Abstract
Extremely thermophilic bacteria of the genus Caldicellulosiruptor utilize carbohydrate components of plant cell walls, including cellulose and hemicellulose, facilitated by a diverse set of glycoside hydrolases (GHs). From a biofuel perspective, this capability is crucial for deconstruction of plant biomass into fermentable sugars. While all species from the genus grow on xylan and acid-pretreated switchgrass, growth on crystalline cellulose is variable. The basis for this variability was examined using microbiological, genomic, and proteomic analyses of eight globally diverse Caldicellulosiruptor species. The open Caldicellulosiruptor pangenome (4,009 open reading frames [ORFs]) encodes 106 GHs, representing 43 GH families, but only 26 GHs from 17 families are included in the core (noncellulosic) genome (1,543 ORFs). Differentiating the strongly cellulolytic Caldicellulosiruptor species from the others is a specific genomic locus that encodes multidomain cellulases from GH families 9 and 48, which are associated with cellulose-binding modules. This locus also encodes a novel adhesin associated with type IV pili, which was identified in the exoproteome bound to crystalline cellulose. Taking into account the core genomes, pangenomes, and individual genomes, the ancestral Caldicellulosiruptor was likely cellulolytic and evolved, in some cases, into species that lost the ability to degrade crystalline cellulose while maintaining the capacity to hydrolyze amorphous cellulose and hemicellulose.
INTRODUCTION
Interest in cellulosic biofuels (29) has sparked efforts to isolate microorganisms capable of both hydrolysis and fermentation of plant biomass, a process referred to as consolidated bioprocessing (CBP) (49, 50). Since plant biomass deconstruction could be accelerated at elevated temperatures, thermophilic microorganisms have been considered catalysts for CBP (8). Of particular note in this regard are members of the extremely thermophilic genus Caldicellulosiruptor that inhabit globally diverse, terrestrial hot springs (12, 27, 56, 57, 61, 69, 80, 98) and thermally heated mud flats (31). Caldicellulosiruptor species are Gram-positive bacteria and typically associate with plant debris; consequently, all isolates characterized to date hydrolyze certain complex carbohydrates characteristic of plant cell walls (8, 97). As such, members of the genus Caldicellulosiruptor are excellent genetic reservoirs of enzymes for plant biomass degradation and, pending the development of functional genetics systems, are potential metabolic hosts for CBP (9).
Currently, there are two main paradigms described for microbial degradation of crystalline cellulose: cellulosomal (3) and noncellulosomal (48, 54). Enzymatically, both systems require the concerted efforts of cellobiohydrolases, endocellulases, and β-glucosidases (49). Crystalline cellulose deconstruction via cell membrane-bound cellulosomes was first described in the thermophile Clostridium thermocellum and has since been described in other mesophilic Firmicutes, such as Clostridium cellulolyticum, Acetivibrio cellulolyticus, and Ruminococcus flavefaciens (3). Analysis of genome sequence data from biomass-degrading microorganisms has helped to identify noncellulosomal bacteria that also lack identifiable cellobiohydrolases, such as Cytophaga hutchinsonii (96) and Fibrobacter succinogenes (77), both of which require close attachment to cellulose for efficient hydrolysis, and Sacharophagus degradans (95), which uses processive endocellulases (94), indicating that there is great diversity in strategies used for crystalline cellulose hydrolysis. As members of the phylum Firmicutes, Caldicellulosiruptor species are distinct from the thermophilic, anaerobic clostridia in that they secrete free and S-layer-bound cellulases and hemicellulases (9, 23, 24, 43, 44, 58, 60, 63, 75, 84, 89, 90) that are not assembled into cellulosomes (85, 89). In this respect, their strategy for crystalline cellulose deconstruction is similar to that for noncellulosomal biomass-degrading aerobic fungi, such as Trichoderma reesei, (54), the thermophilic fungi Myceliophthora thermophila and Thielavia terrestris (7), or the thermophilic aerobe Thermobifida fusca (48).
The noncellulosomal strategy used by the genus Caldicellulosiruptor for plant biomass deconstruction involves novel, multidomain, carbohydrate-active enzymes (23, 24, 58, 60, 75, 84, 89). However, while all Caldicellulosiruptor species hydrolyze hemicellulose, not all can degrade crystalline cellulose. This gives rise to significant disparity across the genus with respect to the capacity to deconstruct plant cell walls. To date, only limited information is available on the genus Caldicellulosiruptor, especially with respect to the diversity within the genus and the characteristics of individual species. Given the variability within the genus for cellulose deconstruction, insights into this important environmental and biotechnological capability could be obtained by comparative examination of weakly to strongly cellulolytic Caldicellulosiruptor species. To this end, here we examine the core genomes and pangenomes of eight members of this genus, in conjunction with exoproteomics analysis, to identify common and distinctive determinants that drive plant biomass deconstruction.
MATERIALS AND METHODS
Cultivation of Caldicellulosiruptor spp.
Seven Caldicellulosiruptor species were revived from freeze-dried cultures provided by the German Collection of Microorganisms and Cell Cultures (DSMZ [http://www.dsmz.de]) in the recommended culture medium, after which they were transferred to modified DSMZ medium 640 (Trypticase, resazurin, cysteine-HCl, and FeCl3 · 6H2O were not added; the reducing agent was 10% [wt/vol] Na2S · 9H2O added to a final concentration of 0.5% in prepared medium) (9). The eighth strain of the species examined in this study, C. obsidiansis, was isolated from the Obsidian Pool, Yellowstone National Park (27). Complex substrates used as carbon sources for growth included microcrystalline cellulose (Avicel; PH-101; FMC), birchwood xylan (Sigma), and acid-pretreated switchgrass (Panicum virgatum [70]), all added to growth medium at 5 g/liter and, in the case of biomass, 5 g/liter wet weight. Cell density measurements at 24 h were averages from two biological replicates in 50-ml cultures. Enumerations of cell densities were conducted under epifluorescence microscopy using acridine orange (Kodak) as a fluorescent dye (30). The qualitative rating of cellulose hydrolysis ability was by the organism's ability to shred Whatman no. 1 filter paper while being grown in Hungate tubes, as described previously (9). Those species capable of shredding filter paper were designated cellulolytic. Those that were noted to grow on microcrystalline cellulose (Avicel) but not shred filter paper were designated weakly cellulolytic.
Genomic DNA isolation and quality control.
High-molecular-weight genomic DNA (gDNA) from five Caldicellulosiruptor species was harvested as described before (9). Overall, the cultures were grown to early stationary phase on DSMZ culture medium as recommended by DSMZ (without resazurin), using DSMZ medium 671 with cellobiose (for C. hydrothermalis, C. kristjanssonii, C. kronotskyensis, and C. lactoaceticus) and DSMZ medium 144 with glucose (for C. owensensis). Cultures were harvested by centrifugation at 5,000 rpm for 15 min, and gDNA was isolated as previously described (20), except with an additional step requiring lysozyme (100 mg/ml), and the final precipitation of gDNA in isopropanol was collected by a flamed glass hook and then gently washed in 70% (vol/vol) ethanol. Dried gDNA was resuspended in Tris-EDTA (TE) buffer to roughly 1 μg/μl and checked for quality on a 1% [wt/vol] agarose gel in 1× Tris-acetate-EDTA (TAE) buffer. Molecular size standards and the protocol for assessing gDNA quality using agarose gel electrophoresis were both provided by the DOE Joint Genome Institute (JGI) (http://my.jgi.doe.gov/general/protocols/20100809-Genomic-DNA-QC.doc).
Genome sequencing.
The finished genome sequences of C. bescii (60, 98), C. obsidiansis (17), and C. saccharolyticus (89) were completed prior to this project. For C. hydrothermalis, C. kristjanssonii, C. kronotskyensis, C. lactoaceticus, and C. owensensis, a combination of 454 Titanium (51) and Illumina (5) technologies was used (10), similar to the sequencing strategy for C. obsidiansis. Detailed protocols explaining these methods can be found at http://www.jgi.doe.gov/.
Genome assembly and annotation.
For the five genome sequences that were completed for this project, assembly has been previously described (10). Genes were identified using Prodigal (33) as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline (64). The predicted open reading frames (ORFs) were translated and used to search the National Center for Biotechnology Information (NCBI) nr database (6), UniProt (88), TIGRFAM (26), Pfam (68), PRIAM (14), KEGG (34), COG (83), and InterPro (100) databases. These data sources were combined to assert a product description for each predicted protein. Noncoding genes and miscellaneous features were predicted using tRNAscan-SE (46), RNAmmer (39), Rfam (25), TMHMM (36), and SignalP (v3.0) (4). The C. saccharolyticus annotation was updated using the same pipeline, except that manual curation was done without GenePRIMP. Further annotation of selected proteins included molecular size and isoelectric point (pI) prediction (19), signal peptide prediction (SignalP v4.0) (67), and transmembrane (TMHMM) (36) prediction.
Phylogenetic analysis.
All three copies of 16S rRNA genes were used in the construction of a phylogenetic tree. ClustalW (86) was used to align 16S sequences from all sequenced Caldicellulosiruptor species, plus one copy of a 16S rRNA gene from three distantly related species. Pairwise distance calculations were done using the MEGA4 software package (82) with the Tajima-Nei substitution model. These distance calculations were then used to construct dendrograms based on neighbor joining and assessed with 1,000 bootstraps. Average nucleotide identity was used to assess the relatedness of species, taking their whole-genome sequences into consideration. All eight sequenced Caldicellulosiruptor species and the same three outliers mentioned above were uploaded into the Jspecies package using the ANIb BLASTn option (71). Average nucleotide identity (ANI), reported as percent identity, was represented using the cellplot feature of JMP (v9) to create a heat plot. ANIb percentages can be found in Table S1 in the supplemental material.
Prediction of orthologous and functional groups of proteins.
Using all eight finished genomes, orthologous groups of proteins were predicted by OrthoMCL (42). Parameters were selected at a P value of 1E−5, percent identity cutoff of 0, percent match cutoff of 0, Markov clustering algorithm (MCL) inflation of 1.5, and molecular weight of 316. OrthoMCL output (see Data Set S1 in the supplemental material), based on protein-protein homology, was used to compute the core genome and pangenome according to Tettelin et al. (85). Top-ranked similarity searches against genomes in the KEGG database (34) used BLASTp (1). Functional classification of proteins was determined based on searches against databases from NCBI (COG) (83), CAZy (13), integrated microbial genomes (IMG) (53), and InterProScan sequence search (100). Predictions of carbohydrate transporters were done as previously described (91) and also utilized the Find Functions database of the IMG portal (53).
Fractionation of substrate-bound proteins, extracellular proteins, and intracellular proteins.
