Abstract
The growth of Salmonella enterica serovar Typhimurium mutants lacking the ProP and ProU osmoprotectant transport systems is stimulated by glycine betaine in high-osmolarity media, suggesting that this organism has an additional osmoprotectant transport system. Bioinformatic analysis revealed that the genome of this organism contains a hitherto-unidentified operon, designated osmU, consisting of four genes whose products show high similarity to ABC-type transport systems for osmoprotectants in other bacteria. The osmU operon was inactivated by a site-directed deletion, which abolished the ability of glycine betaine to alleviate the inhibitory effect of high osmolarity and eliminated the accumulation of [14C]glycine betaine and [14C]choline-O-sulfate in high-osmolarity media in a strain lacking the ProP and ProU systems. Although the OsmU system can take up glycine betaine and choline-O-sulfate, these two osmoprotectants are recognized at low affinity by this transporter, suggesting that there might be more efficient substrates that are yet to be discovered. The transcription of osmU is induced 23-fold by osmotic stress (0.3 M NaCl). The osmU operon is present in the genomes of a number of Enterobacteriaceae, and orthologs of the OsmU system can be recognized in a wide variety of Bacteria and Archaea. The structure of the periplasmic binding protein component of this transporter, OsmX, was modeled on the crystallographic structure of the glycine betaine-binding protein ProX of Archaeoglobus fulgidus; the resultant model indicated that the amino acids that constitute substrate-binding site, including an “aromatic cage” made up of four tyrosines, are conserved between these two proteins.
INTRODUCTION
Cells with stress-bearing walls, such as those found in most bacteria, fungi, and plants, generate turgor pressure by maintaining the concentration (strictly speaking, activity) of solutes in the cytoplasm higher than that found externally (13). This inequality between the internal and external solute activity favors the net influx of water into the cells. Most bacteria use K+ and glutamate for the regulation of the intracellular osmolarity. However, there is a set of special solutes, called osmoprotectants, that are used in preference to K+ and glutamate by a variety of species. These compounds can be accumulated to high levels by transport from the medium (12) or by intracellular synthesis (5, 51) and can dramatically stimulate the growth rate of cells in media of high osmolarity. Figure 1 shows the structures of several osmoprotectants used in this work, each of which is found in nature (46).
Fig 1.
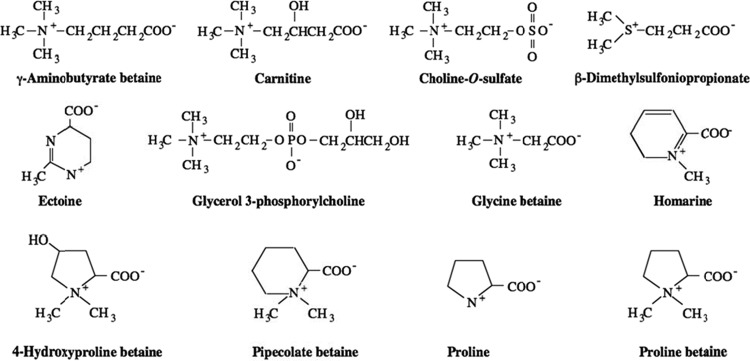
Osmoprotectants used in the present study.
Salmonella enterica serovar Typhimurium and Escherichia coli K-12 accumulate osmoprotectants by the ProP and ProU transport systems (12). ProP, which is a proton symporter (40), is a member of the major facilitator superfamily (MFS) of permeases, and ProU, which uses ATP hydrolysis to drive transport, belongs to the ATP-binding cassette (ABC) superfamily of transporters (http://www.tcdb.org/search/result.php?tc=3.A.1). Bacterial and archaeal ABC-type uptake systems are made up of three different types of functional units: ATPases that supply the energy for the transport, integral membrane proteins that form the substrate-conducting pores, and extracytoplasmic substrate-binding components that present the substrates to the pore proteins (4). In a frequent arrangement of ABC type transport systems, the ATPases are homodimeric proteins, the pore proteins are dimers of two similar or identical polypeptides, and the substrate-binding proteins are monomers, although there are examples of ABC systems in which the membrane-bound pore domains are fused in a single polypeptide to the ATPases or to the substrate-binding components (4). In Gram-negative bacteria, the substrate-binding components of ABC transporters are soluble proteins located in the periplasm, and in Gram-positive bacteria and archaebacteria, they are attached to the cytoplasmic membrane by a lipid or a hydrophobic polypeptide. For the ProU system of Escherichia coli and Salmonella, the ATPase, pore, and substrate binding proteins are called ProV, ProW, and ProX, respectively.
Despite their seemingly diverse structures, most osmoprotectants are transported by both the ProP and ProU systems. ProP has a relatively low affinity for proline and glycine betaine, with an apparent Km of ∼0.1 mM for both (40). ProU has much higher affinity for glycine betaine, with an apparent Km of ∼1 μM (6). However, despite ample physiological and genetic evidence that ProU can also take up proline (11, 20, 26), it has not been possible to measure the activity of this permease with proline as a substrate in standard transport assays (6, 11) suggesting that it has a poor affinity for proline. The substrate preference of the ProU system is reflected by the binding-affinity of the ProX protein, which has an apparent KD (equilibrium dissociation constant) of 1 and 5 μM for glycine betaine and proline betaine (48), respectively, but shows no detectable binding of proline (3, 29, 31).
There are other, less extensively characterized osmoprotectant transporters in Salmonella and E. coli. Choline, which needs to be oxidized to glycine betaine in order to function as an osmoprotectant, is taken up by the proton motive force-driven BetT system, which is a member of the betaine/carnitine/choline transporter (BCCT) superfamily and is highly specific for choline over glycine betaine (http://www.tcdb.org/search/result.php?tc=2.A.15). This permease is present in E. coli but not in S. enterica (35). Several clinical E. coli isolates possess another BCCT-type glycine betaine transporter BetU, which is absent from the laboratory E. coli K-12 strain (39) and from all Salmonella strains. Finally, bioinformatic analysis revealed that both E. coli and S. Typhimurium contain the yehZYXW operon, which specifies an ABC-type transporter, whose ATPase component YehX and pore components YehW/Y show 50 to 56% sequence similarities, respectively, to the ProV and ProW proteins of the E. coli ProU system (9, 39). However, the predicted binding protein component YehZ does not share significant similarity with the ProX protein. The yehZYWV operon is induced by osmotic shock and stationary phase by the σS (RpoS) RNA polymerase (9), but the substrates and the function of this transporter remain to be determined.
Simultaneous mutations in the ProP and ProU systems abolish the ability of S. Typhimurium LT2 and E. coli K-12 to use proline as an osmoprotectant (11, 26), suggesting that these two porters are responsible for high level accumulation of this osmoprotectant in media of elevated osmolarity in these two species. Although E. coli K-12 proP proU double mutants are also unable to use glycine betaine as an osmoprotectant (14), S. Typhimurium proP proU double mutants exhibit residual stimulation in media of high osmolarity by glycine betaine (21), choline-O-sulfate, and β-alanine betaine (A. Hanson, unpublished data). These observations suggest that S. Typhimurium has an additional osmoprotectant transporter that is absent from E. coli K-12. We describe both the identification and the physiological and bioinformatic characterization of this transporter, which we designate the OsmU system.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used, which were all derived from S. Typhimurium LT2, are shown in Table 1. The rich medium used was Luria-Bertani (LB) medium (16), and the minimal medium was M63 (10) or morpholinepropanesulfonic acid (MOPS) (41), containing 10 mM glucose as carbon and energy source. The osmolarity of these minimal media was increased with NaCl, added to final concentrations of 0.3 or 0.65 M, and the pH was adjusted to 7.2 with KOH. The osmolarity of M63 or M63 plus 0.3 M NaCl was 0.22 or 0.83 osM, respectively (20), and that of MOPS or MOPS plus 0.3 M NaCl was 0.1 or 0.65 osM, respectively (8). Solid media contained an additional 20 g of agar (Difco). Cells were grown with aeration at 37°C. Generalized transductions were carried out using bacteriophage P22 HT105/1 int-201 (called P22 hereafter) according to the procedures of Davis et al. (16). Antibiotics, when used, were added at the following concentrations: chloramphenicol (Cm) and kanamycin sulfate (Km) at 25 μg/ml, sodium ampicillin at 100 μg/ml, and tetracycline (Tc) at 20 μg/ml. Sensitivity and resistance to antibiotics is indicated by a superscript “s” and “r”, respectively.
Table 1.
