Abstract
Streptococcus oligofermentans is an oral commensal that inhibits the growth of the caries pathogen Streptococcus mutans by producing copious amounts of H2O2 and that grows faster than S. mutans on galactose. In this study, we identified a novel eight-gene galactose (gal) operon in S. oligofermentans that was comprised of lacABCD, lacX, and three genes encoding a galactose-specific transporter. Disruption of lacA caused more growth reduction on galactose than mutation of galK, a gene in the Leloir pathway, indicating that the principal role of this operon is in galactose metabolism. Diauxic growth was observed in cultures containing glucose and galactose, and a luciferase reporter fusion to the putative gal promoter demonstrated 12-fold repression of the operon expression by glucose but was induced by galactose, suggesting a carbon catabolite repression (CCR) control in galactose utilization. Interestingly, none of the single-gene mutations in the well-known CCR regulators ccpA and manL affected diauxic growth, although the operon expression was upregulated in these mutants in glucose. A double mutation of ccpA and manL eliminated glucose repression of galactose utilization, suggesting that these genes have parallel functions in regulating gal operon expression and mediating CCR. Electrophoretic mobility shift assays demonstrated binding of CcpA to the putative catabolite response element motif in the promoter regions of the gal operon and manL, suggesting that CcpA regulates CCR through direct regulation of the transcription of the gal operon and manL. This provides the first example of oral streptococci using two parallel CcpA-dependent CCR pathways in controlling carbohydrate metabolism.
INTRODUCTION
The human oral cavity harbors between 700 and 19,000 operative taxonomic units (16, 19, 45), among which streptococci are the most abundant microorganisms, contributing about 80% of the total microbial population in the dental biofilm (5, 31). Complex interspecies competitions occur among the oral streptococci due to limited space and shared nutritional requirements (30, 37, 38, 50). The outcome of these interspecies competitions may determine the health status of the oral cavity.
Carbohydrates are common carbon and energy sources for all streptococci, and as such, differences in uptake and metabolic efficiency among the species affect each species' competitiveness in the oral biofilm. The carbohydrate metabolism of bacteria is usually under the control of carbon catabolite repression (CCR) (13, 34, 35, 42), in which rapidly metabolizable carbohydrates, usually glucose, repress the utilization of nonpreferred carbohydrates. It is generally believed that catabolite control protein A (CcpA) (8, 15) and the histidine phosphocarrier protein HPr (9) function in the regulation of CCR in most low-G+C Gram-positive bacteria. HPr phosphorylation (HPr-Ser-P) is triggered by fructose-1,6-bisphosphate (9), a glycolysis intermediate. In the presence of a preferred carbohydrate, i.e., glucose, HPr-Ser-P serves as a cofactor to assist CcpA binding to the promoter regions of the CCR-regulated genes (9).
CCR also occurs in carbohydrate utilization in oral streptococci. Interestingly, CcpA does not play a dominant role in CCR of the dental caries pathogen Streptococcus mutans (14, 20, 33, 44, 46, 47, 49). Instead, HPr-Ser-P and three phosphoenolpyruvate-dependent sugar-phosphotransferase system (PTS) EII complexes, EIIABMan, FruI, and EIILev, assume primary control over the transport and catabolism of nonpreferred carbohydrates (1, 46, 47, 49). The “mitis group” streptococci are major competitors of S. mutans; however, how their CCR is regulated is not well understood.
Our previous studies have shown that Streptococcus oligofermentans, an oral commensal of the mitis group, can outcompete S. mutans by producing abundant H2O2 when protein or glucose is present (37, 38). Further, S. oligofermentans, although it utilizes fewer saccharides than S. mutans, metabolizes galactose more efficiently and yields greater cell mass on galactose than on glucose. To further understand this highly efficient galactose metabolism, in this study, we identified and characterized a galactose operon in S. oligofermentans and showed that it is under the control of CCR. However, unlike in S. mutans and other lactic acid bacteria, galactose utilization in S. oligofermentans is regulated by both CcpA and EIIMan (ManL).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
S. oligofermentans wild-type strain AS 1.3089T (39) and its derivatives were routinely cultured in brain heart infusion (BHI) broth (BD Difco, Franklin Lakes, NJ) at 37°C as static cultures, and the plate cultures were incubated in a candle jar. Transformants were selected on BHI agar plates supplemented with kanamycin (1 mg/ml), chloramphenicol (17 μg/ml), or spectinomycin (800 μg/ml). Growth measurements of S. oligofermentans strains were carried out in TY (1% tryptone-1% yeast extract) or TV broth (7) containing glucose and galactose. Escherichia coli strain DH5α was used for cloning, as well as plasmid amplification, and was grown in Luria-Bertani broth or on agar plates at 37°C. Transformants were selected with ampicillin (100 μg/ml), spectinomycin (250 μg/ml), chloramphenicol (17 μg/ml), or kanamycin (50 μg/ml).
DNA manipulation.
Standard recombinant DNA techniques were used for plasmid constructions. All restriction and ligation enzymes were purchased from New England BioLabs (Beverly, MA). The genomic DNA of S. oligofermentans was extracted and purified using the method of Marmur (24) with slight modifications (11). All primers were designed according to the unreleased draft genome of S. oligofermentans and synthesized by the Sangon Company (Shanghai, China). PCR amplifications were performed with KOD-Plus-Neo (Toyobo, Japan), and purification of the PCR products was carried out with a Qiagen QIAquick PCR Purification Kit (Valencia, CA). DNA extracted from agarose gel was purified with a Tiangen (Beijing, China) Tiangel Midi Purification Kit, and plasmids were extracted and purified with a Tiangen (Beijing, China) Tianprep Mini Plasmid Kit.
Construction of deletion mutants.
lacA, galK, EIIABC, manL, ccpA, and hprK allelic-replacement mutants were constructed using the PCR-ligation method (18). Briefly, DNA fragments corresponding to about 600 bp of the upstream and downstream sequences of each gene were amplified by PCR with the primer pairs listed in Table S1in the supplemental material. BamHI was used to digest the purified PCR products and to cut the nonpolar kanamycin resistance gene cassette from plasmid pALH124 (3). The three fragments were then purified, mixed at a molar ratio of 1:1:1, and ligated by T4 DNA ligase at 16°C for 30 min. The ligation mixture was transformed into the S. oligofermentans wild-type strain using a method described previously (36). Transformants were selected on BHI agar plates containing 1 mg/ml kanamycin, and the gene deletion was confirmed by PCR and sequencing. To construct ccpA-manL and ccpA-hprK double mutants, each 600-bp upstream and downstream sequence of the ccpA gene was amplified with PCR using primer pairs Bi-ccpA-upF/Bi-ccpA-upR and Bi-ccpA-dnF/Bi-ccpA-dnR (see Table S1in the supplemental material) and digested with HindIII and SmaI. A nonpolar spectinomycin resistance gene cassette was released from vector pFW5-luc (28) by digestion with the same enzyme. After gel purification, the three fragments were ligated with DNA ligase, and the ligation mixture was transformed into the manL and hprK mutants. The positive recombinants were selected on BHI agar plates supplemented with kanamycin (1 mg/ml) and spectinomycin (800 μg/ml) and verified by PCR and sequencing.