Seven Caldicellulosiruptor species, four cellulolytic and three weakly cellulolytic, were selected for proteomic analysis. Samples were transferred on Avicel PH-101 three times prior to inoculation of two independent 500-ml cultures, each in 1,000-ml, 45-mm-diameter screw-top Pyrex bottles. A starting inoculum of 1 × 106 cells/ml was used for all cultures, and growth proceeded in batch for 24 h. After 24 h, biological repeats were combined for processing. Spent Avicel with substrate-bound (SB) proteins was isolated by decanting the supernatant (SN) and whole cells (WC) and washing the SB fraction twice with ice-cold DSMZ medium 640, following which the medium was decanted and combined with the SN-plus-WC fraction. Further washing of the SB fraction was done, as described previously (72), with TBS-Ca-T buffer (25 mM Tris-HCl, pH 7.0, 150 mM NaCl, 1 mM CaCl2, and 0.05% [vol/vol] Tween 20). Cell-free SN fraction was obtained by centrifugation at 4°C and 5,000 rpm for 15 min, followed by bottle-top filtration through a 0.22-μm-pore-size filter (Millipore). The resulting WC pellet after centrifugation was washed once with ice-cold DSMZ medium 640 and collected by centrifugation as described above.
Proteomic measurements of Avicel-induced protein fractions.
Each fraction for proteomic analysis (WC, SN, and SB) was independently prepared for bottom-up, two-dimensional liquid chromatography-tandem mass spectrometry (2D-LC-MS/MS) to retain fractional protein localization. Proteins in each fraction were first isolated and denatured by one of the following related methodologies. (i) Cells in the WC fraction were lysed by a combination of boiling and sonication (Branson sonifier) in SDS lysis buffer (4% SDS, 100 mM Tris-HCl, 50 mM dithiothreitol [DTT]). Released proteins (2 mg of crude lysate as measured by bicinchoninic acid [BCA] assay) were then isolated via 20% trichloroacetic acid (TCA) and resuspended in 250 μl 8 M urea as previously described (21). (ii) SN proteins were concentrated to 1 ml by centrifugal membrane filtration (Vivaspin 20 PES; 5-kDa cutoff; GE Healthcare), TCA precipitated, acetone washed, and resuspended in 250 μl of 8 M urea. (iii) Proteins bound to Avicel (SB) were first stripped from the spent substrate (∼10 ml) with 10 ml SDS lysis buffer plus boiling and sonication. Samples were then centrifuged at 4,500 × g and supernatant collected. Proteins in this crude SB fraction were then concentrated, precipitated, washed, and resuspended as described for method ii. Following isolation and denaturation, proteins obtained from each fraction, now in 250 μl of 8 M urea, were reduced (dithiothreitol), alkylated (iodoacetamide), digested (two additions of trypsin), and prepared for 2D-LC-MS/MS analysis as previously described (21). Peptide concentrations were measured by BCA assay.
To reveal the protein complement of each fraction, 25, 50, or 100 μg peptides (SB, SN, and WC, respectively) was bomb loaded onto a biphasic MudPIT back column (55, 93) packed with ∼5 cm strong cation exchange (SCX) resin, followed by ∼3 cm reversed-phase (RP) resin (Luna and Aqua resins, respectively; Phenomenex). Loaded peptides were then washed/desalted offline, placed in line with an in-house pulled nanospray emitter packed with 15 cm RP resin, and analyzed via MudPIT 2D-LC-MS/MS analysis as previously described (21). Briefly, for WC analysis, a full 24-h MudPIT was employed (11 salt cuts at 5, 7.5, 10, 12.5, 15, 17.5, 20, 25, 35, 50, and 100% of 500 mM ammonium acetate followed by a 100-min organic gradient). For both SN and SB peptide fractions, a mini-MudPIT was utilized (4 salt cuts at 10, 20, 35, and 100% of 500 mM ammonium acetate followed by a 100-min organic gradient). Peptide fragmentation data were collected via a hybrid LTQ XL-Orbitrap mass spectrometer (Thermo Fisher Scientific) operating in data-dependent mode. Full MS1 scans (2 microscans; 5 MS/MS per MS1) were obtained using the Orbitrap mass analyzer set to 15K resolution, while MS/MS scans (2 microscans) were obtained/performed in the LTQ mass spectrometer.
Resultant peptide fragmentation data (MS/MS) obtained from each fraction/organism were scored against their respective annotated proteomes, which were downloaded from NCBI (Table 1) using the SEQUEST database-searching algorithm (18). Peptide-sequenced MS/MS spectra were filtered (XCorr [cross-correlation score], +1, 1.8; +2, 2.5; +3, 3.5; DeltCN [normalized difference between first and second match scores], 0.08) and assembled into protein loci by DTASelect (81). Peptide spectral counts (SpC) resulting from intraspecies, nonunique peptides were balanced across their shared protein source (21) to prevent overestimation of protein abundance that could occur between proteins with high degrees of homology, i.e., glycoside hydrolases. Once balanced, SpC for each fraction (SB, SN, and WC) were converted to normalized spectral abundance factors (NSAF) (101) by applying a fractional SpC shift (0.33) to all proteins as described in reference 43. Normalized data from each species and fraction were grouped together based on OrthoMCL to identify trending by orthologous proteins. Using the NSAF values, enrichment scores for both SB (SBE = NSAFSB/[NSAFWC + NSAFSN]) and EC (ECE = [NSAFSB+NSAFSN]/NSAFWC) fractions were calculated. Subcellular and extracellular partitioning was calculated (50% indicates equal partitioning) to visualize in Excel how the NSAF was split between fractions.
Table 1.
General Caldicellulosiruptor genome characteristics
| Species | Accession no. |
Genome size (Mb) | No. of protein-coding genes | G+C content (%) | Reference | |
|---|---|---|---|---|---|---|
| Culture | Genome | |||||
| C. bescii | DSM 6725 | CP001393 | 2.93 | 2,776 | 35.2 | 15 |
| C. hydrothermalis | DSM 18901 | CP002219 | 2.77 | 2,625 | 36.5 | 10 |
| C. kristjanssonii | DSM 12137 | CP002326 | 2.80 | 2,648 | 36.0 | 10 |
| C. kronotskyensis | DSM 18902 | CP002330 | 2.84 | 2,583 | 35.0 | 10 |
| C. lactoaceticus | DSM 9545 | CP003001 | 2.62 | 2,492 | 36.1 | 10 |
| C. obsidiansis | ATCC BAA2073 | CP002164 | 2.53 | 2,331 | 35.2 | 17 |
| C. owensensis | DSM 13100 | CP002216 | 2.43 | 2,264 | 35.4 | 10 |
| C. saccharolyticus | DSM 8903 | CP000679 | 2.97 | 2,760 | 35.2 | 89 |
RESULTS AND DISCUSSION
General genome characteristics.
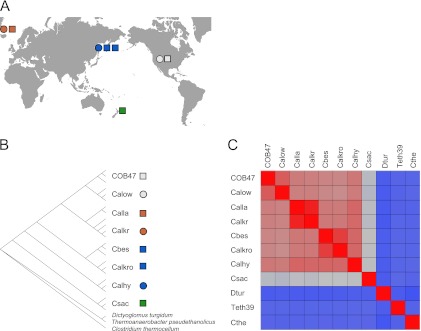
Eight closed Caldicellulosiruptor genome sequences were examined to seek out determinants for the capacity to degrade plant biomass, defined by the ability to hydrolyze the various polysaccharide components of biomass, including crystalline cellulose (Table 1). These species represent globally diverse, terrestrial isolation sites (North America, Iceland, Russia, and New Zealand) (Fig. 1A). Genome sizes for the Caldicellulosiruptor species range from 2.43 to 2.97 Mb, with an average genome size of 2.74 Mb and average G+C content of 35.5% across the genus (Table 1). Previous work has demonstrated a range in cellulolytic capacity for this genus of closely related members (9). No one feature of the Caldicellulosiruptor genomes appears to correlate with location or phenotype; however, the two North American strains have the smallest genomes, both by nucleotide number and ORF count (Table 1). Phylogenetic analysis based on the three 16S rRNA loci found in each genome (Fig. 1B) confirms previous reports that the genera are closely related to each other, with C. saccharolyticus, an isolate from New Zealand, being the most divergent among this group. Dendrograms were built based on 16S rRNA phylogeny of species sharing common biogeography form location-specific clades, regardless of phenotype, such as the isolates from North America, Iceland, and Russia (Fig. 1A and B). Using members from the orders Clostridiales, Thermoanaerobacterales, and Dictyoglomales as outgroups, C. saccharolyticus appears to be the oldest member of its genus due to greater divergence from the other species, having branched off earliest in the Caldicellulosiruptor clade (Fig. 1A).
Fig 1.
Biogeography of sequenced Caldicellulosiruptor species. (A) Global distribution of cellulolytic and weakly cellulolytic species. Squares denote cellulolytic species and circles denote weakly cellulolytic species. Colors shading the shapes indicate common isolation locations. (B) Phylogenetic tree using 16S rRNA sequences from sequenced species plus related outliers. MEGA4 was used to calculate distances and build the phylogenetic tree (82). (C) Phylogenomic heat plot using ANI as a measure of relatedness. Red indicates more closely related species, while gray to blue indicates more distantly related species; the percentages of homology for each pairing of species can be found in Table S1 in the supplemental material. Abbreviated species names are after the assigned locus tags and are the following: Cbes, C. bescii; Calhy, C. hydrothermalis; Calkr, C. kristjanssonii; Calkro, C. kronotskyensis; Calla, C. lactoaceticus; COB47, C. obsidiansis; Calow, C. owensensis; Csac, C. saccharolyticus; Cthe, Clostridium thermocellum; Dtur, Dictyoglomus turgidum; and Teth39, Thermoanaerobacter pseudethanolicus.
The ancestral nature of C. saccharolyticus is reinforced by considering the whole genome using average nucleotide identity (ANI) (Fig. 1C) (71). Since location-specific clades formed when we used 16S rRNA sequences, we explored whether or not this same trend would hold true once entire genomes were considered. This proved to be the case with the Icelandic species, which are highly related (∼98% shared identity), and the North American species (∼92% shared identity) (Fig. 1C; also see Table S1 in the supplemental material). Interestingly, one species isolated from Russia, C. hydrothermalis, is slightly more related to an Icelandic species, C. lactoaceticus, when ANI is considered (Fig. 1C; also see Table S1). Furthermore, when the closed genome sequences are aligned based on geographical location, areas of macrosynteny are apparent, again regardless of cellulolytic phenotype (see Fig. S1). These areas of macrosynteny are not apparent when all eight genomes are aligned due to genetic rearrangement during evolution of the genus (data not shown). While 16S phylogeny and ANI are widely used for taxonomic classification of species, they are not appropriate metrics to assign phenotypes within the genus Caldicellulosiruptor, especially with respect to cellulolytic capability.
Growth on plant biomass and complex carbohydrates differentiates between weakly and strongly cellulolytic Caldicellulosiruptor species.