Bacterial strains used
| Strain | Genotype | Construction or sourcea |
|---|---|---|
| TL1 | proP+ proU+ osmU+ (wild-type S. enterica serovar Typhimurium LT2) | B. Ames via S. Kustu |
| TL188 | proP+ proU1655::Tn10 osmU+ | This laboratory |
| TL3463 | proP-4::Tn10dCm proU+ osmU+ | This laboratory |
| TL3465 | proP-4::Tn10dCm proU1655::Tn10 osmU+ | This laboratory |
| TL4072-4075 | osmU::kan zfa-9223::kan metA22 metE551 trpD2 ilv-452 leu pro (leaky) hsdLT6 hsdSA29 hsdB strA120/pKD46 (oriR101 repA101(Ts) araC+ bla λred [gam+ bet+ exo+]) | osmU replaced by kan by λgam-mediated recombination in TT22971b |
| TL4003 | pntBAB::cam | This laboratory |
| TL4093 | osmU::kan proP+ proU+ | P22(TL4073) → TL1 = Kmr |
| TL4095 | osmU::kan proP+ proU1655::Tn10 | P22(TL4073) → TL188 = Kmr [Tcr] |
| TL4097 | osmU::kan proP-4::Tn10dCm proU+ | P22(TL4073) → TL3463 = Kmr [Cmr] |
| TL4099 | osmU::kan proP-4::Tn10dCm proU1655::Tn10 | P22(TL4073) → TL3465 = Kmr [Cmr Tcr] |
| T22971 | zfa-9223::kan metA22 metE551 trpD2 ilv-452 leu pro (leaky) hsdLT6 hsdSA29 hsdB strA120/pKD46 (oriR101 repA101(Ts) araC+ bla λred [gam+ bet+ exo+]) | Eric Kofoid and J. Roth |
| TT23691 | zfa-9223::kan metA22 metE551 trpD2 ilv-452 leu pro (leaky) hsdLT6 hsdSA29 hsdB strA120 | Eric Kofoid and J. Roth |
The notation P22(X) → Y = phenotype 1 [phenotype(s)] indicates that P22 grown on strain X was used to transduce strain Y, selecting for phenotype 1 and scoring the phenotype(s) indicated in brackets. Phenotypes: Cmr, chloramphenicol resistance; Tcr, tetracycline resistance; Kmr, kanamycin resistance.
As described in Materials and Methods.
Osmoprotectants.
Glycine betaine, d-carnitine, l-carnitine, glycerol 3-phosphorylcholine, and proline were purchased from Sigma Chemical Co. β-Dimethylsulfoniopropionate was obtained from Research Plus, Inc., and ectoine was purchased from Bitop AG. γ-Aminobutyrobetaine, homarine, 4-hydroxyproline betaine, pipecolate betaine, and proline betaine were synthesized as described by Fletcher et al. (25) and were generous gifts from David Rhodes (Department Horticulture and Landscape Architecture, Purdue University). [14C-methyl]glycine betaine was synthesized as described by Ladyman et al. (34), and unlabeled choline-O-sulfate and [14C-methyl]choline-O-sulfate were synthesized as described by Hanson et al. (30).
Construction of the osmU deletion.
The four genes of the osmU operon, STM1491 to STM1494, are located on the minus strand of the S. Typhimurium LT2 sequence (GenBank accession no. AE006468.1) between nucleotides 1568523 and 1571969. The osmU operon was deleted by linear recombination (recombineering) (52). The kan gene in strain TT23691, which is flanked with “universal primer sequences” (http://rothlab.ucdavis.edu/protocols/Lin.Transform.html) was amplified by PCR with the primers kanfor-40 (5′-CCTAAGCCGCATAAGGGCAAAGCATAATTGTTAATGATGTTCACCAAACACCCCCCAAAACC-3′) and kanrev-40 (5′-GTGAATATTTAGCGCCCGACGCGTGCGGCCTGAGCGAGATAGTCCACACAACCACACCACACCAC-3′). In these two primers, the underlined nucleotides hybridize, respectively, to the left and right universal primer sites flanking the kan gene; the first 41 nucleotides of kanfor-40 hybridize to nucleotides 1572109 to 1572150 of the S. Typhimurium LT2 sequence, and the first 44 nucleotides of kanrev-40 hybridize to nucleotides 1568535 to 1568579 of the chromosome, respectively. The amplicon was electroporated into strain TT2291 and recombined into the chromosome with the phage λ recombinase expressed from plasmid pKD46 (15) after induction with arabinose, selecting Kmr progeny. We obtained four Kmr derivatives, designated TL4072 to TL4075. This construction was designed to delete sequences from nucleotide 139 upstream of the first gene of the osmU operon (STM1494) to 56 nucleotides from the 3′ end of the last gene of the operon (STM1491). The 3′ end of the clcB (STM1495) gene, which is the next gene upstream of the osmU operon, is at nucleotide 1572273 and therefore would not be predicted to be affected by the deletion. As an initial test of whether the deletion has been targeted to osmU, we determined whether the kan element in the four isolates was near the pntAB genes, which are located 10 to 16 kbp from the four genes of the osmU operon. Phage P22 lysates obtained of TL4072 to TL4075 were used to transduce TL4003 (pntAB::cam) Kmr, and ∼30% of the transductants proved to have lost the cam determinant, indicating that the kan gene is linked to the pntAB locus. The expected structure of the construct was confirmed by two PCR amplifications using two pairs of primers: betPU1 (5′-CATACCCGCCAGCCCCAATAAAAT-3′) plus UTFRD (5′-GGTTTTGGGGGGTGTTTGGTG-3′), which hybridize to nucleotides 1567275 to 1567298 of the chromosome downstream of osmU and to the left “universal primer” pointing outward, respectively, and betPL2 (5′-GCATCAAGCGGCCACT-3′) plus UTRer (5′-GTGGTGTGGTGTGGTTGTTTG-3′, which hybridize to nucleotides 1572740 to 1572756 of the chromosome upstream of osmU and to the right “universal primer” pointing outward, respectively. We obtained identical amplicons with both pairs of primers from TL4072 to 4075 that were consistent with the osmU operon having been replaced by the kan gene (data not shown). The osmU::kan insertion was transduced from TL4073 into strains TL1 (proP+ proU+), TL188 (proP+ proU::Tn10), TL3463 (proP::Tn10dCm proU+), and TL3465 (proP::Tn10dCm proU::Tn10), generating strains TL4093, TL4095, TL4097, and TL4099, respectively, that carry the osmU::kan insertion, together with the four combinations of wild-type and mutant alleles of proP and proU.
Growth studies.
The efficacy of osmoprotectants was determined in liquid cultures. Cells from single colonies on LB plates were inoculated into liquid LB and, after overnight growth, subcultured at a 1:100 dilution into M63 glucose. After growth to saturation, the cells were inoculated at a 1:20 dilution (to an optical density at 600 nm [OD600] of ∼0.05 to 0.07 [ca. 5 × 107 to 7 × 107 cells/ml]) into 20 ml of M63 plus 10 mM glucose and 0.65 M NaCl, without or with a 1 mM concentration of the indicated osmoprotectants, and grown to stationary phase. The growth of the cultures was monitored by periodic measurement of the light scattering at 600 nm in a Shimadzu UV-Vis spectrophotometer.
Long-term accumulation of 14C-labeled osmoprotectants.
Cells from single colonies on LB medium were inoculated into liquid LB medium grown overnight, and then subcultured at 1:100 dilution into M63 plus 10 mM glucose plus 0.3 M NaCl. After overnight growth, 0.05-ml portions of the cultures were inoculated into 1.0 ml of M63 plus 10 mM glucose plus 0.3 M NaCl plus 1 nCi of [14C-methyl]glycine betaine or [14C-methyl]choline-O-sulfate added along with 1 mM unlabeled glycine betaine or choline-O-sulfate, respectively. After overnight growth, the cells were sedimented by centrifugation in an Eppendorf microfuge, washed three times in 1 ml of M63 plus 10 mM glucose plus 0.3 M NaCl, and suspended in 0.1 ml of H2O, and the radioactivity in the cells was measured by liquid scintillation counting. The biomass at the end of the growth was determined as the OD600 using parallel cultures that were grown in nonradioactive media. The accumulation of radioactive substrates was calculated as the nmol/OD600 and converted to nmol/mg of protein, where an OD600 of 1.0 corresponds to 0.22 ± 0.04 mg of protein/ml for cultures grown in M63 plus 0.3 M NaCl plus 1 mM glycine betaine. In the competition experiments, 10 μM glycine betaine or choline-O-sulfate containing 1 nCi of the respective radioactively labeled compounds was added along with a 1.0 mM concentration of the unlabeled competitor test compounds, unless otherwise indicated.
Quantitative real-time reverse transcription-PCR (qRT-PCR) analysis of the osmotic induction of osmU.
