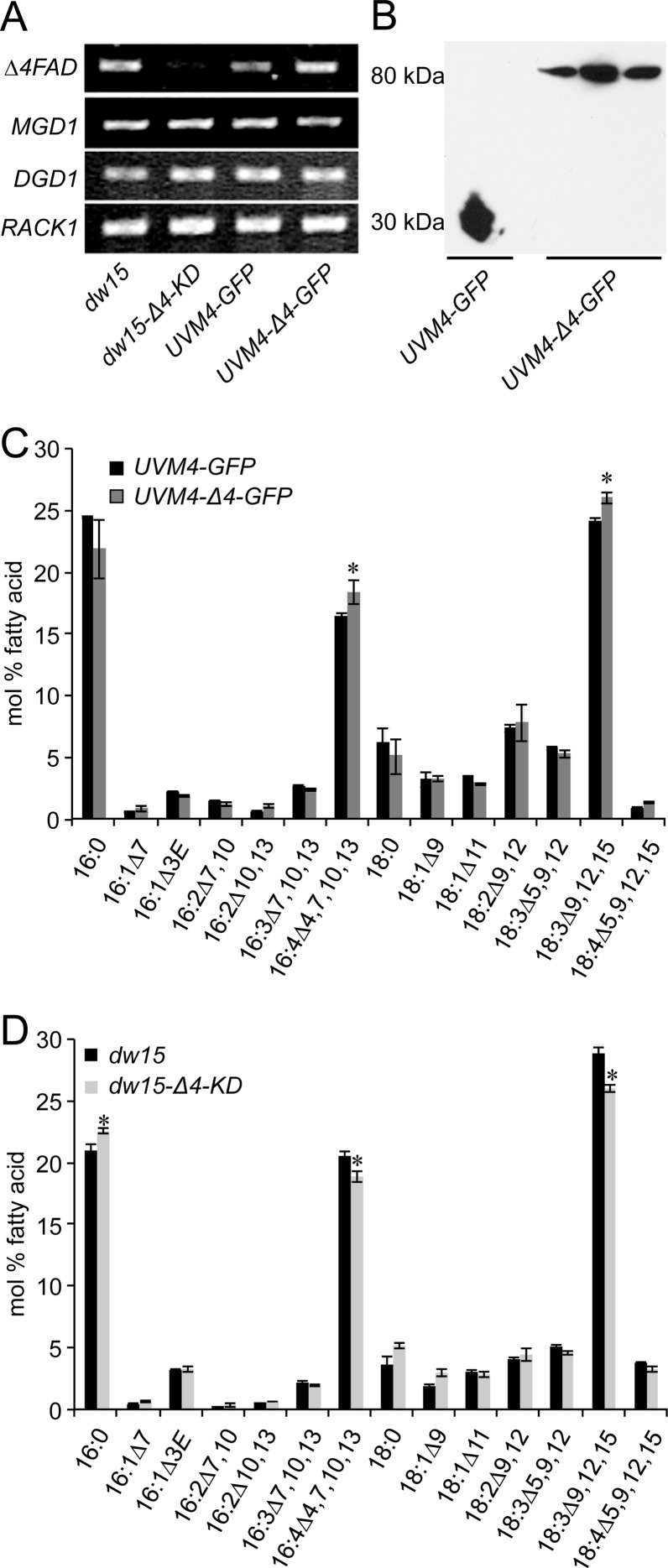
A CrΔ4FAD-GFP fusion construct was introduced into Chlamydomonas strain UVM4, which is optimized for the expression of nuclear transgenes (29). Transgenic lines were screened for enhanced CrΔ4FAD transcript levels by PCR, as well as for increased protein abundance by immunoblotting using an anti-GFP antibody (Fig. 4B).
Fig 4.