Construction of luciferase reporter strains.
The promoter regions of the gal operon and manL, which include approximately 320 and 267 bp of the sequence upstream of the start codon of each gene, respectively, were PCR amplified from the chromosome DNA of S. oligofermentans with the primer pairs listed in Table S1in the supplemental material. The purified PCR product was subsequently digested with BamHI and NheI, gel purified, and ligated to the compatible sites on the pFW5-luc (28) vector using DNA ligase. Correct recombinants (pFW5-luc-Pgal and pFW5-luc-PmanL) were confirmed by restriction analysis, PCR, and sequencing. pFW5-luc-Pgal was then transformed into the S. oligofermentans wild type, and pFW5-luc-PmanL was transformed into the wild-type strain and the ccpA mutant. Transformants were selected on BHI agar plates containing 800 μg/ml spectinomycin and confirmed by PCR and luciferase activity.
To construct luciferase reporter strains of ccpA-manL and ccpA-hprK double mutants, a fragment of pFW5-luc-PmanL vector without the spectinomycin resistance cassette was amplified by PCR using primer pair pFW5-luc-ScaI/pFW5-luc-SalI (see Table S1in the supplemental material), and a chloramphenicol acetyltransferase (CAT)-encoding gene fragment was amplified from the vector PlevD-cat (46) with CAT-ScaI/CAT-SalI (see Table S1in the supplemental material); then, both fragments were digested with ScaI and SalI, gel purified, and ligated to the compatible sites using DNA ligase. The correct recombinant pFW5-luc-cat-manL was verified by restriction analysis, PCR, and sequencing and then was transformed into ccpA-manL and ccpA-hprK double mutants. Transformants were selected on BHI agar plates containing 800 μg/ml spectinomycin, 1 mg/ml kanamycin, and 17 μg/ml chloramphenicol and confirmed by PCR and luciferase activity assay.
Luciferase activity assay.
Twenty-five microliters of 1 mM d-luciferin (Sigma-Aldrich, St. Louis, MO) solution (suspended in 1 mM citrate buffer, pH 6.0) was added to 100-μl samples, and luciferase activities were measured essentially as previously described (21) using a TD 20/20 luminometer (Turner Biosystems, Sunnyvale, CA). The optical densities at 600 nm (OD600) of the samples were determined with a 2100 visible spectrophotometer (Unico, Shanghai, China) and used to normalize the luciferase activity. All the measurements were done with duplicate samples, and all experiments were repeated at least three times.
Overexpression and purification of S. oligofermentans CcpA.
An amino-terminal fusion of His6 to CcpA of S. oligofermentans was constructed as follows. A 1,005-bp region containing the entire ccpA gene was PCR amplified from S. oligofermentans genomic DNA (gDNA) using the primer pair ccpA-28a-F/ccpA-28a-R (see Table S1in the supplemental material). The resultant product was digested with NdeI/BamHI and ligated into the compatible sites on pPET-28a (Novagen, Madison, WI) to produce pPET-28a-ccpA. Following verification by DNA sequencing, pPET-28a-ccpA was transformed into E. coli BL21(DE3)(pLysS) (Novagen, Madison, WI) cells and cultured in Luria-Bertani medium supplemented with 50 mg/ml kanamycin. The cells were grown at 37°C to an OD600 of 0.4 to 0.6. Overproduction of the CcpA protein was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (Sigma-Aldrich, St. Louis, MO). The culture was allowed to grow for an additional 3 to 4 h before being harvested. Cells were collected by centrifugation at 8,200 × g for 10 min, resuspended in a 1/10 volume of binding buffer (20 mM sodium phosphate, 0.5 M NaCl, 30 mM imidazole, pH 7.4), and then lysed by sonication for 10 min. The cell lysate was centrifuged at 13,400 × g for 15 min, and the supernatant was filtered through a 0.22-nm polyvinylidene difluoride membrane (Millipore, Billerica, MA) and then applied to a Ni2+-charged chelating column (GE Healthcare, Piscataway, NJ) previously equilibrated with binding buffer. Proteins were eluted by elution buffer (20 mM sodium phosphate, 0.5 M NaCl, 500 mM imidazole, pH 7.4), and the elution fractions were analyzed by electrophoresis on a 12% sodium dodecyl sulfate-polyacrylamide gel. The fractions with the desired protein were pooled and dialyzed against TKED buffer (100 mM Tris-HCl, 150 mM KCl, 1 mM EDTA, 0.1 mM dithiothreitol) (4) and stored in aliquots in 10% glycerol at −80°C until they were used.
EMSA.
Double-stranded DNA (dsDNA) probes, corresponding to the 320 and 267 bp of the promoter fragments of the gal operon and manL, respectively, were generated by PCR using two pairs of biotin-labeled primers, galp-F EMSA/galp-R EMSA and manLp-F EMSA/manLp-R EMSA (see Table S1in the supplemental material), respectively. A gal promoter fragment lacking the putative cre sequence (denoted gal-p-cre) was also generated by overlap extension PCR. Briefly, two DNA fragments with overlapping ends were generated by PCR using two pairs of primers, Ngalp-F EMSA/galp-cre-R EMSA and Ngalp-R EMSA/galp-cre-F EMSA (see Table S1in the supplemental material). The two fragments were then mixed at a 1:1 ratio and used as a template for a second-round PCR amplification with the biotin-labeled primer pair galp-F EMSA/galp-R EMSA. PCR products amplified with the unlabeled primer pair Ngalp-F EMSA/Ngalp-R EMSA and NmanLp-F EMSA/NmanLp-R EMSA were used as competitive probes. An electrophoretic mobility shift assay (EMSA) was performed using a LightShift Chemiluminescent EMSA Kit (Pierce, Rockford, IL) as recommended by the supplier with minor modifications. Briefly, a constant amount of biotin-labeled dsDNA probe (0.2 nM) and increasing amounts of purified His6-CcpA (2 to 100 nM) were used in each reaction. The reaction buffer was as previously described (25) and contained 12 mM HEPES (pH 7.5), 12% glycerol, 1 mM EDTA, 0.6 mM dithiothreitol, 60 mM KCl, 5 mM MgCl2, 40 ng/μl poly(dI-dC). Competition assays were performed by addition of increasing amounts of the unlabeled dsDNA probe to the binding reaction mixtures. After incubation for 30 min at 30°C, the reaction mixtures were mixed in 1% (vol/vol) Ficoll, 0.02% (wt/vol) bromophenol blue and were separated on a 10% polyacrylamide gel at room temperature. The DNA-protein complex was transferred onto a nylon membrane and cross-linked by using a GS Gene Linker UV Chamber (Bio-Rad Laboratories, Hercules, CA). The biotin-labeled DNA was detected by chemiluminescence.
RNA extraction and quantitative real-time RT-PCR.