Strain TL1 was grown to saturation in MOPS plus glucose and then inoculated into four independent cultures composed of MOPS plus glucose or MOPS plus glucose plus 0.3 M NaCl to an OD600 of 0.08. The cells were grown to mid-exponential phase to an OD600 of ∼0.5, and 0.5-ml samples were rapidly mixed with 1 ml of RNAprotect reagent (Qiagen). Total RNA was isolated with an RNeasy minikit (Qiagen) according to the manufacturer's protocol. DNA was removed using a Turbo DNA-free kit (Invitrogen). The concentration of RNA was determined with a NanoDrop 1000 (Thermo Scientific), and 1,000 ng of RNA was used for random hexamer-primed synthesis of cDNA with the iScript cDNA synthesis kit (Bio-Rad).
In order to correct for variability in the handling of the samples, the level of the gnd (gluconate-6-phosphate dehydrogenase) mRNA was also determined for each culture, along with the osmU mRNA, and the results were expressed as the level of osmU mRNA/gnd mRNA. The gnd gene was used for normalization because its expression is unaffected by growth rate in Salmonella (53) and varies by <1.4-fold in cells that are adapted to low versus high osmolarity (2). Amplification primers, which were designed with the Primer3 software version 4.0 (47), had the followings sequences (5′ to 3′), with their nucleotide positions downstream from the translation start site of the genes shown in parentheses: gene STM1494 of the osmU operon, GCACACCCTCACCCTAAAAC (positions 3 to 22) and GCAGGTCGGCTGAGTAAAAT (positions 215 to 196), and gnd, CAACATCGAAAGCCGTGGTT (positions 58 to 77) and GGCGTTTCGAGGGATTCAA (positions 198 to 180). qRT-PCR amplifications were performed using an AB 7300 RT-PCR thermocycler (Applied Biosystems) with FastStart Universal SYBR green Master (Rox) (Roche). After an initial denaturation step (95°C for 15 min), the reactions involved up to 40 cycles of denaturation (95°C for 30 s), annealing (59° for 1 min), and elongation (72° for 30 s). For each of the eight independent cultures, the osmU mRNA was analyzed in two duplicate reactions, and the gnd mRNA was analyzed in a single determination.
The mRNA levels for the STM1494 gene of the osmU operon and gnd were quantified as the threshold cycle (CT) value, which is the number of PCR cycles in the exponential phase of amplification that result in an equal signal for the two genes. The osmotic induction ratio of the osmU operon, normalized relative to the abundance of the gnd mRNA, was calculated as described by Livak and Schmittgen (38). The duplicate osmU CT values were averaged for each culture, and the corresponding gnd CT value was subtracted from the average osmU CT, generating a ΔCT for each culture. The ratio of the abundance of the osmU mRNA per gnd mRNA was calculated as 2−ΔCT and averaged (± the standard deviation) separately for the four cultures grown with 0.3 M NaCl or without NaCl. The osmotic induction ratio of osmU was calculated by dividing the average ratio (± the variability) of the osmU/gnd mRNA levels for both types of cultures by the average osmU/gnd ratio of the cultures grown without NaCl (setting the ratio for the culture grown without NaCl to 1).
Modeling of OsmX protein.
The sequences of OsmX and Archaeoglobus fulgidus ProX proteins were aligned with CLUSTAL W (http://www.ebi.ac.uk/Tools/msa/clustalw2) (36). Residues 153 to 161 of OsmX were omitted because this region is an apparent insertion on the surface of the C-terminal lobe of OsmX and remote from the substrate binding site. The sequence alignment and the structure coordinates of ProX bound to glycine betaine (Protein Data Bank identifier, 1SW2) were entered into Modeler (23) to produce a model of OsmX bound to the substrate.
RESULTS AND DISCUSSION
Evidence for the third glycine betaine transport system, the OsmU system, in Salmonella.
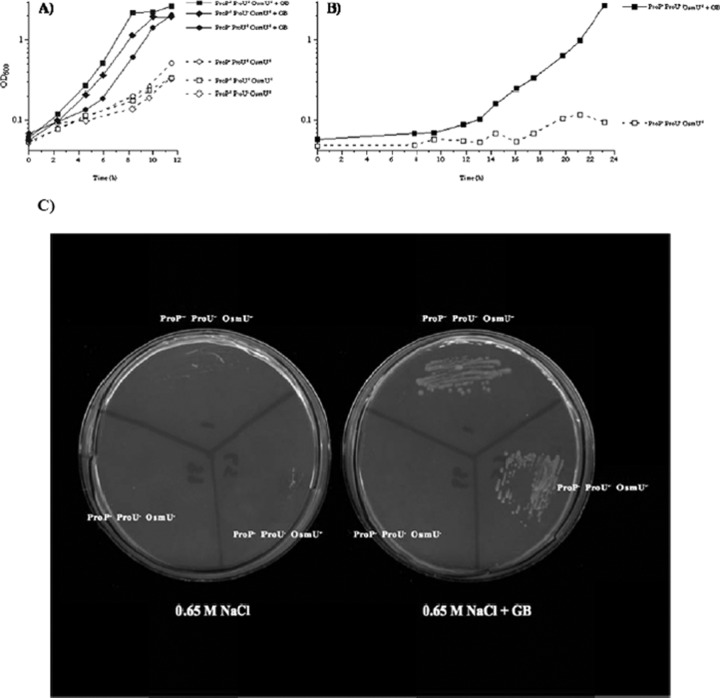
The observation that S. Typhimurium proP proU double mutants are still able to use glycine betaine as an osmoprotectant is reproduced in Fig. 2. Panel A of this figure shows the well-documented ability of glycine betaine to alleviate the inhibitory effects of high osmolarity in strains that have a functional ProP and/or ProU system. As previously observed (27), elimination of both the ProP and the ProU systems resulted in increased sensitivity to high osmolarity (Fig. 2B); the reason for this phenotype is not understood. However, the ProP− ProU− double mutant responded to residual growth rate stimulation by glycine betaine in both liquid and solid media containing 0.65 M NaCl (Fig. 2B and C, respectively). Although glycine betaine stimulated the growth rate of the ProP− ProU− double mutant in high-osmolarity media, it was not as effective for the double mutant as for ProP+ or ProU+ strains. In high-osmolarity liquid media, the growth-stimulatory effect of glycine betaine for the ProP− ProU− double mutant was generally observable after a variable lag phase, lasting between 2 and 12 h. The fact that glycine betaine can still function as an osmoprotectant for the ProP− ProU− double mutant suggests that there is a third transporter for glycine betaine in S. enterica, which we designate the OsmU system (for Osmoprotectant Uptake).
Fig 2.
Glycine betaine can alleviate the inhibitory effects of high osmolarity in a ProP− ProU− OsmU+ strain. (A and B) The effect of glycine betaine in liquid M63 glucose plus 0.65 M NaCl was determined as described in Materials and Methods. Open symbols, strains grown in the absence glycine betaine; closed symbols, strains grown with of 1 mM glycine betaine (GB). Results for strains TL1 (wild type; ProP+ ProU+ OsmU+), TL188 (ProP+ ProU− OsmU+), and TL3463 (ProP− ProU+ OsmU+) (A) and strain TL3465 (ProP− ProU− OsmU+) (B) are shown. (C) Osmoprotective effect of glycine betaine on solid medium containing M63 glucose plus 0.65 M NaCl (left petri dish) or with 1 mM glycine betaine (right petri dish). Cells from single colonies grown on M63 glucose plates were streaked onto these plates, followed by incubation at 37°C for 4 days. The test strains were as follows (clockwise from the top): TL1 (ProP+ ProU+ OsmU+), TL3465 (ProP− ProU− OsmU+), and TL4099 (ProP− ProU− OsmU−).
To identify the gene(s) for this novel permease, we carried out Blastp searches (blast.ncbi.nlm.nih.gov/Blast.cgi) (1) of the predicted proteome of S. Typhimurium LT2, using the sequences of the S. Typhimurium ProVWX and ProP proteins as queries. In addition to the expected perfect matches of these proteins to their own sequences, the sequences of both permeases resemble a number of other proteins with high statistical significance. As has been already noted for E. coli (9, 39), ProV matches YehX, and ProW matches YehW and YehY with 54 to 56% similarity in S. Typhimurium also. However, ProV also shows 57% sequence similarity (with an expect value [E] of 6e-64) to the predicted S. Typhimurium protein STM1491, and ProW matches STM1494 and STM1492 with 50 and 54% similarities, respectively (E ≤ 6e-15). The close clustering of the STM1491, STM1492, and STM1494 genes suggests that they are components of ABC-type transport systems, whose ATP binding component is STM1491 and whose membrane-bound transport pores are made up of heterodimeric proteins (STM1492 and STM1494). Both the yehYXW and the STM491, STM1492, STM1494 clusters also include genes encoding potential periplasmic binding proteins (YehZ and STM1493), but the latter two proteins do not have meaningful similarity to ProX (E ≥ 0.039), indicating that the predicted periplasmic binding proteins of these two ABC transporters are not closely related to ProX. Because, in contrast to E. coli K-12, the ability to use glycine betaine is not abolished by proP proU double mutations in Salmonella, we hypothesized that the transport system consisting of STM1491 to STM1494 is more likely to be the additional osmoprotectant transporter in Salmonella than the transporter made up of YehW, YehY, YehX, and YehZ, since the former set of proteins is absent from E. coli K-12, whereas there are faithful orthologs for the latter set in both organisms. Therefore, we targeted a deletion to STM1491 to STM1494. In analogy with the ProU system, we renamed the STM1491, STM1492, STM1493, and STM1494 genes osmV, osmW, osmX, and osmY, respectively. These four genes are arranged as a potential operon, transcribed in the direction osmYXWV, whose protein products constitute the OsmU system.
