Abstract
Preprolipopoprotein diacylglyceryl transferase (Lgt) is the gating enzyme of lipoprotein biosynthesis, and it attaches a lipid structure to the N-terminal part of preprolipoproteins. Using Lgt from Escherichia coli in a BLASTp search, we identified the corresponding Lgt homologue in Mycobacterium tuberculosis and two homologous (MSMEG_3222 and MSMEG_5408) Lgt in Mycobacterium smegmatis. M. tuberculosis lgt was shown to be essential, but an M. smegmatis ΔMSMEG_3222 mutant could be generated. Using Triton X-114 phase separation and [14C]palmitic acid incorporation, we demonstrate that MSMEG_3222 is the major Lgt in M. smegmatis. Recombinant M. tuberculosis lipoproteins Mpt83 and LppX are shown to be localized in the cell envelope of parental M. smegmatis but were absent from the cell membrane and cell wall in the M. smegmatis ΔMSMEG_3222 strain. In a proteomic study, 106 proteins were identified and quantified in the secretome of wild-type M. smegmatis, including 20 lipoproteins. All lipoproteins were secreted at higher levels in the ΔMSMEG_3222 mutant. We identify the major Lgt in M. smegmatis, show that lipoproteins lacking the lipid anchor are secreted into the culture filtrate, and demonstrate that M. tuberculosis lgt is essential and thus a validated drug target.
INTRODUCTION
Mycobacteria belong to the group of GC-rich actinobacteria among the Gram-positive bacteria. Comprising more than 130 species, the genus Mycobacterium is rather diverse. Members of this genus are, among others, the slow-growing, pathogenic Mycobacterium tuberculosis, the causative agent of tuberculosis, Mycobacterium bovis bacillus Calmette-Guérin, the live attenuated vaccine applied to protect against tuberculosis, Mycobacterium leprae, the causative agent of leprosy, and the fast-growing Mycobacterium smegmatis, a nonpathogenic, saprophytic mycobacterial model organism. Although classified as Gram-positive bacteria, the cellular envelope of mycobacteria resembles the cell envelope of Gram-negative bacteria, having an outer membrane-like structure (15). Mycobacteria interact with their environment by secreted and surface-localized proteins. Lipoproteins are a heterogeneous subgroup of membrane-associated proteins universally present in bacteria. One to 3% of bacterial genomes encode lipoproteins (2, 16). The common feature of lipoproteins is a universally conserved N-terminal cysteine modified with a lipid structure functioning as a membrane anchor. Synthesized as precursors in the cytoplasm, lipoproteins are translocated across the cytoplasmic membrane by either the Sec translocation machinery or the twin-arginine translocation (Tat) system (21, 28, 42, 48). Lipoprotein maturation subsequently occurs on the periplasmic side of the cytoplasmic membrane by the consecutive action of the three enzymes Lgt (preprolipoprotein diacylglyceryl transferase), LspA (prolipoprotein signal peptidase), and Lnt (apolipoprotein N-acyltransferase). These posttranslational modifications are directed by a lipobox motif comprising four amino acids, including the invariant cysteine (LVI)(ASTVI)(GAS)C (2). As a first step, Lgt attaches a diacylglycerol residue to the thiol group of the universally conserved cysteine within the lipobox. Second, LspA cleaves off the signal peptide N-terminal of the modified cysteine, followed by the attachment of a third acyl residue to the free amino group of the cysteine mediated by Lnt (22). The membrane anchor of mycobacterial lipoproteins has been resolved at the molecular level recently (45). Mycobacterial lipoproteins are modified with a thioether-linked diacylglyceryl residue composed of an ester-linked tuberculostearic and an ester-linked palmitic acid as well as an additional palmitic acid amide linked to the N-terminal α-amino group. Diacylglycerol modification and the signal sequence cleavage are prerequisites for N-acylation (5, 45, 48).
The functional diversity of lipoproteins is manifold; among others, they have direct virulence-related functions, such as invasion of host cells, evasion from host defense, and immunomodulation in Gram-positive and Gram-negative bacterial pathogens (18). All three enzymes of the lipoprotein biosynthesis pathway are essential in Gram-negative but not in Gram-positive bacteria. In M. tuberculosis, targeted deletion of lspA demonstrated a role of the lipoprotein biosynthesis pathway in pathogenesis. An M. tuberculosis lspA mutant is unable to cleave the signal peptide of lipoproteins, and this was associated with a 3- to 4-log-reduced number of CFU in an animal model of tuberculosis. Additionally, this strain induced hardly any lung pathology and did not spread to the secondary organs spleen and liver (27, 33).
Lipoproteins (from different bacteria, including mycobacteria) are potent agonists of Toll-like receptor 2 (TLR2). TLR2 agonist activity has been shown for several M. tuberculosis lipoproteins, including LpqH, LprA, LprG, and PstS1 (10). Successful immune evasion of M. tuberculosis has been partly attributed to TLR2-dependent inhibition of antigen processing and presentation (10, 12). Although TLR signaling enhances both innate and adaptive immune responses, it can also downregulate some immune functions.
Virulence assays indicated an important role of the second enzyme (LspA) of the lipoprotein biogenesis in the pathogenesis of tuberculosis, and functional investigations elucidated that the mycobacterial lipoprotein anchor carries three fatty acids and thus is similar to the membrane anchor of Gram-negative bacteria. However, the physiological role of mycobacterial Lgt, the gating enzyme of lipoprotein biosynthesis, remains to be demonstrated. Of note, a high-density mutagenesis study suggested that M. tuberculosis Lgt is essential (36). There is an urgent need for novel drugs and verification of drug targets, since the antituberculosis drug pipeline is not sufficiently filled and more and more drug-resistant M. tuberculosis strains emerge (31). Essential genes, particularly those which are restricted to bacteria, encode drug targets that have great potential. Therefore, we here investigated the prolipoprotein diacylglyceryl transferase in mycobacteria.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
M. smegmatis was grown on LB (Luria-Bertani) agar or on Middlebrook 7H10 agar supplemented with oleic acid albumin dextrose (OADC; Difco). M. tuberculosis was grown on Middlebrook 7H10 agar supplemented with OADC. Tween 80 (0.05%, vol/vol) was added to liquid broth LB, 7H9, and 7H9-OADC to avoid clumping. When appropriate, antibiotics were added at the following concentrations: kanamycin, 50 μg ml−1; streptomycin, 100 μg ml−1; hygromycin, 25 μg ml−1; and gentamicin, 10 μg ml−1. Strain designations were the following: Δlgt, lgt knockout mutant; Δlgt-lgt, M. smegmatis Δlgt transformed with complementing vector pMV361-hyg-lgt expressing M. tuberculosis lgt; Δlgt-MSMEG_3222, M. smegmatis Δlgt transformed with complementing vector pMV361-hyg-MSMEG_3222 expressing M. smegmatis lgt.
Disruption of lgt in M. smegmatis.
For disruption of M. smegmatis lgt (MSMEG_3222), a 1,330-bp fragment upstream and a 1,415-bp fragment downstream of the predicted ORF were amplified by PCR. XbaI/EcoRI linker sequences were added to the upstream fragment and EcoRI/MluI linker sequences were added to the downstream fragment to facilitate oriented cloning. The resulting fragments were cloned into pMCS5-rpsL, resulting in pMCS5-rpsL-lgt. A fragment containing the aph cassette was cloned into the EcoRI site between the upstream and downstream fragments, resulting in plasmid pMCS5-rpsL-lgt::aph. Using the rpsL counterselection strategy (32), the Δlgt allele was substituted for lgt in M. smegmatis, deleting 959 bp from the open reading frame (ORF) coding for Lgt. Substitution was confirmed by Southern blot analysis by probing with a 967-bp ApaI lgt gene fragment. For complementation, an 8,024-bp SfiI/PvuII fragment of the M. tuberculosis chromosome, encompassing the complete lgt (Rv1614) under the control of its own promoter, was cloned in pMV361-hyg, and the resulting plasmid was transformed into Δlgt. A corresponding complementation vector, carrying a 2.5-kbp M. smegmatis lgt fragment, was also constructed and transformed. A strategy similar to that for generating an M. smegmatis deletion mutant was applied to generate corresponding M. tuberculosis mutants.
Whole-genome sequencing, data analysis, and single-nucleotide polymorphism (SNP) confirmation.
Genomic DNA of M. smegmatis Smr5, a streptomycin-resistant derivative of M. smegmatis mc2155 (37) whose sequence has been published, and M. smegmatis Δlgt was prepared as follows. Bacteria were grown for 2 to 3 days on plates. Bacteria were resuspended in 340 μl Tris-EDTA (TE) buffer and heat inactivated for 20 min at 80°C. After cooling down to room temperature, 2 μl 20% Tween 80 and 10 μl lysozyme (80 mg ml−1; Roche) were added, followed by incubation for 2 h at 37°C. After addition of 20 μl 20% SDS and 20 μl proteinase K (2 mg ml−1; Roche), samples were incubated for 1 h at 50°C. Four hundred μl phenol-chloroform-isoamylalcohol (25:24:1, vol/vol) was added, and samples were shaken for 1 h. Subsequently, samples were centrifuged (16,000 × g for 20 min at 4°C), and the supernatant was transferred into a fresh 1.5-ml tube. Eight μl 5 M NaCl and 2.5 volumes (1 ml) of ethanol were added, and the mixtures were incubated overnight at −20°C. After centrifugation of the samples at 16,000 × g for 20 min at 4°C, the pellet was washed twice with 70% ethanol, dried under vacuum, and resuspended in 100 to 300 μl water.
The strains were sequenced using the Illumina Genetic Analyzer (Illumina, Saffron Walden, United Kingdom) to produce paired-end fragment reads of 35 bp. Sequencing was performed at GATC Biotech Ltd. (Constance, Germany). Reads of both strains were mapped against M. smegmatis mc2155 (37) (NC_008596) using CLC Genomics Workbench 4.8 (CLCbio). SNP detection tool (CLCbio) parameters were set as the following: window length, 11; maximum number of gaps and mismatches, 2; minimum average quality of surrounding bases, 15; minimum quality of central base, 20; minimum coverage, 4; and minimum variant frequency, 35%. SNP confirmation was performed by Sanger sequencing with an ABI Prism 310 Genetic Analyzer (Applied Biosystems).