Knockdown and overexpression of CrΔ4FAD in Chlamydomonas. (A) Knockdown lines (dw15-Δ4-KD) show lower CrΔ4FAD expression than the empty-vector control (dw15). CrΔ4FAD expression is elevated in strain UVM4-Δ4-GFP, expressing CrΔ4FAD-GFP. UVM4-GFP (UVM4 expressing GFP alone) was used as a control. The expression of the control genes MGD1, DGD1, and RACK1 was not affected in any of the lines. cDNA was prepared from equal amounts of total RNA and was used as the PCR template with differing cycle numbers depending on the gene: 34 for CrΔ4FAD, 30 for MGD1 and DGD1, and 24 for the control gene RACK1. (B) Immunoblot showing the expression of the CrΔ4FAD-GFP fusion construct in three independent lines. Chlamydomonas cells were harvested, mixed with sodium dodecyl sulfate sample buffer, and directly loaded onto a denaturing gel. Proteins were then transferred to a polyvinylidene difluoride membrane and were subjected to immunoblot analysis using an anti-GFP antibody. (C) The fatty acid composition of Chlamydomonas cells overexpressing CrΔ4FAD is changed: the levels of 16:4 and 18:3Δ9,12,15 are higher in overexpression lines (shaded bars) than in the empty-vector control (filled bars). (D) Total fatty acid composition in CrΔ4FAD knockdown lines. Knockdown lines (shaded bars) show lower levels of 16:4 and 18:3Δ9,12,15 acyl groups than does the empty-vector control (filled bars). For panels C and D, total fatty acids were converted to their FAMEs and were quantified by GC-flame ionization detection. Significant differences in fatty acid composition between the two lines are marked by asterisks (P, <0.05 by a nonpaired two-sample t test).
To repress CrΔ4FAD activity in Chlamydomonas, an artificial microRNA construct targeting specifically the 5′ part of the coding sequence identified was generated and was introduced into the cell wall-less strain dw15. Transformants had reduced CrΔ4FAD transcript levels (Fig. 4A).
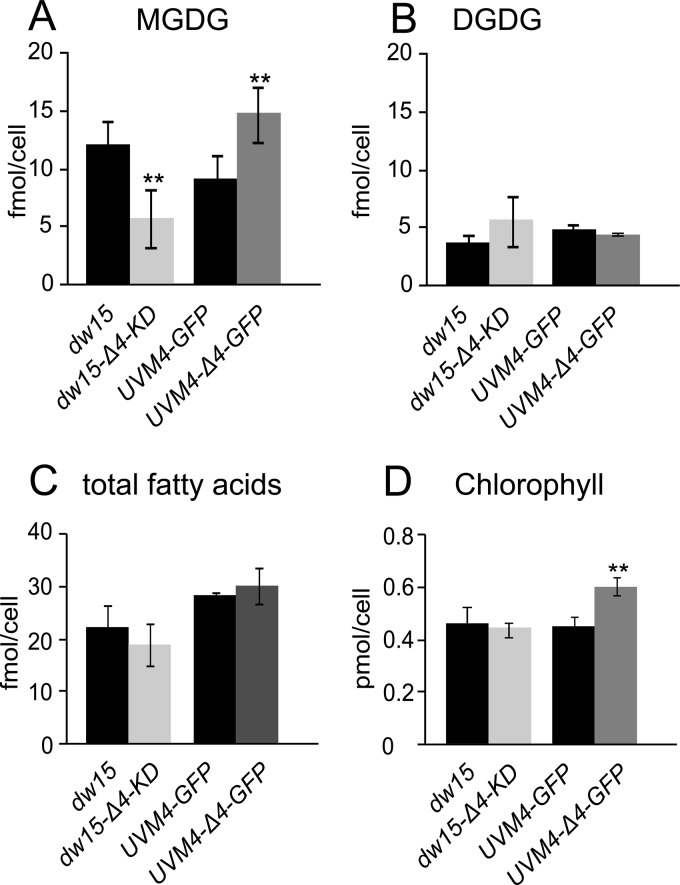
Because in the polar lipid fraction, the 16:4 acyl group is found almost exclusively in MGDG, we initially analyzed only the fatty acid composition of this lipid for decreased or increased 16:4 levels in CrΔ4FAD knockdown and overexpression lines. No obvious alterations in the fatty acid composition of MGDG were observed. Interestingly, the total fatty acid profile of the overexpression lines showed a statistically significant increase in 16:4 and 18:3Δ9,12,15 acyl groups (with a concomitant decrease in the corresponding saturated acyl groups, e.g., 16:0) (Fig. 4C). Vice versa, the knockdown lines had decreases in the 16:4 and 18:3Δ9,12,15 acyl groups (Fig. 4D). The decrease in the 18:3Δ9,12,15 acyl groups was initially surprising. However, this is explained by the decreased amount of MGDG present in knockdown lines (Fig. 5A). Vice versa, overexpression of CrΔ4FAD led to an increase in total-MGDG levels and corresponding increases in the predominant acyl groups present in this lipid.