In the Blastp analysis with ProP, we found six ORFs—YhjE, ShiA, TcuT, KgtP, NanT, and YhjB—that exhibited significant amino acid sequence similarities (E < 1e-15) to this member of the MFS family of transporters. Of these, only TcuT, which encodes a known tricarboxylate transport protein, is unique to Salmonella, whereas the other five are also found in E. coli K-12. Furthermore, ShiA, KgtP, and NanT have been shown to transport shikimate, α-ketoglutarate, and sialic acid, respectively. Therefore, we considered it unlikely that an additional glycine betaine transporter in Salmonella would be a ProP paralog.
The osmU deletion eliminates the residual osmoprotection by glycine betaine in a proP proU double mutant.
The effect of inactivation of the OsmU system on the ability of the strain to use glycine betaine as an osmoprotectant was tested by growth studies in Fig. 3: deletion of the osmU in a mutant that is lacking the ProP and the ProU systems abolished the residual ability of glycine betaine to stimulate growth in medium containing 0.65 M NaCl (compare Fig. 3A to Fig. 2B). The result that the osmU deletion abolished the residual response of the proP proU mutant to glycine betaine as an osmoprotectant was reproduced on solid media of high osmolarity (Fig. 2C). These observations demonstrate that the OsmU is the third transport system, which in addition to ProP and ProU, can import glycine betaine in S. enterica. In strains in which ProP or ProU are functional, the osmU deletion had no discernible effect on the utilization of glycine betaine as an osmoprotectant (compare Fig. 3B and Fig. 2A).
Fig 3.
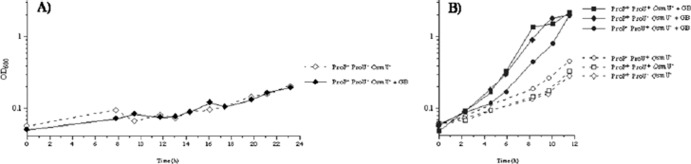
Deletion of the osmU operon eliminates the accumulation of glycine betaine as an osmoprotectant in a ProP− ProU− strain. The effect of glycine betaine on the growth of strains was determined in M63 glucose plus 0.65 M NaCl, as described in Materials and Methods. Open and closed symbols: strains grown in the absence or presence of 1 mM glycine betaine, respectively. (A) Strain TL4099 (ProP− ProU− OsmU−); (B) strains TL4093 (ProP+ ProU+ OsmU−), TL4095 (ProP+ ProU− OsmU−), and TL4097 (ProP− ProU+ OsmU −).
The osmU deletion abolishes the accumulation of glycine betaine and choline-O-sulfate in a proP proU double mutant.
Although data presented above support the conclusion that the OsmU system can transport glycine betaine, our laboratory (28) and others (6) have not been able to detect any glycine betaine transport activity via this permease by standard radioactive assays. This situation is reminiscent of the problem in the initial characterization of ProU, which has proven to be recalcitrant to measurement by radioactive proline transport assays, despite strong physiological evidence that it can take up this osmoprotectant (11, 20). The reason for the inability to assay the glycine betaine transport activity of OsmU is not clear, but one possibility could be that this transporter has a high Km (>1 mM) for glycine betaine, as is the case for ProU and proline (48).
In lieu of short-term uptake assays, we carried out long-term studies of the accumulation of [14C]glycine betaine and [14C]choline-O-sulfate, similar to the experiments that demonstrated the long-term accumulation of proline by the ProU system (11). Since Salmonella is unable to catabolize glycine betaine and choline-O-sulfate (27, 30, 33), the radioactivity recovered in the cells represents the accumulation of these substrates in the cytoplasm, rather than the incorporation of their breakdown products into macromolecules. Table 2 shows the results of these experiments. The wild-type strain TL1 accumulated ∼6 × 102 nmol of glycine betaine/mg of protein and ∼1.6 × 103 nmol of choline-O-sulfate/mg of protein after overnight growth in M63 containing 0.3 M NaCl plus 1 mM concentrations of the respective radioactive substrates. The introduction of single proU and proP mutations diminished the accumulation of glycine betaine by 51 and 29% and the accumulation of choline-O-sulfate by 59 and 17%, respectively. Although accumulation of glycine betaine and choline-O-sulfate was reduced by 82 and 66%, respectively, by the proP proU double mutations, the osmU+ strain TL3465 was still able to accumulate both of these osmolytes. The accumulation of the two osmoprotectants in the proU proP osmU triple mutant TL4099 was reduced to <1% of the level seen in the wild type. These results provide more direct evidence for the conclusion that the OsmU system is able to take up both glycine betaine and choline-O-sulfate. The fact that the ProU−, ProP−, and OsmU− single mutants contained less glycine betaine and choline-O-sulfate than the wild type indicates that each of the three transporters contributes to the accumulation of these quaternary amines. The long-term accumulation of glycine betaine and choline-O-sulfate is dependent on osmotic stress because the wild-type strain accumulated <4 and <1% of these compounds, respectively, in M63 without NaCl than it did in M63 plus 0.3 M NaCl. Finally, the results in Table 2, together with the growth data in Fig. 2, show that despite its similarity to OsmU or ProU, the YehWXYZ transport system is not effective in accumulating glycine betaine or choline-O-sulfate.
Table 2.
Long-term accumulation of glycine betaine and choline-O-sulfate in strains carrying various combinations of mutations in osmoprotectant transport systemsa
| Strain | Presence or absence of functional osmoprotectant transporter |
NaCl concn (M) | Mean accumulation (nmol/mg of protein) ± SEM |
|||
|---|---|---|---|---|---|---|
| ProP | ProU | OsmU | Glycine betaine | Choline-O-sulfate | ||
| TL1 | + | + | + | 0.3 | 627 ± 244 | 1,612 ± 46 |
| TL188 | + | – | + | 0.3 | 307 ± 145 | 661 ± 45 |
| TL3463 | – | + | + | 0.3 | 446 ± 125 | 1,338 ± 116 |
| TL3465 | – | – | + | 0.3 | 116 ± 29 | 547 ± 163 |
| TL4093 | + | + | – | 0.3 | 422 ± 203 | 963 ± 123 |
| TL4095 | + | – | – | 0.3 | 265 ± 99 | 283 ± 22 |
| TL4097 | – | + | – | 0.3 | 504 ± 176 | 874 ± 15 |
| TL4099 | – | – | – | 0.3 | 5.2 ± 3.4 | 1.3 ± 0.7 |
| TL1 | + | + | + | 0.0 | 23 ± 2 | 34 ± 20 |
| TL4099 | – | – | – | 0.0 | 4.6 ± 2.4 | <2.5 |
Strains were grown in M63, glucose, the indicated NaCl concentration, and either 1 mM glycine betaine plus 1 nCi of [14C]glycine betaine or 1 mM choline-O-sulfate plus 1 nCi of [14C]choline-O-sulfate. The long-term accumulation of the radioactive substrates was determined as described in Materials and Methods. The data are the averages of the results of three independent measurements.
Competition between glycine betaine and other osmoprotectants for accumulation by OsmU.
Because the OsmU system appears to have a low affinity for glycine betaine, we tested whether it might prefer some other osmoprotectant as a substrate. Accordingly, we determined the efficacies of various osmoprotectants as competitors for the long-term accumulation of glycine betaine by the OsmU+ ProP− ProU− mutant in M63 plus 0.3 M NaCl containing 10 μM [14C]glycine betaine and 1 mM concentrations of the test compounds. It should be emphasized that these experiments do not measure transport rates because the net accumulation of osmoprotectants is not only determined by the affinity of the transport system, but it also depends on excretion by efflux systems (37) and on the final biomass. Therefore, these experiments can be expected to give at most qualitative insights into the substrate specificity of the transporter.