Microscopy.
For electron microscopy, bacteria were centrifuged and fixed for 30 min at room temperature in 3% paraformaldehyde—0.1% glutaraldehyde. The cells were washed in phosphate-buffered saline (PBS) and postfixed for 30 min in 2% OsO4 before dehydration in ethanol and embedding in Epon. Thin sections were stained with uranyl acetate and lead citrate and examined in a Phillips CM12 electron microscope. Standard laboratory techniques were used for Ziehl-Neelsen and auramineO/rhodamine staining. For the study of microcolonies, bacteria were grown for 3 days on 7H10 agar plates.
Cloning of Mpt83 and LppX.
Plasmid pMV261-Gm, a derivative of pMV261, is a shuttle vector replicating in E. coli as well as in mycobacteria (39). M. tuberculosis LppX and Mpt83 were amplified by PCR from genomic DNA and fused to the M. tuberculosis 19-kDa (lpqH) promoter. Two sequences encoding a hemagglutinin and a hexa-His epitope were fused to the 3′ part of the genes to facilitate subsequent detection by Western blotting. The insert was cloned into the EcoRI site, resulting in pMV261-Gm-FusLppX and pMV261-Gm-FusMpt83, respectively.
Western blotting.
Bacteria from liquid cultures were harvested, resuspended in PBS containing Complete EDTA-free tablets (Roche) to inhibit protein degradation, and subjected to 15 to 30 min of ultrasonication in an ice bath. Soluble and insoluble fractions were separated by centrifugation at 15,000 × g for 20 min at 4°C. Extracts corresponding to 1 to 5 μg of total protein were separated by SDS-PAGE (12%) and analyzed by Western blotting. Antiserum against hemagglutinin (HA) epitope (Roche) and LprG (provided by H. Bercovier) were used in dilutions of 1:300 or 1:10,000, respectively. Appropriate secondary antibodies conjugated with horseradish peroxidase were used at dilutions of 1:10,000. The blots were developed using enhanced chemiluminescence (Bio-Rad).
[14C]palmitic acid incorporation.
Lipoprotein labeling was performed as described previously (38, 50).
Subcellular fractionation of cells.
Subcellular fractionation was performed as described in reference 29. Briefly, cells of 1-liter cultures were harvested, washed in 0.16 M NaCl, and resuspended in lysis buffer (0.05 M potassium phosphate, 0.022% [vol/vol] β-mercaptoethanol, pH 6.5). Cell lysis was performed by a French press (American Instrument Company) followed by low-speed centrifugation at 1,000 × g to remove unbroken cells. Centrifugation was repeated 3 to 5 times for 40 min at 27,000 × g to pellet outer lipid layer material. The supernatant was called SN27.1. The pellet was resuspended in lysis buffer and disrupted by a French press. Subsequent centrifugation at 27,000 × g resulted in a pellet enriched for cell wall components (mycolic acids) and supernatant SN27.2. The supernatants SN27.1 and SN27.2 were pooled and centrifuged at 100,000 × g for 1 h. The resulting pellet was enriched in cytoplasmic membrane and the supernatant in cytosolic components.
Preparation of the extracellular protein fraction.
M. smegmatis cells were grown in 1 liter of LB broth (Difco), and 500 ml cells was harvested during exponential growth (optical density at 600 nm of 0.4 to 0.7) at the transient phase (t0), as well as after the entry into the stationary phase (t1), by centrifugation for 10 min at 4°C (1 mM phenylmethylsulfonyl fluoride [PMSF] was added immediately after harvesting). The extracellular proteins of the supernatant were precipitated with ice-cold 10% (wt/vol) trichloroacetic acid (TCA) overnight on ice and centrifuged for 45 min at 13,500 × g and 4°C. The resulting protein pellet was scraped with a spatula from the wall of the centrifuge tube, washed with 96% (vol/vol) ethanol 5 times, and dried.
Extracellular proteome analysis and image analysis.
The TCA-precipitated extracellular proteins of the M. smegmatis wild type and lgt mutant were washed extensively with ethanol, dried in a speed vacuum, and resolved in a solution containing 2 M thiourea and 8 M urea. Insoluble material was removed by centrifugation. The protein content was determined using the Bradford assay (4). For two-dimensional polyacrylamide gel electrophoresis (2D PAGE), 200 μg of the protein extracts was separated using the nonlinear immobilized pH gradients (IPG; pH range, 4 to 7; Amersham Biosciences) and a Multiphor II apparatus (Amersham Pharmacia Biotech) as described previously (1). The resulting 2D gels were fixed in 40% (vol/vol) ethanol, 10% (vol/vol) acetic acid and stained with colloidal Coomassie brilliant blue (Amersham Biosciences). The image analysis and quantification were performed with Decodon Delta 2D software.
Identification of proteins in the secretome using MALDI-TOF-TOF tandem mass spectrometry (MS/MS).
Proteins were cut manually from the Coomassie-stained proteome and tryptically in-gel digested using the Ettan spot handling platform as described previously (7). The matrix-assisted laser desorption ionization—tandem time-of-flight (MALDI-TOF-TOF) measurement of spotted peptide solutions was carried out on a Proteome-Analyzer 4800 (Applied Biosystems, Foster City, CA) as described previously (7). The spectra were recorded in reflector mode in a mass range from 900 to 3,700 Da with a focus mass of 2,000 Da. For one main spectrum, 25 subspectra with 100 shots per subspectrum were accumulated using a random search pattern. If the autolytic fragment of trypsin with the monoisotopic (M+H)+ m/z at 2,211.104 reached a signal-to-noise ratio (S/N) of at least 10, an internal calibration was automatically performed using this peak for one-point calibration. The peptide search tolerance was 50 ppm, but the actual RMS value was between 10 and 20 ppm. After calibration, the peak lists were created by using the “peak to mascot” script of the GPS Explorer software, v.3.6, with the following settings: mass range from 900 to 3,700 Da, peak density of 50 peaks per range of 200 Da, minimal area of 100, maximal 200 peaks per protein spot, and minimal S/N ratio of 6. The peak lists were searched against an M. smegmatis database extracted from UniprotKB release 12.7 (46) using the Mascot search engine, v.2.1.04 (Matrix Science Ltd., London, United Kingdom).
MALDI-TOF-TOF MS/MS analysis was performed for the three strongest peaks of the TOF spectrum. For one main spectrum, 20 subspectra with 125 shots per subspectrum were accumulated using a random search pattern. The internal calibration was automatically performed as one-point calibration if the monoisotopic arginine (M+H)+ m/z at 175.119 or lysine (M+H)+ m/z at 147.107 reached an S/N ratio of at least 5. The peak lists were created by using the “peak to mascot” script of the GPS Explorer software, v.3.6, with the following settings: mass range from 60 Da to a mass that was 20 Da lower than the precursor mass, peak density of 5 peaks per 200 Da, minimal area of 100, maximal 20 peaks per precursor, and a minimal S/N ratio of 5. Peptide mixtures that yielded a molecular weight search (MOWSE) score of at least 50 in the reflector mode and a sequence coverage of at least 30% that were confirmed by subsequent MS/MS analysis were regarded as positive identification.
RESULTS
Generation of mycobacterial lgt deletion mutants.
Using E. coli Lgt as a query in a BLASTp search, we identified Rv1614, annotated as Lgt, in M. tuberculosis as a preprolipoprotein diacylglyceryl transferase. We applied the rpsL counterselection strategy to generate an M. tuberculosis lgt deletion mutant. rpsL has been shown to be a powerful tool to generate deletion mutants (32, 33). The vector-carried wild-type rpsL confers a streptomycin-sensitive phenotype in a streptomycin-resistant rpsL mutant strain due to the dominance of the wild-type allele. Transformation of a streptomycin-resistant M. tuberculosis H37Rv derivative with a suicide plasmid for targeted deletion of lgt resulted in single-crossover recombinants which had integrated the suicide plasmid at the lgt locus. Upon counterselection, an lgt deletion mutant could not be generated. Only spontaneously streptomycin-resistant single-crossover mutants, which had not undergone second recombination, were obtained. However, the construction of an M. tuberculosis lgt deletion mutant was successful in an lgt-complemented strain, indicating that replacement of lgt is feasible in principle but that a functional copy elsewhere is required. The data from our targeted gene deletion confirm the findings of Sassetti et al. predicting lgt to be essential for growth based on the high-density mutagenesis of M. tuberculosis (36). We previously succeeded in generating an lspA as well as an lnt deletion mutant in M. smegmatis (45). Therefore, we used this nonpathogenic mycobacterial model organism, which is fast growing and amenable to genetic manipulation, to investigate the function of lgt.
Using E. coli Lgt as a query in a BLASTp search, we identified two putative paralogous open reading frames, i.e., MSMEG_5408 and MSMEG_3222. We used pairwise sequence alignment with a Needleman-Wunsch algorithm (http://www.ebi.ac.uk/Tools/psa/emboss_needle/) with default settings to compare both M. smegmatis ORFs to M. tuberculosis, E. coli, and Bacillus subtilis Lgt sequences (see Table S1 in the supplemental material). MSMEG_5408 shows higher percentages of identities/similarities to E. coli or B. subtilis Lgt than does MSMEG_3222. Of note, MSMEG_3222 has an extended C-terminal sequence which is missing from E. coli or B. subtilis Lgt. When both M. smegmatis ORFs were compared to M. tuberculosis Lgt, MSMEG_3222 shows the highest identities/similarities (see Table S1). A recent study from Pailler et al. identified residues essential and important for the function of E. coli Lgt (24). We also used pairwise sequence alignment with the Needleman-Wunsch algorithm (http://www.ebi.ac.uk/Tools/psa/emboss_needle/) with default settings to analyze the conservation of these residues in MSMEG_3222 and MSMEG_5408 (see Table S2). MSMEG_3222 showed conservation of all four essential residues, the Lgt signature motif, and 10 of 17 important residues. Seven of 17 important residues were different from E. coli Lgt, but only four residues were different from M. tuberculosis Lgt. In contrast, comparison of MSMEG_5408 and E. coli Lgt revealed two essential residues which were not conserved in MSMEG_5408, i.e., Y26 and N146 (E. coli numbering). At position Y26 a histidine was found, and at position N146, located in the Lgt signature motif, a cysteine was found in MSMEG_5408. Moreover, six important residues were not conserved in MSMEG_5408 in addition to alteration in two essential residues. Because of the conservation of all residues essential for Lgt function and higher homology to M. tuberculosis Lgt, we focused on MSMEG_3222 for further characterization. MSMEG_3222 localizes at positions 3,299,959 to 3,301,809 in the genome sequence of M. smegmatis that is available from NCBI (http://www.ncbi.nlm.nih.gov/genome/?term=NC_008596). The ORF encodes a protein of 616 amino acids. The six conserved domains observed in Lgt (26) are all present in MSMEG_3222. Following transformation with the suicide plasmid pMCS5-rpsL-lgt::aph, single-crossover recombinants were isolated and subjected to counterselection. Intramolecular homologous recombination occurs with a frequency of about 10−4 in mycobacteria (25). Counterselection of the MSMEG_3222 single-crossover recombinants was much less frequent (<10−8). However, a single-mutant strain resulting from allelic exchange was found after counterselection and is referred to as M. smegmatis Δlgt. Replacement of lgt with a kanamycin resistance marker was verified by Southern blot analysis and PCR (Fig. 1). The mutant strain was complemented by introducing plasmid pMV361-hyg-lgt expressing M. tuberculosis lgt, resulting in strain M. smegmatis Δlgt-lgt. In addition, the mutant strain was complemented with a corresponding vector carrying M. smegmatis lgt (MSMEG_3222) (data not shown).
