Abstract
Putrescine as the sole carbon source requires a novel catabolic pathway with glutamylated intermediates. Nitrogen limitation does not induce genes of this glutamylated putrescine (GP) pathway but instead induces genes for a putrescine catabolic pathway that starts with a transaminase-dependent deamination. We determined pathway utilization with putrescine as the sole nitrogen source by examining mutants with defects in both pathways. Blocks in both the GP and transaminase pathways were required to prevent growth with putrescine as the sole nitrogen source. Genetic and biochemical analyses showed redundant enzymes for γ-aminobutyraldehyde dehydrogenase (PatD/YdcW and PuuC), γ-aminobutyrate transaminase (GabT and PuuE), and succinic semialdehyde dehydrogenase (GabD and PuuC). PuuC is a nonspecific aldehyde dehydrogenase that oxidizes all the aldehydes in putrescine catabolism. A puuP mutant failed to use putrescine as the nitrogen source, which implies one major transporter for putrescine as the sole nitrogen source. Analysis of regulation of the GP pathway shows induction by putrescine and not by a product of putrescine catabolism and shows that putrescine accumulates in puuA, puuB, and puuC mutants but not in any other mutant. We conclude that two independent sets of enzymes can completely degrade putrescine to succinate and that their relative importance depends on the environment.
INTRODUCTION
Polyamines are ubiquitous and important biological molecules. The major polyamine in Escherichia coli is the diamine putrescine (1,4-diaminobutane), although the triamine spermidine (aminopropyl-putrescine) and the diamine cadaverine (1,5-diaminopentane) are also present (6). Intracellular polyamine concentrations positively correlate with growth rate (2, 51). A variety of functions have been attributed to the polyamines. Polyamines affect ribosomal structure, protein and nucleic acid elongation rates, translational fidelity, chromosomal structure, and interactions between mRNA and ribosomes (8, 9, 15, 53). In Escherichia coli, polyamines, especially putrescine, affect the expression of several hundred genes, often affecting translation of specific genes (3, 52, 53). A polyamine-deficient mutant still grows aerobically, albeit about twice as slowly, but does not grow anaerobically or in 95% oxygen and is more sensitive to reactive oxygen species (5, 33). An appropriate level of polyamines would seem to be important for a variety of functions during growth. In addition, polyamine accumulation can be detrimental (1, 11, 28). Polyamines are also involved in several stress responses, and not surprisingly stress responses can affect intracellular polyamine concentrations (1, 34, 49, 50).
Putrescine can also be used a the sole nitrogen source. Growth with a nitrogen source other than ammonia limits growth and is considered nitrogen limited. Such growth induces at least 100 genes whose products assimilate ammonia and scavenge nitrogen-containing compounds (40, 54). This coordinated expression is called the nitrogen-regulated or Ntr response. Twenty Ntr proteins transport and degrade arginine (which can be degraded to putrescine), γ-aminobutyric acid (GABA) (a product of putrescine catabolism), and possibly putrescine (54).
Two pathways have been proposed to degrade putrescine via GABA to succinate (Fig. 1). Transaminase-dependent deamination initiates one pathway (48), while glutamylation of putrescine initiates a second pathway (23). The glutamylated putrescine (GP) pathway is essential for utilization of putrescine as the sole carbon source (22–26). Putrescine as the sole nitrogen source may involve the transaminase-dependent pathway, since nitrogen limitation appears to induce proposed enzymes of the transaminase-dependent pathway but not enzymes of the GP pathway (54). Furthermore, genetic analyses have not established which enzymes catalyze which reactions in vivo. We characterized utilization of putrescine as the sole nitrogen source by examining growth in mutants lacking genes of both pathways. We show that both pathways contribute to putrescine catabolism, redundant enzymes catalyze several reactions, and one enzyme, PuuC, catalyzes three separate reactions. We also characterize regulation of the GP pathway. A separate communication will describe the complex stress-induced regulation of the transaminase pathway. We conclude that two independent pathways can degrade putrescine, and their relative importance depends on growth conditions.
Fig 1.
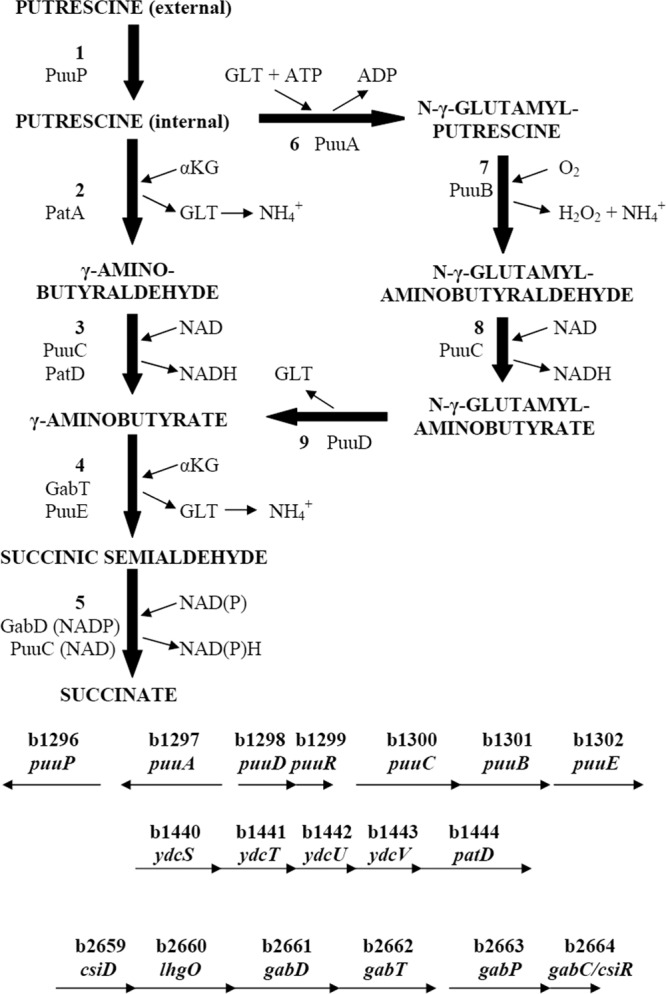
Pathways and genes of putrescine catabolism. The transaminase pathway is reactions 2 to 5 (PatA, PatD, GabT, and GabD), and the glutamylated putrescine (GP) pathway is reactions 6 to 9, 4, and 5 (PuuA, PuuB, PuuC, PuuD, PuuE, and PuuC). Reactions 2, 4, and 7 generate ammonia for assimilation. The abbreviations are GLT for glutamate and α-KG for α-ketoglutarate. All the genes considered in this work are indicated in the bottom half, except for patA (b3073). The solid arrows indicate the genes and the direction of transcription, drawn to scale. There are documented promoters preceding puuA, puuD, ydcS, csiD, and gabD. Other promoters may exist. Evidence for a puuAP operon is presented in this work. Evidence for a nitrogen-regulated gabDTPC operon has been presented (46). Evidence for the larger csiD-lhgO-gabDTPC operon controlled by σS has been described (30). The patA gene (not shown) is monocistronic.
MATERIALS AND METHODS
Strains and plasmids.
All strains used for growth rate determinations and enzyme assays are derivatives of E. coli K-12 strain W3110. Strains and plasmids used in this study are listed in Table 1. Deletions were constructed and verified as described previously (7). All deletions are in-frame deletions if the antibiotic resistance gene is removed. The extent of deletions in individual genes is shown in Table 2. The deletion of ydcSTUV left codons 1 to 5 of ydcS and codons 228 to 264 of ydcV. The deletion of ydcSTUV-patD left codons 1 to 5 of ydcS and 468 to 474 of patD. A derivative of PKD13 (7), pKD13cat, in which the chloramphenicol marker replaced the original kanamycin marker, was used to construct strains with chloramphenicol resistance. P1 phage transductions were done as described previously (32). The strains were tested for the appropriate antibiotic resistance, and the deletion or insertion was verified by PCR.
Table 1.