Total RNA was extracted from S. oligofermentans cultures harvested at mid-log phase (OD600 = 0.4 to 0.5) using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the recommended protocol. The RNA extract was subsequently treated with RNase-free DNase (Promega, Madison, WI) and analyzed by PCR for possible chromosomal-DNA contamination. cDNA templates were then generated from 2 μg total RNA with random primers using Moloney murine leukemia virus reverse transcriptase (Promega) according to the supplier's instructions. Transcript quantification was performed using an ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA) with SYBR green SuperMix (TaKaRa, Dalian, China). Transcript levels of the following genes were probed using gene-specific primers: qPCR-EIIB-F/qPCR-EIIB-R for EIIB, qPCR-16S-F/qPCR-16S-R for the 16S rRNA gene, and qPCR-manL-F/qPCR-manL-R for manL (see Table S1in the supplemental material). DNA used for generating the standard curve was 10-fold (10−2 to 10−8) serially diluted, and PCRs were performed in triplicate. Each cDNA sample was processed in triplicate. The copy numbers in each cDNA sample were calculated according to the calibration curve generated by the PCR products of the genes, and the transcript abundance of each gene was calculated as a ratio to the 16S rRNA gene copies and normalized.
RESULTS
Identification of a galactose operon for galactose metabolism in S. oligofermentans.
Compared to other carbohydrates utilized by S. oligofermentans, galactose supported a higher growth rate (doubling time, 127.6 min in galactose versus 147.0 min in glucose) and a higher yield for S. oligofermentans (data not shown). As galactose can be metabolized through both the Leloir and tagatose-6-phosphate pathways (10), we used the Streptococcus gordonii genes galK (SGO_0932), encoding galactokinase involved in the Leloir pathway, and lacA (SGO_1519), encoding a galactose-6-phosphate isomerase subunit in the tagatose-6-phosphate pathway, as queries to search the draft genome sequence of S. oligofermentans. Two open reading frames (ORFs) were identified. ORF 1555 is a homolog of galK and resides as the first gene in a three-gene cluster encoding proteins of the classic Leloir pathway. ORF 440 is a homolog of lacA located in an eight-gene operon (Fig. 1A) consisting of the putative PTS EIIABC genes (annotated as the galactitol-specific PTS EIIABC complex), lacX, lacA, lacB, lacC, and lacD. Each of these genes exhibited >90% identity in the protein sequences with its counterparts in other streptococci. The operon also resembled the streptococcal lac operon (22, 48) in regard to gene composition, except that it lacked the galactosidase gene lacG. This suggested that the putative S. oligofermentans lac operon does not encode lactose metabolism but encodes galactose metabolism, which also agrees with our previous finding that S. oligofermentans is unable to use lactose as a carbon source. Hence, we designated the operon a gal operon. A putative regulatory gene, lacR (with 99% identity to the lactose PTS repressor of Streptococcus cristatus), is located upstream of this eight-gene operon and is transcribed in the same orientation.
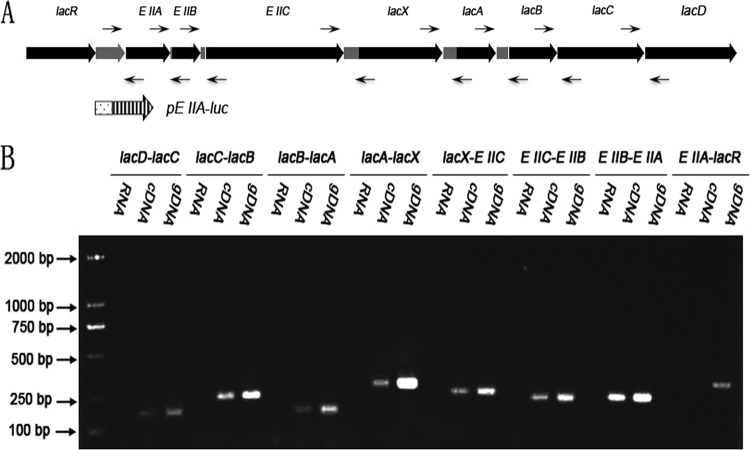
Fig 1.
The gal operon of S. oligofermentans. (A) Schematic representation of the gal operon. lacAB encode a heteromeric galactose-6-phosphate isomerase; lacC encodes a tagatose-6-phosphate kinase; lacD encodes a tagatose-1,6-bisphosphate aldolase; EIIA, EIIB, and EIIC encode the IIA, IIB, and IIC components of a putative galactitol-specific PTS enzyme II, respectively; lacX encodes a predicted aldolase I epimerase; and an adjacent gene, lacR, encodes a putative regulator of the operon. The promoterless luciferase gene (striped arrow) was fused to the putative promoter (dotted rectangle) of the operon to monitor operon expression. (B) RT-PCR amplification of mRNA in the intergenic regions. The primers used are listed in Table 1. Total RNA was generated from galactose-grown cultures of the wild-type strain AS 1.3089T. All reactions were performed in triplicate.
To verify that the 8 genes are indeed located on the same operon, reverse transcription (RT)-PCR was performed to measure the transcript in the intergenic regions. As shown in Fig. 1B, positive RT-PCR products were obtained from all intergenic regions except one between EIIA and lacR, suggesting that lacR is transcribed independently while the rest are highly likely to be cotranscribed.
Relative contributions of the gal operon and galK in galactose utilization.
To further determine the functions of the gal operon and galK in galactose metabolism in S. oligofermentans, deletion mutants were constructed for lacA and galK. The mutants were then analyzed for growth in galactose. As shown in Fig. 2, the lacA mutation severely reduced the growth rate, as well as the final yield, compared with the wild type, as shown by an 8-fold-lower final cell density (OD600). In contrast, the galK mutation reduced the final yield only 2-fold compared with the wild type, and the reduction in the growth rate was much less than for the lacA mutation. These results indicate that the tagatose-6-phosphate pathway encoded by the gal operon is the primary pathway in galactose metabolism in S. oligofermentans, while the classic Leloir pathway plays a relatively minor role in galactose utilization.
Fig 2.
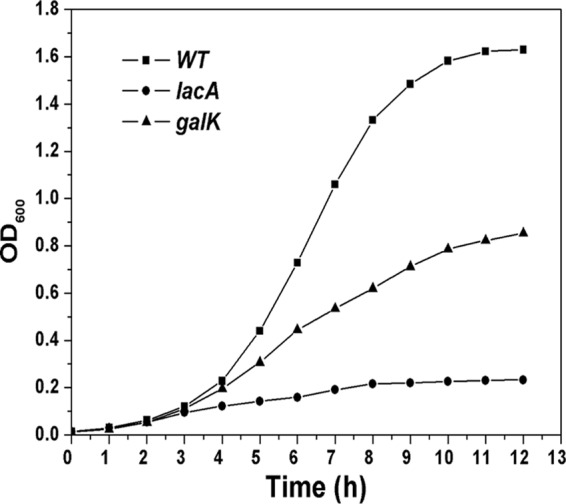
Growth curves of the wild-type (WT) strain and the lacA and galK mutants. Cells were cultured overnight in BHI and then diluted 1:50 in TY broth containing 0.5% galactose. Experiments were performed at least 3 times with similar results. A typical result is presented.
Identification of PTS transporters for galactose in S. oligofermentans.
Although annotated as galactitol-specific PTS permease-encoding genes, the function of EIIABC in galactose metabolism in S. oligofermentans is unknown. Therefore, we constructed an EIIABC deletion mutant and analyzed its growth in glucose and galactose. As shown in Fig. 3A, the EIIABC mutation significantly reduced the growth rate (a doubling time of 205.4 ± 4.0 min versus 127.6 ± 6.4 min) in galactose compared with the wild type. In contrast, no significant difference in growth rate and final yield was observed between the two strains when glucose was used as the carbon source (Fig. 3B). Additionally, neither the wild-type nor the mutant strain grew on galactitol (data not shown), indicating that this EIIABC gene functions as a galactose transporter, not a galactitol transporter, in S. oligofermentans.