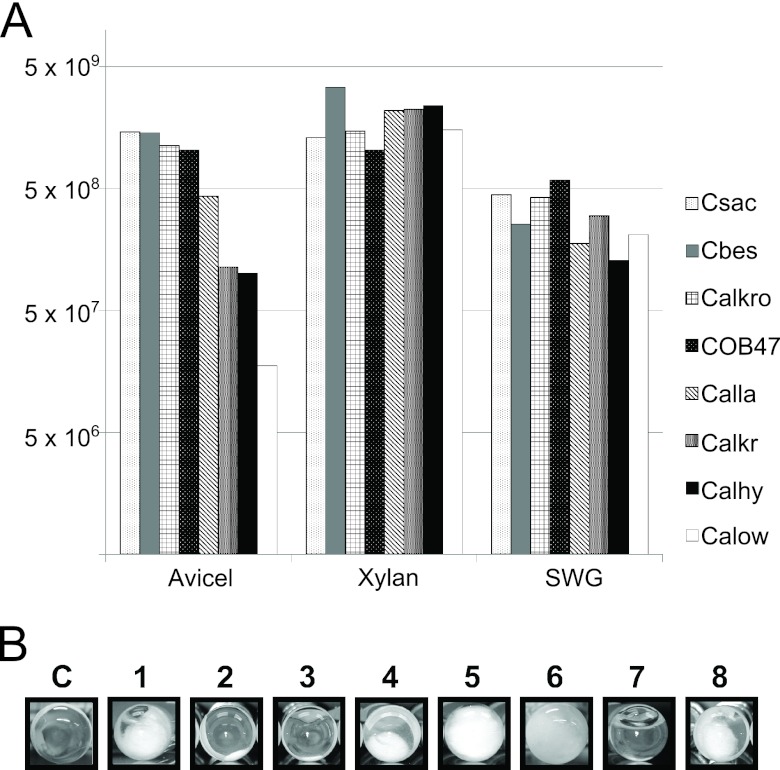
To explore the relationship between genome content and growth on complex carbohydrates, the eight Caldicellulosiruptor species were cultured on crystalline cellulose (Avicel), xylan, acid-treated switchgrass, and filter paper (Fig. 2). While all species grew well on acid-pretreated SWG, which contains hemicellulose, cellulose, and pectin (70), more variability was noted for growth on Avicel (Fig. 2A). All species also grew well on xylan, as expected, based on the core genome (Fig. 2). However, growth on Avicel (Fig. 2A) and filter paper (Fig. 2B) differentiated the strongly from weakly cellulolytic species across the genus. In general, C. bescii, C. kronotskyensis, C. saccharolyticus, and C. obsidiansis grew best on Avicel and filter paper, with C. lactoaceticus growth being at a somewhat lower level. C. hydrothermalis, C. kristjanssonii, and C. owensensis, however, grew less on these substrates and did not break down filter paper to any visible extent (Fig. 2B). These growth experiments provided a perspective for comparative genomic analysis with respect to crystalline cellulose hydrolyzing capability.
Fig 2.
Capacity for crystalline cellulose deconstruction and growth of Caldicellulosiruptor species on complex substrates. (A) Cell density (cells/ml) for each species after 24 h of growth on the following: Avicel, microcrystalline cellulose; Xylan, birchwood xylan; and SWG, acid-treated switchgrass. Standard deviations are equal to one-third or less of the cell density. Abbreviations are after the assigned locus numbering system and are the following: C, control; 1, Cbes, C. bescii; 2, Calhy, C. hydrothermalis; 3, Calkr, C. kristjanssonii; 4, Calkro, C. kronotskyensis; 5, Calla, C. lactoaceticus; 6, COB47, C. obsidiansis; 7, Calow, C. owensensis; and 8, Csac, C. saccharolyticus. (B) Microbial deconstruction of Whatman number 1 filter paper during growth. Fibers released from the substrate at the bottom of the Hungate culture tube are indicative of enzymatic activity against crystalline cellulose.
Caldicellulosiruptor core and pangenomes.
To identify specific determinants among the Caldicellulosiruptor genomes that would enable some species and not others to hydrolyze crystalline cellulose, a baseline view of the genomes is required. The Caldicellulosiruptor core and pangenomes (see Fig. S1 and Data Set S1 in the supplemental material), based on these eight sequenced species, contain 1,580 and 4,009 genes, respectively (42, 85). The pangenome was found to be open, such that the projected number of orthologous genes discovered with each new species sequenced reaches an asymptote of 125 genes. This result is not surprising, given that these species are isolated from dynamic environments, specifically environments with variable nutrient types for organotrophic growth (27). Functional characterization of the core Caldicellulosiruptor genome using COG analysis indicated that while translation and amino acid transport families are enriched in the core versus pangenome, carbohydrate metabolism and transport remain the major features of the genus Caldicellulosiruptor (see Fig. S2 and Table S2). For the core genome, approximately 120 genes are involved in carbohydrate transport and metabolism according to COG classification (see Table S2), which includes the main glycolysis pathway, 6 ABC transporters, and 30 CAZy-related proteins (see Fig. S3). This suggests that the core Caldicellulosiruptor genome is capable of extracellular xylan, glucan, and starch hydrolysis, xylan deacetylation, and the import of the resulting oligosaccharides and their catabolism through central metabolism (Fig. 3; also see Fig. S4). Interestingly, all six of the core ABC transporters are from the CUT1 group (see Table S4) (91), which forms the basis for Caldicellulosiruptor organotrophic import of oligosaccharides (74, 76), which are then further processed to their respective monosaccharides. Of additional interest is the colocalization of glycoside hydrolases and carbohydrate ABC transporters, especially among those included in the Caldicellulosiruptor core genome (see Fig. S3). A previous study (91) also observed this phenomenon in C. saccharolyticus and may be indicative of synergy between centralized carbohydrate-hydrolyzing and import systems. However, the core genome suggests that not all Caldicellulosiruptor species are capable of crystalline cellulose hydrolysis, given that GHs belonging to families known to exhibit these biocatalytic capabilities are not identifiable in several genomes.
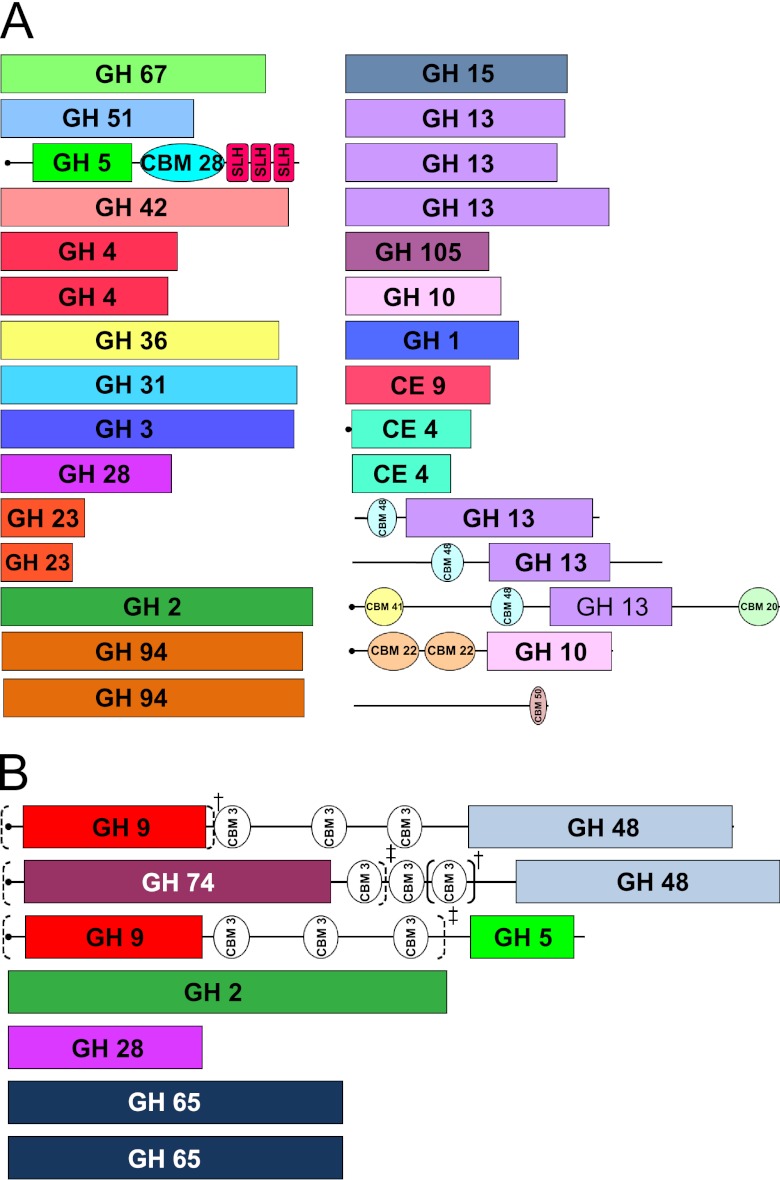
Fig 3.
Core carbohydrate-active enzymes and carbohydrate-binding motif-containing proteins from all eight Caldicellulosiruptor species. (A) Core glycoside hydrolases (GH), polysaccharide lyases (PL), carbohydrate esterases (CE), and carbohydrate-binding motifs (CBM). Numbers refer to protein families established and curated by CAZy (http://www.cazy.org) (13). (B) Core glycoside hydrolases for strongly cellulolytic species. Dashed-line parentheses indicate gene truncations in C. lactoaceticus (†) and C. saccharolyticus (‡); solid-line parentheses indicate an additional CBM family 3 domain in C. lactoaceticus.
The convergence of the number of orthologs in the core genome and the open nature of the Caldicellulosiruptor pangenome indicates that each species is endowed with a set of specific genes beyond the core that relate to the types of carbohydrates present in their environment. Comparisons between the frequencies of the unique Caldicellulosiruptor KEGG BLASTp hits in the Caldicellulosiruptor core genome versus pangenome showed an increase in unique proteins in the pangenome versus the core genome, with C. bescii possessing the largest number and highest frequency of unique Caldicellulosiruptor proteins (see Table S3 in the supplemental material). Analysis of the top-ranked BLASTp hits from strongly cellulolytic versus weakly cellulolytic species did not exhibit any trends based on cellulolytic capability. Top-ranked KEGG BLASTp hits do highlight the major phyla with homologs to proteins from the genus Caldicellulosiruptor, including Firmicutes, Dictyoglomi, Thermotogae, Proteobacteria, and Euryarchaeota. Since the genus Caldicellulosiruptor is classified under the phylum Firmicutes, identifying the majority of best BLASTp hits from Firmicutes was expected (47). Members of the phyla Dictyoglomi, Thermotogae, Proteobacteria, and Euryarchaeota are often isolated or identified from the same locations as the genus Caldicellulosiruptor (32, 37). As such, Caldicellulosiruptor proteins that are direct homologs to proteins from the above-mentioned phyla are likely the result of historical horizontal gene transfer (HGT) in their environment (52). Common biogeography influencing 16S rRNA and ANI-based phylogenetic analyses was not necessarily observed in the context of the number of distinct phyla from KEGG best BLASTp hits, indicative of HGT that is not otherwise detected by phylogenetic analyses. For example, the highly related species C. kristjanssonii and C. lactoaceticus (ANI, 98 to 98.1%; see Table S1) share similar frequencies of best BLASTp hits from the major related phyla (see Table S3); however, C. kristjanssonii had BLASTp best hits to a total of 31 phyla, while C. lactoaceticus had best hits to 22 phyla. Due to the open nature of the Caldicellulosiruptor pangenome, HGT events are important for the evolution of Caldicellulosiruptor species capable of succeeding in their dynamic environments. Increasing the number of Caldicellulosiruptor genome sequences available would also further identify unique genes acquired through HGT, a fraction of which map back to specific aspects of carbohydrate hydrolysis, transport, and metabolism.