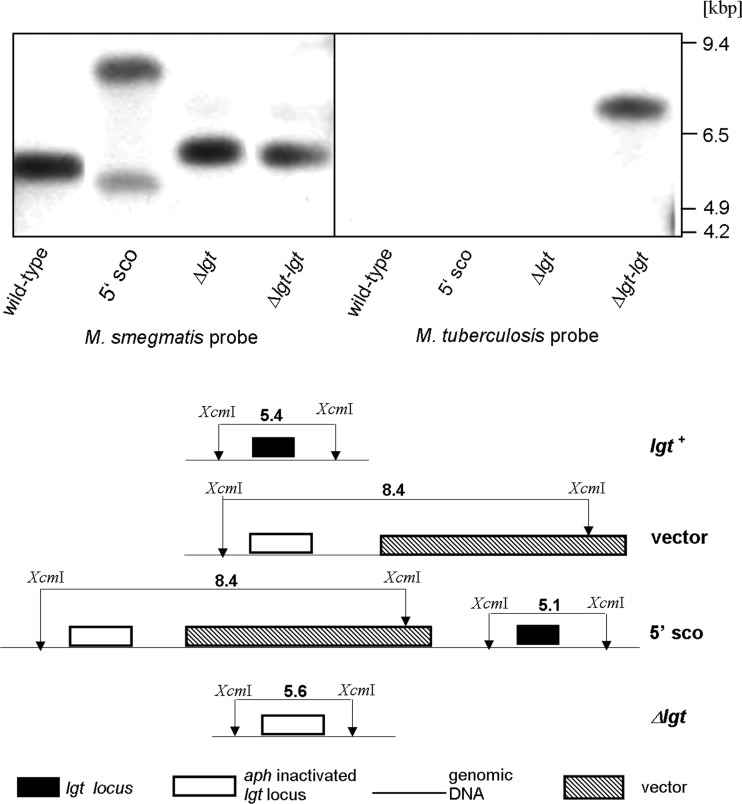
Fig 1.
Disruption of M. smegmatis lgt (MSMEG_3222). (Left) Genomic DNAs from M. smegmatis (lane 1), lgt single-crossover (5′sco) mutant (lane 2), Δlgt mutant (lane 3), and Δlgt-lgtMtb mutant (lane 4; lgtMtb indicates lgt from M. tuberculosis) were digested with XcmI and probed with a 967-bp ApaI lgt gene fragment. The presence of a single 5.6-kbp fragment in the Δlgt knockout strain compared to the single 5.4-kbp fragment in the parental strain demonstrates inactivation of lgt. The shift in fragment size in the Δlgt knockout strain results from replacement of a 959-bp lgt fragment with a 1.2-kbp kanamycin resistance cassette. (Right) Genomic DNAs of M. smegmatis (lane 5), lgt-5′ single-crossover mutant (lane 6), Δlgt mutant (lane 7), and Δlgt-lgtMtb mutant (lane 8) were digested with BamHI and probed with a 491-bp SacI M. tuberculosis lgt gene fragment to demonstrate complementation. Complementation is indicated by a hybridization signal with genomic DNA derived from strain Δlgt-lgtMtb (the M. tuberculosis lgt probe does not hybridize with M. smegmatis lgt).
Whole-genome comparison of M. smegmatis Smr5 and Δlgt.
Isolation of a single mutant only and the apparent discrepancy with respect to the essentiality of lgt in M. tuberculosis H37Rv and M. smegmatis Smr5 prompted us to compare the whole genome of parental M. smegmatis Smr5 and M. smegmatis Δlgt by using Illumina sequencing technology. We reasoned that suppressor mutations in the knockout strain might explain the success of lgt deletion in M. smegmatis. The genome of M. smegmatis mc2155 (37) contains 6,988,209 nucleotides (90% coding) with a GC content of 67%. It comprises 6,938 genes, with 6,717 encoding proteins (http://www.ncbi.nlm.nih.gov/genome?term=nc_008596). M. smegmatis Smr5, the parental strain of the Δlgt mutant, is a direct derivative of strain mc2155 and is therefore assumed to differ from strain mc2155 at least by an rpsL mutation that renders the strain streptomycin resistant (32). The average sequence coverage was 122.90-fold (12.8 million paired-end reads) for M. smegmatis Smr5 and 71.84-fold (7.4 million paired-end reads) for M. smegmatis Δlgt. Using M. smegmatis mc2155 (NC_008596) as a reference, the genomes were mapped with the CLC Genomics Workbench. Results indicated that 99.9266% (Smr5) and 99.3704% (Δlgt) of the genomes were covered with at least one read. Neither insertions nor deletions were detected in the Smr5 wild type and the Δlgt mutant, except for the genetically engineered lgt deletion in the Δlgt mutant. Parental Smr5 and Δlgt had 35 single-nucleotide polymorphisms in common that were not found in the reference strain mc2155. One of these SNPs was the rpsL mutation conferring streptomycin resistance. The other putative SNP found in both strains located to GC-rich regions with low coverage. One SNP, a missense mutation, was unique to the Δlgt mutant. The SNP was identified in the ORF MSMEG_3278, a gene with unknown function. An ORF (MSMEG_3280) encoding a lipoprotein is located in that region. Sanger sequencing confirmed this SNP.
In vitro growth characteristics of M. smegmatis Δlgt.
The in vitro growth characteristics of parental M. smegmatis, Δlgt mutant, and complemented strain expressing M. tuberculosis lgt were investigated. Growth retardation of the Δlgt mutant strain was observed in liquid Middlebrook-Tween broth (7H9) supplemented with OADC. Generation times of the strains at maximum growth rate were the following: wild-type, 3 h 20 min; Δlgt, 5 h 10 min; and Δlgt-lgt, 3 h 50 min. A 1.5-fold growth retardation of the Δlgt mutant was also observed in nutrient-poor medium, i.e., in liquid Middlebrook-7H9-Tween broth without OADC (Fig. 2A). Generation times were 4 h 17 min for the wild type, 6 h 30 min for Δlgt, and 4 h 40 min for Δlgt-lgt.
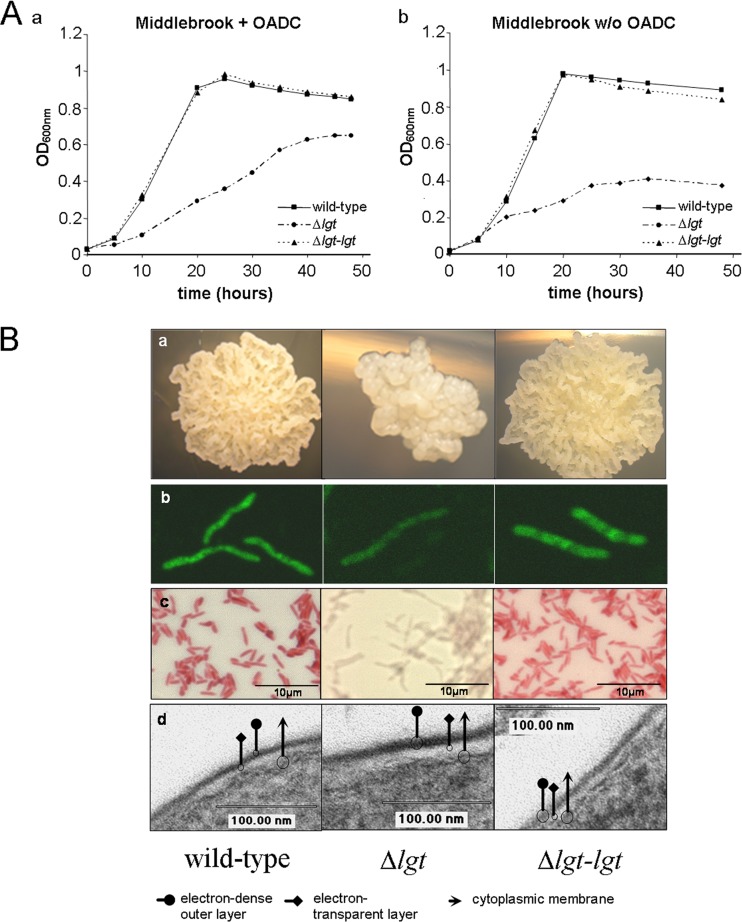
Fig 2.
Growth characteristics of M. smegmatis Δlgt. (A) M. smegmatis wild type, M. smegmatis Δlgt, and M. smegmatis Δlgt-lgtMtb were grown in rich medium (a) or in nutrient-poor medium (b). (B) Inactivation of lgt affects cell and colony morphology. (a) Microcolonies of M. smegmatis grown on 7H10 agar supplemented with OADC for 3 days (total magnification, ×10); (b) auramine-stained M. smegmatis grown in 7H9-Tween (total magnification × 2000); (c) Ziehl-Neelsen-stained M. smegmatis grown in 7H9-Tween (total magnification, ×1,000); (d) electron microscopic photographs of M. smegmatis (total magnification, ×200,000). Column 1, parental M. smegmatis; column 2, Δlgt mutant; column 3, Δlgt-lgtMtb complemented strain; arrow, cytoplasmic membrane; square, electron-translucent layer; circle, electron-dense outer layer.