Fig 3.
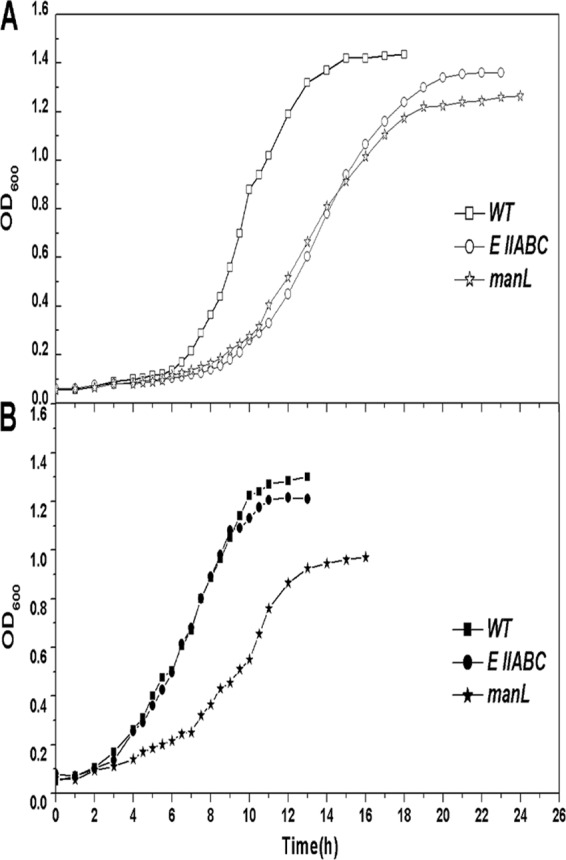
Growth of the wild type, the EIIABC mutant, and the manL mutant in galactose (A) and glucose (B). Cells were cultured overnight in BHI and then diluted 1:50 in TY broth containing either 0.5% galactose or 0.5% glucose. Experiments were performed at least 3 times with similar results. A typical result is presented.
The EIIABC mutation caused only delayed growth on galactose (Fig. 3A); thus, other galactose transporters might exist in S. oligofermentans. One candidate is the glucose/mannose-specific PTS EIIABC complex encoded by manLMN, which has been shown to be a galactose transporter in S. mutans and S. gordonii (1, 40, 46–48). We searched for the manL homolog in the draft genome of S. oligofermentans using the manL gene (SGO_1679) of S. gordonii as a query. A putative manL (ORF 787) was identified, and a deletion mutant was constructed and tested for growth in glucose or galactose. As shown in Fig. 3, the manL mutation reduced the growth rate in both galactose (doubling time, 211.4 ± 4.5 min versus 127.6 ± 6.4 min) and glucose (doubling time, 167.1 ± 8.6 min versus 147.0 ± 3.2 min) compared with the wild type. The mutant also showed a 15% reduction in the final yield when grown in galactose (Fig. 3A) and an ∼30% reduction in the final yield when grown in glucose (Fig. 3B) compared with the wild type. These results suggest that this EIIABMan (ManL) homolog in S. oligofermentans might serve as both a galactose and a glucose transporter, although its role in glucose transport appeared to be more dominant, as judged by the larger reduction in the final yield in glucose-grown mutant cells (Fig. 3B).
Galactose utilization is subjected to CCR in S. oligofermentans.
As the metabolism of most nonpreferred carbohydrates in bacteria is subjected to CCR (13, 34, 35, 42), we analyzed CCR in galactose metabolism in S. oligofermentans by growing the cells in 0.5% galactose plus 0.05% glucose. A diauxic growth curve was observed (Fig. 4), demonstrating that glucose suppressed galactose utilization in S. oligofermentans. Interestingly, S. oligofermentans grew faster on galactose (doubling time, 127.6 ± 6.4 min) than on glucose (doubling time, 147.0 ± 3.2 min).
Fig 4.
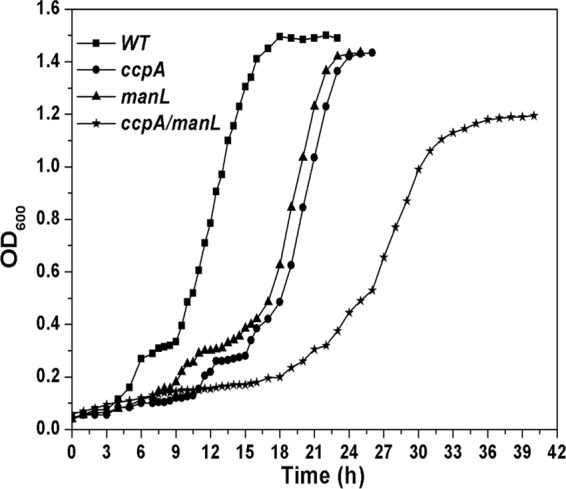
Diauxic growth of the wild type, the ccpA and manL mutants, and the ccpA-manL double mutant in TY broth supplemented with 0.05% glucose and 0.5% galactose. Experiments were performed as described in the legend to Fig. 3.
To see whether this glucose suppression of galactose metabolism is at the transcription level, we used quantitative PCR (qPCR) to determine the gal operon expression when the strain was grown in TY broth containing glucose (0.5%), galactose (0.5%), or glucose plus galactose (each 0.5%) (wt/vol). As shown in Table 1, the EIIB gene transcript was reduced 7- to 13-fold in cells grown on glucose or glucose plus galactose versus cells grown on galactose. This result was confirmed by a luciferase reporter strain, which was constructed by fusing the luc gene to the promoter of the gal operon (Fig. 1A). As shown in Table 2, luciferase activity was >10 times lower in cells grown on glucose or glucose plus galactose than in cells grown on galactose, suggesting that glucose suppressed gal operon transcription.
Table 1.
Relative transcript abundances of EIIB and manL in the wild-type and mutant strains grown in different sugarsa
| Strain or genotype |
EIIBb |
manLc |
||||
|---|---|---|---|---|---|---|
| Glu | Glu + Gal | Gal | Glu | Glu + Gal | Gal | |
| WTd | 0.64 ± 0.12 | 1.32 ± 0.03 | 8.77 ± 1.38 | 0.92 ± 0.12 | 0.85 ± 0.09 | 0.71 ± 0.02 |
| ccpA | 3.66 ± 0.37f | 6.68 ± 0.19f | 35.27 ± 3.61e | 1.29 ± 0.13 | 1.73 ± 0.15e | 1.28 ± 0.14e |
| manL | 6.34 ± 0.92f | 6.58 ± 0.29f | 1.45 ± 0.10e | NDg | ND | ND |
| ccpA-manL | 8.17 ± 0.24f | 10.25 ± 0.54f | 12.32 ± 1.27e | ND | ND | ND |
| hprK | 0.86 ± 0.02 | 4.71 ± 0.35f | 3.58 ± 0.10f | ND | ND | ND |
| ccpA-hprK | 3.25 ± 0.13f | 131.97 ± 2.31f | 56.63 ± 2.56f | ND | ND | ND |
Data are the averages of three independent cultures.
