Abstract
The model archaeon Pyrococcus furiosus grows optimally near 100°C on carbohydrates and peptides. Its genome sequence (NCBI) was determined 12 years ago. A genetically tractable strain, COM1, was very recently reported, and here we describe its genome sequence. Of 1,909,827 bp in size, it is 1,571 bp longer (0.1%) than the reference NCBI sequence. The COM1 genome contains numerous chromosomal rearrangements, deletions, and single base changes. COM1 also has 45 full or partial insertion sequences (ISs) compared to 35 in the reference NCBI strain, and these have resulted in the direct deletion or insertional inactivation of 13 genes. Another seven genes were affected by chromosomal deletions and are predicted to be nonfunctional. In addition, the amino acid sequences of another 102 of the 2,134 predicted gene products are different in COM1. These changes potentially impact various cellular functions, including carbohydrate, peptide, and nucleotide metabolism; DNA repair; CRISPR-associated defense; transcriptional regulation; membrane transport; and growth at 72°C. For example, the IS-mediated inactivation of riboflavin synthase in COM1 resulted in a riboflavin requirement for growth. Nevertheless, COM1 grew on cellobiose, malto-oligosaccharides, and peptides in complex and minimal media at 98 and 72°C to the same extent as did both its parent strain and a new culture collection strain (DSMZ 3638). This was in spite of COM1 lacking several metabolic enzymes, including nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase and beta-glucosidase. The P. furiosus genome is therefore of high plasticity, and the availability of the COM1 sequence will be critical for the future studies of this model hyperthermophile.
INTRODUCTION
The genus Pyrococcus represents a group of obligate anaerobic archaea that grow optimally near 100°C and utilize a wide range of poly- and oligosaccharides and peptides (36). They are found in marine hydrothermal vents and are some of the best-studied archaea with potential biotechnological applications (2). Complete genome sequences are available for five Pyrococcus species (9, 26, 28, 34, 49). Their evolution and genomic diversity have been linked to a high degree of DNA recombination efficiency and the presence of mobile genetic elements (58). These insertion sequences (ISs) are small (<2.5-kb), self-directed segments of DNA capable of inserting at many nonhomologous sites in the target DNA (38). Numerous reports have indicated that IS element transposition has led to extensive chromosomal rearrangements and lateral gene transfer (LGT) in the Pyrococcus genus (13, 16, 22, 27, 58, 60). For example, environmental isolates of Pyrococcus differ in their distribution of IS elements, suggesting a high degree of mobility (58), as has been reported for other hyperthermophilic archaea from freshwater vents (40). These genomic features are thought to play a role in the adaptation to rapidly changing environmental conditions (58).
The best characterized of the Pyrococcus species is P. furiosus, the first to be isolated (19) and the first to have its genome sequenced (49). This organism has been studied by numerous “-omics”-based approaches, including transcriptomics by tiling (59) and DNA microarrays (14, 35, 51, 54, 57), comparative genomics (8, 33, 60), proteomics (32, 42), metallomics (11), and structural genomics (25). In many ways, it has become one of the model hyperthermophiles, a status recently sustained by the development of a genetic system for the organism (37). This was based on isolation of a variant in a lab strain population, designated COM1, which was highly efficient in taking up and recombining exogenous DNA in both circular and linear forms. The COM1 strain was obtained by targeted gene disruption of the pyrF locus (PF1114) using a plasmid designed for double-crossover recombination (37).
The COM1 strain has now been used to generate marked or markerless deletions of a number of genes, either singly or iteratively in the same strain, including those encoding two hydrogenases, a sulfur-reducing system and an iron-sulfur biosynthesis system (4, 37). A stable replicating shuttle vector is also available (18). In addition, recombinant forms that contain inserted DNA have been generated, such as highly active promoters or forms encoding affinity tags for rapid protein purification (7, 24). For example, an affinity-tagged, catalytically active, H2 gas-producing subcomplex of P. furiosus hydrogenase was produced at approximately 100 times the level of the native hydrogenase (24). A more robust genetic method that allows for selection on complex growth media for agmatine auxotrophy has also been developed, involving a thermostable compound required for polyamine biosynthesis (50). This was used to produce an affinity-tagged (Strep) version of the native form of the cytoplasmic hydrogenase of P. furiosus (7).