On 7H10 agar, microcolonies of M. smegmatis typically showed rough, dry, and irregular surfaces. In contrast, colonies of the Δlgt mutant were smaller and less ruffled (Fig. 2B). In contrast to the parental strain, M. smegmatis Δlgt was barely stained by auramine O/rhodamine and Ziehl-Neelsen, respectively (Fig. 2B). These morphological alterations had no correlation at the ultrastructural level as revealed by electron microscopy. Both the wild-type and Δlgt strains showed a multilayered cell envelope typical for mycobacteria, i.e., the plasma membrane and the electron-dense, presumably peptidoglycan layer are covered by an electron-transparent layer and an irregular electron-dense outer layer (Fig. 2B). Wild-type-like growth on solid agar and staining with auramine O/rhodamine and Ziehl-Neelsen was restored by introducing a wild-type copy of M. tuberculosis lgt, demonstrating that morphological and staining alterations in the mutant are due to the deletion of lgt and that M. tuberculosis lgt is functional in M. smegmatis. Growth rate and colony morphology of the Δlgt mutant were also restored when complemented with MSMEG_3222 (see Fig. S2 in the supplemental material).
Impact of Lgt depletion on mycobacterial lipoproteins.
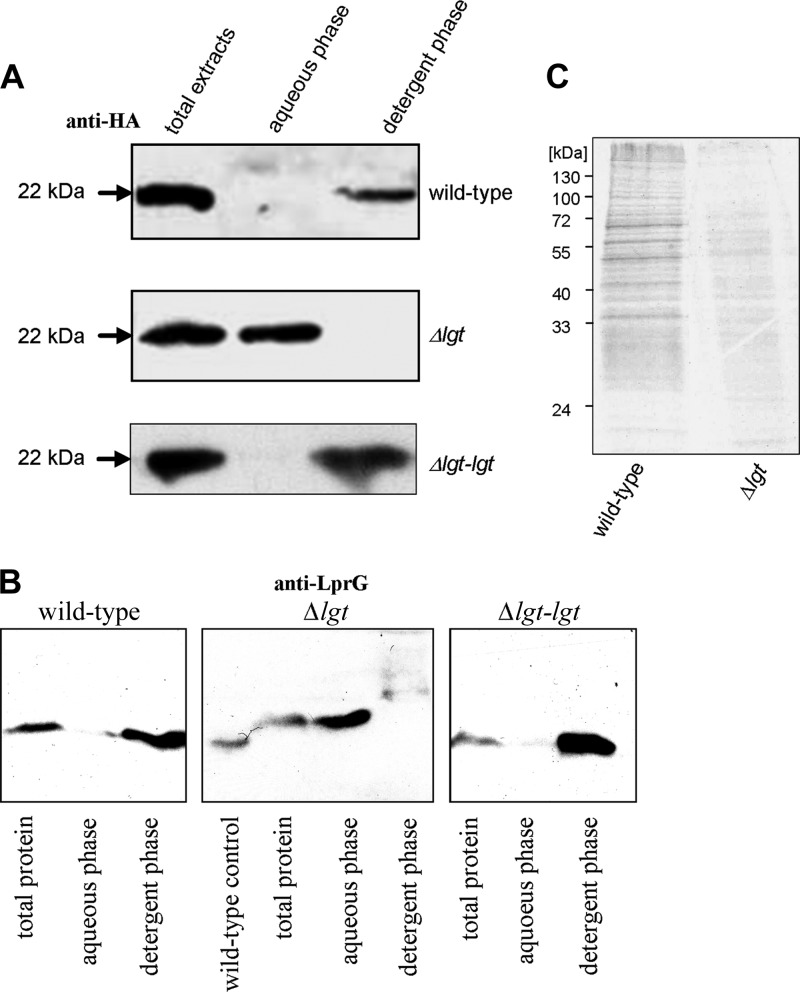
To verify that MSMEG_3222 is the gating enzyme of lipoprotein biosynthesis, we performed Triton-X114 extraction using Mpt83 and LppX, two well-characterized M. tuberculosis lipoproteins, as reporters to monitor the fate of representative lipoproteins. Phase partition of protein extracts with the detergent Triton-X114 leads to the accumulation of lipophilic proteins in the detergent phase, whereas hydrophilic proteins accumulate in the aqueous phase. The diacylglyceryl residue, composed of a glyceryl and two ester-bound fatty acids, renders hydrophilic proteins lipophilic. Mpt83 and LppX, extracted from the parental M. smegmatis, accumulated in the detergent phase, whereas both lipoproteins extracted from the Δlgt mutant accumulated in the aqueous phase, suggesting the absence of the lipid anchor from the Δlgt mutant. Complementation with M. tuberculosis lgt reverted the phenotype of the Δlgt mutant (Fig. 3A and B). To corroborate these findings obtained with representative lipoproteins, metabolic labeling experiments with [14C]palmitic acid were performed for a broader set of proteins. The same amount of total protein from the parental strain and Δlgt mutant was loaded on SDS gels. Autoradiography of protein extracts separated by SDS-PAGE revealed multiple 14C-labeled proteins in the wild-type extract. In contrast, the Δlgt mutant showed almost no incorporation of [14C]palmitic acid (Fig. 3C). The accumulation of Mpt83 and LppX from the Δlgt mutant in the aqueous phase indicates a lack of the diacylglyceryl anchor. The almost-abolished [14C]palmitic acid incorporation supports this indication. This demonstrates that MSMEG_3222 encodes an M. smegmatis prolipoprotein diacylglyceryl transferase Lgt.
Fig 3.
M. smegmatis Δlgt mutant fails to attach the lipid anchor. (A) Triton X-114 phase partition of M. smegmatis strains expressing recombinant lipoprotein Mpt83. (B) Triton X-114 phase partition of M. smegmatis parental strain, Δlgt mutant, and Δlgt-lgtMtb complemented strain expressing recombinant LppX using anti-LprG antibody. Of note, anti-LprG antibody (provided by H. Bercovier) cross-reacts with LppX (see Fig. S1 in the supplemental material). (C) [14C]palmitic acid incorporation in the wild type and Δlgt mutant.
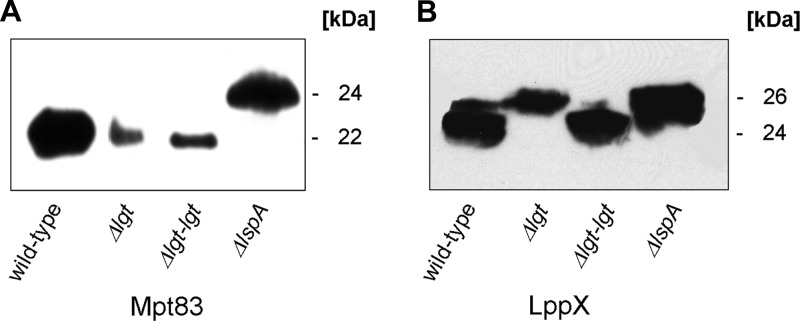
Lgt is the gating enzyme for lipoprotein biosynthesis. It catalyzes the first step in the biosynthesis of lipoproteins by forming a thioether linkage between a diacylglyceryl residue and the sulfhydryl group of the +1 cysteine, thereby attaching the first building block of the membrane anchor. It is assumed that afterwards, in a sequential manner, lipoprotein-specific signal peptidase A (LspA) then cleaves off the signal sequence, which in turn is a prerequisite for N-acylation. The lack of the diacylglyceryl modification should abolish recognition of immature lipoproteins by LspA and subsequent modifications. Western blot analyses with protein extracts from the M. smegmatis parental strain, lgt mutant, and lgt mutant complemented with the M. tuberculosis lgt (Rv1614) homologue were performed to investigate the processing of lipoproteins in the M. smegmatis Δlgt mutant. An M. smegmatis lspA mutant served as a control (45). Temperature-sensitive lspA mutants of E. coli and lspA knockout mutants of Gram-positive bacteria accumulate diglyceride prolipoprotein, since conversion of prolipoprotein to apolipoprotein is inhibited due to the inability to cleave the signal sequence from the diglyceride prolipoprotein (35, 47). Mpt83 expressed in the lgt mutant showed the same apparent molecular mass as Mpt83 from the parental strain, as demonstrated by Western blot analysis (Fig. 4A). This indicates Lgt-independent cleavage of the signal sequence of Mpt83. In contrast, an increased molecular mass of Mpt83 derived from the lspA mutant was observed. The difference in size of about 2 to 3 kDa corresponds to the mass of the signal sequence. The absence of smaller forms of Mpt83 in the lspA mutant indicates that Mpt83 is not processed by alternative signal peptidases when lgt is functional. Lgt-independent LspA cleavage has been shown for a limited number of lipoproteins in Listeria monocytogenes (3). Therefore, we also analyzed the M. smegmatis parental strain, Δlgt mutant, and complemented strain expressing a second heterologous lipoprotein, namely, M. tuberculosis LppX. Western blot analysis of whole-cell extracts revealed an increased molecular mass of recombinant LppX of about 2 kDa in the lgt mutant compared to wild-type cells (Fig. 4B). The increased molecular size corresponds to the size of the N-terminal signal sequence of LppX. The same increase in size was also observed in the lspA mutant. LppX from the complemented lgt mutant showed the same molecular mass as that from the wild type. The LppX signal peptide was also cleaved when Δlgt mutant was complemented with MSMEG_3222 (see Fig. S2 in the supplemental material). This demonstrates that accumulation of pro-LppX in the Δlgt mutant is related to Lgt depletion. The accumulation of prolipoproteins in the lspA mutant shows that the cloned proteins are recognized as lipoproteins. The accumulation of the prolipoprotein form of LppX in the lgt mutant clearly demonstrates that the lack of Lgt abolishes the signal sequence cleavage for LppX but not for Mpt83.
Fig 4.