Strains and growth rates in glucose-putrescine mediuma
| Line no. | Strain | Descriptionb | Generation time (min ± SEM) |
|---|---|---|---|
| 1 | W3110 | lacL8 lacIq | 225 ± 7 (18) |
| 2 | CP2 | ΔpatA | 220 ± 11 (6) |
| 3 | CP6 | ΔgabT | 221 ± 14 (4) |
| 4 | CP14 | ΔpuuE::cat | 183 ± 10 (3) |
| 5 | CP36 | ΔgabT ΔpuuE::cat | 305 ± 8 (3) |
| 6 | BLS71 | ΔpatA Δ(ydcSTUV-patD)::kan | 240 ± 14 (9) |
| 7 | BLS73 | Δ(ydcSTUV-patD)::kan | 228 ± 7 (24) |
| 8 | BLS74 | ΔpatA ΔpatD::cat | 227 ± 8 (9) |
| 9 | BLS75 | ΔpatD::cat | 241 ± 13 (10) |
| 10 | BLS77 | ΔpuuR::cat | NG, S |
| 11 | BLS79 | ΔpuuP::cat | NG |
| 12 | BLS80 | ΔpuuA::cat | NG |
| 13 | BLS81 | ΔpuuA | 348 ± 9 (6) |
| 14 | BLS82 | ΔpatA ΔpatD | 210 ± 9 (9) |
| 15 | BLS83 | ΔpatD | 241 ± 9 (12) |
| 16 | BLS84 | ΔpuuR | 121 ± 6 (12) |
| 17 | BLS85 | ΔpuuP | NG |
| 18 | BLS88 | ΔpatA ΔpuuA | NG |
| 19 | BLS89 | ΔpuuC::cat | 305 ± 15 (8) |
| 20 | BLS90 | ΔpuuC | 315 ± 12 (10) |
| 21 | BLS93 | ΔpuuC ΔpatD | VSG |
| 22 | BLS96 | ΔpuuB::cat | 320 ± 10 (6) |
| 23 | BLS97 | ΔpuuB | 367 ± 18 (13) |
| 24 | BLS98 | ΔpuuB patA::kan | NG |
| 25 | BLS99 | ΔpuuC Δ(ydcSTUV-patD)::kan | VSG |
| 26 | BLS100 | ΔpuuC ΔpatD::cat | VSG |
| 27 | BLS102 | ΔgabD::cat ΔpatD | 235 ± 10 (3) |
| 28 | BLS103 | ΔpuuC ΔpatD ΔgabD::cat | NG |
| 29 | BLS104 | ΔydcSTUV::cat | 234 ± 7 (6) |
| 30 | BLS105 | ΔydcSTUV | 225 ± 9 (6) |
| 31 | BLS106 | ΔpuuC ΔgabD::cat | 443 ± 24 (3) |
| 32 | BLS108 | ΔcsiD ΔpuuA | 344 ± 16 (3) |
| 33 | BLS109 | ΔpuuA ΔpatD | 507 ± 33 (3) |
| 34 | BLS110 | ΔpatA ΔpuuC | NG |
| 35 | PAJ2 | ΔpuuD | NG |
| 36 | SR4 | ΔgabD::cat | 184 ± 23 (4) |
NG, no growth; S, suppressors accumulate frequently on plates; VSG, very slow growth (10% increase per day) for several days, before a growth burst, which is assumed to be from accumulation of suppressors.
Deletions are assumed to be polar for genes with an antibiotic resistance gene indicated. In-frame deletions are assumed to be nonpolar, and there is no associated antibiotic resistance gene.
Table 2.
Extent of deletions in genes
| Gene | Size (no. of residues) | Deleted residues |
|---|---|---|
| gabD | 482 | 109–482 |
| gabT | 426 | 47–391 |
| patD | 474 | 2–467 |
| patA | 497 | 1–488 |
| puuA | 472 | 6–462 |
| puuB | 426 | 9–387 |
| puuC | 495 | 5–485 |
| puuD | 254 | 8–233 |
| puuE | 421 | 2–391 |
| puuP | 461 | 5–452 |
| puuR | 185 | 6–181 |
Plasmids from the ASKA collection were used for complementation studies. These plasmids contained puuA, puuB, puuC, puuD, puuP, puuR, patA, patD, and gabD under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter. These plasmids code for chloramphenicol resistance. When the mutant was also chloramphenicol resistant, the cat gene from the ASKA plasmid was deleted by digesting with SspI and replacing it with the pUC18-derived ampicillin resistance marker bla. The AlwNI-BsrBI fragment from pUC18 was blunt-ended using T4 DNA polymerase before ligation into the ASKA plasmid. The ampicillin-resistant plasmid was constructed for plasmids containing gabD, puuC, puuD, and puuR. Detectable, and often abundant, protein was observed for PuuB, PuuC, PuuD, PuuE, PatD, and GabD but not PuuA, PuuC, PuuP, and PatA (http://ecoli.naist.jp/GB8). Despite the failure to observe PuuC and PuuP, each complemented the appropriate mutant. Plasmids containing puuA and patA did not complement the appropriate mutant, but there is no evidence that these plasmids code for a functional protein.
The plasmid pBLS17 was used to construct lacZ translational fusions (20). pBLS18, a derivative of pBLS17 with lacZ from pAH125 with the trp terminator deleted, was used to construct lacZ transcriptional fusions (20). To construct the puuA-lacZ and puuD-lacZ fusions, a PCR product containing 53 bases upstream from the start codon of puuA to 59 bases upstream from the start codon of puuD was inserted into plasmids pBLS17 and pBLS18. The lacZ fusions were integrated into the chromosomal lambda attachment site and checked for single insertion by PCR (12). pBLS17 and pBLS18 were also inserted into the chromosome as negative controls.
Media and growth conditions.
W salts minimal medium contained 0.4% glucose as the carbon source and 0.2% of each nitrogen source (41). All cultures for growth studies and assays were grown at 30°C and aerated at 220 rpm. Growth was monitored with a Klett colorimeter, model 800-3, using a 42 filter. Luria-Bertani broth and agar plates were supplemented with 100 μg/ml ampicillin, 50 μg/ml kanamycin, or 20 μg/ml chloramphenicol where appropriate. The complementation analysis required determining the correct concentration of inducer for expression of the complementing gene from the appropriate ASKA plasmid. Initially, experiments were performed with 100 μM IPTG, and this worked well for the plasmids with puuB, puuD, puuP, puuR, patD, and gabD. If this failed, higher and lower concentrations were tested. The plasmid with puuC required a higher inducer concentration (200 μM) and did not generate detectable PuuC protein after induction and purification (http://ecoli.naist.jp/GB8). These results imply poor expression.
Enzyme assays.
For all assays, 10-ml cultures were harvested in late exponential phase (∼100 Klett units or A600 of ∼0.6), pelleted at 4°C, washed twice with cold 150 mM NaCl, and frozen at −80°C. For β-galactosidase assays, thawed cell pellets were resuspended in 1 ml Z buffer (31) with 1 mM β-mercaptoethanol and sonicated on ice in three 5-s bursts. After centrifugation at 4°C, the supernatants were assayed as described previously (31). For dehydrogenase assays, thawed cells were resuspended in 0.1 M KPO4 buffer (pH 7.5)-1 mM dithiothreitol and disrupted with three 5-s bursts of sonication. Succinic semialdehyde dehydrogenase activity was assayed in 0.1 M KPO4 buffer (pH 7.8), 0.28 mM NADP or NAD, and 0.6 mM succinic semialdehyde at 37°C. The reaction was started with the addition of succinic semialdehyde, and the A340 was monitored. For the γ-aminobutyraldehyde dehydrogenase assay, crude extracts were ultracentrifuged for 90 min at 120,000 × g to remove NADH-oxidizing activity. This dehydrogenase was assayed in glycine buffer (pH 9.5), 0.28 mM NADP or NAD, and 0.5 mM γ-aminobutyraldehyde at 37°C. The reaction was started with the addition of γ-aminobutyraldehyde, and the A340 was monitored. The γ-aminobutyraldehyde substrate was freshly prepared from 4-amino-butyraldehyde diethylacetal (Sigma-Aldrich) by boiling for 10 min in 0.2 M HCl (47). All protein concentrations were determined using bovine serum albumin as a standard (29). Activities were expressed as nmol minute−1 mg protein−1.