The development of additional genetic tools for COM1 and its use as a platform for genetic manipulation and metabolic engineering can be anticipated. For such studies, the complete genome sequence of this strain is obviously essential for all molecular manipulations. It was also important to determine if the genome sequence of COM1 could reveal any unanticipated phenotypic changes, particularly those affecting its ability to metabolize growth substrates or its requirement for vitamins or cofactors. The genome sequence of COM1 has now been determined, and comparison with the published sequence of the P. furiosus reference strain (49) (NC_003413) revealed a surprisingly large number of changes. The results of this study have implications not only for the utility of this new genetic system for a well-studied hyperthermophile but also for our understanding of the dynamics of genomic change and chromosome maintenance in P. furiosus and of the function of two carbohydrate metabolism enzymes, nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) and β-glucosidase (CelB).
MATERIALS AND METHODS
Strains and growth conditions.
The three P. furiosus strains used in this study are termed COM1, which is the genetically tractable strain (37); Parent, which is the lab strain from which COM1 was derived; and the DSMZ control strain (3638), which was acquired in October 2010 from the German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany). For genomic DNA isolation (and routine growth studies), cells were grown from a single colony of the COM1 strain, the Parent lab strain population, and the minimally passaged DSMZ control strain. They were grown in a complex cellobiose medium as described previously (37) with uracil (20 μM) added to the growth medium of the auxotrophic COM1 strain. Growth experiments to evaluate phenotypes were carried out in biological triplicate in 100-ml serum bottles with 50 ml of complex or defined medium with cellobiose, maltose, or malto-oligosaccharides as carbon sources at 98°C or 72°C. To obtain cells for enzyme assays, cultures were grown on cellobiose, peptides were grown in a 20-liter DCI-Biolafitte BioPro Evo Series SIP fermentor, and cells were harvested at the end of exponential growth.
DNA sequencing.
Cells for genomic DNA isolation were harvested from a 400-ml stationary-phase culture and suspended in 2 ml buffer A (25% sucrose, 50 mM Tris-HCl, 40 mM EDTA, pH 7.4) followed by incubation at 37°C for 1 h with 0.6 mg/ml RNase, 0.2 mg/ml proteinase K, and 0.25 M EDTA, pH 8.0, and then incubation at 65°C for 45 min with 1% SDS, Triton X-100, cetyltrimethylammonium bromide (CTAB), and 0.7 M NaCl. Genomic DNA was extracted using phenol-chloroform-isoamyl alcohol (25:24:1, buffered at pH 8), ethanol precipitated, and suspended in 10 mM Tris buffer, pH 8.0, to a concentration of ∼0.1 μg/μl. All genomic sequencing libraries were prepared according to the manufacturer's guidelines. Single-end 454 sequencing of COM1 was performed using a Roche GS FLX titanium pyrosequencer, with a quarter-plate. Illumina sequencing was performed using a single lane on a GAIIx sequencer. The mate-pair library was constructed with a 2- to 5-kb protocol. Sanger sequencing of PCR products was used to confirm mutations and genome rearrangements. PCR targets were amplified using PfuTurbo DNA polymerase (Stratagene) on a Bio-Rad C1000 thermal cycler. Products were analyzed on an agarose gel and purified using a StrataPrep PCR purification kit (Agilent Technologies).
Genome sequences, assembly, and analysis.