Relationship between ABC carbohydrate transporter inventory and growth substrate range.
Since noncore genes appear to be involved in a species' ability to hydrolyze crystalline cellulose, the inventory of carbohydrate transporters was first considered. Overall, the genus Caldicellulosiruptor has 6 core ATP-binding cassette (ABC) transporters out of 45 in the pangenome (see Table S4 in the supplemental material). Substrate preferences for five of these core transporters have previously been assigned based on transcriptomic analysis of C. saccharolyticus (91). Only C. hydrothermalis, C. kronotskyensis, and C. saccharolyticus contain unique transporters not found in the other sequenced Caldicellulosiruptor species. As mentioned above, all of the core ABC transporters are of the CUT1 type, which are typically involved in oligosaccharide import (74, 76), although some of these CUT1 transporters from C. saccharolyticus will respond to monosaccharides (91). These transporters appear to import some, but not all, oligosaccharides that are generated by plant biomass hydrolysis. As there is a wide variety of CAZy-related genes found in the Caldicellulosiruptor genomes, there are also particular ABC transporters used by individual species to support growth on various types of plant biomass.
A connection between ABC transporter number, diversity, and substrate range was evident in examining the genomes. C. lactoaceticus has the most restricted carbohydrate preferences (9, 57) and also has the fewest carbohydrate-related ABC transporters, one-third of those found in C. hydrothermalis. This further supports the point that C. lactoaceticus has evolved as a specialist on higher-chain plant polysaccharides and cannot use glucose to support growth due to the lack of a transporter for glucose. The next closest related species to C. lactoaceticus, C. kristjanssonii, has only three more transporters than C. lactoaceticus and is capable of growth on glucose (9, 12), strongly implicating one of those three additional transporters in glucose uptake. Two of these transporters have previously been implicated in glucose import for C. saccharolyticus, and this finding further confirms that result (91).
C. hydrothermalis contains the most transporters of any member of the genus, with 39 ABC transporters predicted to be involved in carbohydrate transport. On the whole, the G+C content of C. hydrothermalis is higher than that of the rest of the genus (Table 1), implying that it has obtained genes through HGT. Indeed, seven ABC transporters from C. hydrothermalis are unique to the genus and could be the result of HGT. Interestingly, C. hydrothermalis grows weakly on Avicel (Fig. 2A) and does not visibly deconstruct filter paper (Fig. 2B), indicating that transporter inventory does not correlate with the ability to hydrolyze crystalline cellulose. Instead, it appears that C. hydrothermalis has evolved by either importing diverse types of carbohydrates into the cell or using multiple transporters to maximize the uptake of specific carbohydrates.
Overall, no common transporter set could be identified that was only present in cellulolytic but not weakly cellulolytic Caldicellulosiruptor species (see Table S4 in the supplemental material). This finding seems reasonable, given that all isolated species have been described as having the ability to grow on cellobiose (12, 27, 31, 61, 69, 80, 98). Since these bacteria are assumed to live in plant biomass-degrading communities, even if a species is lacking strong cellulolytic machinery it would be beneficial to maintain the ability to import cellulose hydrolysis products. In addition, no correlation between the number of transporters and cellulolytic ability was evident (Table 2). However, the diversity of carbohydrate transporters in weakly cellulolytic species merits further consideration for the design of a biocatalyst for CBP. By incorporating a large number of diverse carbohydrate transporters, flux through many different catabolic pathways could be maintained, supported by the fact that the genus does not exhibit carbon catabolite repression (CCR) (89, 91).
Table 2.
Carbohydrate-related domains and transporter inventory
| Species | No. of ORFs witha: |
Totalb | SigPc | CTd | ||||
|---|---|---|---|---|---|---|---|---|
| GH | CBM | PL | CE | GT | ||||
| C. bescii | 52 | 22 | 4 | 7 | 29 | 68 | 23 | 20 |
| C. hydrothermalis | 62 | 17 | 1 | 6 | 28 | 74 | 15 | 39 |
| C. kristjanssonii | 37 | 15 | 3 | 5 | 31 | 48 | 14 | 15 |
| C. kronotskyensis | 77 | 28 | 4 | 9 | 35 | 93 | 32 | 28 |
| C. lactoaceticus | 44 | 18 | 4 | 4 | 27 | 54 | 17 | 12 |
| C. obsidiansis | 47 | 18 | 2 | 5 | 29 | 59 | 16 | 20 |
| C. owensensis | 51 | 16 | 4 | 8 | 31 | 67 | 19 | 18 |
| C. saccharolyticus | 59 | 17 | 1 | 6 | 30 | 70 | 17 | 25 |
GH, glycoside hydrolase; CBM, carbohydrate binding module; PL, polysaccharide lyase; CE, carbohydrate esterase; GT, glycosyl transferase. Numbers of carbohydrate-active protein domains were retrieved from the CAZy database at http://www.cazy.org (13).
Indicates the total number of ORFs that contain either glycoside hydrolases, carbohydrate-binding modules, polysaccharide lyases, or carbohydrate esterases.
SigP, number of signal peptides.
CT, number of ATP binding cassette (ABC) carbohydrate transporters.
Similarities and subtle differences in core metabolism influence carbohydrate preferences.
Since carbohydrate transporter diversity did not appear to correlate with specific determinants for cellulolytic ability, an examination of the metabolic capacity should be considered. However, based on the information reported here and for the previously sequenced Caldicellulosiruptor genomes (15, 89), the core metabolic pathways across the genus appear to be highly conserved. All species are capable of glycolysis through the Embden-Meyerhof-Parnas (EMP) pathway, fermentation of xylose through a nonoxidative pentose phosphate pathway (PPP), uronic acid metabolism, oxidation of acetate-coenzyme A (CoA), and reduction of pyruvate through an incomplete citric acid cycle (TCA) (see Fig. S4 in the supplemental material). The highly conserved EMP pathway would be responsible for oxidation of glucose liberated from cellulose or starch and highlights the importance of both α-d- and β-d-glucose as energy sources.
Aside from the metabolism of cellulose and pectin, there are some differences between Caldicellulosiruptor species with respect to various monosaccharide metabolic pathways involved in hemicellulose metabolism. One subtle difference concentrates on the xylose isomerase (XI) of C. saccharolyticus, which is a class I XI, in contrast to the other species, which use a class II XI (28, 40). The significance of this is unknown; however, all Caldicellulosiruptor species grow well on xylose (9) and the analogous complex polysaccharide xylan (Fig. 2A), indicating that both types of XI are able to catalyze efficient xylose metabolism for the genus Caldicellulosiruptor.
Three other alternative pathways that feed into the PPP involve other aldopentoses: d-ribose, l-arabinose, and d-arabinose. To metabolize l-arabinose, a component of pectin and arabinoxylan, a putative l-fucose isomerase (MCL group 1847; see Data Set S1 in the supplemental material) appears to be used by most species (see Fig. S4). In contrast, the Icelandic Caldicellulosiruptor species lack the genes to metabolize any of these aldopentoses, which also explains their inability to grow on d/l-arabinose and ribose (12, 57). This apparent lack of d/l-arabinose-specific isomerases and kinases would then theoretically reduce their capacity to metabolize a portion of the hydrolysis products from arabinoxylan.
Another example of upstream carbohydrate conversion pathways influencing carbohydrate growth profiles is the metabolism of deoxy-sugars, such as l-fucose and l-rhamnose. The plant cell wall component pectin can contain l-rhamnose, and xyloglucans can also be fucosylated (99), making the catabolism of deoxy-sugars important for the complete metabolism of all biomass-related carbohydrates. While some species possess complete pathways to metabolize deoxy-sugars, not all species have been described as being capable of growth on them; for example, C. bescii was described as being unable to grow on fucose (80). In addition, other species with incomplete deoxy-sugar pathways have been described as being capable of growth on rhamnose, with C. owensensis being one such example (31). This highlights the overall need for a better understanding of the alternate upstream carbohydrate conversion pathways.
GH inventory reflects the capacity for crystalline cellulose hydrolysis.
Ultimately, the answer to what makes an organism weakly or strongly cellulolytic rests to a large extent on its enzymatic inventory. As discussed above, the inventory of carbohydrate transporters and metabolic pathways only gives clues about the metabolic capacity of the organism after deconstruction of plant biomass. Therefore, a comparative analysis of their glycoside hydrolase (GH) inventory should highlight distinct determinants for cellulose deconstruction. The pangenome of the genus Caldicellulosiruptor encodes 134 carbohydrate-active enzymes (CAZy) (13), here classified as GHs, carbohydrate esterases (CEs), polysaccharide lyases (PLs), and carbohydrate binding modules (CBMs); 48 of these contain signal peptides and are predicted to be extracellular (Table 2). Carbohydrate-active enzyme inventory of the pangenome constitutes the collective capacity of the genus to metabolize complex and simple carbohydrates, including various types of plant biomass. In a preliminary screen of carbohydrate-active enzyme inventory from the genus based on draft genome sequence data, GH family 48 and CBM family 3 were implicated as essential elements for crystalline cellulose hydrolysis by Caldicellulosiruptor species (9). With eight finished genome sequences, a more complete assessment can be done.
As might be expected of microorganisms capable of plant biomass degradation, each Caldicellulosiruptor species contains a significant number of GH domains and CBM modules in their genomes, from 38 and 26, respectively, for C. kristjanssonii up to 84 and 63, respectively, for C. kronotskyensis (Table 2). These numbers are higher than those for other thermophilic anaerobes but are smaller than those for fungi, such as Trichoderma reesei (∼200) (15, 54). The genome of C. kronotskyensis contains 84 GH domains that represent 38 different GH families, which is also the highest diversity of GH domains found in an anaerobic thermophile (13, 60). This is about 50% more GH domains than many other Caldicellulosiruptor species (Table 2). However, the diversity of GH families does not necessarily map back to cellulolytic capability, as C. hydrothermalis and C. saccharolyticus possess 60 and 61 families, respectively, and have vastly different plant biomass deconstruction capabilities (Fig. 2B).