RESULTS
The genetics of putrescine catabolism to GABA.
Our previous studies of GABA and arginine as the sole nitrogen sources were conducted for cells grown at 30°C (21, 45, 46). Therefore, we studied putrescine as the sole nitrogen source at this temperature. In contrast, analysis of putrescine as the sole carbon source was conducted at 20°C (22–26). These differences are significant and will be discussed later.
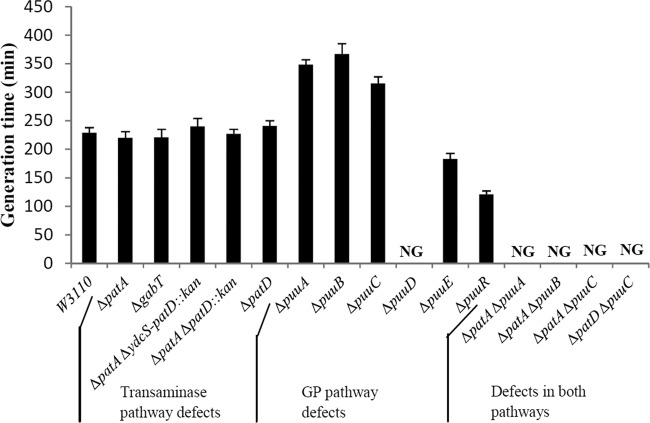
We first disrupted putrescine catabolic genes of the transaminase pathway, which are known to be nitrogen regulated (54). The transaminase pathway converts putrescine to γ-aminobutyrate by a transaminase-dependent deamination, catalyzed by PatA (also called YgjG), and an NAD-dependent aminobutyraldehyde dehydrogenase (ABDH), catalyzed by YdcW (also called Prr) (43, 44). Mutants with in-frame deletions of their genes have not been described. We will show that they are the major enzymes that catalyze their respective reactions. We redesignated YdcW as PatD (putrescine aminotransferase pathway, dehydrogenase). Colonies of CP2 (ΔpatA strain) were smaller than those of W3110 (wild type) on agar plates containing either broth or glucose-putrescine minimal medium (not shown). However, CP2 in liquid minimal medium grew normally with putrescine, ammonia, or alanine as the sole nitrogen sources, slightly faster (≤20%) with ornithine or GABA, and slightly slower (≤20%) with agmatine (Fig. 2 and results not shown). A ΔpatD strain grew normally with all nitrogen sources tested, including putrescine (Fig. 2 and results not shown). Two different ΔpatA ΔpatD double mutants also grew normally with putrescine as the nitrogen source (Fig. 2). The patD gene appears to be part of a larger ydcSTUV-patD operon. The first four genes of the operon specify a proposed transporter for putrescine/spermidine. Growth with putrescine was not affected by deletion of these four genes in wild-type, ΔpatD, or ΔpatA ΔpatD backgrounds (Fig. 2 and Table 1, lines 6, 7, 29 and 30). These results indicate that PatA and PatD are not required for putrescine catabolism.
Fig 2.
Growth with putrescine as the nitrogen source. The strains and doubling times are listed in Table 1. NG indicates no growth.
The enzymes of the GP pathway catalyze ATP-dependent glutamylation of putrescine (PuuA), oxygen-dependent deamination (PuuB), dehydrogenase-dependent oxidation (PuuC), and deglutamylation of N-γ-glutamyl-γ-GABA (PuuD) (Fig. 1). Strains with in-frame deletions of puuA, puuB, or puuC had a doubling time 50% greater than the wild type with putrescine as the sole nitrogen source (Fig. 2). ASKA plasmids containing puuB and puuC restored wild-type growth to ΔpuuB and ΔpuuC strains, respectively (Fig. 3). (The ASKA plasmid containing puuA did not restore wild-type growth to a ΔpuuA strain. This is a problem with the ASKA plasmid, which does not produce detectable PuuA after induction and purification [http://ecoli.naist.jp/GB8]. A different puuA plasmid has been shown to complement the defect in a ΔpuuA strain [25]. We did not reconstruct the plasmid.) Unlike the puuA, puuB, and puuC mutants, a ΔpuuD mutant did not grow with putrescine (Fig. 2). The ASKA plasmid containing puuD not only restored growth to a ΔpuuD strain but also resulted in faster growth than a wild-type strain (Fig. 3). We conclude that the enzymes of the GP pathway (except for PuuD) are not required for putrescine catabolism as the nitrogen source, although their absence detectably impaired growth.
Fig 3.
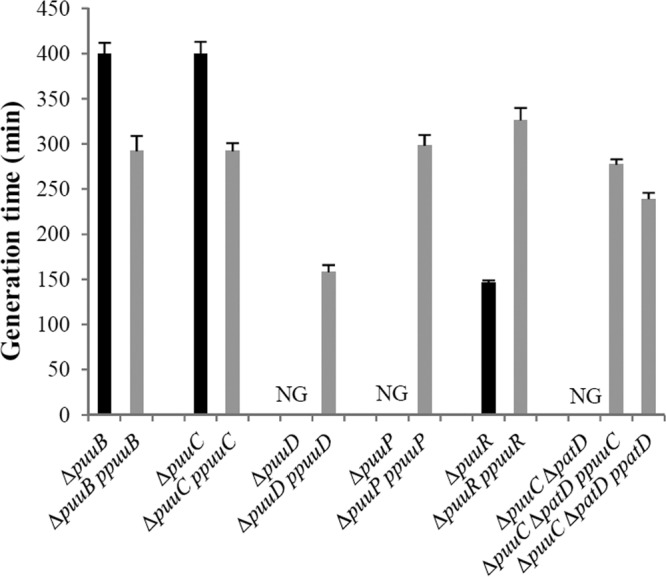
Complementation analysis of mutants with defects in putrescine catabolism. The strains are listed in Table 1. The designation of ppuuB, etc., indicates an ASKA plasmid with the indicated gene, e.g., puuB. NG indicates no growth. The black bar indicates a mutant, while the gray bar indicates a complemented strain.
We then examined mutants with defects in both pathways. Strains with deletions of patA and any of the following—puuA, puuB, or puuC—did not grow with putrescine as the nitrogen source (Fig. 2). Putrescine supplementation in either glucose-ammonia (nitrogen-rich) or glucose-glutamine (nitrogen-limited) minimal medium did not affect growth of these double mutants, which suggests that putrescine toxicity cannot account for the mutant phenotype (results not shown). A ΔpuuA ΔpatD strain had a doubling time that was 50% greater than that of a ΔpuuA strain (Table 1, lines 13 and 33). This suggests that PatD is not the only enzyme with ABDH activity in the transaminase pathway. A ΔpuuC ΔpatD strain, which lacks the proposed aldehyde dehydrogenases of both pathways, failed to grow (Fig. 2). This suggests that PuuC may be the second enzyme with ABDH activity. ASKA plasmids containing either puuC or patD restored a wild-type growth rate to the ΔpuuC ΔpatD mutant (Fig. 3). Defects in both pathways generally prevent putrescine utilization, which implies that either pathway is sufficient for putrescine utilization.
It is possible that other pathways may contribute to putrescine catabolism, although not sufficiently for putrescine utilization as the sole nitrogen source. We specifically tested whether CsiD (also called YgaT) contributes to putrescine catabolism. CsiD is one product of a complex csiD-lhgO-gabDTPC operon (30). The last four genes specify proteins involved in GABA metabolism (30, 46). Homology and structural analysis suggests that CsiD is a nonheme iron II-dependent oxygenase, possibly with α-ketoglutarate as one substrate (4). LhgO (previously called YgaF) is a flavin adenine dinucleotide-dependent l-2-hydroxyglutarate oxidase (18). Together, CsiD and LhgO could conceivably degrade putrescine to GABA by an oxygen-dependent pathway that is known to exist in some bacteria (27). We transferred a deletion of csiD into a ΔpuuA mutant, which is detectably impaired in putrescine utilization. The deletion of csiD did not further impair the growth of a ΔpuuA mutant (Table 1, lines 13 and 32), which suggests that CsiD does not contribute to putrescine catabolism.