Fig 5.
CrΔ4FAD expression specifically affects the MGDG content (A), but not the DGDG content (B), of Chlamydomonas. The total fatty acid content remains unchanged (C), but the higher total-galactolipid content in the overexpression lines leads to an increased chlorophyll content (D). Lipids for panels A to C were isolated from a defined amount of cells, separated by TLC, and quantified as FAMEs. Pigments (D) were isolated, and their absorbance spectra were detected in 80% acetone. Asterisks indicate significant differences between knockdown or overexpression lines and their corresponding controls (P, <0.05 by a nonpaired two-sample t test). Data were obtained from at least six independent experiments. Strains are explained in the legend to Fig. 4.
DGDG, the second prominent galactolipid, is derived from MGDG but contains only trace amounts of 16:4 (10). The fatty acid composition of DGDG was not affected by changes in CrΔ4FAD expression, but we observed a slight increase in the total DGDG amount in the knockdown lines (Fig. 5B). Transcript levels of the galactolipid synthesis genes MGD1 and DGD1 were not altered (Fig. 4A).
The increase in the total-galactolipid amount in the overexpression lines was accompanied by an increase in the chlorophyll content per cell (Fig. 5D) but did not affect the chlorophyll a/b ratio (see also Fig. S4 in the supplemental material).
The cytochrome b5 domain of CrΔ4FAD is active in vitro.
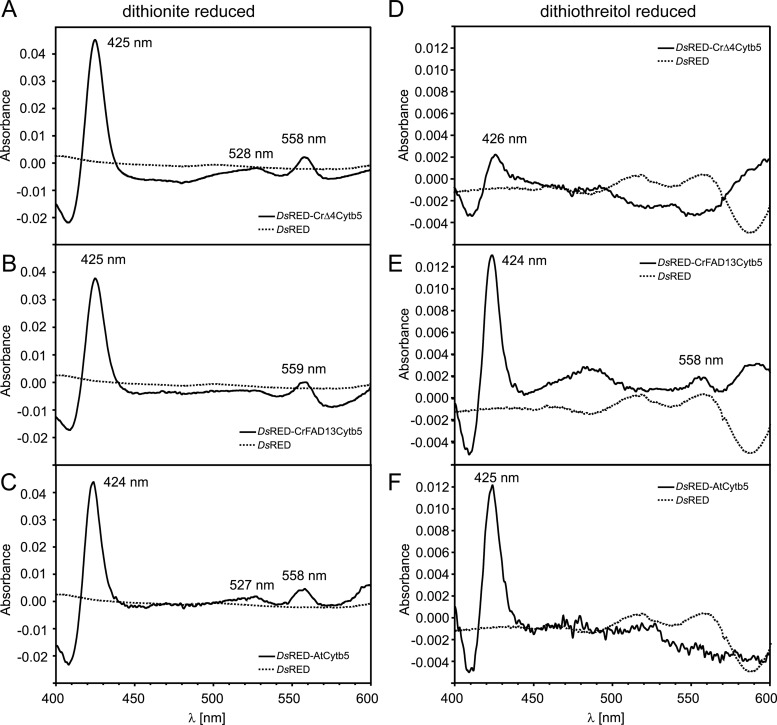
To test the functionality of the cytochrome b5 domain, DsRED fusion constructs were expressed in E. coli, and the proteins were purified by their C-terminal His tag. Redox difference spectra were calculated from the absorbance of the air-oxidized and dithionite-reduced samples (redox potential [ΔE], −660 mV). The observed absorbance difference maxima at 425, 527, and 557 nm (Fig. 6A) were similar to those of other fatty acid desaturase cytochrome b5 domains (12, 40). They were also detected in protein extracts from cells producing DsRED fused to free cytochrome b5 from Arabidopsis and the cytochrome b5 domain of the ER-localized desaturase FAD13 from Chlamydomonas, but not in the DsRED control (Fig. 6B and C). The weaker reducing agent dithiothreitol (DTT) (ΔE, −330 mV) was still able to reduce DsRED-AtCytb5 and DsRED-CrFAD13Cytb5, though at lower levels, but not DsRED-CrΔ4Cytb5 (Fig. 6D to F).