Approximately one-fourth of the 121 CAZy-related ORFs are conserved across all eight sequenced Caldicellulosiruptor genomes and constitute the core. These 30 ORFs include 26 enzymes containing GH domains, three containing CE domains, and one with only a single CBM domain (Fig. 3A). Four ORFs from this core are predicted to be extracellular, including Csac_0678 and its orthologs, a bifunctional GH5 domain enzyme (63), a putative xylanase, a putative pullulanase, and a carbohydrate esterase (Fig. 3A). In theory, these genes represent the minimal set of CAZy-related genes required for biomass deconstruction by a Caldicellulosiruptor species. While it may be tempting to use this list as an indication of the minimal set of extracellular enzymes required by the genus to support a plant biomass-degrading lifestyle, functional homology of non-core enzymes must also be considered. Indeed, C. kristjanssonii has 11 GH domain-containing enzymes above the core Caldicellulosiruptor set, the lowest number of total GH domain-containing enzymes in the genus. Note that the minimal set of carbohydrate-active enzymes in the genus does not equip the microbe for crystalline cellulose hydrolysis, although the GH5-containing enzyme does allow for random cleavage of amorphous cellulose (63). C. lactoaceticus, a species closely related to C. kristjanssonii, is cellulolytic and possesses only 6 more CAZy-related ORFs above that of C. kristjanssonii (Table 2). Comparison to core cellulolytic enzymes will highlight which of these six additional CAZy-related ORFs are important for cellulose hydrolysis. It appears that both species isolated from Iceland are minimalists with respect to gene inventory for carbohydrate hydrolysis.
Seven additional genes conserved among the cellulolytic species comprise the core cellulolytic carbohydrate-active enzyme list (Fig. 3B). This set includes full or partial homologs of enzymes with GH9 and GH48 domains linked by CBM3 modules, GH74 and GH48 domains linked by CBM3 modules, and GH9 and GH5 domains linked by CBM3 modules (Fig. 3B). Indeed, as a previous preliminary analysis suggested, those species that are strongly cellulolytic also possess enzymes with GH9 and GH48 catalytic domains and CBM3 modules (9) (see Tables S5 and S6 in the supplemental material). In fact, these enzymes are colocalized in loci that contain anywhere from four to seven modular multidomain enzymes, all of which possess CBM3 modules (Fig. 4). One weakly cellulolytic species, C. kristjanssonii, also has some CBM3-linked enzymes; however, none also has a GH48 domain, which appears to be the absolute determinant for crystalline cellulose hydrolysis in the genus (see Table S5). In the comparison between C. kristjanssonii and C. lactoaceticus, where six additional ORFs are present in the cellulolytic C. lactoaceticus, three are conserved among cellulolytic species and only two multidomain multifunctional ORF products are encoded by cellulolytic Caldicellulosiruptor species, the GH74:GH48 enzyme and CelA, a GH9:GH48 enzyme (Fig. 3B). Family 48 GHs are often characterized as cellobiohydrolases (2), supporting the theory that this particular family is responsible for the strong cellulolytic phenotype. Indeed, mutations in GH48-containing enzymes have disrupted the cellulolytic ability of Ruminococcus albus 8 (16) and reduced the cellulolytic efficiency of Clostridium thermocellum (60) and Clostridium cellulolyticum (66). There are cases, however, where the sole presence of a GH48 domain is not enough to promote a strong cellulolytic phenotype, as is the case for cellulosomal Clostridium acetobutylicum (59, 73), even though the GH48 enzyme was expressed and secreted as part of the cellulosome (45). Evidently, even in the case of the strongly cellulolytic Caldicellulosiruptor species, additional determinants beyond the presence of GH domains and CBM modules most likely exist that promote crystalline cellulose hydrolysis.
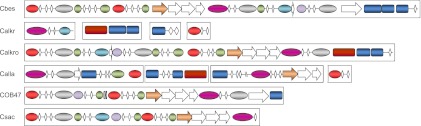
Fig 4.
Gene clusters of CBM3-containing glycoside hydrolases. Locus tags are the following: Cbes, Athe_1867-Athe_1853; Calkr, Calkr_0017, Calkr_1847∼Calkr_1849, Calkr_2455, and Calkr_2522; Calkro, Calkro_0850∼Calkro_0864; Calla, Calla_0015∼Calla_0018, Calla_1251∼Calla_1249, Calla_2311∼Calla_2308, and Calla_2385; COB47, COB47_1673∼COB47_1662; Csac, Csac_1076∼Csac_1085. CBM3 modules are denoted by white diamonds, and dashed lines mean that orthologs possess the CBM3 module; green ovals, GH5; red ovals, GH9; lilac ovals, GH10; blue ovals, GH44; gray ovals, GH48; purple ovals, GH74; blue rectangles, polysaccharide lyase; beige arrow, GT39; and brown rectangle, AraC transcriptional regulator.
Identification of secreted proteins provides insights into substrate attachment and hydrolysis.
To further probe what determinants exist beyond the cellulolytic GH family containing enzymes in the genus Caldicellulosiruptor, Avicel-induced proteins were identified via bottom-up proteomics. Avicel was used as a model plant biomass substrate due to the large proportion of cellulose in plant cell walls and previous studies on T. reesei cellulase systems demonstrating strong affinity of cellulases for Avicel (38, 62). A strong, potentially irreversible interaction between Caldicellulosiruptor proteins and Avicel would be ideal for proteomic screening to identify substrate-bound proteins, since their affinity for Avicel would have to withstand washing steps to remove cells. Previous proteomic screens from members of the genus focused on the cell-free extracellular and whole-cell fractions of cellulolytic Caldicellulosiruptor species (15, 43, 44). We previously reported on differential two-dimensional SDS-PAGE profiles of cell-free supernatant from cells grown on Avicel in an attempt to capture protein-level differences of weakly to strongly cellulolytic Caldicellulosiruptor species (9). To fully capture differential protein expression between weakly and strongly cellulolytic Caldicellulosiruptor species, an expanded proteomic screen was employed. Here, we describe the first comprehensive genus-wide screen of Avicel-induced proteins identified not only from SN and WC but also from the Avicel-bound (SB) fraction from four selected strongly cellulolytic and three weakly cellulolytic Caldicellulosiruptor species.
Overall, between 36 and 48% of total protein-coding sequences predicted from Caldicellulosiruptor genomes was detected as peptides from the SB, SN, and WC fractions using mass spectrometry (see Data Set S2 in the supplemental material). This is lower than the 54% detection for C. bescii (15) or 65% detection for C. obsidiansis (44); however, previous experiments included two or more growth substrates analyzed and/or measurements at various growth stages, whereas this study only included one growth substrate, Avicel. Peptides identified in the SB fraction ranged from 16 to 24% of total protein-coding sequences detected; however, the numbers could be inflated by the presence of intercellular proteins released by cells adhered irreversibly to Avicel. Weakly cellulolytic Caldicellulosiruptor species had the lowest frequency (16.7 and 20.1% for C. owensensis and C. hydrothermalis, respectively) of proteins detected in the SB fraction. This result is not unexpected. A weakly cellulolytic species would not be expected to produce many proteins that interact with cellulose, including the above-mentioned multidomain modular enzymes with CBM family 3 motifs. However, another weakly cellulolytic species, C. kristjanssonii, had the largest frequency of protein-coding sequences detected in the SB fraction, again potentially from intercellular leakage. In fact, the average substrate-bound enrichment score (SBE) for C. kristjanssonii is lower than the average SBE for the entire genus, indicative of intercellular protein contamination of the SB fraction.
Identification of glycoside hydrolases bound to cellulose.
Peptides classified as CAZy-related ORFs were detected at higher frequencies than the complete proteome, ranging from 54 to 83% detection (see Data Set S2 in the supplemental material). As expected, one of the most detected fractions of extracellular peptides corresponded to proteins encoded by the gene cluster containing enzymes with CBM3 modules (MCL cluster 4; see Data Set S2). These GHs were also enriched in the SB fraction more so than in the SN fraction (weighted percentages of 88, 3, and 9% total NSAF for SB, SN, and WC, respectively), agreeing with the cellulose-binding function of CBM family 3 modules (87). One particular group, orthologs of CelA (GH9-CBM3-CBM3-CBM3-GH48) (Fig. 3B), was the most abundant CBM3-containing enzyme detected in the SB fraction. Previous studies identifying extracellular proteins in C. bescii and C. obsidiansis grown on cellulose also found that CelA is the most abundant CBM3-containing enzyme produced by cellulolytic Caldicellulosiruptor species (44). Enrichment of cellobiohydrolases bound to Avicel has been noted before; in competitive binding assays using T. reesi cellulases, including cellobiohydrolases and endoglucanases, the cellobiohydrolases bound with a higher affinity to Avicel (38). This observation further highlights the association of modular multidomain enzymes containing both GH48 and CBM3 domains to crystalline cellulose and emphasizes their important role in its hydrolysis.
One benefit of identifying proteins in the SB fraction is the discovery of previously overlooked enzymes, such as the enrichment of a modular multidomain mannanase (GH26) enzyme on cellulose (22). This cellulolytic enzyme contains two CBM families, CBM27 and CBM35, which are found in the genomes of C. hydrothermalis, C. kristjanssonii, C. lactoaceticus, and C. obsidiansis (MCL group 2116; see Data Set S2 in the supplemental material). Enrichment of this enzyme in the SB fraction was significantly higher in two weakly cellulolytic species, C. hydrothermalis and C. kristjanssonii (NSAF of 1.57 × 10−2 and 4.82 × 10−3, respectively), and significantly lower (almost nonexistent) in the cellulolytic C. lactoaceticus (NSAF of 2.35 × 10−4). Furthermore, there was no detection of this protein in the SN or WC fractions of strongly cellulolytic C. obsidiansis grown on cellobiose, cellulose, or switchgrass, as shown in another study (43). At a minimum, this indicates that there are different regulatory mechanisms for weakly versus strongly cellulolytic species; those species lacking CBM3 protein loci are likely compensating for other enzymes. As mentioned above, previous reports using an orthologous enzyme from Caldicellulosiruptor sp. Rt8B.4 (22) characterized this enzyme as a mannanase, and there has been no further description of enzyme activity beyond that on gluco- and galactomannans (78). However, when the carbohydrate-binding specificity of the CBM motifs was investigated, it was noted that the N terminus of the protein, comprised of the CBM motifs, demonstrated affinity for not only mannan but also glucans, such as soluble cellulose and β-glucan (79). It is not unusual for noncellulolytic enzymes to be targeted to cellulose to decouple cellulose from surrounding polysaccharides, as is the case for some of the multimodular enzymes containing CBM3 motifs (MCL group 4; see Data Sets 1 and 2 in the supplemental material).
Noncatalytic proteins bound to cellulose.