Lipoproteins without a lipid anchor are not cleaved by LspA. Western blot analysis of whole-cell extracts of M. smegmatis strains expressing Mpt83 (A) or LppX (B). The signal peptide of lipoproteins Mpt83 and LppX accounts for approximately 2 kDa. The signal peptide is not cleaved in the lgt mutant in the case of LppX. The signal peptide of Mpt83 is cleaved in the Δlgt mutant, indicating Lgt-independent signal sequence cleavage. Western blot analyses were performed using anti-HA antibody and corresponding secondary antibody conjugated with horseradish peroxidase.
Taken together, Triton X-114, [14C]palmitic acid labeling, and Western blot analyses strongly indicate that lipoproteins derived from the Δlgt mutant lack the diacylglyceryl residue. The lack of the lipid anchor results in a failure of LspA-mediated signal sequence cleavage for some lipoproteins. In contrast, the signal sequence of Mpt83 derived from the lgt mutant is cleaved off. Lipoproteins can be released into the culture filtrate by shedding or signal peptide processing (shaving). In a secretome analysis of B. subtilis, seven lipoproteins were found to be shed into the extracellular medium in the wild type, and more than 20 lipoproteins were released into the medium in large amounts in the B. subtilis lgt mutant (1). Lipoproteins lacking the lipid anchor were reported to be released into the extracellular medium after signal sequence cleavage by Lsp, SpaseI, or alternative proteases or without further processing in Streptococcus species, L. monocytogenes, and B. subtilis, respectively (1, 3, 8, 9, 43).
Lipoproteins devoid of the lipid anchor are released into culture filtrate.
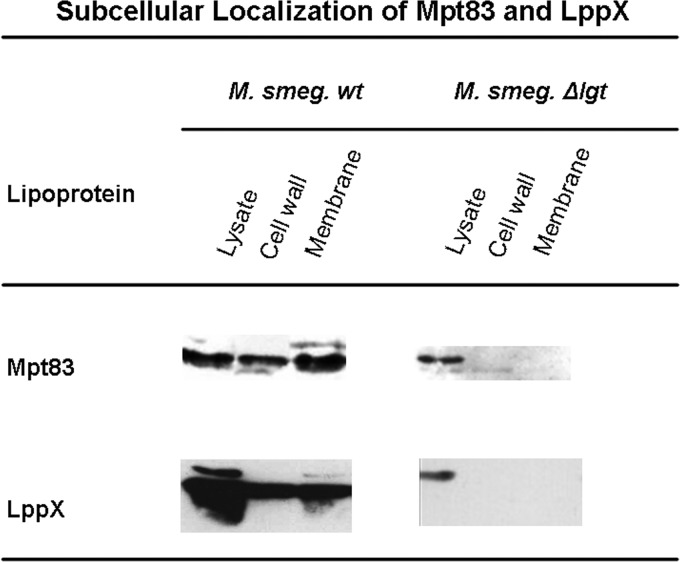
We performed subcellular fractionations (29) of the parental strain and lgt mutant expressing LppX and Mpt83, respectively, to localize mature lipoproteins and lipoproteins lacking the lipid anchor. In the parental strain, both lipoproteins, LppX and Mpt83, were present in the cell envelope fractions (Fig. 5). In the Δlgt mutant, both lipoproteins were absent from the cell wall fraction and from the cytoplasmic membrane fraction. Taken together, this demonstrates that the lack of lipid modification affects subsequent processing and localization. For some lipoproteins, e.g., Mpt83, signal sequence cleavage may occur without prior Lgt-mediated lipid modification, but the absence of a membrane anchor leads to the failure to transport and retain lipoproteins in the mycobacterial cell envelope (Fig. 5).
Fig 5.
Subcellular localization of Mpt83 and LppX in M. smegmatis Δlgt mutant and parental strains. Western blot analyses of fractionated M. smegmatis extracts are shown. Lipoproteins Mpt83 and LppX localize in the cell wall fraction in the parental strain but are absent from the cytoplasmic membrane and the cell wall fraction in the Δlgt mutant. Absence of lipoproteins in the cell envelope fractions suggests that lipoproteins in the Δlgt mutant are released into the supernatant (see Fig. 6 in the supplemental material).
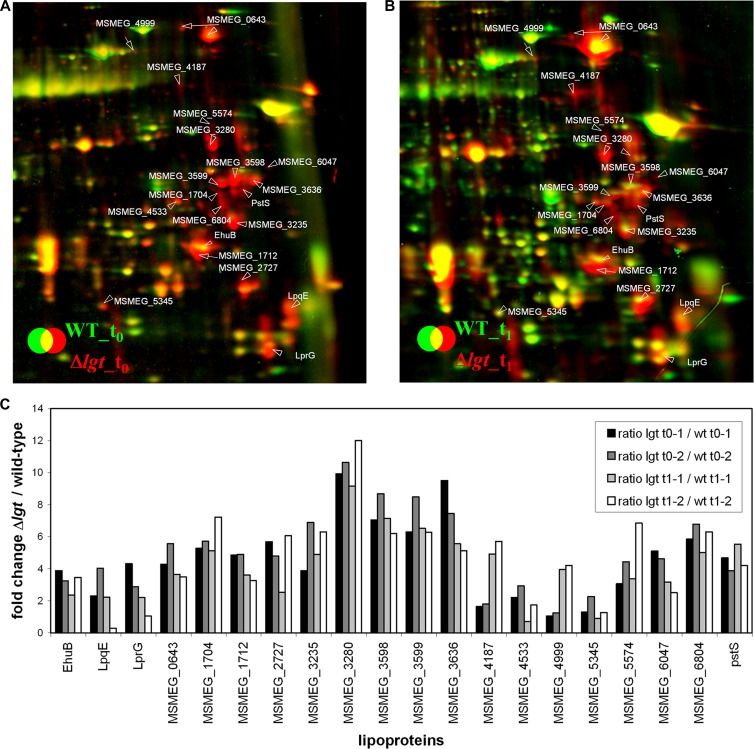
Lipoproteins without the lipid anchor either may be secreted into the culture supernatant because they lack a hydrophobic structure which incorporates them into the membrane, or they may be degraded by proteases. So far, we have focused on the two recombinant M. tuberculosis lipoproteins, Mpt83 and LppX. To get a more complete view of the fate of endogenous lipoproteins, we used a proteomic approach. We analyzed the composition of the secretome of the parental strain and the lgt mutant by two-dimensional gel electrophoresis (2D PAGE) and subsequent protein identification by MALDI-TOF mass spectrometry as described previously (1) (see Fig. S3 in the supplemental material). Experiments were performed in duplicate, and samples were taken during exponential growth (t0) and after entry into the stationary phase (t1). We identified 129 protein spots in the culture supernatant (see Table S1). The identified extracellular proteins are involved in protein transport and solute binding, protein folding and degradation, cell envelope maintenance, intermediary and energy metabolism, transcription and translation, and defense mechanisms. The differences in the levels of 106 secreted proteins between wild-type and lgt mutant cells are quantified in Table S2. In total, 54 proteins were found at 2- to 10-fold larger amounts in the lgt mutant secretome than in the wild-type secretome (Table 1). In the stationary phase, the cytoplasmic proteins Tkt and EF-Tu were more abundant in the lgt mutant secretome than in the secretome of the parental strain, which is probably due to increased cell lysis in t1. We then analyzed all identified proteins for the presence of signal peptides and signal peptidase I and II cleavage sites using the LipoP 1.0 server (http://www.cbs.dtu.dk/services/LipoP/). In total, 45 extracellular proteins were predicted to be synthesized with N-terminal signal peptides, including 25 proteins with type I signal peptides and 20 proteins with lipoprotein-specific type II signal peptides (Table 1). These 45 predicted secretory proteins include 35 proteins that are more abundant in the lgt mutant secretome. In particular, all 20 lipoproteins identified in the culture supernatant were present in larger amounts in the Δlgt mutant secretome (Fig. 6). Tryptic peptide fragments corresponding to signal peptides of lipoproteins were not detected (data not shown). These lipoproteins function mainly as ABC transporter substrate-binding proteins. Other lipoproteins are involved in energy and intermediary metabolism, protein folding and stabilization, transcriptional regulation, and unknown functions. Some proteins with predicted signal peptides were found to be present in the secretome in smaller amounts in the Δlgt mutant. Among these are two secreted proteins (MSMEG_0066 and MSMEG_3493) and a 28-kDa antigen (MSMEG_6919). The decreased amount of these proteins could be due to overrepresentation of lipoproteins in the secretome of the Δlgt mutant, since the secretome of the two strains was normalized with respect to total protein concentration.
Table 1.