Fig 6.
Redox difference spectra of dithionite-reduced (A to C) and dithiothreitol-reduced (D to F) recombinant cytochrome b5. (A to C) Spectra were calculated from the absorbance of air-oxidized and dithionite-reduced DsRED fusion proteins. DsRED-CrΔ4Cytb5 (A), DsRED-CrFAD13Cytb5 (B), and DsRED-AtCytb5 (C) show difference maxima at 425 (424), 528 (527), and 558 (559) nm, respectively, in line with previous observations (12, 40). (D to F) Spectra were calculated from the absorbance of air-oxidized and DTT-reduced DsRED fusion proteins. The characteristic shape of the cytochrome b5 difference spectrum is still maintained in DsRED-CrFAD13Cytb5 (E) and DsRED-AtCytb5 (F), although the difference between oxidized and DTT-reduced forms is much less pronounced than that with dithionite. In contrast, DsRED-CrΔ4Cytb5 cannot be reduced with DTT (D), indicating that CrΔ4Cytb5 requires a stronger reductant than DTT [ΔE, −330 mV, comparable with that for NADH; ΔE(dithionite), −660 mV].
DISCUSSION
Although a large number of different fatty acid desaturases have been characterized from a variety of organisms, including algae, some of the more specialized enzymes are still being discovered. One example is the recent identification of the plastid phosphatidylglycerol-specific desaturase FAD4 from Arabidopsis, which was shown to fall into a distinct class of fatty acid desaturases (9) (see Fig. S3 in the supplemental material). The MGDG-specific Δ4 desaturase from Chlamydomonas described here, CrΔ4FAD, is another example. While we were able to predict it by phylogenetic analysis in the genome of Chlamydomonas, it contained surprising features setting it apart from other known desaturases. Notably, an N-terminal cytochrome b5 domain was present in what we determined is a plastid-localized fatty acid desaturase. Plastid localization of the enzyme was predicted based on the presence of a transit peptide and due to the fact that the enzyme appeared to be highly specific for the exclusively plastid localized galactolipid MGDG.
Different approaches to confirm the activity by expression of the cDNA in Saccharomyces cerevisiae or Arabidopsis were not productive at this time, and modification of CrΔ4FAD abundance in Chlamydomonas did not alter the fatty acid composition of MGDG per se but affected the amount of MGDG in the cells. Apparently, in Chlamydomonas, the availability of the 16:4 acyl group in MGDG limits overall MGDG content in the thylakoid membrane. This result provides evidence for the presence of a regulatory or compensatory system in Chlamydomonas that governs membrane lipid homeostasis. A different compensatory effect was observed in previous attempts to suppress the activity of the betaine lipid synthase Bta1 of Chlamydomonas using an RNA interference (RNAi) approach (34). Rather than reducing the relative amount of betaine lipid, the cells reduced growth to maintain a constant overall lipid composition. Considering both observations, it appears that strong compensatory mechanisms are in place in Chlamydomonas that maintain membrane lipid homeostasis at the level of the lipid class, as observed for betaine lipid, and the molecular species composition of individual lipids, as is the case for MGDG, shown here.