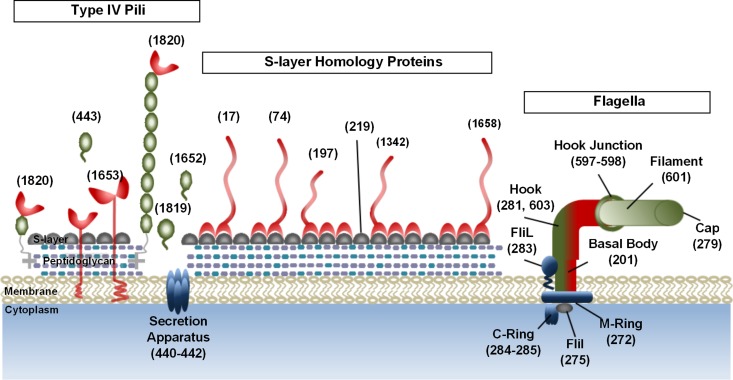
Other proteins that have been theorized to be involved in microbe-substrate interactions were also enriched in the substrate-bound fraction (Fig. 5). The major protein that forms the S layer (MCL group 219; see Data Set S2 in the supplemental material) was found in the extracellular fractions of all species in significant amounts. In fact, this protein alone constituted more than 9% of the total spectra collected across all organisms, with overall fractional partitioning of 35, 53, and 12% (SB, SN, and WC, respectively). However, as previously observed with 2-dimensional SDS-PAGE analysis (9), the supernatants of C. saccharolyticus and C. owensensis are enriched with the S-layer protein (see Data Set S2) relative to the other Caldicellulosiruptor spp. The recently characterized, S-layer-located cellulolytic enzyme Csac_0678 (63) was also enriched in the SB fraction (MCL group 1342; see Data Set S2), as expected. Interestingly, only the ortholog from C. owensensis was strongly enriched in the SN fraction, potentially as a result of the truncated CBM28 motif, highlighting the importance of this particular CBM family in adherence to noncrystalline portions of Avicel (11). A role for four other S-layer-associated proteins in substrate attachment also can be assigned from their observed binding to Avicel (Fig. 5). Although the majority of proteins do not have identifiable carbohydrate-binding modules, they all strongly partition toward the SB fraction (86% of their total SpC collected overall).
Fig 5.
Extracellular, cell membrane-bound proteins involved in microbe-cellulose interactions of strongly cellulolytic Caldicellulosiruptor. Highlighted proteins were detected in the supernatant or substrate-bound proteome. Proteins found enriched in the substrate proteome are shaded red, those enriched in the supernatant are shaded green, and proteins shaded blue indicate enrichment in the cell lysate. Noted proteins shaded gray were detected in all three protein fractions and were not determined to be enriched in one fraction over another. Numbers in parentheses above proteins are nominal labels given to orthologous families of proteins as determined by the OrthoMCL program (42). Exact locus tag numbers for each orthologous protein family are found in Data Set S1 in the supplemental material, and NSAF for each MCL group are found in Data Set S2.
Another group of proteins potentially involved in substrate attachment are those assembled into flagella (Fig. 5). Surprisingly, proteins comprising the flagella were detected primarily in the SN fraction for strongly cellulolytic species (22, 67, and 11% for SB, SN, and WC, respectively, based on total NSAF), while for the weakly cellulolytic species the proteins were enriched in the SB fraction (54, 37, and 9% for SB, SN, and WC, respectively). Enrichment of the flagella in the SN fraction of strongly cellulolytic species indicates that cellulose will induce expression of flagellar genes, although in this case the flagella were not detected to play a role in cellular adhesion. In contrast, enrichment of flagellum components in the SB fraction indicates a more important role for flagella in cellulose adhesion for weakly cellulolytic species. A two-stage mechanism for cell surface attachment has been proposed for the proteobacterium Caulobacter crescentus, with the reversible primary surface attachment mechanism involving the flagella, followed by attachment by type IV pili prior to biofilm formation in the irreversible secondary phase (41). Clearly, there are differing mechanisms for cellulose attachment even within the genus Caldicellulosiruptor, and the enrichment of flagellum-related proteins in the SB fraction from weakly cellulolytic species may be indicative of an extended reversible attachment phase.
Formation of a cellulolytic biofilm by the strongly cellulolytic species C. obsidiansis on cellulose surfaces has been shown previously (92). Since this irreversible secondary stage of cell surface attachment occurs with a strongly cellulolytic species, we looked at the abundance of type IV pilus-related proteins to determine if these structures play a role. Indeed, fewer prepilin subunits were detected for two of the weakly cellulolytic species than for strongly cellulolytic species. In addition, the prepillin subunits were enriched in the SN fraction for all species (5, 93, and 2% for SB, SN, and WC, respectively). However, almost 7-fold fewer of these proteins were detected for the weakly cellulolytic species (MCL groups 443, 1652, and 1819; Fig. 5; also see Data Set S2 in the supplemental material).
Proteins from the type IV pilus genomic region that were enriched in the SB fraction (82% of total NSAF for MCL group 1820 and 97% of total NSAF for MCL group 1653) belonged almost exclusively to the strongly cellulolytic species (Table 3). Annotated as hypothetical proteins, they have no significant homology to proteins outside the genus. Orthologs from highly cellulolytic species (C. bescii, C. kronotskyensis, C. obsidiansis, and C. saccharolyticus) had identity scores ranging from 81 to 95% (MCL group 1820) and 85 to 99% (MCL group 1653), whereas orthologs from species isolated in Iceland were highly homologous to each other (99% identity) yet were much more divergent from the highly cellulolytic group, with identity scores ranging from 36 to 37% (MCL group 1820) and 40% (MCL 1653). Indeed, when predicted parameters such as molecular size and isoelectric point are compared within MCL groups 1820 and 1653, orthologs from C. lactoaceticus and C. kristjanssonii are the smallest proteins, and in the case of MCL group 1820 they are the most positively charged, with a predicted pI of more than 8 (Table 3).
Table 3.
Caldicellulosiruptor adhesins located downstream of type IV pilus gene clusters
| MCL groupa and/or gene locus | Protein property |
Protein abundancee in: |
||||||
|---|---|---|---|---|---|---|---|---|
| Length (aa) | Sizeb (kDa) | pIb | SigPc | TMDd | SB | SN | WC | |
| 1820 | ||||||||
| Athe_1871 | 642 | 70.1 | 5.37 | N | Y | 2.26E−03 | 1.07E−04 | 3.16E−06 |
| Calkr_0826 | 634 | 68.9 | 8.3 | N | Y | 5.37E−03 | 8.54E−04 | 7.07E−05 |
| Calkro_0844 | 642 | 69.6 | 5.18 | N | Y | 4.41E−03 | 8.77E−06 | 2.83E−06 |
| Calla_1507 | 634 | 69.0 | 8.02 | N | Y | 9.80E−03 | 2.32E−03 | 5.45E−04 |
| COB47_1678 | 642 | 69.8 | 5.13 | N | Y | NAf | NA | NA |
| Csac_1073 | 642 | 69.9 | 5.13 | N | Y | 4.29E−03 | 2.49E−03 | 5.99E−05 |
| 1653 | ||||||||
| Athe_1870 | 649 | 70.3 | 6.37 | N | Y | 2.04E−03 | 1.05E−05 | 3.13E−06 |
| Calhy_0908 | 638 | 71.0 | 5.8 | Y | Y | NDg | ND | ND |
| Calkr_0827 | 622 | 68.9 | 5.7 | Y | N | ND | ND | ND |
| Calkro_0845 | 649 | 70.5 | 7.02 | N | Y | 6.95E−04 | 8.67E−06 | 2.80E−06 |
| Calla_1506 | 628 | 69.5 | 6.01 | N | Y | ND | ND | ND |
| COB47_1675 | 649 | 70.3 | 5.72 | N | Y | NA | NA | NA |
| Csac_1074 | 649 | 70.4 | 5.58 | N | Y | 1.84E−04 | 1.75E−05 | 4.80E−05 |
| Calow_1589 | 667 | 71.7 | 9.23 | Y | N | 4.70E−03 | 1.60E−02 | 2.07E−04 |
| Calow_1590 | 900 | 100.2 | 5.12 | N | Y | 2.93E−04 | 7.64E−04 | 7.79E−05 |
OrthoMCL group numbers for orthologous Caldicellulosiruptor proteins (see Data Set S1 in the supplemental material). No orthologous groups were assigned to the two proteins detected from C. owensensis.
Predictions for molecular size and pI used the ExPASy Compute pI/Mw tool (http://web.expasy.org/compute_pi/) (19).
SigP, number of signal peptides; predicted using SignalP (http://www.cbs.dtu.dk/services/SignalP/) (67).
TMD, transmembrane domain; predicted using the TMHMM server (http://www.cbs.dtu.dk/services/TMHMM/) (36).
Protein abundance is reported as NSAF for each fraction screened. SB, substrate bound; SN, supernatant; WC, whole-cell lysate.
NA, protein abundance not available.
ND, not detected in protein fractions using proteomics.
Orthologous MCL group 1820 is expressed by all species examined and was enriched in the SB fraction, in some cases being more than 90% of total NSAF. Since an ortholog in MCL group 1820 is also expressed and found enriched in the SB fraction from the weakly cellulolytic C. kristjanssonii, these proteins do not impart a strong cellulolytic phenotype. However, the ORF directly downstream, represented by orthologous MCL group 1653, was only detected by proteomic screening in the highly cellulolytic species examined and was also enriched in the SB fraction (Table 3). The demonstrated differential expression of this MCL group during growth on cellulose implicates MCL group 1653 in a Caldicellulosiruptor species' ability to hydrolyze crystalline cellulose. Based on genomic proximity of the ORFs to the type IV pilus locus and the enrichment of these proteins in the SB fraction, we propose that these proteins are novel adhesins that mediate attachment of type IV pili to cellulose (MCL groups 1820 and 1653; Fig. 5; also see Data Set S2 in the supplemental material). Gram-positive species sequenced so far generally have one genomic locus that contains the cluster of type IV pilus assembly genes, including hypothetical proteins located adjacent to the locus (65). In the genomic neighborhood of type IV pilus genes, it appears that the adhesins and the type IV pilus locus also reside directly upstream of the cellulase gene cluster in strongly cellulolytic species, lending evidence to a synergistic expression pattern (see Table S7). No orthologs of these adhesins are found in the genome of C. owensensis, a weakly cellulolytic species, which instead possesses other adhesin-like proteins located downstream of the type IV pilus locus (Table 3; also see Table S7). However, both adhesins from C. owensensis were enriched in the SN fraction, and the sole adhesin from C. hydrothermalis was not detected in any of the protein fractions (Table 3). A potential role for those adhesin-like proteins cannot be ruled out, and indeed low levels of mRNA for Calhy_0908 were detected when C. hydrothermalis was grown on cellobiose or switchgrass (data not shown). In the case of C. owensensis, while type IV pilus-proximate proteins were not enriched in the SB fraction, these proteins are expressed in response to the detection of cellooligosaccharides and may mediate attachment to other polysaccharides found in biomass, such as xylan, pectin, or mannans. Determination of the polysaccharide specificity of these putative adhesins, as well as further characterization of the interplay between neighboring adhesins, are the subjects of ongoing experiments.
Was the ancestral Caldicellulosiruptor cellulolytic?
The genomic neighborhoods of type IV pilus- and CBM3-containing enzymes present an interesting case of presumed genomic rearrangement of cellulases in a weakly cellulolytic species, C. kristjanssonii, and the closely related strongly cellulolytic species, C. lactoaceticus. Since the CBM3-containing enzymes of C. kristjanssonii and C. lactoaceticus are found in blocks throughout their respective genomes instead of a single locus, genomic rearrangement can explain the separation of the type IV locus and CBM3-containing enzymes. Genomic rearrangement in this locus could explain the lack of GH48-containing enzymes in C. kristjanssonii and, hence, weak growth on crystalline cellulose (Fig. 2A). Since the genomic identity is very close (ANI of ∼98%; see Table S1 in the supplemental material) between these two species with vastly different phenotypes on cellulose, it begs the question of which phenotype came first in the Caldicellulosiruptor lineage, strongly or weakly cellulolytic?