The transcription abundance is the mean ± standard deviation of the EIIB copy number/0.0001 16S rRNA gene copies.
The transcription abundance is the mean ± standard deviation of the manL copy number/0.01 16S rRNA gene copies.
WT, wild type.
Values with significant difference from the wild type (P < 0.05; Student's t test).
Values with significant difference from the wild type (P < 0.01; Student's t test).
ND, not done.
Table 2.
manL-luc and gal-luc reporter gene expression in various S. oligofermentans strains growing in different sugars
| Fusion | RLU (105)a |
||
|---|---|---|---|
| Glu | Glu + Gal | Gal | |
| WT-PmanL-luc | 15.13 ± 0.02 | 17.19 ± 2.94 | 26.44 ± 1.23 |
| manL-PmanL-luc | 33.43 ± 2.86 | 33.12 ± 2.65 | 32.59 ± 2.02 |
| ccpA-PmanL-luc | 25.73 ± 2.88 | 30.48 ± 3.15 | 35.83 ± 3.30 |
| ccpA-manL-PmanL-luc | 154.68 ± 5.34 | 149.60 ± 2.06 | 160.67 ± 2.17 |
| hprK-PmanL-luc | 34.99 ± 2.09 | 33.69 ± 1.31 | 37.29 ± 0.83 |
| ccpA-hprK-PmanL-luc | 64.56 ± 2.70 | 70.83 ± 2.00 | 38.76 ± 0.60 |
| WT-Pgal-luc | 10.57 ± 1.18 | 9.30 ± 1.85 | 125.61 ± 19.92 |
RLU, relative light units. The data are the averages from three independent batches of cultures.
To test whether other saccharides exert a CCR on galactose utilization, we determined the gal operon expression in S. oligofermentans cells growing in all of its utilizable saccharides, i.e., fructose, mannose, maltose, or sucrose and their mixtures with galactose. Table S2 in the supplemental material shows that the luciferase activity of the Pgal-luc reporter strain was reduced 2.7- to 16-fold in the other four sugars or their mixtures with galactose compared with that in galactose alone (see Table S2 in the supplemental material). However, the activity increased a maximum of 3.6-fold in response to different concentrations of galactose (0.025 to 0.5% [wt/vol]). This demonstrated that galactose metabolism is subject to CCR in S. oligofermentans, while galactose induces the expression of the gal operon.
Involvement of CcpA and ManL in CCR of S. oligofermentans.
CcpA, the catabolite control protein, is believed to play a key role in CCR regulation in Gram-positive bacteria (8, 15, 17), whereas ManL, the glucose/mannose PTS permease, is also reported to be involved in CCR in S. mutans and S. gordonii (1, 40, 46, 47). To further understand the regulatory mechanism of CCR in S. oligofermentans, we searched for ccpA homologs in the draft genome of S. oligofermentans using the S. cristatus ccpA gene (ZP_08059449.1) as a query and found a putative ccpA gene (ORF 247). A deletion mutation was made in the putative ccpA, and both the ccpA and the manL mutants were analyzed for growth in 0.5% galactose plus 0.05% glucose. Unexpectedly, both mutant strains still exhibited diauxic growth, although the lag phase appeared to be longer than in the wild type (Fig. 4). This suggested that deletion of either gene is not sufficient to overcome the CCR effect. To further test this supposition, we created a ccpA-manL double mutation and measured its growth effect. As shown in Fig. 4, this double mutation eliminated diauxic growth, suggesting that in S. oligofermentans, both ccpA and manL are involved in CCR of galactose utilization.
Gene expression of the gal operon in the ccpA and manL single- and double-mutant backgrounds was also measured by qPCR. As shown in Table 1, compared with that of the wild type, gal operon expression, as represented by the EIIB gene, was upregulated ∼5-fold in the ccpA or manL single mutant and ∼7.7-fold in the double-mutant background when cells grew in a mixture of glucose and galactose. When cells were grown in glucose alone, the expression levels of the gal operon in the ccpA and manL mutants and the ccpA-manL double mutants were 6-, 10-, and 12-fold higher than in the wild-type strain, respectively. These results confirmed the involvement of both CcpA and ManL in the CCR of galactose metabolism in S. oligofermentans. Notably, EIIB expression in the manL mutant was about 6-fold lower than that in the wild-type strain when it was grown on galactose, which might be due to decreased galactose transport via ManL.
In addition, to determine the effect of HPr (Ser-P), the cofactor of CcpA, on CCR, we constructed a deletion mutation of the gene encoding HPrK, a kinase that specifically phosphorylates the HPr protein at serine residue 46. A double gene mutation of ccpA-hprK was constructed, as well. qPCR determination (Table 1) showed that gal operon expression was 3.6-fold higher in the hprK mutant than in the wild-type strain when it was grown on glucose plus galactose but that expression was reduced in galactose. Whereas the expression levels of the gal operon in the ccpA-hprK double mutant were about 5-, 100-, and 6-fold higher than in the wild-type strain when it was grown on glucose, a mixture of glucose and galactose, and galactose, respectively. This result implies a direct involvement of HPr (Ser-P) in CCR, in addition to acting as a cofactor of CcpA.
CcpA negatively regulates manL gene expression.
It has been reported that CcpA negatively regulates manL in other streptococci (40, 46). We determined the manL gene expression in the wild-type strain of S. oligofermentans and the ccpA mutant using both qPCR and a luciferase reporter, which was constructed by fusion of the luc gene to the promoter of the manL gene. As shown in Tables 1 and 2, manL expression in the ccpA mutant was about 1.5- to 2-fold higher than in the wild-type strain when it was grown on glucose, galactose, and their mixture. This indicates that CcpA negative regulation of manL is most likely to be glucose independent, as the level of upregulation was higher in galactose.
To investigate the possible autoregulation of manL in S. oligofermentans, as reported in the oral commensal S. gordonii (40), we determined manL gene expression in the manL mutant and the ccpA-manL double mutant. As shown in Table 2, when grown on glucose or a mixture of glucose and galactose, the activity of the PmanL-luc fusion in the manL mutant was about 1.9- to 2.2-fold higher than in the wild-type strain, while it was increased 14- to 16-fold in the ccpA-manL double mutant. Though manL expression in the manL mutant was about the same as in the wild-type strain on galactose, it increased 6-fold in the ccpA-manL double mutant. This suggests that though autoregulation is present, manL expression is also under the control of CcpA.
Furthermore, to see the effect of HPr (Ser-P) on manL expression, manL gene expression was measured in hprK and ccpA-hprK mutants. As shown in Table 2, manL expression was enhanced about 2-fold in the hprK mutant when it was grown on glucose or a mixture of glucose and galactose, while it increased by about 4-fold in the ccpA-hprK double mutant. However, manL expression was only slightly elevated in either hprK or ccpA-hprK mutants when they were grown on galactose. This indicates that HPr might be involved in the direct regulation of manL expression.
CcpA directly binds to promoters of the gal operon and manL.