While plastid localization for CrΔ4FAD was expected, the presence of a cytochrome b5 domain was not. In fact, the deduced protein sequence contained all structural features of a membrane-bound “front end” desaturase typically associated with the ER: besides an N-terminal cytochrome b5 domain, three histidine box motifs were identified, which have been shown to be essential for activity in comparable enzymes (38). As in all “front end” desaturases identified so far, one of the histidines in the third conserved box is replaced by a glutamine (motif QXXHH). Replacement of the corresponding glutamine residue with a histidine in the Δ6-desaturase from borage abolished activity (37). Cytochrome b5 fusions typically occur at the N terminus in “front end” desaturases and at the C terminus of Δ9-desaturases in fungi and red algae. Only a few “front end” desaturases without cytochrome b5 are known, e.g., the Δ6-desaturase from the cyanobacterium Synechocystis sp. strain PCC 6802 (32).
The exclusive presence of 16:4 acyl groups in Chlamydomonas under nutrient-replete conditions in the sn-2 position of MGDG suggests strong substrate specificity for CrΔ4FAD in vivo or high specificity of substrate channeling toward the active site of the enzyme. Another example of a highly specific plastid desaturase is FAD4, responsible for the synthesis of 16:1Δ3(E) esterified to the sn-2 position of phosphatidylglycerol (9). In a second study involving transgenic plants producing bacterial diacylglycerol kinase in the plastid, thereby causing channeling of different lipid molecular species into the FAD4 substrate phosphatidylglycerol, the Δ3(E) double bond was also found in C18 fatty acids (8). This demonstrates that FAD4 activity is lipid specific but independent of the length of the acyl chain. On the other hand, Heilmann et al. (13) forced a new substrate on the normally plastid localized desaturase FAD5 by altering subcellular targeting to the ER. Instead of the native substrate MGDG, the double bonds were introduced into the acyl chains of phosphatidylcholine, albeit with altered regioselectivity.
We identified CrΔ4FAD by sequence comparison with a known Δ4-desaturase involved in DHA biosynthesis. The activity of the Δ4-desaturase from Euglena was shown to be dependent on a preexisting Δ7 double bond but, like FAD4, independent of the acyl chain length (25). Given that the Δ4 double bond is found in small amounts also in 16:2Δ4,7 and 16:3Δ4,7,10 (Fig. 1), CrΔ4FAD may also be dependent on Δ7 unsaturated substrates. Elucidation of substrate preferences was not possible due to the lack of an in vitro system. It seems possible that the major limitation may be the availability of a suitable electron donor. In the ER, electrons are transferred to cytochrome b5 by a specific oxidoreductase. This leads to the question of the nature of the primary electron donor of CrΔ4FAD. We did not find evidence for a cytochrome b5 oxidoreductase, which could be imported into the plastid of Chlamydomonas. Furthermore, our in vitro expression data for the cytochrome b5 domain suggest that CrΔ4FAD requires a stronger reducing agent than free cytochrome b5 in the ER. Of CrΔ4Cytb5, CrFAD13Cytb5, and AtCytb5, all three could be fully reduced by dithionite (ΔE, −660 mV), but only AtCytb5 and CrFAD13Cytb5 could also be fully reduced by DTT (ΔE, −330 mV). The redox potential of DTT is within the same range as that of NADH (ΔE, −315 mV). Taking into account the fact that the redox potentials of ferredoxins from Chlamydomonas range from −390 to −425 mV (5), it seems likely that CrΔ4Cytb5 receives electrons from a ferredoxin rather than NAD(P)H.
The presence of this unusual desaturase in Chlamydomonas chloroplasts and the apparent correlation of 16:4 acyl group content and MGDG amounts raise the question of the function of this particular molecular species of MGDG. Absent an efficient system for the complete inactivation of genes in Chlamydomonas, it will be difficult to answer this question by creating a null mutant. But it seems clear, based on the analysis of the transgenic lines with altered CrΔ4FAD abundance, that exquisite compensatory mechanisms are in place in Chlamydomonas to adjust the abundance of MGDG and its molecular species composition.