Two clusters of CAZy-related enzymes exist in the pangenome; one cluster includes primarily glucan-degrading enzymes (GDL) with CBM3 domains (Fig. 4), and the other contains xylan-degrading enzymes (XDL) and xylooligosaccharide transporters (91). Since Caldicellulosiruptor species from more than one continental location contain one or both clusters, it is likely that the ancestral Caldicellulosiruptor species contained both clusters. This also suggests that the ancestral Caldicellulosiruptor species was strongly cellulolytic and capable of crystalline cellulose deconstruction, and that weakly cellulolytic species have lost that ability through gene deletion events.
Members of the genus, except C. hydrothermalis and C. owensensis, have at least one homolog contained within the GDL, which means that C. hydrothermalis and C. owensensis either branched off from the Caldicellulosiruptor lineage prior to acquisition of those genes by the ancestral species or that they lost the entire region after speciation. As mentioned before, the type IV pilus operon is also located directly upstream of the GDL in strongly cellulolytic species. The separation of these colocated regions, in addition to further genomic rearrangements in the GDL of Icelandic species, makes it likely that C. hydrothermalis and C. owensensis lost the GDL after speciation. In addition to the loss of the GDL, these two species also lost one or both cellulose-associating adhesins from the type IV pilus loci, indicating that gene loss occurred further upstream than just the GDL. Furthermore, if the weakly cellulolytic Caldicellulosiruptor species were the result of a separate lineage in the genus, one would expect the weakly cellulolytic species to be more genetically similar to one another, which 16S phylogeny and ANI both disprove (Fig. 1; also see Table S1 in the supplemental material). It is also interesting that many genes located in the GDL cluster of the strongly cellulolytic Caldicellulosiruptor species appear to be the result of various recombination events after gene duplication of glycoside hydrolase domains with CBM3 domains (23, 35, 60) (Fig. 4). Microsynteny in the GDL between C. saccharolyticus and C. kronotskyensis, two geographically distinct species (Fig. 4), indicates that there has been additional rearrangement in the GDL of C. bescii after speciation.
Conclusions.
Eight whole-genome sequences from the genus Caldicellulosiruptor, ranging from weakly to strongly cellulolytic species (Fig. 2A), were assessed for determinants of cellulolytic capability. While biogeography was determined to play a role in the level of relatedness between species based on 16S phylogeny and ANI (Fig. 1), it was not a reliable metric to predict phenotype. Using detailed comparative analysis of the genomes, carbohydrate transport and catabolic pathways were indicative of carbohydrate metabolic capabilities. However, genomic analysis is not enough to predict cellulolytic capability. This is not to say that there is no benefit of such an analysis, since metabolic engineering of a CBP organism will require detailed characterization of the import and metabolism of carbohydrates.
Further analysis of the CAZy-related gene inventory did reaffirm previously predicted determinants for cellulolytic ability, namely, enzymes possessing GH family 48 domains with CBM family 3 modules. Indeed, when the GDL for the cellulolytic species C. lactoaceticus is compared to that of the highly related C. kristjanssonii, the presence of a GH48-containing enzyme, a GH5-containing enzyme, and an additional GH9 enzyme in C. lactoaceticus are the main differences. Since C. kristjanssonii already possesses a GH9 linked to CBM3 modules and other GH5-containing enzymes in its genome, it is unlikely that these were the determinants for a cellulolytic phenotype. Most likely, it is the presence of a GH48-containing enzyme that makes the difference, since GH family 48 members are most often characterized as cellobiohydrolases (13). Additionally, when species that grow better than C. lactoaceticus on cellulose are considered (Fig. 2A), the enzyme CelA, which links a GH9 and GH48 with three CBM3 modules (Fig. 4B), appears to be the determinant for strong cellulolytic growth. Lastly, proteomic-based identification of substrate-bound extracellular proteins revealed additional determinants for a strong cellulolytic phenotype, including two type IV pilus-associated adhesins. As more Caldicellulosiruptor species genomes become available, the insights discussed here can be further evaluated.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Bioenergy Science Center (BESC), Oak Ridge National Laboratory, a U.S. Department of Energy Bioenergy Research Center funded by the Office of Biological and Environmental Research in the DOE Office of Science (contract no. DE-PS02-06ER64304 [DOE 4000063512]).
We gratefully acknowledge the efforts of Lynne Goodwin (JGI-LANL) and Karen Walston Davenport (LANL) on the Caldicellulosiruptor sequencing project. We also thank Dhaval Mistry and Dustin Nelson (NCSU) for their technical assistance in gathering physiological data.
Footnotes
Published ahead of print 25 May 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barr BK, Hsieh YL, Ganem B, Wilson DB. 1996. Identification of two functionally different classes of exocellulases. Biochemistry 35:586–592 [DOI] [PubMed] [Google Scholar]
- 3. Bayer EA, Lamed R, White BA, Flint HJ. 2008. From cellulosomes to cellulosomics. Chem. Rec. 8:364–377 [DOI] [PubMed] [Google Scholar]
- 4. Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 5. Bennett S. 2004. Solexa Ltd. Pharmacogenomics 5:433–438 [DOI] [PubMed] [Google Scholar]
- 6. Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2011. GenBank. Nucleic Acids Res. 39:D32–D37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Berka RM, et al. 2011. Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris. Nat. Biotechnol. 29:922–927 [DOI] [PubMed] [Google Scholar]
- 8. Blumer-Schuette SE, Kataeva I, Westpheling J, Adams MW, Kelly RM. 2008. Extremely thermophilic microorganisms for biomass conversion: status and prospects. Curr. Opin. Biotechnol. 19:210–217 [DOI] [PubMed] [Google Scholar]
- 9. Blumer-Schuette SE, Lewis DL, Kelly RM. 2010. Phylogenetic, microbiological, and glycoside hydrolase diversities within the extremely thermophilic, plant biomass-degrading genus Caldicellulosiruptor. Appl. Environ. Microbiol. 76:8084–8092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blumer-Schuette SE, et al. 2011. Complete genome sequences for the anaerobic, extremely thermophilic plant biomass-degrading bacteria Caldicellulosiruptor hydrothermalis, Caldicellulosiruptor kristjanssonii, Caldicellulosiruptor kronotskyensis, Caldicellulosiruptor owensensis, and Caldicellulosiruptor lactoaceticus. J. Bacteriol. 193:1483–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boraston AB, Ghaffari M, Warren RAJ, Kilburn DG. 2002. Identification and glucan-binding properties of a new carbohydrate-binding module family. Biochem. J. 361:35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bredholt S, Sonne-Hansen J, Nielsen P, Mathrani IM, Ahring BK. 1999. Caldicellulosiruptor kristjanssonii sp. nov., a cellulolytic, extremely thermophilic, anaerobic bacterium. Int. J. Syst. Bacteriol. 49 991–996 [DOI] [PubMed] [Google Scholar]
- 13. Cantarel BL, et al. 2009. The Carbohydrate-Active enZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Claudel-Renard C, Chevalet C, Faraut T, Kahn D. 2003. Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 31:6633–6639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dam P, et al. 2011. Insights into plant biomass conversion from the genome of the anaerobic thermophilic bacterium Caldicellulosiruptor bescii DSM 6725. Nucleic Acids Res. 39:3240–3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Devillard E, et al. 2004. Ruminococcus albus 8 mutants defective in cellulose degradation are deficient in two processive endocellulases, Cel48A and Cel9B, both of which possess a novel modular architecture. J. Bacteriol. 186:136–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Elkins JG, et al. 2010. Complete genome sequence of the cellulolytic thermophile Caldicellulosiruptor obsidiansis OB47T. J. Bacteriol. 192:6099–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eng JK, McCormack AL, Yates JR. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976–989 [DOI] [PubMed] [Google Scholar]
- 19. Gasteiger E, et al. 2003. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31:3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Geslin C, et al. 2003. PAV1, the first virus-like particle isolated from a hyperthermophilic euryarchaeote, Pyrococcus abyssi. J. Bacteriol. 185:3888–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Giannone RJ, et al. 2011. Proteomic characterization of cellular and molecular processes that enable the Nanoarchaeum equitans-Ignicoccus hospitalis relationship. PLoS One 6:e22942 doi:10.1371/journal.pone.0022942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gibbs MD, Elinder AU, Reeves RA, Bergquist PL. 1996. Sequencing, cloning and expression of a beta-1,4-mannanase gene, manA, from the extremely thermophilic anaerobic bacterium, Caldicellulosiruptor Rt8B. 4. FEMS Microbiol. Lett. 141:37–43 [DOI] [PubMed] [Google Scholar]
- 23. Gibbs MD, et al. 2000. Multidomain and multifunctional glycosyl hydrolases from the extreme thermophile Caldicellulosiruptor isolate Tok7B.1. Curr. Microbiol. 40:333–340 [DOI] [PubMed] [Google Scholar]
- 24. Gibbs MD, Saul DJ, Luthi E, Bergquist PL. 1992. The beta-mannanase from “Caldocellum saccharolyticum” is part of a multidomain enzyme. Appl. Environ. Microbiol. 58:3864–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. 2003. Rfam: an RNA family database. Nucleic Acids Res. 31:439–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haft DH, Selengut JD, White O. 2003. The TIGRFAMs database of protein families. Nucleic Acids Res. 31:371–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hamilton-Brehm SD, et al. 2010. Caldicellulosiruptor obsidiansis sp. nov., an anaerobic, extremely thermophilic, cellulolytic bacterium isolated from Obsidian Pool, Yellowstone National Park. Appl. Environ. Microbiol. 76:1014–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hartley BS, Hanlon N, Jackson RJ, Rangarajan M. 2000. Glucose isomerase: insights into protein engineering for increased thermostability. Biochim. Biophys. Acta 1543:294–335 [DOI] [PubMed] [Google Scholar]
- 29. Himmel ME, et al. 2007. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315:804–807 [DOI] [PubMed] [Google Scholar]
- 30. Hobbie JE, Daley RJ, Jasper S. 1977. Use of nucleopore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang CY, Patel BK, Mah RA, Baresi L. 1998. Caldicellulosiruptor owensensis sp. nov., an anaerobic, extremely thermophilic, xylanolytic bacterium. Int. J. Syst. Bacteriol. 48:91–97 [DOI] [PubMed] [Google Scholar]
- 32. Hugenholtz P, Pitulle C, Hershberger KL, Pace NR. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hyatt D, et al. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. 2012. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40:D109–D114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kataeva I, Li X-L, Chen H, Choi S-K, Ljungdahl LG. 1999. Cloning and sequence analysis of a new cellulase gene encoding CelK, a major cellulosome component of Clostridium thermocellum: evidence for gene duplication and recombination. J. Bacteriol. 181:5288–5295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 37. Kublanov IV, et al. 2009. Biodiversity of thermophilic prokaryotes with hydrolytic activities in hot springs of Uzon Caldera, Kamchatka (Russia). Appl. Environ. Microbiol. 75:286–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kyriacou A, Neufeld RJ, MacKenzie CR. 1989. Reversibility and competition in the adsorption of Trichoderma reesei cellulase components. Biotechnol. Bioeng. 33:631–637 [DOI] [PubMed] [Google Scholar]