As a LacI family member, CcpA is believed to control CCR through direct regulation of the target genes by binding to the catabolite response elements (cre) of their promoters (8, 15, 17). Inspection of the promoter regions of the gal operon and manL in S. oligofermentans identified cre-like motifs (TGACAGCGCTATCA; located 57 to 70 bp upstream of the ATG start codon) and TGAAAACGTTTTAT (67 to 80 bp upstream of the ATG codon), respectively. These sequence motifs are slightly different from the consensus cre sequence (TGWNANCGNTNWCA, where W = A/T) in Bacillus subtilis (43). To determine if CcpA binds to the two promoters, CcpA was overexpressed in E. coli, and the purified protein was used to perform an EMSA with the gal operon and manL promoters as targets. A labeled 50-bp DNA fragment containing the putative binding site of the response regulator ComE (com box), which was located in the promoter region of the comC gene encoding a competence signal peptide, was included as the negative control. As shown in Fig. 5A, lanes 1 to 6, CcpA bound to the gal promoter in a dose-dependent manner. To test the specificity of this binding, an unlabeled gal promoter fragment was used to compete with the labeled promoter fragment. As shown in lanes 7 to 10, as the concentration of the unlabeled probe increased, the intensity of the shifted bands decreased, indicating that CcpA binding to the gal promoter is specific. As a further confirmation of the binding specificity, a gal promoter fragment lacking the putative cre sequence (denoted gal-p-cre) did not show mobility shifts until the CcpA concentration reached 40 nM (lanes 11 to 14). Of note, a shifted band appeared when the CcpA concentration increased to 100 nM (lane 15). This might be caused by the presence of the cre motif-like sequence upstream of the cre knockout site in the gal promoter. To further verify whether the binding was specific, increased concentrations of unlabeled gal-p-cre were added to the reaction mixtures. It was shown that unlabeled gal-p-cre could not compete with the labeled gal-p for CcpA binding (data not shown). These results demonstrate that CcpA specifically binds to the gal promoter and that the cre sequence is required for high-affinity binding.
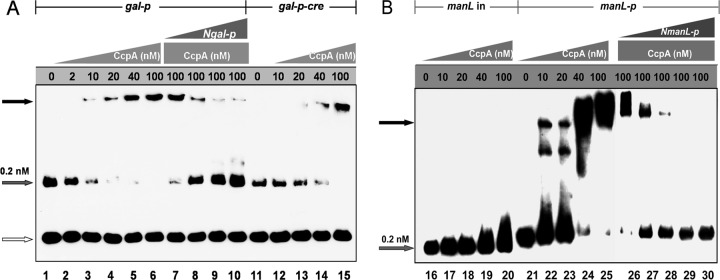
Fig 5.
EMSA of His6-CcpA with the promoters of the gal operon (A) and manL (B). DNA fragments of the gal operon promoter carrying intact cre (gal-p) (lanes 1 to 10) or without cre (gal-p-cre) (lanes 11 to 15) and the manL coding region (manL in) (lanes 16 to 20) and promoter (manL-p) (lanes 21 to 30) were PCR amplified and end labeled with biotin. The added CcpA concentrations are indicated above the lanes. Lanes 7 to 10 (A) and lanes 26 to 30 (B) contain the same amount (100 nM) of CcpA, but with increasing concentrations (2, 10, 20, and 40 nM) of unlabeled gal promoter fragment (Ngal-p) or manL promoter fragment (NmanL-p), respectively, as a competing substrate for CcpA. The black arrows indicate the shifted migration positions of the CcpA-bound promoter fragment, the gray arrows indicate the free DNA fragments, and the open arrow (A) indicates the negative-control DNA (com box).
CcpA binding to the manL promoter was also determined by EMSA. A fragment from the coding region of manL (denoted manL in) with a similar length of the manL promoter was used as the negative control. As shown in Fig. 5B, CcpA bound to the manL promoter with an affinity comparable to that to the gal promoter (lanes 21 to 25). Similarly, the intensity of the shifted bands decreased in response to the increased concentration of the unlabeled promoter fragment (lanes 26 to 30), indicating that the binding of CcpA to the manL promoter is specific. Taken together, these results support the hypothesis that CcpA regulates CCR through direct transcriptional repression of the gal operon and manL.
DISCUSSION
Lactose and galactose, which are abundant sugars in milk and yogurt, are important carbon sources for the microorganisms in the dental biofilm. The relative efficiency of uptake and metabolism of these saccharides by oral commensals and pathogens may determine the outcome of interspecies competition and, thus, the status of oral health. Previously, we found that S. oligofermentans, an oral commensal, utilized galactose more efficiently than the cariogenic S. mutans (data not shown). In this study, a novel gal operon was identified for galactose metabolism in the microorganism. Further studies demonstrated that S. oligofermentans has two galactose transporters, EIIGal, which is encoded by the gal operon and transports only galactose, and EIIMan, which is encoded by manL and transports both glucose and galactose. The presence of two transporters for galactose may partially explain the faster growth of S. oligofermentans on galactose. We further demonstrated that galactose metabolism in S. oligofermentans is subject to CCR and that CcpA is directly involved in regulating CCR by binding to the promoter regions of the gal operon and manL. Furthermore, we revealed a unique CCR regulatory mechanism in S. oligofermentans in which both CcpA and ManL are involved, but their regulatory pathways appear to be parallel rather than hierarchical. Based on our results and a previous model for CcpA-mediated CCR regulation (13), we propose a modified model for CcpA regulation of manL and gal operon expression in S. oligofermentans (Fig. 6). In this model, the preferred sugar, glucose, is transported by the manL gene product, EIIMan, and fermented through glycolysis, resulting in the activation of the kinase activity of HPrK and phosphorylation of HPr to HPr (Ser-P). HPr (Ser-P) or other cofactors then interact with CcpA, which increases the affinity of CcpA for cre in the promoters of the gal operon and manL. Binding of CcpA to the two promoters represses transcription of manL and the gal operon, thus suppressing the transport and utilization of galactose. Meanwhile, transport of glucose leads to dephosphorylation of ManL at its conserved His residue, and dephosphorylated ManL negatively regulates expression of the gal operon and manL, further repressing galactose utilization. Although many details of this model need to be experimentally demonstrated in S. oligofermentans, the model could serve as a working map to guide our future studies. The highlighted aspects of this study are discussed below.
Fig 6.
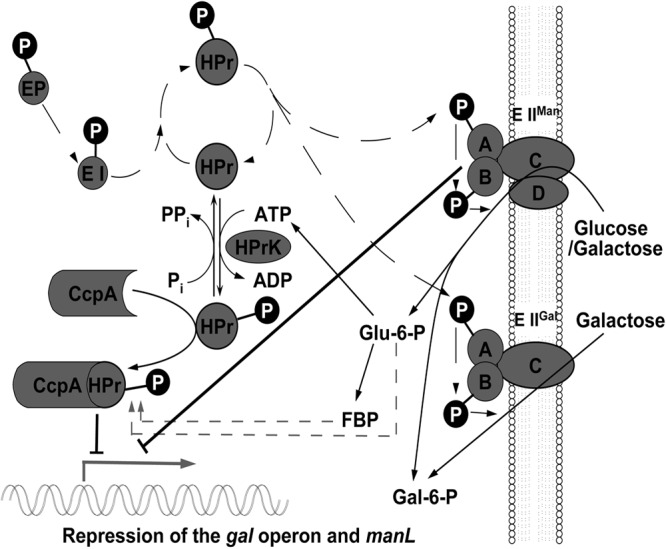
Modified model of CcpA-mediated repression of gal operon expression in S. oligofermentans. Details are provided in Discussion. FBP, fructose-1,6-bisphosphate. manL encodes the A and B components of EIIMan. EIIABC encodes the A, B, and C components of galactitol-specific PTS permease, denoted EIIGal, which is part of the gal operon. Adapted by permission from Macmillan Publishers Ltd. 13.