Supplementary Material
ACKNOWLEDGMENTS
We thank Melinda Frame (Center for Advanced Microscopy at MSU) for help with the confocal imaging and Beverley Chamberlin (Mass Spectrometry Facility, MSU) for the GC-flame ionization detection-MS analyses.
This work was funded by a DAAD Postdoc Research Fellowship (http://www.daad.de) to S.Z.; by the National Science Foundation, supporting T.B. during a summer internship program; and by a grant from the Air Force Office of Scientific Research, to C.B.
W. Jochum was a participant in the 2010 DAAD Rise in North America Undergraduate Program (http://www.DAAD.de). T. Bigorowski was a participant in the 2010 Michigan State University Plant Genomics Summer Undergraduate Program (http://www.plantgenomics.msu.edu).
Footnotes
Published ahead of print 4 May 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Abramoff M, Magelhaes P, Ram S. 2004. Image processing with ImageJ. Biophotonics Int. 11:36–42 [Google Scholar]
- 2. Allen MD, Kropat J, Tottey S, del Campo JA, Merchant SS. 2007. Manganese deficiency in Chlamydomonas results in loss of photosystem II and MnSOD function, sensitivity to peroxides, and secondary phosphorus and iron deficiency. Plant Physiol. 143:263–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 4. Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917 [DOI] [PubMed] [Google Scholar]
- 5. Cammack R, et al. 1977. Midpoint redox potentials of plant and algal ferredoxins. Biochem. J. 168:205–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chi X, et al. 2008. Fatty acid biosynthesis in eukaryotic photosynthetic microalgae: identification of a microsomal Δ12 desaturase in Chlamydomonas reinhardtii. J. Microbiol. 46:189–201 [DOI] [PubMed] [Google Scholar]
- 7. Fan J, Andre C, Xu C. 2011. A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii. FEBS Lett. doi: 10.1016/j.febslet.2011.05.018 [DOI] [PubMed] [Google Scholar]
- 8. Fritz M, et al. 2007. Channeling of eukaryotic diacylglycerol into the biosynthesis of plastidial phosphatidylglycerol. J. Biol. Chem. 282:4613–4625 [DOI] [PubMed] [Google Scholar]
- 9. Gao J, et al. 2009. FATTY ACID DESATURASE4 of Arabidopsis encodes a protein distinct from characterized fatty acid desaturases. Plant J. 60:832–839 [DOI] [PubMed] [Google Scholar]
- 10. Giroud C, Gerber A, Eichenberger W. 1988. Lipids of Chlamydomonas reinhardtii: analysis of molecular species and intracellular site(s) of biosynthesis. Plant Cell Physiol. 29:587–595 [Google Scholar]
- 11. Guy JE, Whittle E, Kumaran D, Lindqvist Y, Shanklin J. 2007. The crystal structure of the ivy Δ4-16:0-ACP desaturase reveals structural details of the oxidized active site and potential determinants of regioselectivity. J. Biol. Chem. 282:19863–19871 [DOI] [PubMed] [Google Scholar]
- 12. Haak D, Gable K, Beeler T, Dunn T. 1997. Hydroxylation of Saccharomyces cerevisiae ceramides requires Sur2p and Scs7p. J. Biol. Chem. 272:29704–29710 [DOI] [PubMed] [Google Scholar]
- 13. Heilmann I, Mekhedov S, King B, Browse J, Shanklin J. 2004. Identification of the Arabidopsis palmitoyl-monogalactosyldiacylglycerol Δ7-desaturase gene FAD5, and effects of plastidial retargeting of Arabidopsis desaturases on the fad5 mutant phenotype. Plant Physiol. 136:4237–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Joyard J, et al. 2010. Chloroplast proteomics highlights the subcellular compartmentation of lipid metabolism. Prog. Lipid Res. 49:128–158 [DOI] [PubMed] [Google Scholar]