De novo hybrid assembly of COM1 single-end 454 and mate-pair Illumina data was performed using the MIRA assembler (version 3.2.1). Illumina library insert size was estimated using Bowtie version 0.12.7 on the NCBI reference P. furiosus sequence (NC_003413). Scaffolding of the resulting MIRA-produced contigs was performed using SSPACE version 1.1 in extension mode with the mate-pair Illumina reads. Scaffold regions with ambiguous base calls were PCR amplified and sequenced. Genome block alignments were performed using Progressive Mauve version 2.3.1. Fuzznuc, part of the Emboss toolkit 6.3.1 (48), was used to locate open reading frames (ORFs) with exact matches to NCBI-annotated sequences and to sequences derived from a mapped assembly using the P. furiosus NCBI genome as a reference. Additional ORFs present in the NCBI annotation that did not have exact matches in the de novo-assembled chromosome were located using BLAST version 2.2.24, and coordinates were determined by visual inspection. In addition to the NCBI gene names, InterPro's Iprscan version 4.8 was used to assign functional annotations to ORFs. Transmembrane domains and signal peptides were predicted using the optional TMHMM and SignalP packages. IS elements were determined using the ISFinder BLAST analysis tool (http://www.is.biotoul.fr/) (52). The COM1 assembled genome was visualized, and annotations were corrected using the CLC Genomics Workbench version 6.2. Annotated ORFs were translated using Transeq (EMBOSS toolkit 6.3.1) with Table 11. Needleman-Wunsch global alignments of the translated ORFs with NCBI reference protein sequences were performed using Needle (EMBOSS toolkit 6.3.1). An alignment of the NCBI reference genome (NC_003413) with COM1 was performed using Nucmer version 3.0.7 with a maximum gap size of 500 and a minimum cluster length of 100. A dot plot of the alignment was created using Mummerplot version 3.5.
Enzyme activities.
Cytoplasmic extracts of each strain were prepared as previously described (42). All steps were carried out under strict anaerobic conditions unless otherwise noted. Nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) activity was measured at 70°C, monitoring the rate of NAD(P)H formation at 340 nm (41). β-Glucosidase (CelB) activity was measured aerobically in a discontinuous assay with β-d-glucopyranoside-p-nitrophenyl as the substrate at 80°C (29). Reaction mixtures contained 50 mM Tris-HCl (pH 7.4) for GAPN and 50 mM sodium phosphate (pH 6.0) for CelB. The Bradford method (3) was used to estimate protein concentrations in cell extracts using bovine serum albumin as the standard.
Nucleotide sequence accession number.
The fully assembled DNA sequence of COM1 has been deposited in GenBank (accession number CP003685).
RESULTS
Genome sequence.
De novo assembly of 5,213,746 total reads of the COM1 genomic DNA resulted in one scaffold of 1,909,827 bp. This is 1,571 bp longer (0.1%) than the NCBI reference sequence for P. furiosus (1,908,256 bp) (49). The average coverage of the COM1 scaffold was 32× and 89× for 454 and Illumina reads, respectively.
Genome organization.
Alignment of the COM1 genome sequence with the P. furiosus NCBI reference (49) revealed a high overall degree of synteny, with two major inversions (Fig. 1). Sequence blocks, labeled A to E, were assigned to the COM1 genome based on the organization of the reference sequence (Fig. 2) with block A (red arrow) starting at PF0001. Block boundaries are located at the sites of IS elements (black tick marks), with the exception of the boundary between blocks A and E, which represents the boundary between the first and last annotated ORFs in the NCBI reference sequence (PF0001 and PF2065). Block A (red) begins at position 1495214 in the COM1 genome sequence and continues through PF0189, followed by block D (blue, PF0390 to PF1603′), block C (green, PF0349 to PF0388), block B (yellow, PF0190 to PF0347), and block E (purple, PF1603′ to PF2065). Relative to the NCBI reference sequence, blocks AE and C are inverted in COM1. The IS elements identified at the boundaries flanking block C were part of the originally annotated sequence (PF0348 and PF0389), whereas the other sites of rearrangement are due to recent IS activity in the intergenic region upstream of PF0190 and within PF1603.