The results of these competition studies are shown in Table 3. When glycine betaine was used as the competitor at a 100-fold excess, it diminished the accumulation of radioactivity only by 81%. If the accumulation of glycine betaine were determined only by the Km of the OsmU system for this substrate, it would be expected that this competitor would have reduced the intracellular radioactivity 100-fold, proportionally to the reduction in the radioactive specific activity. However, the cells that were grown in 1 mM glycine betaine had a 13-fold-higher level of glycine betaine than the cells that were grown in the presence of 10 μM glycine betaine (data not shown), and therefore the higher total accumulation of glycine betaine in the former culture can account for the apparent excess of radioactivity in those cells. The most important result in Table 3 is that choline-O-sulfate, added at a 100-fold excess over [14C]glycine betaine, reduced the accumulation radioactivity by 99%. d-Carnitine, l-carnitine, and proline betaine caused a qualitatively similar decrease in the accumulation of radioactivity to that resulting from glycine betaine, suggesting that these compounds are probably not better substrates for OsmU than glycine betaine. Ectoine, dimethylsulfoniopropionate, 4-hydroxyproline betaine, glycerol 3-phosphorylcholine, pipecolate betaine, γ-aminobutyrobetaine, homarine, and proline were even less effective competitors. Thus, the major insight from these experiments is that of the 13 compounds tested, only choline-O-sulfate is a better substrate for OsmU than glycine betaine.
Table 3.
Competition between glycine betaine by other osmoprotectants for uptake via the OsmU systema
| Competitor compound | Mean amt of GB taken up (nmol/mg protein) ± SEM | % Reduction in GB accumulation |
|---|---|---|
| None | 100 ± 13 | 0 |
| GB at: | ||
| 1 mM | 19 ± 7 | 81 |
| 0.5 mM | 34 ± 16 | 66 |
| Choline-O-sulfate | 0.7 ± 0.3 | 99 |
| d-Carnitine | 19 ± 12 | 81 |
| l-Carnitine | 19 ± 12 | 81 |
| Proline betaine | 25 ± 6 | 75 |
| Ectoine | 54 ± 14 | 46 |
| β-Dimethylsulfoniopropionate | 60 ± 12 | 40 |
| 4-Hydroxyproline betaine | 76 ± 14 | 24 |
| Glycerol 3-phosphorylcholine | 78 ± 24 | 22 |
| Pipecolate betaine | 79 ± 13 | 21 |
| γ-Aminobutyrobetaine | 85 ± 27 | 15 |
| Homarine | 95 ± 18 | 5 |
| Proline | 95 ± 21 | 5 |
Strain TL3465 (osmU+ proP-4::Tn10dCm proU1655::Tn10) was grown in M63, glucose, 0.3 M NaCl, 10 μM glycine betaine (GB) containing 1 nCi of [14C]glycine betaine, and the indicated unlabeled competitor compounds added at 1 mM, except for glycine betaine, which was also used at 0.5 mM. The efficacy of the compounds as competitors of glycine betaine uptake was determined as described in Materials and Methods. The data are averages of the results of three independent measurements.
As a further test of the substrate specificity of OsmU, we carried out additional competition experiments between 10 μM [14C]glycine betaine and unlabeled choline-O-sulfate and between [14C]choline-O-sulfate and unlabeled glycine betaine (Table 4). The addition of 0.05, 0.1, and 0.2 mM unlabeled choline-O-sulfate to the cultures grown with 10 μM [14C]glycine betaine reduced the accumulation of radioactivity by 46, 72, and 96%, respectively. In contrast, glycine betaine did not cause a significant reduction in the intracellular level of [14C]choline-O-sulfate even at a 100-fold excess. These results confirm that choline-O-sulfate is a better substrate for OsmU than glycine betaine. However, choline-O-sulfate is not more potent an osmoprotectant than glycine betaine, because it conferred a 2.5-fold stimulation of the growth rate of the OsmU+ ProP− ProU− strain in M63 glucose plus 0.65 M NaCl, similar to the 2.6-fold stimulation seen with glycine betaine (data not shown). Furthermore, despite the higher preference of the OsmU system for choline-O-sulfate over glycine betaine, we have been also unsuccessful in measuring the activity of this permease with choline-O-sulfate in the standard short-term radioactive transport assays (data not shown). As is the case for glycine betaine, choline-O-sulfate cannot stimulate the growth rate of the proP proU osmU triple mutant in high-osmolarity media (data not shown), indicating that there is no additional efficient uptake system for choline-O-sulfate besides ProP, ProU, and OsmU.
Table 4.
Competition between glycine betaine and choline-O-sulfate for accumulation via the OsmU system
| Radioactive substrate (10 μM) | Unlabeled competitor | Concn (mM) of unlabeled competitor | Mean amt of radioactive substrate accumulated (nmol/mg of protein) ± SEMa | % Radioactive substrate accumulated in the presence of the competitorb |
|---|---|---|---|---|
| Choline-O-sulfate | Glycine betaine | 0 | 183 ± 33 | 100 |
| 0.5 | 223 ± 93 | 124 | ||
| 2 | 190 ± 15 | 104 | ||
| 10 | 216 ± 29 | 116 | ||
| Glycine betaine | Choline-O-sulfate | 0 | 205 ± 12 | 100 |
| 0.05 | 110 ± 23 | 54 | ||
| 0.1 | 58 ± 18 | 28 | ||
| 0.2 | 8.7 ± 1.8 | 4 |
Strain TL3465 (osmU+ proP-4::Tn10dCm proU1655::Tn10) was grown in M63, glucose, 0.3 M NaCl, and either 1 nCi of [14C]choline-O-sulfate or 1 nCi of [14C]glycine betaine in 10 μM carrier plus unlabeled competitor glycine betaine or choline-O-sulfate, respectively, at the indicated concentrations. The accumulation of the radioactive substrate was determined as described in Materials and Methods. The data are averages of the results of three independent measurements.
The percent radioactive substrate accumulated in the presence of the competitor at the indicated concentrations was calculated relative to the accumulation in the absence of competitor set to 100%.
The osmU operon is induced by osmotic stress.
We carried out qRT-PCR analysis to probe whether the transcription of osmU is regulated by the osmolarity of the medium. This analysis indicated that the level of the osmV mRNA, normalized to the level of the constitutive gnd mRNA, was 23-fold higher in cells grown in MOPS plus glucose plus 0.3 M NaCl than in cells grown in MOPS plus glucose (Fig. 4). The fact that the osmU operon is induced by osmotic stress provides additional support for the conclusion that OsmU is a transport system for osmoprotectants. However, it will be necessary to carry out more extensive characterization of the transcriptional control of osmU by various factors, including the NaCl concentration, osmoprotectants, and growth phase.
Fig 4.
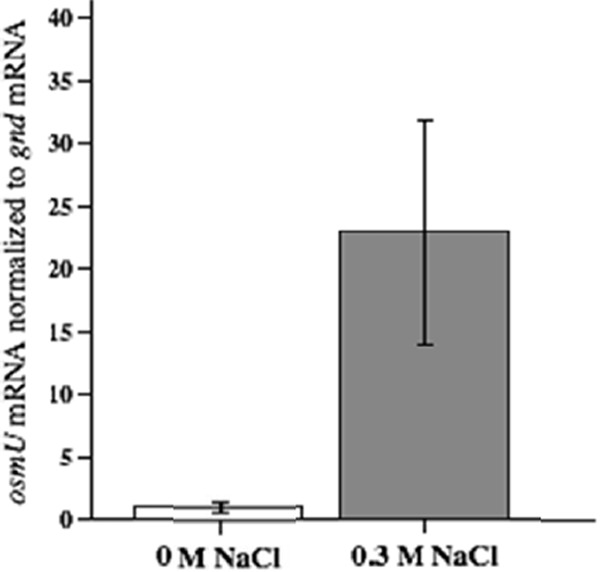
The osmU operon is induced by osmotic stress. Strain TL1 was grown in MOPS medium plus glucose with 0 M or 0.3 M NaCl, and the osmU and gnd transcript levels were determined by qRT-PCR as described in Materials and Methods. The data are shown as the ratios of osmU and gnd mRNAs, with the ratio for the cells grown with 0 M NaCl set to 1. For both media, the results are the averages of results obtained with four independent mid-exponential phase cultures. In a two-sample, one-tailed t test, the difference in the ratios of osmU/gnd expression at 0.3 M NaCl and 0 M NaCl was significant (P < 0.01).
Phylogenetic distribution of OsmU.