- 15. Kachroo A, Kachroo P. 2009. Fatty acid-derived signals in plant defense. Annu. Rev. Phytopathol. 47:153–176 [DOI] [PubMed] [Google Scholar]
- 16. Kajikawa M, et al. 2006. A front-end desaturase from Chlamydomonas reinhardtii produces pinolenic and coniferonic acids by ω13 desaturation in methylotrophic yeast and tobacco. Plant Cell Physiol. 47:64–73 [DOI] [PubMed] [Google Scholar]
- 17. Kriechbaumer V, et al. 2009. Subcellular distribution of tail-anchored proteins in Arabidopsis. Traffic 10:1753–1764 [DOI] [PubMed] [Google Scholar]
- 18. Kwan AL, Li L, Kulp DC, Dutcher SK, Stormo GD. 2009. Improving gene-finding in Chlamydomonas reinhardtii: GreenGenie2. BMC Genomics 10:210 doi: 10.1186/1471-2164-10-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lindqvist Y, Huang W, Schneider G, Shanklin J. 1996. Crystal structure of Δ9 stearoyl-acyl carrier protein desaturase from castor seed and its relationship to other di-iron proteins. EMBO J. 15:4081–4092 [PMC free article] [PubMed] [Google Scholar]
- 20. Lu B, Benning C. 2009. A 25-amino acid sequence of the Arabidopsis TGD2 protein is sufficient for specific binding of phosphatidic acid. J. Biol. Chem. 284:17420–17427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lumbreras V, Stevens DR, Purton S. 1998. Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J. 14:441–447 [Google Scholar]
- 22. Maggio C, Barbante A, Ferro F, Frigerio L, Pedrazzini E. 2007. Intracellular sorting of the tail-anchored protein cytochrome b5 in plants: a comparative study using different isoforms from rabbit and Arabidopsis. J. Exp. Bot. 58:1365–1379 [DOI] [PubMed] [Google Scholar]
- 23. Merchant SS, et al. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318:245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Metz JG, et al. 2001. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science 293:290–293 [DOI] [PubMed] [Google Scholar]
- 25. Meyer A, et al. 2003. Biosynthesis of docosahexaenoic acid in Euglena gracilis: biochemical and molecular evidence for the involvement of a Δ4-fatty acyl group desaturase. Biochemistry 42:9779–9788 [DOI] [PubMed] [Google Scholar]
- 26. Miller R, et al. 2010. Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol. 154:1737–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moellering ER, Benning C. 2010. RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot. Cell 9:97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Molnar A, et al. 2009. Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 58:165–174 [DOI] [PubMed] [Google Scholar]
- 29. Neupert J, Karcher D, Bock R. 2009. Generation of Chlamydomonas strains that efficiently express nuclear transgenes. Plant J. 57:1140–1150 [DOI] [PubMed] [Google Scholar]
- 30. Porra RJ, Thompson WA, Kriedemann PE. 1989. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975:384–394 [Google Scholar]
- 31. Qiu X, Hong H, MacKenzie SL. 2001. Identification of a Δ4 fatty acid desaturase from Thraustochytrium sp. involved in the biosynthesis of docosahexanoic acid by heterologous expression in Saccharomyces cerevisiae and Brassicajuncea. J. Biol. Chem. 276:31561–31566 [DOI] [PubMed] [Google Scholar]
- 32. Reddy AS, Nuccio ML, Gross LM, Thomas TL. 1993. Isolation of a Δ6-desaturase gene from the cyanobacterium Synechocystis sp. strain PCC 6803 by gain-of-function expression in Anabaena sp. strain PCC 7120. Plant Mol. Biol. 22:293–300 [DOI] [PubMed] [Google Scholar]