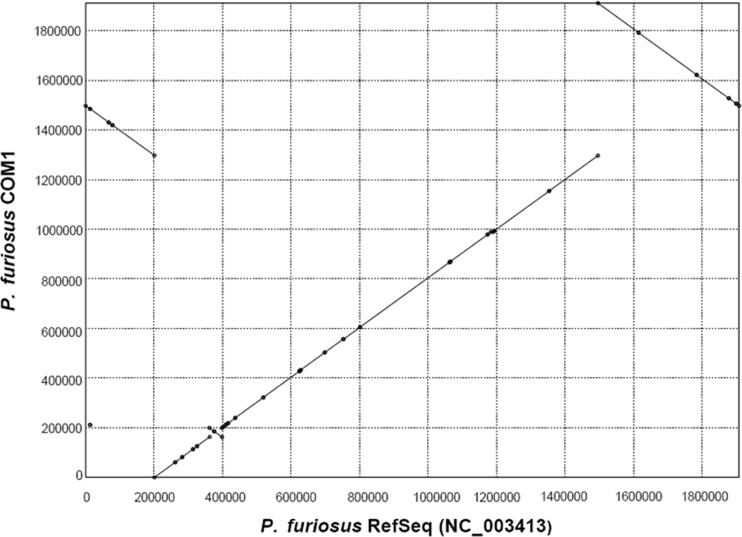
Fig 1.
Dot plot of the alignment of the P. furiosus NCBI reference sequence (NC_003413) on the x axis and the P. furiosus COM1 strain on the y axis indicates high overall synteny with two major inversions. Inverted segments in COM1 synonymous with the circular diagram in Fig. 2 are block A (positions 1495214 to 1295163), block E (positions 1909827 to 1495215), and block C (positions 199132 to 162245). The small segment (positions 210431 to 211213) is a result of an IS element insertion between PF0401 and PF0402 in COM1.
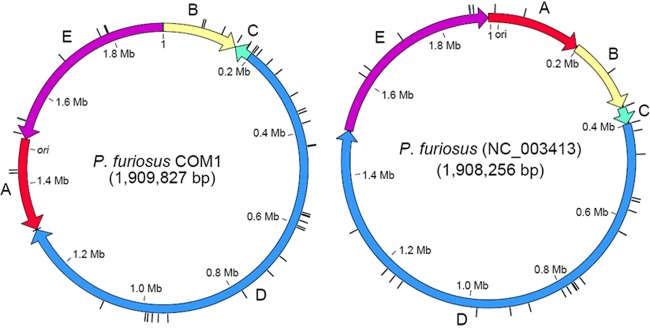
Fig 2.
COM1 genome organization (left) compared to the P. furiosus reference sequence (NC_003413, right). Black tick marks on the outside of the circular diagrams represent full-length IS elements in both genomes, with two additional IS elements located at the boundaries on the COM1 genome between blocks A and D and blocks B and E. The block boundary between A and E indicates the start/end of the reference sequence and is just upstream of the origin of replication (ori) in both genomes. PF numbers associated with each block are ordered in the 5′-to-3′ direction of each arrow: A (red), PF0001 to PF0189; B (yellow), PF0190 to PF0347; C (green), PF0349 to PF0388; D (blue), PF0390 to PF1603′; E (purple), PF1603′ to PF2065. PF1603 is disrupted by an IS element in the COM1 genome.
Confirmation of the COM1 genome organization (Fig. 1 and 2) was obtained by amplifying and sequencing PCR products that spanned the boundaries of each genome block (see Fig. S1 and S2 in the supplemental material). Care was taken to design unique primers not within transposable elements. The chromosomal orientations of the Parent and DSMZ strains were also analyzed. Surprisingly, both the NCBI reference order (Fig. 2) and the inverted orientations of blocks AE and C were observed in the COM1 and Parent strains. Only the inverted order of block C and the reference order of block AE were observed in the DSMZ control strain.
IS element activity.