We carried out a bioinformatic survey of the database for occurrence of OsmU orthologs in bacterial and archaeal genomes, using the Blastn discontinuous megablast algorithm (http://blast.ncbi.nlm.nih.gov/Blast.cgi [default settings]). The osmVWXY genes are present in 63 S. enterica genomes, with 96 to 100% conservation over ≥3,324 of the 3,447 nucleotides of the S. enterica serovar Typhimurium osmU operon. Orthologs of the osmU operon that show high nucleotide sequence similarity are also evident in the following genera or species in the family of Enterobacteriaceae for which complete genomic sequences are available: Citrobacter (two species), Enterobacter (three species), Escherichia fergusonii (three strains), Cronobacter (two species), Pantoea (three species), and Salmonella bongori, Serratia marcescens Db11, and Erwinia billingiae (one species, each). The genomes of these organisms contain matches for each of the osmVWXY genes (in that order) and the intergenic spacers with Blastn sequence conservation ranging from 91% (in S. bongori) to 71% (in Pantoea ananatis) throughout ≥96% of the entire operon (E < 1e-99).
The only other completely sequenced organism that has a good nucleotide sequence ortholog for osmU is Chromobacterium violaceum (betaproteobacterium, member of the Neisseriaceae), which contains a potential operon of four genes that matches the S. Typhimurium LT2 osmU operon with 53% Blastn identity (E < 1e-99) over 85% of the sequence. The lower sequence similarity of the osmU operon of C. violaceum compared to all of the enterobacterial versions leads us to speculate that the betaproteobacterium may have acquired this operon by a horizontal gene transfer event that was independent of the introduction of this operon into the Enterobacteriaceae.
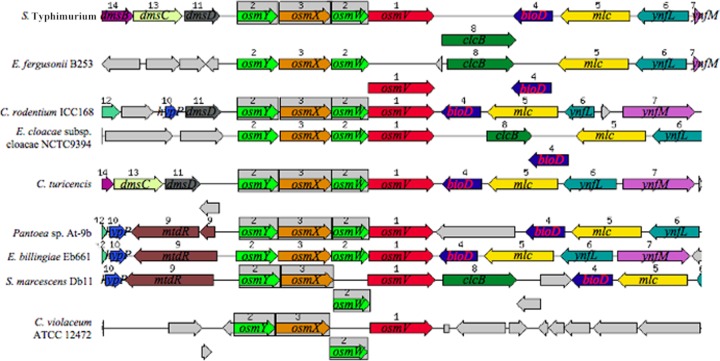
We used the SEED database (http://pubseed.theseed.org/seedviewer.cgi) (43) to analyze the genetic neighborhoods of the osmU orthologs in the chromosomes of the Enterobacteriaceae that contain significant nucleotide sequence matches to this operon (Fig. 5). In all S. enterica serovars, as well as in S. bongori, the osmU operon is located between the dms operon and the clcB, bioD, mlc, and yntL genes. Although this grouping of genes is not perfectly conserved in other Enterobacteriaceae that have osmU, all of these organisms have orthologs in common near osmU (especially on the 3′ side of the operon). The conservation of the arrangement of genes suggests that the osmU operon probably has been derived by these organisms from a chromosomal segment that was present in a common ancestor. Other members of the Enterobacteriaceae that do not have an osmU operon, e.g., all other species of E. coli, Klebsiella, and Yersinia, have some or all of the flanking genes in this neighborhood but without the intervening osmU operon (data not shown). Interestingly, although S. marcescens Db11 has an osmU operon, S. odorifera, S. proteamaculans, and five other Serratia species lack these genes, even though other genes that are found in the proximity of the osmU operon in other Enterobacteriaceae are also conserved in Serratia species. In C. violaceum, the osmU ortholog is in a chromosomal region that does not share any significant conservation of genetic neighborhood with the arrangement in Enterobacteriaceae, in accord with the suggestion that the osmU operon may have been acquired by this strain by horizontal transfer independently of the event that introduced this operon into a subset of the Enterobacteriaceae. None of the bacteria that have orthologs of the four genes of the osmU operon has a closely situated gene that specifies a predicted transcriptional regulatory gene. Therefore, the osmotic induction of osmU is mediated either by a regulatory protein that is encoded in a distal gene or by differential interaction of RNA polymerase with the osmU promoter at low and high osmolarity.
Fig 5.
Conservation of orthologous genes flanking the osmU operon in Enterobacteriaceae. The synteny of genes was analyzed and displayed using the SEED database and algorithm (http://pubseed.theseed.org/seedviewer.cgi) (43). Sets of orthologous genes that are conserved around the osmU operon are shown in the same colors, with the direction of transcription indicated by the arrows; genes that are gray are not conserved across these organisms. Conserved orthologs encode the proteins as follows: dmsBCD, three subunits of dimethylsulfate reductase; clcB, Cl− channel; bioD, dethiobiotin synthetase; mlc, maltose regulon repressor; ynfL, putative LysR family repressor; ynfM, major superfamily transporter; mldR, multidrug resistance protein B; hypP, hypothetical protein. The organisms shown are S. Typhimurium, Escherichia fergusonii B253, Citrobacter rodentium ICC168, Enterobacter cloacae subsp. cloacae NTC9394, Cronobacter turicensis, Pantoea sp. strain At-9b, Erwinia billingiae Eb661, Serratia marcescens DB11, and Chromobacterium violaceum ATCC 12472.
We also carried out a wider survey of the occurrence of OsmU orthologs in Bacteria and Archaea, based on amino acid sequence conservation, using the Blastp algorithm and the SEED database. The assignment of transport systems as orthologs of OsmU was based on two criteria: (i) that the protein components of the putative ortholog should show highly significant sequence match to each of the Salmonella OsmV, OsmW/Y, and OsmX proteins (E ≤ 5e-20) and (ii) that genes for these proteins be directly adjacent to each other on the chromosome of the host organism. However, it should be borne in mind that high amino acid sequence conservation of the subunits of such systems alone is not sufficient evidence for the fact that they transport the same or related substrates in different organisms. Furthermore, we cannot make any inferences from bioinformatic analysis alone as to whether or not these transport systems are necessarily functional in their host organisms. With these caveats in mind, we made the following observations in this analysis.
Except for the Mollicutes, there are good orthologs of the four proteins of the OsmU system encoded by genes arranged in a potential operon organization in other eubacterial phyla in the National Center for Biotechnology Information Taxonomy database (24) (Actinobacteria, Bacteroides, Chlorobi, Chlamydiae, Cyanobacteria, Firmicutes, Spirochaetes, and five subdivisions of Proteobacteria). Mollicutes, which lack peptidoglycan and consequently cannot maintain a turgor pressure, do have a number of ATP-binding components of ABC transport systems that are highly similar to the OsmV protein of Salmonella (with expect value as low as 3e-28), but none of these organisms have meaningful orthologs of the transmembrane pore and the extracellular binding protein components of the OsmU system. In the archaeal domain, there are organisms in the phylum Euryarchaeota that have robust matches to each of the four proteins of OsmU that are specified by a set of directly adjacent genes, but none of the members of the phyla of Crenarchaeota and Nanoarchaeota contain ABC-type transport systems that show significant matches to the OsmW/Z and OsmX proteins.
Among the three types of components of paralogous ABC osmoprotectant transporters, it is generally the extracytoplasmic binding proteins that show the highest sequence divergence, suggesting that the sequence differences might reflect differences in the substrate specificities of these permeases (45, 49). Crystal structures have been obtained for eight binding proteins of ABC-type transport systems for quaternary amine osmoprotectants: the ProX component of the ProU system of E. coli (48), the ProX protein of the thermophilic archaeon Archaeoglobus fulgidus (49), the OpuBC (45), OpuCC (19), and OpuAC (50) proteins of Bacillus subtilis, the ChoX protein of Sinorhizobium meliloti (42), the substrate-binding fragment OpuAC of the OpuABC protein of Lactococcus lactis (54), and the OpuCC protein of Staphylococcus aureus (PDB ID 3O66). Figure 6 shows the phylogenetic relationship of the predicted OsmX and YehZ proteins of Salmonella to these other proteins. In this small sample of proteins, OsmX is most closely related to YehZ, whose substrate specificity is not known, and to the A. fulgidus ProX protein, which has been shown to recognize glycine betaine and proline betaine with apparent KD values of ∼50 nM (49). These three proteins fall into the same clade that also includes the B. subtilis OpuCC, which has broad specificity for osmoprotectants (19, 32); the B. subtilis OpuBC protein, which is highly specific for choline (45); and the protein OpuCC of S. aureus, whose substrate specificity has not been determined. A different clade is formed by the glycine betaine/proline betaine binding fragment OpuAC from Lactococcus lactis (54), the glycine betaine/proline betaine/dimethylsulfonioacetate binding protein OpuAC of B. subtilis (50), the choline/acetylcholine binding protein ChoX of S. meliloti (42), and the ProX component of the ProU system of E. coli, which has high affinity for glycine betaine and proline betaine (KD values of 1 and 5 μM, respectively) but does not bind proline. This analysis suggests that ABC-type osmoprotectant transport systems can be divided into two broad families on the basis of the sequences of the binding proteins, an OsmX family and a ProX family, both of which have representatives in a wide variety of Bacteria and Archaea.
Fig 6.