Quantification of proteins that are present in larger amountsa in the secretome of the M. smegmatis Δlgt mutant than in the parental strain
| Protein group and name | Function or description | Δlgt/wild-type ratio at: |
|||
|---|---|---|---|---|---|
| t0-1 | t0-2 | t1-1 | t1-2 | ||
| Transport and solute binding | |||||
| MSMEG_3247d | Branched-chain amino acid ABC transporter substrate-binding protein | 4.74 | 7.57 | 4.68 | 4.51 |
| MSMEG_3280 Lipc | Polyamine-binding lipoprotein | 9.93 | 10.64 | 9.15 | 12.00 |
| MSMEG_3598 Lip | Periplasmic sugar-binding proteins | 7.06 | 8.68 | 7.14 | 6.21 |
| MSMEG_3235 Lip | ABC-type amino acid transport system, secreted component | 3.87 | 6.90 | 4.91 | 6.31 |
| MSMEG_2727 Lip | Glutamate-binding protein | 5.68 | 4.80 | 2.53 | 6.06 |
| MSMEG_6804 Lip | Sugar ABC transporter substrate-binding protein | 5.85 | 6.78 | 5.02 | 6.31 |
| MSMEG_1704 Lip | ABC transporter | 5.27 | 5.71 | 5.12 | 7.23 |
| MSMEG_3636 Lip | Ferric iron-binding periplasmic protein of ABC transporter | 9.51 | 7.45 | 5.57 | 5.13 |
| MSMEG_0643 Lip | Extracellular solute-binding protein, family protein 5, putative | 4.29 | 5.56 | 3.66 | 3.50 |
| MSMEG_6524 SPb | ABC polyamine/opine/phosphonate transporter, periplasmic ligand binding protein | 3.17 | 1.84 | 5.80 | 4.35 |
| MSMEG_6047 Lip | Cation ABC transporter, periplasmic cation-binding protein, putative | 5.10 | 4.62 | 3.18 | 2.51 |
| PstS (MSMEG_5782) Lip | Periplasmic phosphate-binding protein | 4.68 | 3.88 | 5.53 | 4.21 |
| EhuB (MSMEG_5368) Lip | Ectoine/hydroxyectoine ABC transporter solute-binding protein | 3.88 | 3.25 | 2.36 | 3.46 |
| MSMEG_4533 Lip | Sulfate-binding protein | 2.21 | 2.93 | 0.72 | 1.74 |
| MSMEG_1712 Lip | ABC transporter periplasmic-binding protein YtfQ | 4.85 | 4.90 | 3.62 | 3.26 |
| MSMEG_5574 Lip | Substrate-binding protein | 3.06 | 4.44 | 3.38 | 6.85 |
| MSMEG_4999 Lip | Bacterial extracellular solute-binding protein, family protein 5 | 1.06 | 1.24 | 3.95 | 4.21 |
| Protein fate: folding, stabilization and degradation | |||||
| MSMEG_3070 Lip | LprG protein | 4.31 | 2.88 | 2.20 | 1.06 |
| MSMEG_3903 SP | Low-molecular-wt antigen MTB12 | 5.67 | 1.98 | 6.11 | 3.10 |
| tig (MSMEG_4674) | TF | 1.09 | 1.30 | 0.88 | 0.91 |
| tig-2 (MSMEG_4674)e | TF | 5.35 | 3.42 | 2.62 | 2.97 |
| MSMEG_5664 | Peptidyl-prolyl cis-trans isomerase | 1.30 | 0.70 | 1.25 | 1.28 |
| MSMEG_5015 SP | Secreted protein | 2.98 | 3.46 | 2.06 | 3.04 |
| Defense mechanisms | |||||
| MSMEG_6567 SP | Iron-dependent peroxidase | 1.46 | 1.05 | 2.33 | 9.89 |
| MSMEG_2658 SP | Beta-lactamase | 1.76 | 1.47 | 1.02 | 1.18 |
| MSMEG_3811 | Universal stress protein family protein, putative | 1.79 | 1.44 | 0.68 | 0.51 |
| Energy, intermediary and fatty acid metabolism | |||||
| MSMEG_0806 | Hydrolase | 4.98 | 4.10 | 3.65 | 5.88 |
| MSMEG_0194 SP | Serine esterase, cutinase family protein | 2.64 | 2.64 | 2.39 | 4.67 |
| MSMEG_1403 SP | Cutinase superfamily protein | 1.85 | 0.94 | 0.70 | 1.38 |
| MSMEG_0361 SP | Glycosyl hydrolase family protein 3 | 1.49 | 1.47 | 1.00 | 0.40 |
| MSMEG_0645 SP | Putative beta-1,3-glucanase | 1.82 | 1.89 | 1.36 | 1.56 |
| MSMEG_3962 | Lactate 2-monooxygenase | 1.07 | 1.27 | 0.51 | 0.73 |
| MSMEG_6398 SP | Antigen 85-A | 1.69 | 1.55 | 1.45 | 1.56 |
| MSMEG_5789 | Putative thiosulfate sulfurtransferase (Rhodanese-like protein) | 2.00 | 1.52 | 1.12 | 1.59 |
| MSMEG_3580 | Antigen 85-C | 1.20 | 1.61 | 1.01 | 1.19 |
| MSMEG_5345 Lip | Glycosyl hydrolase family protein 16 | 1.29 | 2.26 | 0.89 | 1.26 |
| MSMEG_0216 | 3-Hydroxyacyl-coenzyme A dehydrogenase | 1.62 | 1.38 | 0.25 | 0.31 |
| glcB (MSMEG_3640) | Malate synthase G | 0.98 | 1.90 | 1.06 | 0.94 |
| tkt (MSMEG_3103) | Transketolase | 0.43 | 0.62 | 0.99 | 1.31 |
| Other and unknown function | |||||
| MSMEG_5617 SP | Immunogenic protein MPT63 | 2.19 | 1.78 | 0.42 | 5.55 |
| MSMEG_1051 SP | Immunogenic protein MPB64/MPT64 | 1.59 | 1.57 | 1.27 | 0.82 |
| MSMEG_3599 Lip | Sugar-binding transcriptional regulator, LacI family protein | 6.31 | 8.48 | 6.52 | 6.28 |
| MSMEG_2408 | Uncharacterized oxidoreductase MSMEG_2408 | 1.67 | 1.92 | 0.84 | 1.56 |
| MSMEG_1038 | GTP cyclohydrolase II | 1.37 | 1.10 | 0.33 | 0.78 |
| tuf (MSMEG_1401) | Elongation factor Tu (EF-Tu) | 0.56 | 0.49 | 0.14 | 5.28 |
| MSMEG_0233 SP | Lipoprotein LppS | 1.79 | 1.67 | 1.33 | 1.71 |
| MSMEG_3528 SP | ErfK/YbiS/YcfS/YnhG family protein | 6.25 | 7.05 | 6.02 | 9.87 |
| MSMEG_2381 | Putative uncharacterized protein | 4.53 | 2.34 | 3.15 | 1.45 |
| MSMEG_6078 Lip | LpqE protein | 2.31 | 4.04 | 2.22 | 0.29 |
| MSMEG_1322 SP | ErfK/YbiS/YcfS/YnhG family protein | 3.00 | 1.36 | 9.71 | 6.74 |
| MSMEG_0035 | FHA domain protein | 1.74 | 0.97 | 4.52 | 8.20 |
| MSMEG_6289 | Trypsin | 1.32 | 1.23 | 0.94 | 0.16 |
| MSMEG_0065 | Putative uncharacterized protein | 1.92 | 1.17 | 0.67 | 1.17 |
| MSMEG_4187 Lip | Putative uncharacterized protein | 1.65 | 1.79 | 4.92 | 5.70 |
Larger amounts indicates that mean values of duplicates of Δlgt/wt ratios are greater than 1.1 for at least one time point (t0 or t1). t0-1 and t0-2 indicate analysis of two biological replicates at t0; t1-1 and t1-2 indicate analysis of two biological replicates at t1.
SP, signal peptidase I cleavage site.
Lip, lipobox motif according to LipoP 1.0.
According to LipoP 1.0, MSMEG_3247 is not a lipoprotein. MSMEG_3247 was identified as a lipoprotein manually based on the lipobox motif IAGC. This gene was not included as a lipoprotein in the calculations.
Trigger factor (TF) was identified in two different protein spots (tig and tig-2). tig-2 is a putative processed form of tig.
Fig 6.
Close-ups of the lipoproteins that are secreted into the medium in the Δlgt mutant due to the missing lipid anchor. Shown are sections of the dual-channel images of the secretome of the M. smegmatis Δlgt mutant (red image) and the wild-type strain (green image) at the transition phase (t0; A) and 1 h after entry into the stationary phase (t1; B). The secretome was precipitated with TCA and separated using 2D PAGE in the pH range of 4 to 7, as described in Materials and Methods. Quantification of the dual-channel image was performed using Decodon Delta 2D software. (C) The induction ratios of the identified lipoproteins in the extracellular proteome of the lgt mutant to those of the wild type are shown in the corresponding diagram. Two biological replicates are used for quantification in panel C. Proteins are annotated with their corresponding MSMEG_ number. Lipoproteins were identified using LipoP 1.0.
The increased amount of lipoproteins in the culture filtrate of the Δlgt mutant clearly demonstrates that the diacylglyceryl residue attached to the +1 cysteine is responsible for anchoring lipoproteins in the cell envelope. Lipoproteins lacking this lipid structure are not retained in the mycobacterial cell envelope; rather, they are secreted into the supernatant. The redistribution of all lipoproteins in the M. smegmatis Δlgt mutant from the cell envelope to the medium most probably is responsible for the severe growth and physiological phenotype.
DISCUSSION
Lipoproteins have been known since the discovery of the major lipoprotein of E. coli by Braun and Rehn in 1969. Later, the three enzymes involved in lipoprotein biosynthesis were identified first in E. coli. Therefore, the biosynthesis pathway of E. coli lipoproteins is well described. Lipoproteins are present in all bacterial species, but their structure and biosynthesis pathways differ, particularly between Gram-negative and Gram-positive bacteria (22). Mycobacteria have features of both Gram-negative and Gram-positive bacteria. Mycobacterial and E. coli lipoproteins have three fatty acids in common, but mycobacterial lipoproteins differ from E. coli lipoproteins with respect to the fatty acids of the diacylglyceryl residue linked to the sulfhydryl group of the +1 cysteine. The mycobacterial diacylglyceryl contains the mycobacterium-specific fatty acid 10-methyl octadecanoic acid (tuberculostaric acid) (45). Lipoproteins and the enzymes involved in their synthesis are virulence factors in bacterial pathogens (18). M. tuberculosis lipoproteins in particular have been shown to suppress innate immune mechanisms. LspA catalyzes the second step of lipoprotein synthesis, and its inactivation already attenuates M. tuberculosis severely (33). Characterization of the step preceding signal peptide cleavage of mycobacterial lipoproteins is therefore of major interest. The enzyme which catalyzes the diacylglycerol attachment and the function of the diacylglyceryl on lipoproteins in mycobacteria is characterized here. Based on Himar-1 transposon mutagenesis, lgt in M. tuberculosis is an essential gene, and this was confirmed by our targeted approach. Lgt and the other lipoprotein biosynthesis enzymes (Lsp and Lnt) are essential in Gram-negative bacteria (49). Interference with lipoprotein synthesis leads to mislocalization of lipoproteins involved in major outer membrane biogenesis pathways (30, 44). In Gram-positive bacteria, lgt is not essential, although some lipoproteins are essential, indicating that lipoproteins are active without their membrane anchor (20, 48). In mycobacteria, few lipoproteins are functionally characterized, but most of the approximately 140 mycobacterial lipoproteins are of unknown function. Seven mycobacterial lipoproteins have been shown to be essential (LppL, LppY, LpqB, LpqF, LpqK, LpqW, and LprB) (36). LpqW (MSMEG_5130, Rv1166) acts at the branching point of phosphatidylinositol and lipoarabinomannan biosynthesis to control the abundance of the two species in the cell (17). Cell wall biogenesis is essential for mycobacteria and is a target of several antimycobacterial drugs. Mislocalization of LpqW and, in turn, failure to synthesize the correct mannose-capped lipoarabinomannan could be a reason for essentiality of Lgt in M. tuberculosis.