- 39. Lagesen K, et al. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35:3100–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lewis D. 2010. Functional genomics analysis of extremely thermophilic fermentative microorganisms from the archaeal genus Pyrococcus and bacterial genus Caldicellulosiruptor. North Carolina State University, Raleigh, NC [Google Scholar]
- 41. Li G, et al. 2012. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol. Microbiol. 83:41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lochner A, et al. 2011. Label-free quantitative proteomics for the extremely thermophilic bacterium Caldicellulosiruptor obsidiansis reveal distinct abundance patterns upon growth on cellobiose, crystalline cellulose, and switchgrass. J. Proteome Res. 10:5302–5314 [DOI] [PubMed] [Google Scholar]
- 44. Lochner A, et al. 2011. Use of label-free quantitative proteomics to distinguish the secreted cellulolytic systems of Caldicellulosiruptor bescii and Caldicellulosiruptor obsidiansis. Appl. Environ. Microbiol. 77:4042–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lopez-Contreras AM, et al. 2004. Substrate-induced production and secretion of cellulases by Clostridium acetobutylicum. Appl. Environ. Microbiol. 70:5238–5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ludwig W, Schleifer K-H, Whitman W. 2009. Revised road map to the phylum Firmicutes. In de Vos P P., et al. (ed), Bergey's manual of systematic bacteriology, vol 3 Springer, New York, NY [Google Scholar]
- 48. Lykidis A, et al. 2007. Genome sequence and analysis of the soil cellulolytic Actinomycete Thermobifida fusca YX. J. Bacteriol. 189:2477–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lynd LR, Wyman CE, Gerngross TU. 1999. Biocommodity engineering. Biotechnol. Prog. 15:777–793 [DOI] [PubMed] [Google Scholar]
- 51. Margulies M, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Markowitz VM, et al. 2010. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 38:D382–D390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Markowitz VM, et al. 2012. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 40:D115–D122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Martinez D, et al. 2008. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol. 26:553–560 [DOI] [PubMed] [Google Scholar]
- 55. McDonald WH, Ohi R, Miyamoto DT, Mitchison TJ, Yates JR., III 2002. Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int. J. Mass Spectrom. 219:245–251 [Google Scholar]
- 56. Miroshnichenko ML, et al. 2008. Caldicellulosiruptor kronotskyensis sp. nov. and Caldicellulosiruptor hydrothermalis sp. nov., two extremely thermophilic, cellulolytic, anaerobic bacteria from Kamchatka thermal springs. Int. J. Syst. Evol. Microbiol. 58:1492–1496 [DOI] [PubMed] [Google Scholar]
- 57. Mladenovska Z, Mathrani IM, Ahring BK. 1995. Isolation and characterization of Caldicellulosiruptor lactoaceticus sp. nov., an extremely thermophilic, cellulolytic, anaerobic bacterium. Arch. Microbiol. 163:223–230 [Google Scholar]
- 58. Morris DD, Gibbs MD, Ford M, Thomas J, Bergquist PL. 1999. Family 10 and 11 xylanase genes from Caldicellulosiruptor sp. strain Rt69B. 1. Extremophiles 3:103–111 [DOI] [PubMed] [Google Scholar]
- 59. Nolling J, et al. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823–4838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Olson DG, et al. 2010. Deletion of the Cel48S cellulase from Clostridium thermocellum. Proc. Natl. Acad. Sci. U. S. A. 107:17727–17732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Onyenwoke RU, Lee YJ, Dabrowski S, Ahring BK, Wiegel J. 2006. Reclassification of Thermoanaerobium acetigenum as Caldicellulosiruptor acetigenus comb. nov. and emendation of the genus description. Int. J. Syst. Evol. Microbiol. 56:1391–1395 [DOI] [PubMed] [Google Scholar]
- 62. Otter DE, Munro PA, Scott GK, Geddes R. 1989. Desorption of Trichoderma reesei cellulase from cellulose by a range of desorbents. Biotechnol. Bioeng. 34:291–298 [DOI] [PubMed] [Google Scholar]
- 63. Ozdemir I, Blumer-Schuette SE, Kelly RM. 2012. S-layer homology (SLH) domain proteins Csac_0678 and Csac_2722 implicated in plant polysaccharide deconstruction by the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Appl. Environ. Microbiol. 78:768–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pati A, et al. 2010. GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat. Methods 7:455–457 [DOI] [PubMed] [Google Scholar]
- 65. Pelicic V. 2008. Type IV pili: e pluribus unum? Mol. Microbiol. 68:827–837 [DOI] [PubMed] [Google Scholar]
- 66. Perret S, Maamar H, Belaich J-P, Tardif C. 2004. Use of antisense RNA to modify the composition of cellulosomes produced by Clostridium cellulolyticum. Mol. Microbiol. 51:599–607 [DOI] [PubMed] [Google Scholar]
- 67. Petersen TN, Brunak S, Heijne Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786 [DOI] [PubMed] [Google Scholar]
- 68. Punta M, et al. 2012. The Pfam protein families database. Nucleic Acids Res. 40:D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rainey FA, et al. 1994. Description of Caldicellulosiruptor saccharolyticus gen. nov., sp. nov: an obligately anaerobic, extremely thermophilic, cellulolytic bacterium. FEMS Microbiol. Lett. 120:263–266 [DOI] [PubMed] [Google Scholar]
- 70. Raman B, et al. 2009. Impact of pretreated switchgrass and biomass carbohydrates on Clostridium thermocellum ATCC 27405 cellulosome composition: a quantitative proteomic analysis. PLoS One 4:e5271 doi:10.1371/journal.pone.0005271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Richter M, Rosselló-Móra R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U. S. A. 106:19126–19131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rincon MT, et al. 2007. A novel cell surface-anchored cellulose-binding protein encoded by the sca gene cluster of Ruminococcus flavefaciens. J. Bacteriol. 189:4774–4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sabathe F, Belaich A, Soucaille P. 2002. Characterization of the cellulolytic complex (cellulosome) of Clostridium acetobutylicum. FEMS Microbiol. Lett. 217:15–22 [DOI] [PubMed] [Google Scholar]
- 74. Saier MH. 2000. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64:354–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Saul DJ, et al. 1990. celB, a gene coding for a bifunctional cellulase from the extreme thermophile “Caldocellum saccharolyticum.” Appl. Environ. Microbiol. 56:3117–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Schneider E. 2001. ABC transporters catalyzing carbohydrate uptake. Res. Microbiol. 152:303–310 [DOI] [PubMed] [Google Scholar]
- 77. Suen G, et al. 2011. The complete genome sequence of Fibrobacter succinogenes S85 reveals a cellulolytic and metabolic specialist. PLoS One 6:e18814 doi:10.1371/journal.pone.0018814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sunna A. 2010. Modular organisation and functional analysis of dissected modular beta-mannanase CsMan26 from Caldicellulosiruptor Rt8B. 4. Appl. Microbiol. Biotechnol. 86:189–200 [DOI] [PubMed] [Google Scholar]
- 79. Sunna A, Gibbs MD, Bergquist PL. 2001. Identification of novel beta-mannan- and beta-glucan-binding modules: evidence for a superfamily of carbohydrate-binding modules. Biochem. J. 356:791–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Svetlichnyi VA, Svetlichnaya TP, Chernykh NA, Zavarzin GA. 1990. Anaerocellum thermophilum gen. nov sp. nov: an extremely thermophilic cellulolytic eubacterium isolated from hot springs in the Valley of Geysers. Microbiology 59:598–604 [Google Scholar]
- 81. Tabb DL, McDonald WH, Yates JR. 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1:21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 83. Tatusov RL, et al. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Te'o VS, Saul DJ, Bergquist PL. 1995. celA, another gene coding for a multidomain cellulase from the extreme thermophile Caldocellum saccharolyticum. Appl. Microbiol. Biotechnol. 43:291–296 [DOI] [PubMed] [Google Scholar]
- 85. Tettelin H, et al. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. U. S. A. 102:13950–13955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Tormo J, et al. 1996. Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J. 15:5739–5751 [PMC free article] [PubMed] [Google Scholar]
- 88. UniProt Consortium 2010. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 39:D214–D219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. van de Werken HJ, et al. 2008. Hydrogenomics of the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Appl. Environ. Microbiol. 74:6720–6729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. VanFossen AL, Ozdemir I, Zelin SL, Kelly RM. 2011. Glycoside hydrolase inventory drives plant polysaccharide deconstruction by the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Biotechnol. Bioeng. 108:1559–1569 [DOI] [PubMed] [Google Scholar]
- 91. VanFossen AL, Verhaart MR, Kengen SM, Kelly RM. 2009. Carbohydrate utilization patterns for the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus reveal broad growth substrate preferences. Appl. Environ. Microbiol. 75:7718–7724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wang Z-W, Lee S-H, Elkins JG, Morrell-Falvey JL. 2011. Spatial and temporal dynamics of cellulose degradation and biofilm formation by Caldicellulosiruptor obsidiansis and Clostridium thermocellum. AMB Express. 1:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Washburn MP, Wolters D, Yates JR. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242–247 [DOI] [PubMed] [Google Scholar]
- 94. Watson BJ, Zhang H, Longmire AG, Moon YH, Hutcheson SW. 2009. Processive endoglucanases mediate degradation of cellulose by Saccharophagus degradans. J. Bacteriol. 191:5697–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Weiner RM, et al. 2008. Complete genome sequence of the complex carbohydrate-degrading marine bacterium, Saccharophagus degradans strain 2-40T. PLoS Genet. 4:e1000087 doi:10.1371/journal.pgen.1000087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Xie G, et al. 2007. Genome sequence of the cellulolytic gliding bacterium Cytophaga hutchinsonii. Appl. Environ. Microbiol. 73:3536–3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Yang SJ, et al. 2009. Efficient degradation of lignocellulosic plant biomass, without pretreatment, by the thermophilic anaerobe “Anaerocellum thermophilum” DSM 6725. Appl. Environ. Microbiol. 75:4762–4769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yang SJ, et al. 2010. Reclassification of 'Anaerocellum thermophilum' as Caldicellulosiruptor bescii strain DSM 6725T sp. nov. Int. J. Syst. Evol. Microbiol. 60:2011–2015 [DOI] [PubMed] [Google Scholar]
- 99. York WS, van Halbeek H, Darvill AG, Albersheim P. 1990. Structural analysis of xyloglucan oligosaccharides by 1H-n.m.r. spectroscopy and fast-atom-bombardment mass spectrometry. Carbohydrate Res. 200:9–31 [DOI] [PubMed] [Google Scholar]
- 100. Zdobnov EM, Apweiler R. 2001. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848 [DOI] [PubMed] [Google Scholar]
- 101. Zybailov B, et al. 2006. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 5:2339–2347 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.