There are several polyamine transport systems, including products of the potFGHI and ydcSTUVW operons, that are induced by nitrogen limitation and the Ntr response (14, 54). Deletions of these operons had no effect on growth with putrescine as the nitrogen source (results not shown). The puuP gene, which is adjacent to puuA, specifies a transport system that is not regulated by nitrogen limitation. The ΔpuuP strains failed to utilize putrescine as a nitrogen source at 30°C (Fig. 3 and Table 1, lines 11 and 17). A puuP plasmid restored a wild-type growth rate to the mutant (Fig. 3). PuuP appears to be the major transporter for putrescine as the sole nitrogen source. PuuP has been previously shown to be required for putrescine utilization as a carbon source at 20°C (26).
In summary, three types of defects prevent utilization of putrescine as the sole nitrogen source: loss of PuuD, loss of PuuP, and defects in both the GP and transaminase pathways.
The redundancy and specificity of enzymes that degrade putrescine to GABA.
Loss of PuuA, PuuB, or PuuC, in a strain without PatA, prevents putrescine utilization. Since these four enzymes catalyze different reactions, the phenotypes imply that each is a nonredundant enzyme. We confirmed this conclusion for PatA. Cell extracts of W3110 (wild type) and CP2 (ΔpatA mutant) grown in glucose-putrescine minimal medium had 2.36 ± 0.07 and 0.09 ± 0.02 units of putrescine transaminase activity, respectively, which implies that PatA is essentially the only putrescine transaminase (39).
The ABDH reaction of the transaminase pathway oxidizes γ-aminobutyraldehyde to GABA. Purified PatD (formerly YdcW) has NAD-specific dehydrogenase activity (44). Genetic results in the preceding section suggested that PuuC might have ABDH activity. Assays of NAD-dependent ABDH activity from cell extracts confirmed this possibility (Fig. 4). A ΔpatD puuC+ strain possessed about 45% of wild-type activity, a patD + ΔpuuC strain had about 70% activity, and a ΔpatD ΔpuuC strain had about 15% activity. These results suggest that PatD and PuuC account for 55% and 30%, respectively, of ABDH activity, and that another enzyme or enzymes have 15% residual activity. A plasmid with either patD or puuC increased ABDH activity from a ΔpatD ΔpuuC strain 2 or 14 times over the background level, respectively (Fig. 5). The puuC gene appears to be poorly expressed from the ASKA plasmid (see Materials and Methods, “Strains and plasmids”). Nonetheless, biochemical and genetic results indicate that PuuC and PatD have ABDH activity in vivo. GabD, the remaining aldehyde dehydrogenase in the two putrescine catabolic pathways, does not contribute to ABDH, since a ΔpatD ΔpuuC ΔgabD strain still possessed residual activity (Fig. 4), and cells with a plasmid with gabD which had 50 times more NADP succinic semialdehyde dehydrogenase (SSDH) activity did not have increased ABDH activity (not shown).
Fig 4.
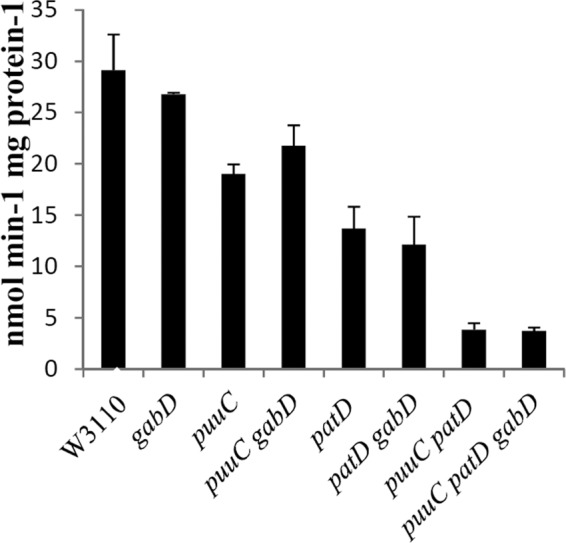
The activity of γ-aminobutyraldehyde dehydrogenase (ABDH). Cells were grown in glucose-aspartate-putrescine minimal medium, which is inducing but eliminates differences in growth rates.
Fig 5.
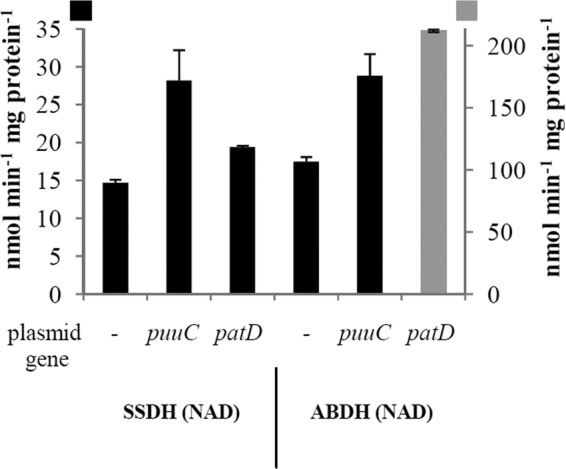
The activity of succinic semialdehyde dehydrogenase (SSDH) and γ-aminobutyraldehyde dehydrogenase (ABDH). Strain BLS93 (ΔpuuC ΔpatD) was grown with ASKA plasmids containing the indicated gene. Cells were grown in LB medium. The black-bar values use the scale on the left, while the gray-bar value uses the scale on the right.
The redundancy and specificity of enzymes degrade GABA.
The transaminase and GP pathways convert putrescine to GABA, which is then metabolized to succinate. GabT converts GABA to succinic semialdehyde, and the extent of homology between GabT and PuuE suggests that PuuE probably catalyzes the same reaction. We examined growth of mutants lacking these enzymes. CP6 (ΔgabT) (Table 1, line 3) and CP14 (ΔpuuE) (Table 1, line 4) grew normally with putrescine as the sole nitrogen source, whereas CP36 (ΔgabT ΔpuuE) (Table 1, line 5) grew 30% slower than the wild type. To determine if CP36 can catabolize the GABA generated from putrescine, we measured growth yields with limiting nitrogen. With 1 mM NH4Cl as the only nitrogen source, the wild-type strain and the three mutants grew to a density of 81 Klett units. With 1 mM putrescine (which has two nitrogens), W3110, CP14, CP6, and CP36 grew to 151 ± 3, 144 ± 2, 176 ± 1, and 78 ± 2 Klett units, respectively. (Each value is the average of three determinations.) We conclude that CP36 (ΔgabT ΔpuuE), which grew to half the level of the other strains, can remove only one nitrogen from putrescine and therefore cannot degrade GABA. Extracts of W3110 (wild type), CP14 (ΔpuuE gabT+), CP6 (ΔgabT puuE+), and CP36 (ΔpuuE ΔgabT) had 430 ± 22, 214 ± 11, 111 ± 8, and −1.0 ± 2.6 units of GABA transaminase activity, respectively. We conclude that (i) PuuE and GabT are the only transaminases that degrade GABA to succinic semialdehyde in vivo and (ii) loss of both enzymes allows utilization of one, but not two, nitrogens of putrescine.
Strains without succinic semialdehyde dehydrogenase (SSDH) activity might be impaired in utilization of one nitrogen from putrescine, or they might grow slower if succinic semialdehyde is toxic. GabD is known to have NADP-dependent SSDH activity. A ΔgabD mutant and a ΔgabD ΔpatD mutant grew as well as the wild type with putrescine as the nitrogen source (Table 1, line 27, and not shown). The doubling time of a ΔpuuC ΔgabD mutant (Table 1, line 31) was 100% greater than that of the wild type, while the doubling time of a ΔpuuC mutant (Table 1, line 20) was only 40% greater. These results are consistent with the possibility that PuuC has SSDH activity in vivo. Therefore, we examined SSDH activity in crude extracts from various mutants. For cells grown in glucose-putrescine minimal medium, GabD accounted for 85% of NADP-dependent SSDH activity (Fig. 6B). Loss of both PuuC and PatD did not diminish NADP-dependent activity (Fig. 7). (Cells for this assay had to be grown in glucose-ammonia-putrescine medium, since the double mutant does not grow in glucose-putrescine medium.) Unexpectedly, PuuC accounted for 75% of NAD-dependent SSDH activity (Fig. 6A). Confirmation of these results was sought from cells containing plasmids with puuC, patD, or gabD. For NADP-dependent SSDH, a ΔpuuC ΔgabD mutant with an empty ASKA plasmid had 11 U of activity, while the mutant with an ASKA-gabD plasmid had 655 U of activity. Plasmids with patD or puuC had no effect on NADP-dependent SSDH (not shown). In other words, only GabD has NADP-dependent activity. For NAD-dependent SSDH, cells with puuC or patD on a plasmid had 100% and 30% more activity than cells with an empty plasmid (Fig. 5). Genetic and biochemical evidence indicates that GabD and PuuC have NADP- and NAD-dependent SSDH activity, respectively.