Differences between the gal operon of S. oligofermentans and the lac operon of other streptococci.
In most lactic acid bacteria, the lac operon encodes enzymes for lactose metabolism through the tagatose-6-phosphate pathway, in which the lactose-specific PTS enzyme II (EIILac) is responsible for lactose transport via the PTS system (29, 41). In oral streptococci, e.g., S. mutans and S. gordonii, transport of galactose is via EIILac and mannose-specific PTS permease (EIIMan) (40, 48). Although genes in the gal operon of S. oligofermentans are homologous to genes of the lac operon in other streptococci (Fig. 1), fundamental differences exist between the gal and lac operons. First, the gal operon lacks lacG, a gene encoding 6-phosphate-β-galactosidase, which makes the operon deficient in a critical gene involved in lactose metabolism. This finding is also consistent with the inability of the microorganism to utilize lactose. Second, the genes for EIILac, present in the lac operon, are replaced by three genes encoding a putative galactitol-specific PTS enzyme IIABC complex (EIIABC), which were shown to be a galactose transporter (Fig. 3). Third, unlike in S. mutans, the tagatose-6-phosphate pathway contributes more than the Leloir pathway to galactose metabolism in S. oligofermentans (Fig. 2). As expected, the lactose repressor gene, lacR, was shown to repress the expression of the gal operon in S. oligofermentans (data not shown).
Differences in CCR control mechanisms between S. oligofermentans and other oral streptococci.
CCR is a common phenomenon in bacteria and controls utilization of the preferred sugar over a secondary carbon source (13, 34, 35, 42). In Gram-positive bacteria, CCR is mostly achieved through a global regulator, CcpA (8, 15). It has been found that CcpA not only acts as a master regulator of catabolite control, but also controls the expression of virulence genes in many pathogens, such as the sagA operon (for streptolysin S production) (17) and the mac gene (encoding immunoglobulin-degrading enzyme) (32) in Streptococcus pyogenes and the pilT and pilD genes (for pilus components) in Clostridium perfringens (26). Similarly, CcpA also regulates virulence-related functions in oral streptococci, e.g., biofilm formation and acid tolerance in S. mutans (2) and penicillin tolerance (6), H2O2 production (50), and the arginine deaminase (12) pathway in S. gordonii. Sugar-specific PTS components have been found to be involved in carbohydrate-dependent regulation of gene expression in many bacteria (23, 27). Furthermore, CCR control of carbohydrate metabolism in oral streptococci has so far been shown to be achieved mainly by PTS permeases and HPr (40, 46, 47), both of which are independent of CcpA. For example, in S. mutans, Abranches et al. (1) and Zeng et al. (46, 47, 49) demonstrated that three PTS permeases, ManL, FruI (IIABCFru), and LevDEFG (EIILev), are involved in CCR of the fruA, levD, and cel operons, while in S. gordonii, EIIABMan regulates fruA and levDEFG expression (40). The mechanism of PTS-mediated CCR appears to involve sugar specificity. For instance, EIIABMan controls CCR only when its transported substrates are present (1, 40), whereas EIILev controls CCR only in the presence of fructose (46). In S. mutans, ManL-mediated CCR was postulated to take place via an allosteric interaction or a phosphorelay with the regulatory proteins (46, 47).
Our study unambiguously demonstrated that both ManL (a PTS permease) and CcpA are involved in CCR of galactose catabolism in S. oligofermentans, where manL and the gal operon are transcriptionally controlled by CcpA through direct binding of CcpA to the cre motifs in their respective promoters. Thus, S. oligofermentans is the first example of an oral streptococcus in which CcpA directly controls CCR, at least for galactose metabolism. Whether CcpA also controls CCR of other carbohydrates in S. oligofermentans has yet to be determined.
The manL gene of S. oligofermentans also seems to be under the control of ManL, in addition to CcpA. It has been suggested that when carbohydrate concentrations reach 3 to 5 mM, CcpA functions to repress the expression of PTS permease by monitoring the carbohydrate flow through the glycolytic pathway (40, 47), whereas at lower carbohydrate concentrations, ManL may adjust the PTS permease level by regulating its own expression. This mechanism may enable S. oligofermentans to adapt to the fluctuating amounts of carbohydrates in the oral environment and thus to be more competitive in the ecological environment.
It is also worth noting that, in addition to galactose metabolism, the ccpA mutant also displayed a significantly longer lag phase than the wild-type strain and the manL mutant when grown in 0.05% glucose plus 0.5% galactose (Fig. 4). This suggested that CcpA likely also regulates other genes, as well. In fact, a genomic analysis of the cre motif in the S. oligofermentans genome identified many genes with a cre-like sequence in their putative promoters (data not shown). We speculate that CcpA, like its counterpart in other streptococci, is likely also a global regulator.
Supplementary Material
ACKNOWLEDGMENT
This study was supported by China NSFC grant 30870042.