In contrast to M. tuberculosis, the generation of an lgt deletion mutant in M. smegmatis eventually was successful due to the presence of a second lgt (MSMEG_5408). Our biochemical analyses demonstrated that MSMEG_3222 is the major mycobacterial prolipoprotein diacylglyceryl transferase. M. smegmatis wild-type cultures pulsed with [14C]palmitic acid incorporated the labeled fatty acid into various proteins of different molecular masses, but in the ΔMSMEG_3222 mutant very little labeling was detected. Using Triton phase partition, Mpt83 and LppX, rather hydrophilic proteins with a GRAVY (grand average of hydropathicity, calculated as the sum of hydropathy values of all amino acids divided by the number of all residues) (19) score of 0.188 and 0.013, respectively, accumulate in the detergent phase when isolated from parental and complemented strains. In contrast, these proteins accumulate in the aqueous phase when isolated from the Δlgt mutant. This clearly demonstrates that a hydrophobic structure is attached to these proteins which is mediated by ORF MSMEG_3222. Third, the lipoprotein LppX was found in prolipoprotein form with a signal sequence in the M. smegmatis Δlgt mutant. Fourth, lipoproteins are abundant in the secretome of the Δlgt mutant. The residual labeling of proteins may be due to low Lgt activity of MSMEG_5408. MSMEG_5408 could be an alternative Lgt with lower efficacy and activity than MSMEG_3222. MSMEG_5408 has two amino acid substitutions in residues essential for Lgt function, i.e., Y26H and N146C (see Table S2 in the supplemental material). While a Y26F alteration does not affect the functionality of Lgt, a Y26A alteration completely abolishes Lgt function in E. coli (24, 34). An N146A mutation also completely inactivates Lgt. N146 is located in the Lgt signature motif. Pailler et al. hypothesized that Y26, located in a putative acyltransferase motif together with the Lgt signature motif, is involved in binding or recognition of phosphatidylglycerol (24). Alterations Y26H and N146C therefore could account for a decreased efficacy or activity of MSMEG_5408. We currently cannot exclude that MSMEG_5408 is a second Lgt. Generation of MSMEG_5408 alone or in combination with MSMEG_3222 would be required to test this hypothesis. Alternatively, in vitro biochemical analysis with purified MSMEG_5408 could be performed (40). It may be worth noting that the closely related actinobacterium Streptomyces coelicolor also has two functional Lgt homologues. Both S. coelicolor lgt homologues rescued a Streptomyces scabies Δlgt mutant (48). Successful deletion of individual genes, but the inability to generate a double mutant, likewise suggested an overlapping function (42). This may also be the case in M. smegmatis. MSMEG_5408 may act only on a small subset of the putative 140 lipoproteins or be particularly active under specific growth conditions. The residual labeling may also be due to other enzymes, such as protein acetyltransferase-modifying proteins in ε-amino groups of lysine residues with degradation products of palmitic acid.
In Western blot analyses, Mpt83 was proteolytically processed in the Δlgt mutant, presumably either by SpaseI or LspA. The two lipoproteins investigated here in more detail not only contain LspA cleavage sites but also have confidently predicted SpaseI cleavage sites in their signal peptides in addition to the lipobox. In the ΔlspA mutant, both lipoproteins, Mpt83 and LppX, were found in the prolipoprotein form. Unspecific proteolysis is not observed for any of the tested lipoproteins in the lspA mutant. While pro-LppX is not cleaved by a signal peptidase, the signal sequence of Mpt83 eventually is cleaved independently of the lipidation by SpaseI or LspA. This phenomenon is well known in L. monocytogenes and Streptococcus spp. (3, 9).
The molecular nature of the diacylglycerol attached to lipoproteins by Lgt was previously determined by MS analyses (5, 45). The lipid structure is thought to anchor proteins to hydrophobic membranes. In B. subtilis, E. coli, and Listeria, failure to attach the lipid anchor results in the loss of precursor lipoproteins (3, 8, 9, 43). The lack of membrane retention results in the accumulation of these proteins in the culture filtrate. The molecular mechanism underlying the release remains obscure, but it was shown in B. subtilis that proteolytic processing by alternative extracellular or cell wall-associated proteases contributes to the release of nonmodified lipoproteins into the medium fraction. This has been revealed by N-terminal sequencing of the lipoproteins secreted in the B. subtilis lgt mutant that all were alternatively processed and lacked the +1 cysteine residue (1). In the parental and the complemented M. smegmatis strain, LppX and Mpt83 were found to accumulate in the cell wall fraction. When we subjected recombinant M. smegmatis Δlgt mutant to subcellular fractionation, we found neither LppX nor Mpt83 in the cytoplasmic membrane and the cell wall fraction, respectively. Cell wall localization of Mpt83 and LppX is consistent with the respective function and predicted localization of the two lipoproteins (41). However, it has also been noted that MPB83, the corresponding M. bovis protein, is released from the cells as both a mature, lipidated, 25- to 26-kDa form and as a hydrophilic 22- to 23-kDa form (14).
The absence of Mpt83 and LppX from the cell wall of the Δlgt mutant suggests that mutant bacteria are unable to retain lipoproteins in their membranes. Therefore, we analyzed the secretome of the Δlgt mutant and parental strain. Overall, 106 different proteins were identified, including 54 proteins that are present in increased amounts in the secretome of the Δlgt mutant. The increased release of some cytoplasmic proteins from M. smegmatis Δlgt mutant such as Tkt and EF-Tu indicates that this strain is more susceptible to cell lysis than the wild type. Furthermore, we identified 20 lipoproteins that were present at higher levels in the Δlgt mutant secretome than in the wild type, including 15 substrate-binding or transport proteins. Substrate-binding proteins are abundant in the genomes of Gram-positive bacteria and mycobacteria as binding components of ABC transport systems (41) and are anchored in the outer leaflet of the cytoplasmic membrane (6). In M. smegmatis, a total of approximately 140 putative lipoproteins are annotated. This increased release of lipid anchor-lacking lipoproteins in the lgt mutant verifies previous secretome results and confirms the function of Lgt in M. smegmatis (1, 3, 8, 9, 43). We also searched the MALDI-TOF data set of the Δlgt mutant secretome results for peptide masses corresponding to lipoprotein-specific signal peptides to analyze whether lipoproteins are released in processed or nonprocessed forms. The MS results showed that none of these released lipoproteins harbored the N-terminal signal peptide, indicating that the majority of released lipoproteins are processed either by Lsp, SpaseI, or alternative proteases. Interestingly, release of nonmodified lipoproteins into the medium was observed in a daptomycin-resistant B. subtilis strain that acquired a point mutation of the pgsA gene encoding the phosphatidyl glycerol synthase (11). In the case of the pgsA mutant, the precursor for the diacylglycerol residue, phosphatidyl glycerol, was missing, which in turn results in the lack of the lipid anchor and an Lgt mutant-like release of lipoproteins.
The release of nonmodified lipoproteins may be particularly deleterious for the pathogen M. tuberculosis, since lipoproteins have important virulence functions and are involved in cell wall synthesis (41). The loss of the lipid anchor leads to redistribution from the membrane to the medium, and in turn the lack of the lipoproteins may result in alteration of the mycobacterial cell envelope. Cell envelope alterations were also observed in lgt mutants of other bacteria. For example, a Bacillus anthracis lgt deletion mutant had a more hydrophilic surface than the wild type, indicating a cell wall defect, and this mutant was markedly attenuated in a mouse spore infection model (23). Growth-deficient phenotypes and lack of immune activation were reported for a Staphylococcus aureus lgt mutant (38). Similarly, the M. smegmatis Δlgt mutant showed severe growth attenuation in liquid broth and lacks acid fastness, also indicating cell wall alteration. The phenotypes of the Δlgt mutant point to a key role of Lgt for correct lipoprotein function and consequently for proper cell wall integrity. Nevertheless, we were able to successfully generate an M. smegmatis lgt deletion strain, perhaps because of the presence of a second lgt homologue. A single-nucleotide polymorphism was found in the gene MSMEG_3278 in the Δlgt mutant. The gene does not have an annotated function and is located in a region encoding proteins involved in polyamine transport. One gene of this transport system (MSMEG_3280) is a lipoprotein and was highly abundant in the secretome of the Δlgt mutant. The role of MSMEG_3278 in the wild type and the Δlgt mutant remains to be investigated.
Lipoteichoic acid (LTA), a cell wall component of S. aureus, has long been assumed to stimulate TLR2. However, isolation of LTA from S. aureus lgt mutant indicated that LTA preparations from wild-type S. aureus are frequently contaminated with lipoproteins (13). Numerous mycobacterial lipoglycans are also supposed to stimulate TLR2. The M. smegmatis Δlgt mutant will be an invaluable tool to investigate lipoprotein-independent TLR2 stimulation by mycobacterial lipoglycans.
Lgt is essential in M. tuberculosis and thus is even more important for M. tuberculosis than LspA, the second enzyme of the lipoprotein biosynthesis pathway, which is not essential but is required for full virulence. Lgt inactivation severely affects growth and cell wall properties of M. smegmatis. M. smegmatis possibly can better compensate for Lgt depletion due to the presence of a second lgt homologue and due to the eventual accumulation of a suppressor mutation, which obviously occurs only at low frequency. There is an urgent need for novel antituberculosis drugs and new potential drug targets, since the antituberculosis drug pipeline is not sufficiently filled and more and more drug-resistant M. tuberculosis strains emerge. Our investigations in M. tuberculosis indicate that Lgt is a valuable target for generation of antituberculosis drugs, while a corresponding M. smegmatis mutant enables us to investigate the physiological role of Lgt in mycobacteria.
ACKNOWLEDGMENT
This work was supported by the Swiss National Foundation (31003A-135705).