Fig 6.
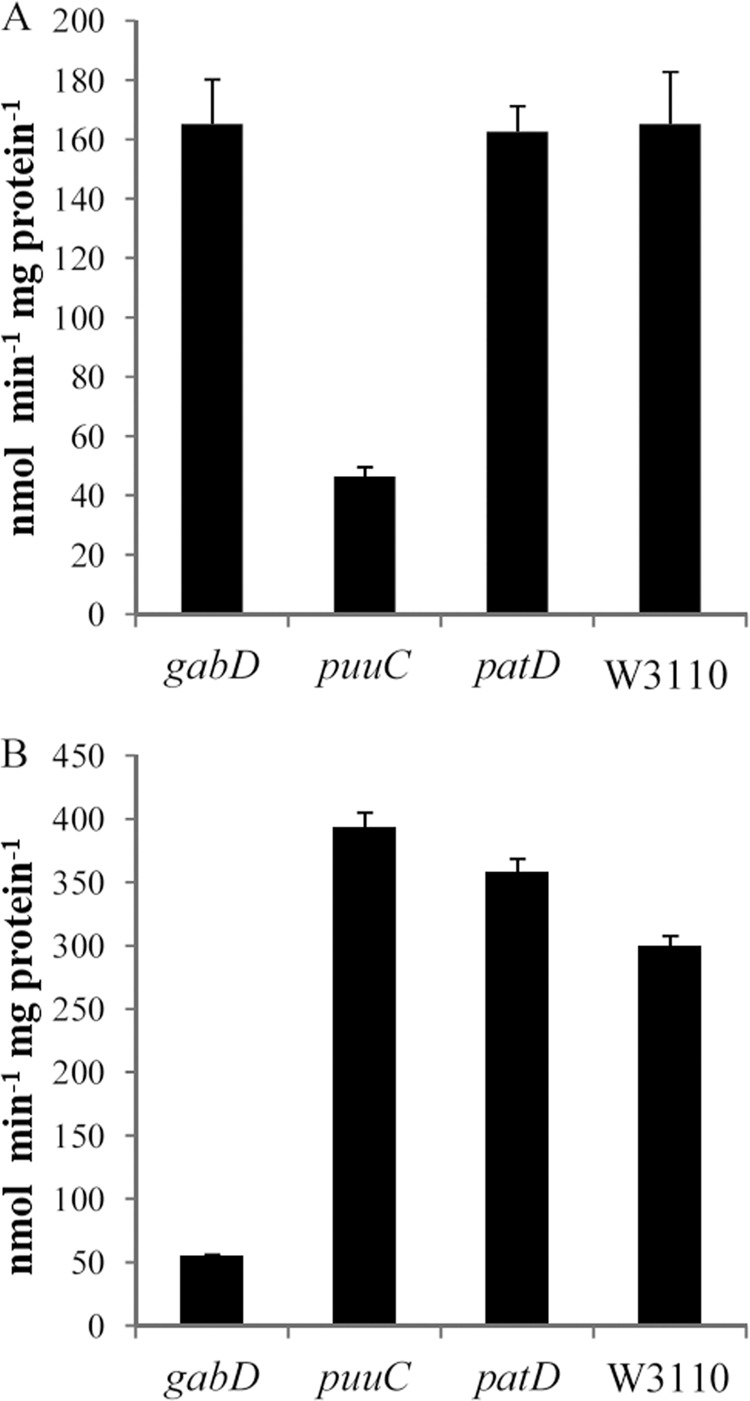
Succinic semialdehyde dehydrogenase activity in various mutants. Cells were grown in glucose-putrescine minimal medium. (A) NAD-dependent activity; (B) NADP-dependent activity.
Fig 7.
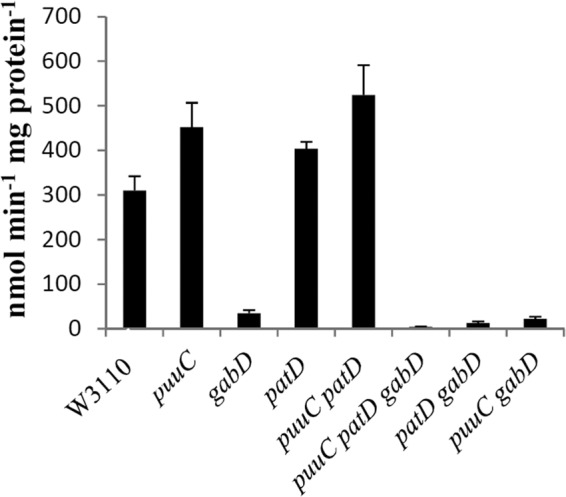
NADP-dependent succinic semialdehyde dehydrogenase activity. Cells were grown in glucose-aspartate-putrescine minimal medium, which allows growth of mutants that cannot grow in glucose-putrescine medium and eliminates growth differences but still allows induction.
Polarity in GP pathway genes.
In the course of mutant construction, we isolated mutants with both insertions, which are presumably polar, and in-frame deletions. There were two examples of differences in growth for the two types of mutants. First, an in-frame deletion of puuA only modestly impaired growth, whereas an insertion in puuA eliminated growth (Table 1, lines 12 and 13). The insertion presumably prevents puuP expression, which is required for putrescine catabolism. This suggests the existence of a puuAP operon. Second, an in-frame deletion of puuR stimulated growth (Table 1, line 16), whereas an insertion in puuR eliminated growth (Table 1, line 10). Loss of PuuD, perhaps by destabilization of puuD mRNA, might explain this phenotype. Another possibility is high expression of the puuAP operon coupled with loss of PuuC, PuuB, and PuuE, resulting in either toxic accumulation of γ-glutamyl-γ-putrescine or depletion of substrates (glutamate and ATP). Strains with insertions in puuB and puuC (Table 1, lines 22 and 19, respectively) grew as well as strains with in-frame deletions of these genes (Table 1, lines 24 and 20, respectively). This is expected, since both the insertions and deletions affect only the GP pathway and not the transaminase pathway. Because of the complex phenotype of the strain with the insertion in puuR, it is difficult to interpret much about polarity in the puuD-puuR-puuC-puuB-puuE region.
Regulation of GP pathway genes: PuuR and putrescine.
We studied expression from the regions preceding puuA, puuC, and puuD with single-copy lacZ transcriptional fusions. (We also constructed puuA-lacZ and puuD-lacZ translational fusions. No differences were observed between the two types of fusions under a variety of conditions, and results are shown only for the transcriptional fusions.) We examined expression from the puuD-lacZ fusion in glucose-ammonia, glucose-ammonia-putrescine, and glucose-putrescine media and observed 1,400, 6,600, and 19,000 units of LacZ, respectively (open boxes, Fig. 8). Virtually identical results were observed for the puuA-lacZ fusions (not shown). The difference in expression from puuD-lacZ strains in glucose-ammonia-putrescine (nitrogen excess) and glucose-putrescine (nitrogen-limiting medium), 6,600 versus 19,000 units, might be due to control by nitrogen limitation. However, there was no difference in LacZ from cells grown in glucose-ammonia (nitrogen excess) versus glucose-glutamine (nitrogen limiting) (not shown), which argues against this possibility. These results suggest induction by putrescine.
Fig 8.