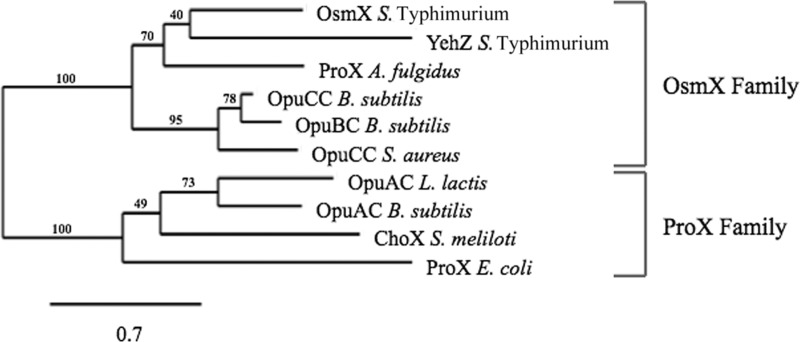
Phylogenetic relationship of the S. Typhimurium OsmX protein to YehZ and the extracellular binding protein components of other osmoprotectant transport systems whose three-dimensional structure has been determined. The phylogenetic tree of the protein sequences (including the signal peptides) was constructed with the PhyML method and displayed by the web service Phylogeny.fr (http://www.phylogeny.fr), using the “advanced mode, bootstrapping procedure” (100 bootstraps), without the Gblocks program (7, 17, 18, 22). The protein sequences (database accession codes) are as follows: OsmX, S. Typhimurium (GI:16764838); YehZ, S. Typhimurium (GI:16765495); ProX, A. fulgidus DSM 4304 (GI:11498587); OpuCC, B. subtilis subsp. subtilis strain 168 (GI:16080434); OpuBC, B. subtilis subsp. subtilis strain 168 (GI:16080424); OpuCC, S. aureus subsp. aureus Mu50 (GI:15925436); OpuAC, L. lactis (GI:296278460); OpuAC, B. subtilis subsp. subtilis strain 168 (GI:16077369); ChoX, S. meliloti 1021 (GI:15966152), and ProX, E. coli strain K-12 substrain MG1655 (GI:16130593).
Predicted three-dimensional structure of the OsmX protein.
Among the osmoprotectant binding proteins for which crystallographic structures are available, the S. enterica OsmX protein shows the highest similarity to the A. fulgidus ProX (Af-ProX) protein (Fig. 6). Therefore, we modeled the OsmX protein on the structure of Af-ProX. The results of this modeling are shown in Fig. 7. Crystallographic analysis of the latter protein revealed that its substrate-binding site contains four tyrosines (Y63, Y111, Y190, and Y214) that form an “aromatic cage” that binds the quaternary ammonium group of betaines with cation-π interactions that is sealed at one end by D109, which also contributes to the binding of the quaternary ammonium group by its main-chain oxygen atom (49). The carboxylate group of glycine betaine interacts via salt bridges and a hydrogen bond with K13, R149, and T66.
Fig 7.
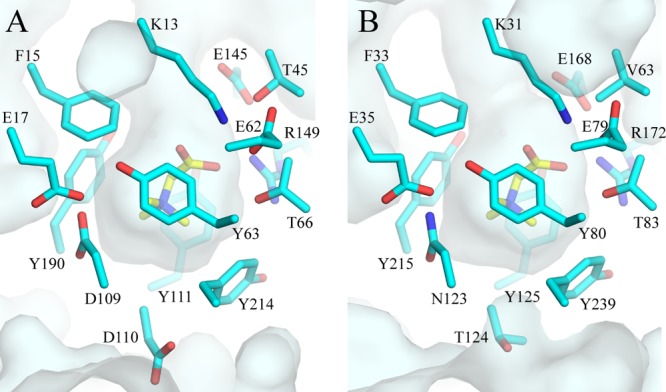
S. enterica OsmX modeled on the glycine betaine-bound conformation of A. fulgidus ProX. A surface slab view is shown for ProX (A) and the modeled OsmX (B) bound to glycine betaine (yellow). Residues within 5 Å of the bound substrate are indicated in cyan. Figure was constructed with PyMOL (www.pymol.org) as described in Materials and Methods. The amino acids are numbered starting from the first residue of the mature protein for ProX (49) and starting from the N-terminal methionine of the predicted full-length protein, including the signal sequence, for OsmX.
Nearly all residues of Af-ProX that are within 5 Å of glycine betaine, including the aromatic cage around the quaternary ammonium moiety, are strictly conserved in OsmX, except for residues V63, N123, and T124 of OsmX (T45, D109, and D110 of Af-ProX). The residue corresponding to N123 of OsmX is also an asparagine in S. Typhimurium YehZ, B. subtilis OpuBC and OpuCC, and S. aureus OpuCC, which are in the same clade with OsmX (Fig. 5); N123 could interact via the main-chain oxygen with the quaternary ammonium moiety of the substrate in the same way as D109 does in Af-ProX. The T124 in OsmX would probably be able to maintain similar van der Waals interactions with the substrate as D110 in Af-ProX. However, the switch of the T45 in At-ProX to the V63 in OsmX would be expected to abolish a hydrogen bond interaction with the substrate carboxylate group in OsmX. This difference might be responsible for the low affinity of the OsmU system for glycine betaine. However, the robustness of the transport of various substrates may not be determined solely by the selectivity of the binding proteins, but it could also be dependent on the interaction of the transmembrane pore proteins with the substrate-binding proteins or with the substrates. Experimental confirmation of the predicted substrate-binding site of OsmX will have to wait until the identification of a molecule that is recognized with high affinity by this protein.
Concluding remarks.
Although we obtained evidence that the OsmU system is an additional transport system besides ProP and ProU that can take up osmoprotectants in Salmonella, our results (Tables 3 and 4) suggest that OsmU has a low affinity for glycine betaine, which is typically one of the most potent osmoprotectants for bacteria. Choline-O-sulfate appears to be a more efficient substrate for OsmU than glycine betaine, but we were not able to demonstrate the uptake of choline-O-sulfate via this transporter in the standard short-term transport assays. Our inability to assay the activity of OsmU with choline-O-sulfate and glycine betaine could be due to the fact that this transporter has a low affinity for these osmoprotectants (Km > 1 mM). This negative result could mean either that the OsmU system is effective in accumulating osmoprotectants only if they are present at high concentrations externally, or it might recognize some uncharacterized osmoprotectants with high affinity. Like all other Salmonella serovars, S. enterica serovar Typhi possesses an OsmU system, but in all sequenced members of this serovar, the ProU system is predicted to be nonfunctional due to a −1 frameshift in the proV gene (44). Therefore, the OsmU system may be of special importance for the accumulation of osmoprotectants in this human pathogen. The fact that the OsmU paralog YehZYXW does not transport glycine betaine or choline-O-sulfate may suggest the existence of other osmoprotectants for Salmonella and other Enterobacteriaceae that have not yet been discovered.
ACKNOWLEDGMENTS
We thank B. J. Gasper and Y. F. Leung for advice and help with the statistical analysis of the qRT-PCR data.
This study was supported in part by U.S. National Science Foundation (NSF) awards MCB-9978253 and IOS-1054977 (to L.N.C.) and IOS-1025398 (to A.D.H.) and by NSF Career Award MCB-99-84919 (to D.A.S.).