Footnotes
Published ahead of print 18 May 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Antelmann H, et al. 2001. A proteomic view on genome-based signal peptide predictions. Genome Res. 11:1484–1502 [DOI] [PubMed] [Google Scholar]
- 2. Babu MM, et al. 2006. A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins. J. Bacteriol. 188:2761–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baumgartner M, et al. 2007. Inactivation of Lgt allows systematic characterization of lipoproteins from Listeria monocytogenes. J. Bacteriol. 189:313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 5. Brulle JK, et al. 2010. Cloning, expression and characterization of Mycobacterium tuberculosis lipoprotein LprF. Biochem. Biophys. Res. Commun. 391:679–684 [DOI] [PubMed] [Google Scholar]
- 6. Chater KF, Biro S, Lee KJ, Palmer T, Schrempf H. 2010. The complex extracellular biology of Streptomyces. FEMS Microbiol. Rev. 34:171–198 [DOI] [PubMed] [Google Scholar]
- 7. Chi BK, et al. 2011. S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol. Cell Proteomics 10:M111.009506. doi:10.1074/mcp.M111.009506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Greeff A, et al. 2003. Lipoprotein signal peptidase of Streptococcus suis serotype 2. Microbiology 149:1399–1407 [DOI] [PubMed] [Google Scholar]
- 9. Denham EL, Ward PN, Leigh JA. 2009. In the absence of Lgt, lipoproteins are shed from Streptococcus uberis independently of Lsp. Microbiology 155:134–141 [DOI] [PubMed] [Google Scholar]
- 10. Drage MG, et al. 2009. TLR2 and its co-receptors determine responses of macrophages and dendritic cells to lipoproteins of Mycobacterium tuberculosis. Cell Immunol. 258:29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hachmann AB, et al. 2011. Reduction in membrane phosphatidylglycerol content leads to daptomycin resistance in Bacillus subtilis. Antimicrob. Agents Chemother. 55:4326–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harding CV, Boom WH. 2010. Regulation of antigen presentation by Mycobacterium tuberculosis: a role for Toll-like receptors. Nat. Rev. Microbiol. 8:296–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hashimoto M, et al. 2006. Not lipoteichoic acid but lipoproteins appear to be the dominant immunobiologically active compounds in Staphylococcus aureus. J. Immunol. 177:3162–3169 [DOI] [PubMed] [Google Scholar]
- 14. Hewinson RG, Michell SL, Russell WP, McAdam RA, Jacobs WR., Jr 1996. Molecular characterization of MPT83: a seroreactive antigen of Mycobacterium tuberculosis with homology to MPT70. Scand. J. Immunol. 43:490–499 [DOI] [PubMed] [Google Scholar]
- 15. Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. 2008. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. U. S. A. 105:3963–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hutchings MI, Palmer T, Harrington DJ, Sutcliffe IC. 2009. Lipoprotein biogenesis in Gram-positive bacteria: knowing when to hold 'em, knowing when to fold 'em. Trends Microbiol. 17:13–21 [DOI] [PubMed] [Google Scholar]
- 17. Kovacevic S, et al. 2006. Identification of a novel protein with a role in lipoarabinomannan biosynthesis in mycobacteria. J. Biol. Chem. 281:9011–9017 [DOI] [PubMed] [Google Scholar]
- 18. Kovacs-Simon A, Titball RW, Michell SL. 2011. Lipoproteins of bacterial pathogens. Infect. Immun. 79:548–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105–132 [DOI] [PubMed] [Google Scholar]
- 20. Leskela S, Wahlstrom E, Kontinen VP, Sarvas M. 1999. Lipid modification of prelipoproteins is dispensable for growth but essential for efficient protein secretion in Bacillus subtilis: characterization of the Lgt gene. Mol. Microbiol. 31:1075–1085 [DOI] [PubMed] [Google Scholar]
- 21. McDonough JA, Hacker KE, Flores AR, Pavelka MS, Jr., Braunstein M. 2005. The twin-arginine translocation pathway of Mycobacterium smegmatis is functional and required for the export of mycobacterial beta-lactamases. J. Bacteriol. 187:7667–7679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Okuda S, Tokuda H. 2011. Lipoprotein sorting in bacteria. Annu. Rev. Microbiol. 65:239–259 [DOI] [PubMed] [Google Scholar]
- 23. Okugawa S, et al. 2012. Lipoprotein biosynthesis by prolipoprotein diacylglyceryl transferase is required for efficient spore germination and full virulence of Bacillus anthracis. Mol. Microbiol. 83:96–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pailler J, Aucher W, Pires M, Buddelmeijer N. 2012. Phosphatidylglycerol::prolipoprotein diacylglyceryl transferase (Lgt) of E. coli has seven transmembrane segments and its essential residues are embedded in the membrane. J. Bacteriol. 194:2142–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pavelka MS, Jr, Jacobs WR., Jr 1999. Comparison of the construction of unmarked deletion mutations in Mycobacterium smegmatis, Mycobacterium bovis bacillus Calmette-Guerin, and Mycobacterium tuberculosis H37Rv by allelic exchange. J. Bacteriol. 181:4780–4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qi HY, Sankaran K, Gan K, Wu HC. 1995. Structure-function relationship of bacterial prolipoprotein diacylglyceryl transferase: functionally significant conserved regions. J. Bacteriol. 177:6820–6824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rampini SK, et al. 2008. LspA inactivation in Mycobacterium tuberculosis results in attenuation without affecting phagosome maturation arrest. Microbiology 154:2991–3001 [DOI] [PubMed] [Google Scholar]
- 28. Rezwan M, Grau T, Tschumi A, Sander P. 2007. Lipoprotein synthesis in mycobacteria. Microbiology 153:652–658 [DOI] [PubMed] [Google Scholar]
- 29. Rezwan M, Laneelle MA, Sander P, Daffe M. 2007. Breaking down the wall: fractionation of mycobacteria. J. Microbiol. Methods 68:32–39 [DOI] [PubMed] [Google Scholar]
- 30. Robichon C, Vidal-Ingigliardi D, Pugsley AP. 2005. Depletion of apolipoprotein N-acyltransferase causes mislocalization of outer membrane lipoproteins in Escherichia coli. J. Biol. Chem. 280:974–983 [DOI] [PubMed] [Google Scholar]
- 31. Sala C, Hartkoorn RC. 2011. Tuberculosis drugs: new candidates and how to find more. Future Microbiol. 6:617–633 [DOI] [PubMed] [Google Scholar]
- 32. Sander P, Meier A, Bottger EC. 1995. rpsL+: a dominant selectable marker for gene replacement in mycobacteria. Mol. Microbiol. 16:991–1000 [DOI] [PubMed] [Google Scholar]
- 33. Sander P, et al. 2004. Lipoprotein processing is required for virulence of Mycobacterium tuberculosis. Mol. Microbiol. 52:1543–1552 [DOI] [PubMed] [Google Scholar]
- 34. Sankaran K, et al. 1997. Roles of histidine-103 and tyrosine-235 in the function of the prolipoprotein diacylglyceryl transferase of Escherichia coli. J. Bacteriol. 179:2944–2948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sankaran K, Wu HC. 1995. Bacterial prolipoprotein signal peptidase. Methods Enzymol. 248:169–180 [DOI] [PubMed] [Google Scholar]
- 36. Sassetti CM, Boyd DH, Rubin EJ. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77–84 [DOI] [PubMed] [Google Scholar]
- 37. Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911–1919 [DOI] [PubMed] [Google Scholar]
- 38. Stoll H, Dengjel J, Nerz C, Gotz F. 2005. Staphylococcus aureus deficient in lipidation of prelipoproteins is attenuated in growth and immune activation. Infect. Immun. 73:2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stover CK, et al. 1991. New use of BCG for recombinant vaccines. Nature 351:456–460 [DOI] [PubMed] [Google Scholar]
- 40. Sundaram S, Banerjee S, Sankaran K. 2012. The first nonradioactive fluorescence assay for phosphatidylglycerol:prolipoprotein diacylglyceryl transferase that initiates bacterial lipoprotein biosynthesis. Anal. Biochem. 423:163–170 [DOI] [PubMed] [Google Scholar]
- 41. Sutcliffe IC, Harrington DJ. 2004. Lipoproteins of Mycobacterium tuberculosis: an abundant and functionally diverse class of cell envelope components. FEMS Microbiol. Rev. 28:645–659 [DOI] [PubMed] [Google Scholar]
- 42. Thompson BJ, et al. 2010. Investigating lipoprotein biogenesis and function in the model Gram-positive bacterium Streptomyces coelicolor. Mol. Microbiol. 77:943–957 doi:10.1111/j.1365–2958.2010.07261.x [DOI] [PubMed] [Google Scholar]
- 43. Tjalsma H, et al. 1999. The role of lipoprotein processing by signal peptidase II in the Gram-positive eubacterium bacillus subtilis. Signal peptidase II is required for the efficient secretion of alpha-amylase, a non-lipoprotein. J. Biol. Chem. 274:1698–1707 [DOI] [PubMed] [Google Scholar]
- 44. Tokuda H. 2009. Biogenesis of outer membranes in Gram-negative bacteria. Biosci. Biotechnol. Biochem. 73:465–473 [DOI] [PubMed] [Google Scholar]
- 45. Tschumi A, et al. 2009. Identification of apolipoprotein N-acyltransferase (Lnt) in mycobacteria. J. Biol. Chem. 284:27146–27156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. UniProt Consortium 2007. The Universal Protein Resource (UniProt). Nucleic Acids Res. 35:D193–D197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Venema R, et al. 2003. Active lipoprotein precursors in the Gram-positive eubacterium Lactococcus lactis. J. Biol. Chem. 278:14739–14746 [DOI] [PubMed] [Google Scholar]
- 48. Widdick DA, et al. 2011. Dissecting the complete lipoprotein biogenesis pathway in Streptomyces scabies. Mol. Microbiol. 80:1395–1412 [DOI] [PubMed] [Google Scholar]
- 49. Wu HC. 1996. Biosynthesis of lipoproteins, p 1005–1014 In Neidhardt FC, et al. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed, vol 2 ASM Press, Washington, DC [Google Scholar]
- 50. Young DB, Garbe TR. 1991. Lipoprotein antigens of Mycobacterium tuberculosis. Res. Microbiol. 142:55–65 [DOI] [PubMed] [Google Scholar]