- 33. Riediger ND, Othman RA, Suh M, Moghadasian MH. 2009. A systemic review of the roles of n-3 fatty acids in health and disease. J. Am. Diet. Assoc. 109:668–679 [DOI] [PubMed] [Google Scholar]
- 34. Riekhof WR, Sears BB, Benning C. 2005. Annotation of genes involved in glycerolipid biosynthesis in Chlamydomonas reinhardtii: discovery of the betaine lipid synthase BTA1Cr. Eukaryot. Cell 4:242–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rossak M, Schäfer A, Xu N, Gage DA, Benning C. 1997. Accumulation of sulfoquinovosyl-1-O-dihydroxyacetone in a sulfolipid-deficient mutant of Rhodobacter sphaeroides inactivated in sqdC. Arch. Biochem. Biophys. 340:219–230 [DOI] [PubMed] [Google Scholar]
- 36. Sato N, Fujiwara S, Kawaguchi A, Tsuzuki M. 1997. Cloning of a gene for chloroplast ω6 desaturase of a green alga, Chlamydomonas reinhardtii. J. Biochem. 122:1224–1232 [DOI] [PubMed] [Google Scholar]
- 37. Sayanova O, et al. 2001. Mutagenesis and heterologous expression in yeast of a plant Δ6-fatty acid desaturase. J. Exp. Bot. 52:1581–1585 [DOI] [PubMed] [Google Scholar]
- 38. Shanklin J, Whittle E, Fox BG. 1994. Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry 33:12787–12794 [DOI] [PubMed] [Google Scholar]
- 39. Sperling P, Heinz E. 2001. Desaturases fused to their electron donor. Eur. J. Lipid Sci. Technol. 103:158–180 [Google Scholar]
- 40. Sperling P, Schmidt H, Heinz E. 1995. A cytochrome-b5-containing fusion protein similar to plant acyl lipid desaturases. Eur. J. Biochem. 232:798–805 [PubMed] [Google Scholar]
- 41. Sperling P, Ternes P, Zank TK, Heinz E. 2003. The evolution of desaturases. Prostaglandins Leukot. Essent. Fatty Acids 68:73–95 [DOI] [PubMed] [Google Scholar]
- 42. Thompson JD, Gibson TJ, Higgins DG. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics Chapter 2:Unit 2.3. doi: 10.1002/0471250953.bi0203s00 [DOI] [PubMed] [Google Scholar]
- 43. Tonon T, Harvey D, Larson TR, Graham IA. 2003. Identification of a very long chain polyunsaturated fatty acid Δ4-desaturase from the microalga Pavlova lutheri. FEBS Lett. 553:440–444 [DOI] [PubMed] [Google Scholar]
- 44. Tonon T, et al. 2005. Fatty acid desaturases from the microalga Thalassiosira pseudonana. FEBS J. 272:3401–3412 [DOI] [PubMed] [Google Scholar]
- 45. Venegas-Caleron M, Sayanova O, Napier JA. 2010. An alternative to fish oils: Metabolic engineering of oil-seed crops to produce omega-3 long chain polyunsaturated fatty acids. Prog. Lipid Res. 49:108–119 [DOI] [PubMed] [Google Scholar]
- 46. von Heijne G. 1992. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487–494 [DOI] [PubMed] [Google Scholar]
- 47. Wada H, Gombos Z, Murata N. 1994. Contribution of membrane lipids to the ability of the photosynthetic machinery to tolerate temperature stress. Proc. Natl. Acad. Sci. U. S. A. 91:4273–4277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang Z, Benning C. 2011. Arabidopsis thaliana polar glycerolipid profiling by thin layer chromatography (TLC) coupled with gas-liquid chromatography (GLC). J. Vis. Exp. 49:2518 doi: 10.3791/2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang ZT, Ullrich N, Joo S, Waffenschmidt S, Goodenough U. 2009. Algal lipid bodies: stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot. Cell 8:1856–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhou XR, et al. 2007. Isolation and characterization of genes from the marine microalga Pavlova salina encoding three front-end desaturases involved in docosahexaenoic acid biosynthesis. Phytochemistry 68:785–796 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.