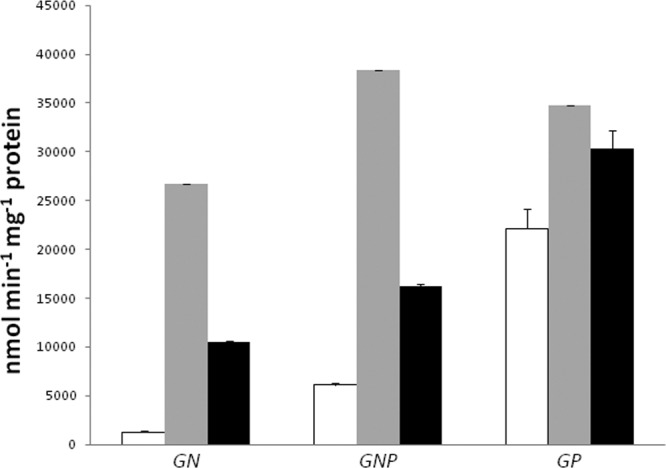
β-Galactosidase activity from a single-copy puuD-lacZ transcription fusion. Cells were grown in the indicated minimal medium: GN, glucose-ammonia; GNP, glucose-ammonia-putescine; and GP, glucose-putrescine. Activities in the wild type (blank), puuR (gray), and puuR strains with complementing plasmid (black) are shown.
The puuC-lacZ fusion contains 365 bases of the C terminus of puuR, the entire 274-base intergenic region, and 32 bases of puuC. The fusion gave 250, 310, and 480 units of LacZ in glucose-ammonia, glucose-ammonia-putrescine, and glucose-putrescine media, respectively. (A promoterless lacZ inserted into the chromosome generated 8 units of activity. All SEMs were ≤15% of the means.) These results are consistent with the possibility of a weak constitutive promoter but do not indicate the presence of a putrescine-inducible promoter. This region was not further analyzed.
The puuR gene codes for a potential 185-residue DNA-binding protein. It is a member of the XRE (xenobiotic response element) family which includes cro and cI of phage λ. The doubling time of a ΔpuuR strain was half that of a wild-type strain in glucose-putrescine medium (Table 1, line 16). An ASKA-puuR plasmid restored a wild-type growth rate (Fig. 3). Loss of PuuR elevated expression from the puuD promoter 22-fold in glucose-ammonia medium, 6-fold in glucose-ammonia-putrescine medium, and 1.6-fold in glucose-putrescine medium (Fig. 8). The ASKA-puuR plasmid restored lower expression in glucose-ammonia and glucose-ammonia-putrescine media (black boxes, Fig. 8). Expression was higher in a puuR mutant than for wild-type cells grown in the most derepressing medium (glucose-putrescine medium) (Fig. 8). Results with the puuA fusion were virtually identical (not shown). These results suggest that PuuR is a repressor for both the puuA and puuD promoters and that PuuR mediates control by putrescine.
To test whether a derivative of putrescine is the inducer, we examined puuA and puuD expression in mutants with different blocks in putrescine catabolism. We grew cells in glucose-ammonia-putrescine medium, which allows growth of putrescine-nonutilizing mutants. In a ΔpuuA ΔpatA background, which does not degrade putrescine, expression of both puuA and puuD was as high as that in a ΔpuuR strain (Fig. 9). These results are sufficient to suggest that putrescine, and not a derivative, is the inducer.
Fig 9.
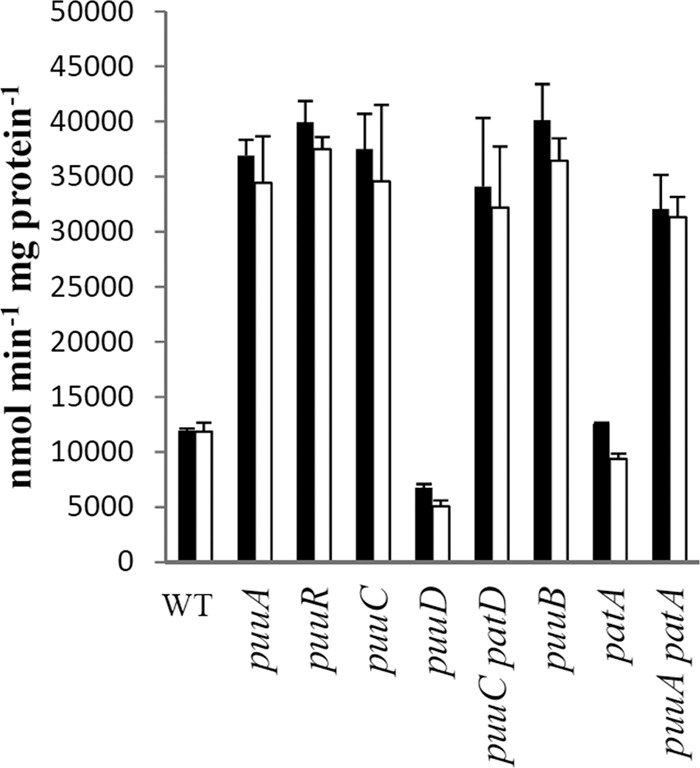
β-Galactosidase activity from strains with puuD-lacZ (black) and puuA-lacZ (white) fusions. Cells were grown in glucose-ammonia-putrescine minimal medium, which shows the largest differential between the wild-type and puuR backgrounds (see Fig. 8). This medium also eliminates the enormous growth rate differences of the various mutants.
This conclusion allows an indirect test for putrescine accumulation in various mutants. Derepressed expression was observed for puuA, puuB, and puuC mutants but not for patA and puuD mutants (Fig. 9). The derepression implies putrescine accumulation in the former mutants. For these experiments, cells had been grown in glucose-aspartate-putrescine medium, which has the inducer but supports the same growth rate for wild-type and mutant strains. For cells grown in glucose-ammonia minimal medium, a ΔpuuA ΔpatA strain derepressed puuA expression about 2-fold but was only about 12% of the level in a wild-type strain grown in glucose-putrescine medium (data not shown). This implies that loss of the catabolic pathways only modestly increases intracellular putrescine in medium without exogenous putrescine.
DISCUSSION
Pathways of putrescine catabolism.
The reactions of PatA, PatD, GabT, and GabD degrade putrescine to succinate (Fig. 1). Mutants lacking any of these enzymes grew normally with putrescine as the nitrogen source. Common regulatory features of these enzymes (discussed below) suggest that these four enzymes constitute a pathway, which we have called the transaminase pathway. The reactions catalyzed by PuuA, PuuB, PuuC, PuuD, and PuuE also completely degrade putrescine to succinate. PuuC catalyzes the third and sixth reactions of this pathway (reactions 8 and 5, Fig. 1). The latter reaction has not been previously described.
Loss of puuA, puuB, and puuC impaired but did not eliminate growth with putrescine as the nitrogen source (Fig. 2). In contrast, loss of puuD prevented growth in glucose-putrescine medium and a complementing plasmid stimulated growth that was faster than wild-type growth (Fig. 2 and 3). The transaminase pathway is intact in all of these mutants, which suggests that an incomplete GP pathway is creating a problem in the puuD mutant. It seems plausible that N-γ-glutamyl-γ-GABA accumulates. This can be a problem if N-γ-glutamyl-γ-GABA is toxic. However, this possibility would suggest impaired growth in glucose-ammonia-putrescine medium, which does not occur. An alternate possibility is that the glutamate in N-γ-glutamyl-γ-GABA is nonproductively sequestered; that is, it is not being recycled. This would create an effective glutamate starvation. This possibility predicts normal growth if glutamate or a precursor for glutamate (in this case ammonia) is provided and this prediction is met.
Specificity and redundancy of the aldehyde dehydrogenases in putrescine catabolism.
PuuC oxidizes γ-aminobutyraldehyde, N-glutamyl-γ-aminobutyraldehyde, and succinic semialdehyde (Fig. 1, reactions 3, 5, and 8). This broad specificity is consistent with two characterizations of PuuC which were published before its role in putrescine catabolism was known. First, an E. coli aldehyde dehydrogenase that oxidizes acetaldehyde was sequenced (13), and current genome resources show that it is PuuC. It has 40% amino acid identity and 60% similarity to nonspecific mammalian aldehyde dehydrogenases (13). Second, a more recent study examined the substrate specificity of purified AldH (an alternate name for PuuC) and showed that it oxidizes a variety of aldehydes, including butyraldehyde (16). The pyridine nucleotide specificity of PuuC has been proposed to be flexible. GenBank and other annotations indicate that PuuC prefers NADP over NAD, which was suggested by the first characterization of PuuC (13). However, the ratio of NADP to NAD activity is not constant with cells from different media (13) or with different substrates (16). Our results show that overexpression of puuC increased NAD-dependent SSDH activity but not NADP-dependent SSDH activity.