Footnotes
Published ahead of print 18 May 2012
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Balaji B, O'Connor K, Lucas JR, Anderson JA, Csonka LN. 2005. Timing of induction of osmotically controlled genes in Salmonella enterica serovar Typhimurium determined with quantitative real-time RT-PCR. Appl. Environ. Microbiol. 71:8273–8283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barron A, Jung JU, Villarejo M. 1987. Purification and characterization of a glycine betaine binding protein from Escherichia coli. J. Biol. Chem. 262:11841–11846 [PubMed] [Google Scholar]
- 4. Biemans-Oldehinkel E, Doeven MK, Poolman B. 2006. ABC transporter architecture and regulatory roles of accessory domains. FEBS Lett. 580:1023–1035 [DOI] [PubMed] [Google Scholar]
- 5. Brill J, Hoffmann T, Bleisteiner M, Bremer E. 2011. Osmotically controlled synthesis of the compatible solute proline is critical for cellular defense of Bacillus subtilis against high osmolarity. J. Bacteriol. 193:5335–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cairney J, Booth IR, Higgins CF. 1985. Osmoregulation of gene expression in Salmonella typhimurium: proU encodes an osmotically induced betaine transport system. J. Bacteriol. 164:1224–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540–552 [DOI] [PubMed] [Google Scholar]
- 8. Cayley S, Lewis BA, Guttman HJ, Record MT., Jr 1991. Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity. J. Mol. Biol. 222:281–300 [DOI] [PubMed] [Google Scholar]
- 9. Checroun C, Gutierrez C. 2004. σS-Dependent regulation of yehZYXW, which encodes a putative osmoprotectant ABC transporter of Escherichia coli. FEMS Microbiol. Lett. 236:221–226 [DOI] [PubMed] [Google Scholar]
- 10. Cohen GN, Rickenberg RH. 1956. Concentration specifique reversible des amino acides chez Escherichia coli. Ann. Inst. Pasteur Paris 91:693–720 [PubMed] [Google Scholar]
- 11. Csonka LN. 1983. A third l-proline permease in Salmonella typhimurium which functions in media of elevated osmotic strength. J. Bacteriol. 151:1433–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Csonka LN, Epstein W. 1996. Osmoregulation, p 1210–1223 In Neidhardt FC, et al. (ed), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed ASM Press, Washington, DC [Google Scholar]
- 13. Csonka LN, Hanson AD. 1991. Prokaryotic osmoregulation: genetics and physiology. Annu. Rev. Microbiol. 45:569–606 [DOI] [PubMed] [Google Scholar]
- 14. Culham DE, et al. 1994. Genes encoding osmoregulatory proline/glycine betaine transporters and the proline catabolic system are present and expressed in diverse clinical Escherichia coli isolates. Can. J. Microbiol. 40:397–402 [DOI] [PubMed] [Google Scholar]
- 15. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Davis RW, Botstein D, Roth JR. 1980. Advanced Bacterial Genetics, vol Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 17. Dereeper A, Audic S, Claverie JM, Blanc G. 2010. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 10:8 doi:10.1186/1471-2148-10-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dereeper A, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36(Suppl 2):W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Du Y, et al. 2011. Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC. Biochem. J. 436:283–289 [DOI] [PubMed] [Google Scholar]
- 20. Dunlap VJ, Csonka LN. 1985. Osmotic regulation of l-proline transport in Salmonella typhimurium. J. Bacteriol. 163:296–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dunlap VJ, Csonka LN. 1985. Regulation of the osmotically stimulated transport of proline and glycine betaine in Salmonella typhimurium, p 115–128 In Key JL, Kosuge T. (ed), Cellular and molecular biology of plant stress. Alan R. Liss, Inc, New York, NY [Google Scholar]
- 22. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 19:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eswar N, et al. 2007. Comparative protein structure modeling using Modeller. Curr. Protoc. Protein Sci. Chapter 2:Unit 2.9 [DOI] [PubMed] [Google Scholar]
- 24. Federhen S. 2012. The NCBI taxonomy database. Nucleic Acids Res. 40:D136–D143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fletcher SA, Rhodes D, Csonka LN. 2001. Analysis of the effects of osmoprotectants on the high osmolality-dependent induction of increased thermotolerance in Salmonella typhimurium. Food Microbiol. 18:345–354 [Google Scholar]
- 26. Grothe S, Krogsrud RL, McClellan DJ, Milner JL, Wood JM. 1986. Proline transport and osmotic stress response in Escherichia coli K-12. J. Bacteriol. 166:253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gutierrez JA. 1993. Adenylate kinase of Salmonella typhimurium is involved in the maintenance of the energy charge during the uptake of the osmoprotectant glycine betaine. Ph.D. thesis Purdue University, West Lafayette, IN [Google Scholar]
- 28. Gutierrez JA, Csonka LN. 1995. Isolation and characterization of adenylate kinase (adk) mutations in Salmonella typhimurium which block the ability of glycine betaine to function as an osmoprotectant. J. Bacteriol. 177:390–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haardt M, Kempf B, Faatz E, Bremer E. 1995. The osmoprotectant proline betaine is a major substrate for the binding-protein-dependent transport system ProU of Escherichia coli. Mol. Gen. Genet. 248:783–786 [DOI] [PubMed] [Google Scholar]
- 30. Hanson AD, Rathinasabapathi B, Chamberlin B, Gage D. 1991. Comparative physiological evidence that alanine betaine and choline-O-sulfate act as compatible solutes in halophytic Limonium species. Plant Physiol. 97:1199–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jebbar M, Talibart R, Gloux K, Bernard T, Blanco C. 1992. Osmoprotection of Escherichia coli by ectoine: uptake and accumulation characteristics. J. Bacteriol. 174:5027–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kappes RM, Bremer E. 1998. Response of Bacillus subtilis to high osmolarity: uptake of carnitine, crotonobetaine, and γ-butyrobetaine via the ABC transport system OpuC. Microbiology 144:83–90 [DOI] [PubMed] [Google Scholar]
- 33. Koo S-P, Higgins CF, Booth IR. 1991. Regulation of compatible solute accumulation in Salmonella typhimurium: evidence for a glycine betaine efflux system. J. Gen. Microbiol. 137:2617–2625 [DOI] [PubMed] [Google Scholar]
- 34. Ladyman JAR, Hitz WD, Hanson AD. 1980. Translocation and metabolism of glycine betaine by barley plants in relation to water-stress. Planta 150:191–196 [DOI] [PubMed] [Google Scholar]
- 35. Lamark T, et al. 1991. DNA sequence and analysis of the bet genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Mol. Microbiol. 5:1049–1064 [DOI] [PubMed] [Google Scholar]
- 36. Larkin MA, et al. 2007. CLUSTAL W and CLUSTAL X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 37. Levina N, et al. 1999. Protection of Escherichia coli cells against extreme turgor by activation of MscS and MscL mechanosensitive channels: identification of genes required for MscS activity. EMBO J. 18:1730–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Livak KJ, Schmittgen TD. 2001. Analysis of the relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 39. Ly A, Henderson J, Lu A, Culham DE, Wood JM. 2004. Osmoregulatory systems of Escherichia coli: identification of betaine-carnitine-choline transporter family member BetU and distributions of betU and trkG among pathogenic and nonpathogenic isolates. J. Bacteriol. 186:296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. MacMillan SV, et al. 1999. The ion coupling and organic substrate specificities of osmoregulatory transporter ProP in Escherichia coli. Biochim. Biophys. Acta 1420:30–44 [DOI] [PubMed] [Google Scholar]
- 41. Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oswald C, Smits SH, Höing M, Bremer E, Schmitt L. 2009. Structural analysis of the choline-binding protein ChoX in a semi-closed and ligand-free conformation. J. Biol. Chem. 390:1163–1170 [DOI] [PubMed] [Google Scholar]
- 43. Overbeek R, et al. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1,000 genomes. Nucleic Acids Res. 33:5691–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Parkhill J, et al. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848–852 [DOI] [PubMed] [Google Scholar]
- 45. Pittelkow M, Tschapek B, Smits SH, Schmitt L, Bremer E. 2011. The crystal structure of the substrate-binding protein OpuBC from Bacillus subtilis in complex with choline. J. Mol. Biol. 411:53–67 [DOI] [PubMed] [Google Scholar]
- 46. Rhodes D, Hanson AD. 1993. Quaternary ammonium and tertiary sulfonium compounds in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44:357–384 [Google Scholar]
- 47. Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365–386 [DOI] [PubMed] [Google Scholar]
- 48. Schiefner A, et al. 2004. Cation-π interactions as determinants for binding of the compatible solutes glycine betaine and proline betaine by the periplasmic ligand-binding protein ProX from Escherichia coli. J. Biol. Chem. 279:5588–5596 [DOI] [PubMed] [Google Scholar]
- 49. Schiefner A, Holtmann G, Diederichs K, Welte W, Bremer E. 2004. Structural basis for the binding of compatible solutes by ProX from the hyperthermophilic archaeon Archaeoglobus fulgidus. J. Biol. Chem. 279:48270–48281 [DOI] [PubMed] [Google Scholar]
- 50. Smits SH, et al. 2008. The compatible-solute-binding protein OpuAC from Bacillus subtilis: ligand binding, site-directed mutagenesis, and crystallographic studies. J. Bacteriol. 190:5663–5671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tempest DW, Meers JL, Brown CM. 1970. Influence of environment on the content and composition of microbial free amino acid pools. J. Gen. Microbiol. 64:171–185 [DOI] [PubMed] [Google Scholar]
- 52. Thomason L, et al. 2005. Recombineering: genetic engineering in bacteria using homologous recombination. Curr. Protoc. Mol. Biol. 70(Suppl):1.16.1–1.16.21 [DOI] [PubMed] [Google Scholar]
- 53. Winkler ME, Roth DJ, Hartman PE. 1979. Promoter- and attenuator-related metabolic regulation of the Salmonella typhimurium histidine operon. J. Bacteriol. 133:830–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wolters JC, et al. 2010. Ligand binding and crystal structures of the substrate-binding domain of the ABC transporter OpuA. PLoS One 5:e10361 doi:10.1371/journal.pone.0010361 [DOI] [PMC free article] [PubMed] [Google Scholar]