Footnotes
Published ahead of print 18 May 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Abranches J, Candella MM, Wen ZT, Baker HV, Burne RA. 2006. Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans. J. Bacteriol. 188:3748–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abranches J, et al. 2008. CcpA regulates central metabolism and virulence gene expression in Streptococcus mutans. J. Bacteriol. 190:2340–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahn SJ, Wen ZT, Burne RA. 2006. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect. Immun. 74:1631–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Almengor AC, Kinkel TL, Day SJ, McIver KS. 2007. The catabolite control protein CcpA binds to Pmga and influences expression of the virulence regulator Mga in the group A Streptococcus. J. Bacteriol. 189:8405–8416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Avila M, Ojcius DM, Yilmaz O. 2009. The oral microbiota: living with a permanent guest. DNA Cell Biol. 28:405–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bizzini A, Entenza JM, Moreillon P. 2007. Loss of penicillin tolerance by inactivating the carbon catabolite repression determinant CcpA in Streptococcus gordonii. J. Antimicrob. Chemother. 59:607–615 [DOI] [PubMed] [Google Scholar]
- 7. Burne RA, Wen ZT, Chen YY, Penders JE. 1999. Regulation of expression of the fructan hydrolase gene of Streptococcus mutans GS-5 by induction and carbon catabolite repression. J. Bacteriol. 181:2863–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Deutscher J. 2008. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 11:87–93 [DOI] [PubMed] [Google Scholar]
- 9. Deutscher J, Francke C, Postma PW. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70:939–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Vos WM, Vaughan EE. 1994. Genetics of lactose utilization in lactic-acid bacteria. FEMS Microbiol. Rev. 15:217–237 [DOI] [PubMed] [Google Scholar]
- 11. Dong X, Xin Y, Jian W, Liu X, Ling D. 2000. Bifidobacterium thermacidophilum sp nov., isolated from an anaerobic digester. Int. J. Syst. Evol. Microbiol. 50:119–125 [DOI] [PubMed] [Google Scholar]
- 12. Dong Y, Chen YY, Burne RA. 2004. Control of expression of the arginine deiminase operon of Streptococcus gordonii by CcpA and Flp. J. Bacteriol. 186:2511–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Görke B, Stülke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6:613–624 [DOI] [PubMed] [Google Scholar]
- 14. Griswold AR, Jameson-Lee M, Burne RA. 2006. Regulation and physiologic significance of the agmatine deiminase system of Streptococcus mutans UA159. J. Bacteriol. 188:834–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Iyer R, Baliga NS, Camilli A. 2005. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 187:8340–8349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Keijser BJ, et al. 2008. Pyrosequencing analysis of the oral microflora of healthy adults. J. Dent. Res. 87:1016–1020 [DOI] [PubMed] [Google Scholar]
- 17. Kinkel TL, McIver KS. 2008. CcpA-mediated repression of streptolysin S expression and virulence in the group A streptococcus. Infect. Immun. 76:3451–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lau PC, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193–205 [DOI] [PubMed] [Google Scholar]
- 19. Lazarevic V, et al. 2009. Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. J. Microbiol. Methods 79:266–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Loesche WJ. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Loimaranta V, Tenovuo J, Koivisto L, Karp M. 1998. Generation of bioluminescent Streptococcus mutans and its usage in rapid analysis of the efficacy of antimicrobial compounds. Antimicrob. Agents Chemother. 42:1906–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Loughman JA, Caparon MG. 2007. Comparative functional analysis of the lac operons in Streptococcus pyogenes. Mol. Microbiol. 64:269–280 [DOI] [PubMed] [Google Scholar]
- 23. Lun S, Willson PJ. 2005. Putative mannose-specific phosphotransferase system component IID represses expression of suilysin in serotype 2 Streptococcus suis. Vet. Microbiol. 105:169–180 [DOI] [PubMed] [Google Scholar]
- 24. Marmur J. 1963. A procedure for the isolation of deoxyribonucleic acid from microorganisms. Methods Enzymol. 6:726–738 [Google Scholar]
- 25. McIver KS, Heath AS, Green BD, Scott JR. 1995. Specific binding of the activator Mga to promoter sequences of the emm and scpA genes in the group A Streptococcus. J. Bacteriol. 177:6619–6624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mendez M, et al. 2008. Carbon catabolite repression of type IV pilus-dependent gliding motility in the anaerobic pathogen Clostridium perfringens. J. Bacteriol. 190:48–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mertins S, et al. 2007. Interference of components of the phosphoenolpyruvate phosphotransferase system with the central virulence gene regulator PrfA of Listeria monocytogenes. J. Bacteriol. 189:473–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Podbielski A, Woischnik M, Leonard BA, Schmidt KH. 1999. Characterization of nra, a global negative regulator gene in group A streptococci. Mol. Microbiol. 31:1051–1064 [DOI] [PubMed] [Google Scholar]
- 29. Postma PW, Lengeler JW, Jacobson GR. 1993. Phosphoenolpyruvate-carbohydrate phosphotransferase system of bacteria. Microbiol. Rev. 57:543–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qi F, Chen P, Caufield PW. 2001. The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Appl. Environ. Microbiol. 67:15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rosan B, Lamont RJ. 2000. Dental plaque formation. Microbes Infect. 2:1599–1607 [DOI] [PubMed] [Google Scholar]
- 32. Shelburne SA, et al. 2008. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc. Natl. Acad. Sci. U. S. A. 105:1698–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Simpson CL, Russell RR. 1998. Identification of a homolog of CcpA catabolite repressor protein in Streptococcus mutans. Infect. Immun. 66:2085–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stülke J, Hillen W. 1999. Carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2:195–201 [DOI] [PubMed] [Google Scholar]
- 35. Titgemeyer F, Hillen W. 2002. Global control of sugar metabolism: a gram-positive solution. Antonie Van Leeuwenhoek 82:59–71 [PubMed] [Google Scholar]
- 36. Tong H, et al. 2006. Establishing a genetic system for ecological studies of Streptococcus oligofermentans. FEMS Microbiol. Lett. 264:213–219 [DOI] [PubMed] [Google Scholar]
- 37. Tong H, et al. 2007. Streptococcus oligofermentans inhibits Streptococcus mutans through conversion of lactic acid into inhibitory H2O2: a possible counteroffensive strategy for interspecies competition. Mol. Microbiol. 63:872–880 [DOI] [PubMed] [Google Scholar]
- 38. Tong H, Chen W, Shi W, Qi F, Dong X. 2008. SO-LAAO, a novel L-amino acid oxidase that enables Streptococcus oligofermentans to outcompete Streptococcus mutans by generating H2O2 from peptone. J. Bacteriol. 190:4716–4721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tong H, Gao X, Dong X. 2003. Streptococcus oligofermentans sp nov, a novel oral isolate from caries-free humans. Int. J. Syst. Evol. Microbiol. 53:1101–1104 [DOI] [PubMed] [Google Scholar]
- 40. Tong H, Zeng L, Burne RA. 2011. The EIIABMan phosphotransferase system permease regulates carbohydrate catabolite repression in Streptococcus gordonii. Appl. Environ. Microbiol. 77:1957–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vadeboncoeur C, Pelletier M. 1997. The phosphoenolpyruvate:sugar phosphotransferase system of oral streptococci and its role in the control of sugar metabolism. FEMS Microbiol. Rev. 19:187–207 [DOI] [PubMed] [Google Scholar]
- 42. Warner JB, Lolkema JS. 2003. CcpA-dependent carbon catabolite repression in bacteria. Microbiol. Mol. Biol. Rev. 67:475–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weickert MJ, Chambliss GH. 1990. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 87:6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wen ZT, Burne RA. 2002. Analysis of cis- and trans-acting factors involved in regulation of the Streptococcus mutans fructanase gene (fruA). J. Bacteriol. 184:126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zaura E, Keijser BJ, Huse SM, Crielaard W. 2009. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 9:259–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zeng L, Burne RA. 2008. Multiple sugar:phosphotransferase system permeases participate in catabolite modification of gene expression in Streptococcus mutans. Mol. Microbiol. 70:197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zeng L, Burne RA. 2010. Seryl-phosphorylated HPr regulates CcpA-independent carbon catabolite repression in conjunction with PTS permeases in Streptococcus mutans. Mol. Microbiol. 75:1145–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zeng L, Das S, Burne RA. 2010. Utilization of lactose and galactose by Streptococcus mutans: transport, toxicity, and carbon catabolite repression. J. Bacteriol. 192:2434–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zeng L, Wen ZT, Burne RA. 2006. A novel signal transduction system and feedback loop regulate fructan hydrolase gene expression in Streptococcus mutans. Mol. Microbiol. 62:187–200 [DOI] [PubMed] [Google Scholar]
- 50. Zheng L, Itzek A, Chen Z, Kreth J. 2011. Environmental influences on competitive hydrogen peroxide production in Streptococcus gordonii. Appl. Environ. Microbiol. 77:4318–4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.