Our results identified the major dehydrogenases in the putrescine catabolism. Other dehydrogenases have been proposed to catalyze reactions in putrescine catabolism, and they may be important in different growth conditions. Sad (also called YneI) oxidizes succinic semialdehyde, and it has been reported that a sad/yneI mutant has a growth defect with putrescine as the carbon source (10, 22). We observed no steady-state growth defect for a ΔyneI strain with putrescine as the sole nitrogen source (unpublished observation).
An enzyme in addition to PatD and PuuC appears to oxidize γ-aminobutyraldehyde, since a double mutant still grew, albeit extremely slowly (Table 1), and this mutant still had 15% residual activity (Fig. 4). We did not specifically identify an enzyme with this residual activity, but several published results suggest that AdhE has ABDH activity. First, E. coli cannot utilize putrescine as a carbon source at ≥30°C. A mutant that could utilize putrescine as a carbon source at 37°C has elevated ABDH activity. This ABDH was purified and shown to have 95-kDa subunits, which excludes PatD (50.7-kDa subunits) and PuuC (53.2-kDa subunits) (38). The unusual size is sufficient to identify this ABDH as AdhE. Its subunits are 70% larger than those of the second largest aldehyde dehydrogenase, which is AldB (56.3-kDa subunits), and AdhE is the only soluble dehydrogenase in E. coli, with subunits of 95 kDa ± 10%. Second, purified AdhE has high activity with butyraldehyde, which suggests that activity with γ-aminobutyraldehyde is likely (42). Third, putrescine induces AdhE (52).
Regulation and relative contributions of the putrescine catabolic pathways.
RNA polymerase complexed with σ54 controls expression of the operons of transaminase pathway genes, gabDTPC, patA (ygjG), and ydcSTUV-patD (40, 54). RNA polymerase complexed with σS also controls these operons. The σS-dependent control for the csiD-lhgO-gabDTPC operon has been described (30), and we will describe the complex σS-dependent control of patA and ydcSTUV-patD in a separate communication. Gene profiling suggests that high temperature increases their expression (17). It appears that these four operons constitute a stress-inducible pathway of putrescine catabolism.
Several factors have been reported to increase expression of GP pathway genes: putrescine, low temperature, and the transition from anaerobic to aerobic growth; other factors lower expression: succinate, high temperature, and low aeration (17, 22, 25, 26, 36). ArcA and PuuR have been implicated in GP pathway control (22, 25, 26, 36) (this paper). ArcA mediates repression during anaerobic growth (36) and probably with low aeration. In addition to these control mechanisms, a metal-catalyzed oxidation initiates posttranslational degradation of PuuA (25). PuuA inactivation is consistent with the importance of polyamines for growth with 95% oxygen (5). These mechanisms do not obviously account for the effects of temperature (described next) and succinate.
Gene profiling shows that expression of GP pathway genes is elevated at lower temperatures, and expression of transaminase pathway genes is repressed (17). This temperature effect can explain some unexpected aspects of putrescine catabolism. First, the GP pathway degrades putrescine as the sole carbon source at 20°C but not at 37°C (22–26). Our wild-type strain cannot use putrescine as the sole carbon source at 30°C. Diminished expression of the GP pathway may explain the failure to utilize putrescine at higher temperatures. Second, mutants that utilize putrescine as a carbon source at 37°C have been isolated, and they have elevated levels of both transaminase pathway enzymes (37). Several rationales can be suggested to account for this temperature effect. First, spermidine, which is derived from putrescine, is toxic at low temperatures (1, 11, 28). Putrescine catabolism could minimize the toxicity. Second, the growth rate of mutants suggests that the GP pathway is quantitatively more important than the transaminase pathway. This would generate more aldehydes, which could be a problem at higher temperatures, as has been proposed for another aldehyde-generating pathway (19, 35). Third, putrescine and other polyamines may be more important at higher temperatures.
We have shown that two pathways catabolize putrescine as the sole nitrogen source. The nonoverlapping regulatory factors and differential effect of temperature suggest that the relative importance of each pathway will depend on the environment. The GP pathway is essential for putrescine as the sole carbon source at low temperatures. It is not essential for putrescine catabolism as the sole nitrogen source, although it appears to be more quantitatively significant than the transaminase pathway.
ACKNOWLEDGMENTS
Grants MCB-0323931 from the National Science Foundation and GM085536 from the National Institutes of Health supported this work.
We thank Christine Pybus and Paul Johnson for construction of some strains.
Footnotes
Published ahead of print 25 May 2012
REFERENCES
- 1. Apirakaramwong A, et al. 1999. Involvement of ppGpp, ribosome modulation factor, and stationary phase-specific sigma factor σS in the decrease in cell viability caused by spermidine. Biochem. Biophys. Res. Commun. 264:643–647 [DOI] [PubMed] [Google Scholar]
- 2. Boyle SM, MacIntyre MF, Sells BH. 1977. Polyamine levels in Escherichia coli during nutritional shiftup and exponential growth. Biochim. Biophys. Acta 477:221–227 [DOI] [PubMed] [Google Scholar]
- 3. Capp MW, et al. 1996. Compensating effects of opposing changes in putrescine (2+) and K+ concentrations on lac repressor-lac operator binding: in vitro thermodynamic analysis and in vivo relevance. J. Mol. Biol. 258:25–36 [DOI] [PubMed] [Google Scholar]
- 4. Chance MR, et al. 2002. Structural genomics: a pipeline for providing structures for the biologist. Protein Sci. 11:723–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chattopadhyay MK, Tabor CW, Tabor H. 2009. Polyamines are not required for aerobic growth of Escherichia coli: preparation of a strain with deletions in all of the genes for polyamine biosynthesis. J. Bacteriol. 191:5549–5552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cohen SS. 1998. A guide to the polyamines. Oxford University Press, New York, NY [Google Scholar]
- 7. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Davis RH, Morris DR, Coffino P. 1992. Sequestered end products and enzyme regulation: the case of ornithine decarboxylase. Microbiol. Rev. 56:280–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Echandi G, Algranati ID. 1975. Defective 30S ribosomal particles in a polyamine auxotroph of Escherichia coli. Biochem. Biophys. Res. Commun. 67:1185–1191 [DOI] [PubMed] [Google Scholar]
- 10. Fuhrer T, Chen L, Sauer U, Vitkup D. 2007. Computational prediction and experimental verification of the gene encoding the NAD+/NADP+-dependent succinate semialdehyde dehydrogenase in Escherichia coli. J. Bacteriol. 189:8073–8078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fukuchi J, Kashiwagi K, Yamagishi M, Ishihama A, Igarashi K. 1995. Decrease in cell viability due to the accumulation of spermidine in spermidine acetyltransferase-deficient mutant of Escherichia coli. J. Biol. Chem. 270:18831–18835 [DOI] [PubMed] [Google Scholar]
- 12. Haldimann A, Wanner BL. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183:6384–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heim R, Strehler EE. 1991. Cloning an Escherichia coli gene encoding a protein remarkably similar to mammalian aldehyde dehydrogenases. Gene 99:15–23 [DOI] [PubMed] [Google Scholar]
- 14. Igarashi K, Ito K, Kashiwagi K. 2001. Polyamine uptake systems in Escherichia coli. Res. Microbiol. 152:271–278 [DOI] [PubMed] [Google Scholar]
- 15. Igarashi K, Kashiwagi K. 2000. Polyamines: mysterious modulators of cellular functions. Biochem. Biophys. Res. Commun. 271:559–564 [DOI] [PubMed] [Google Scholar]
- 16. Jo JE, et al. 2008. Cloning, expression, and characterization of an aldehyde dehydrogenase from Escherichia coli K-12 that utilizes 3-hydroxypropionaldehyde as a substrate. Appl. Microbiol. Biotechnol. 81:51–60 [DOI] [PubMed] [Google Scholar]
- 17. Jozefczuk S, et al. 2010. Metabolomic and transcriptomic stress response of Escherichia coli. Mol. Syst. Biol. 6:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kalliri E, Mulrooney SB, Hausinger RP. 2008. Identification of Escherichia coli YgaF as an l-2-hydroxyglutarate oxidase. J. Bacteriol. 190:3793–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim KS, et al. 2010. The Rut pathway for pyrimidine degradation: novel chemistry and toxicity problems. J. Bacteriol. 192:4089–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim SH, Schneider BL, Reitzer L. 2010. Genetics and regulation of the major enzymes of alanine synthesis in Escherichia coli. J. Bacteriol. 192:5304–5311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kiupakis AK, Reitzer L. 2002. ArgR-independent induction and ArgR-dependent superinduction of the astCADBE operon in Escherichia coli. J. Bacteriol. 184:2940–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kurihara S, Kato K, Asada K, Kumagai H, Suzuki H. 2010. A putrescine-inducible pathway comprising PuuE-YneI in which gamma-aminobutyrate is degraded into succinate in Escherichia coli K-12. J. Bacteriol. 192:4582–4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kurihara S, et al. 2005. A novel putrescine utilization pathway involves γ-glutamylated intermediates of Escherichia coli K-12. J. Biol. Chem. 280:4602–4608 [DOI] [PubMed] [Google Scholar]
- 24. Kurihara S, Oda S, Kumagai H, Suzuki H. 2006. γ-Glutamyl-γ-aminobutyrate hydrolase in the putrescine utilization pathway of Escherichia coli K-12. FEMS Microbiol. Lett. 256:318–323 [DOI] [PubMed] [Google Scholar]
- 25. Kurihara S, et al. 2008. γ-Glutamylputrescine synthetase in the putrescine utilization pathway of Escherichia coli K-12. J. Biol. Chem. 283:19981–19990 [DOI] [PubMed] [Google Scholar]
- 26. Kurihara S, et al. 2009. The putrescine importer PuuP of Escherichia coli K-12. J. Bacteriol. 191:2776–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Large PJ. 1992. Enzymes and pathways of polyamine breakdown in microorganisms. FEMS Microbiol. Rev. 8:249–262 [DOI] [PubMed] [Google Scholar]
- 28. Limsuwun K, Jones PG. 2000. Spermidine acetyltransferase is required to prevent spermidine toxicity at low temperatures in Escherichia coli. J. Bacteriol. 182:5373–5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lowry OH, Rosebrough NJ, Farr L, Randall RJ. 1951. Protein measurement with the folin phenol reagent. J. Biol. Chem. 193:265–275 [PubMed] [Google Scholar]
- 30. Metzner M, Germer J, Hengge R. 2004. Multiple stress signal integration in the regulation of the complex σS-dependent csiD-ygaF-gabDTP operon in Escherichia coli. Mol. Microbiol. 51:799–811 [DOI] [PubMed] [Google Scholar]
- 31. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 32. Miller JH. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 33. Minton KW, Tabor H, Tabor CW. 1990. Paraquat toxicity is increased in Escherichia coli defective in the synthesis of polyamines. Proc. Natl. Acad. Sci. U. S. A. 87:2851–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Munro GF, Hercules K, Morgan J, Sauerbier W. 1972. Dependence of the putrescine content of Escherichia coli on the osmotic strength of the medium. J. Biol. Chem. 247:1272–1280 [PubMed] [Google Scholar]
- 35. Parales RE, Ingraham JL. 2010. The surprising Rut pathway: an unexpected way to derive nitrogen from pyrimidines. J. Bacteriol. 192:4086–4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Partridge JD, Scott C, Tang Y, Poole RK, Green J. 2006. Escherichia coli transcriptome dynamics during the transition from anaerobic to aerobic conditions. J. Biol. Chem. 281:27806–27815 [DOI] [PubMed] [Google Scholar]
- 37. Prieto-Santos MI, Martin-Checa J, Balana-Fouce R, Garrido-Pertierra A. 1986. A pathway for putrescine catabolism in Escherichia coli. Biochim. Biophys. Acta 880:242–244 [DOI] [PubMed] [Google Scholar]
- 38. Prieto MI, Martin J, Balana-Fouce R, Garrido-Pertierra A. 1987. Properties of gamma-aminobutyraldehyde dehydrogenase from Escherichia coli. Biochimie 69:1161–1168 [DOI] [PubMed] [Google Scholar]
- 39. Pybus CA. 2002. Roles of the omega-transaminases of Escherichia coli in nitrogen metabolism and characterization of a putrescine aminotransferase. Master's thesis The University of Texas at Dallas, Richardson, TX [Google Scholar]
- 40. Reitzer L. 2003. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 57:155–176 [DOI] [PubMed] [Google Scholar]
- 41. Rothstein DM, Pahel G, Tyler B, Magasanik B. 1980. Regulation of expression from the glnA promoter of Escherichia coli in the absence of glutamine synthetase. Proc. Natl. Acad. Sci. U. S. A. 77:7372–7376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rudolph FB, Purich DL, Fromm HJ. 1968. Coenzyme A-linked aldehyde dehydrogenase from Escherichia coli. I. Partial purification, properties, and kinetic studies of the enzyme. J. Biol. Chem. 243:5539–5545 [PubMed] [Google Scholar]
- 43. Samsonova NN, Smirnov SV, Altman IB, Ptitsyn LR. 2003. Molecular cloning and characterization of Escherichia coli K-12 ygjG gene. BMC Microbiol. 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Samsonova NN, Smirnov SV, Novikova AE, Ptitsyn LR. 2005. Identification of Escherichia coli K-12 YdcW protein as a γ-aminobutyraldehyde dehydrogenase. FEBS Lett. 579:4107–4112 [DOI] [PubMed] [Google Scholar]
- 45. Schneider BL, Kiupakis AK, Reitzer LJ. 1998. Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli. J. Bacteriol. 180:4278–4286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schneider BL, et al. 2002. The Escherichia coli gabDTPC operon: specific γ-aminobutyrate catabolism and nonspecific induction. J. Bacteriol. 184:6976–6986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sebela M, et al. 2000. Characterisation of a homogeneous plant aminoaldehyde dehydrogenase. Biochim. Biophys. Acta 1480:329–341 [DOI] [PubMed] [Google Scholar]
- 48. Shaibe E, Metzer E, Halpern YS. 1985. Metabolic pathway for the utilization of l-arginine, l-ornithine, agmatine, and putrescine as nitrogen sources in Escherichia coli K-12. J. Bacteriol. 163:933–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tkachenko A, Nesterova L, Pshenichnov M. 2001. The role of the natural polyamine putrescine in defense against oxidative stress in Escherichia coli. Arch. Microbiol. 176:155–157 [DOI] [PubMed] [Google Scholar]
- 50. Tkachenko AG, Pshenichnov MR, Salakhetdinova O, Nesterova L. 1999. The role of putrescine and the energy state of Escherichia coli in regulation of DNA topology during adaptation to oxidative stress. Mikrobiologiia 68:27–32 [PubMed] [Google Scholar]
- 51. Tweeddale H, Notley-McRobb L, Ferenci T. 1998. Effect of slow growth on metabolism of Escherichia coli, as revealed by global metabolite pool (“metabolome”) analysis. J. Bacteriol. 180:5109–5116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yoshida M, et al. 2004. A unifying model for the role of polyamines in bacterial cell growth, the polyamine modulon. J. Biol. Chem. 279:46008–46013 [DOI] [PubMed] [Google Scholar]
- 53. Yoshida M, Meksuriyen D, Kashiwagi K, Kawai G, Igarashi K. 1999. Polyamine stimulation of the synthesis of oligopeptide-binding protein (OppA). Involvement of a structural change of the Shine-Dalgarno sequence and the initiation codon aug in oppA mRNA. J. Biol. Chem. 274:22723–22728 [DOI] [PubMed] [Google Scholar]
- 54. Zimmer DP, et al. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. U. S. A. 97:14674–14679 [DOI] [PMC free article] [PubMed] [Google Scholar]