COM1 and the P. furiosus reference sequences were annotated for IS elements using the ISFinder BLAST analysis tool (52) with an E-value cutoff of ≤4E−17. The reference sequence contains 29 complete and 6 partial IS elements (Table 1), whereas COM1 also contains the same 6 partial sequences in the same relative positions, but the number of complete sequences increased to 39. IS elements represent over 26 kb (1.38%) of the reference genome but over 34 kb (1.79%) of the COM1 genome. Most of the IS elements (80%) identified in both genomes are members of the IS6 family with the remainder representing three other IS families (IS982, IS607, and IS200/IS605 [Table 1]). Species-specific names for the IS elements are based on their origin and include not only ISPfu1 to ISPfu5 but also those derived from Thermococcus species (ISTko2 and ISTsi3 [52]). The full-length copies of the different ISPfu elements (ISPfu1 to ISPfu5) have consistent sizes (781, 782, 933, 1,961, and 779 bp, respectively) and possess properties typical of known transposable elements, i.e., putative transposase genes, terminal inverted repeats, and flanking duplicated target sequences. Locations of these IS elements were mapped (Fig. 2), and it is evident that they are responsible for a large number of genomic changes in COM1 relative to the NCBI sequence. These are summarized in Table 2. They include four excisions resulting in the complete or partial deletion of six neighboring genes, seven insertions within seven genes, and nine intergenic insertions potentially affecting regulatory regions upstream of 11 genes. Of these 24 genes, 10 encode conserved hypothetical proteins, 13 are predicted to be in operons, and the predicted cellular locations of the gene products are divided between the membrane (7 genes), extracellular (4 genes), and cytoplasmic (12 genes) fractions.
Table 1.
Comparison of IS element population in P. furiosus COM1 strain relative to the NCBI reference sequencea
| IS family | Name | No. of IS elements |
|||
|---|---|---|---|---|---|
| COM1 genome |
NCBI reference |
||||
| Full | Partial | Full | Partial | ||
| IS6 | ISPfu1 | 16 | 1 | 8 | 1 |
| ISPfu2 | 13 | 0 | 11 | 0 | |
| ISPfu5 | 4 | 0 | 4 | 0 | |
| IS982 | ISPfu3 | 5 | 0 | 5 | 0 |
| IS607 | ISPfu4 | 1 | 2 | 1 | 2 |
| ISTko2 | 0 | 2 | 0 | 2 | |
| IS200/IS605 | ISTsi3 | 0 | 1 | 0 | 1 |
| Total no. of IS elements | 39 | 6 | 29 | 6 | |
| Total IS length (bp) | 32,406 | 1,798 | 24,592 | 1,798 | |
| Genome (%) | 1.70 | 0.09 | 1.29 | 0.09 | |
The ISFinder BLAST analysis tool (52) was used to annotate IS elements with E-value scores of ≤4E−17 in both the COM1 and NCBI reference genome sequences. Sequence similarity among the members of an IS family and between IS families results in multiple hits at each locus. Only the best BLAST hit was retained for each IS element.
Table 2.
Genes affected by IS activity in the P. furiosus COM1 strain
| IS elementa | Affected gene(s) | Gene annotationb | Length (bp) | Operon positionc | Predicted locationd |
|---|---|---|---|---|---|
| Excisionse | |||||
| PF0013 (ISPfu1) | PF0012 | Hypothetical protein | 39 (3′ end) | Cyt | |
| PF0408 (ISPfu1) | PF0407 | <Carboxypeptidase-like, regulatory domain> | 41 (5′ end) | 1 of 2 | Mem |
| PF0756 (ISPfu2) | PF0755 | Nonphosphorylating GAPDHh (GAPN) | 300 (5′ end) | 4 of 4 | Cyt |
| PF0757 | Hypothetical protein | Complete | Cyt | ||
| PF0758 | <Nucleotide-binding domain (HEPN)> | Complete | Cyt | ||
| PF2035 (ISPfu2) | PF2034 | <Methyltransferase (TYW3)> | 41 (5′ end) | 1 of 2 | Cyt |
| Insertions in ORFf | |||||
| ISPfu2 | PF0061 | Riboflavin synthase subunit alpha | 782 | 1 of 4 | Cyt |
| ISPfu1 | PF0393 | [CRISPR-associated protein, Cas6-2 (21)] | 781 | Cyt | |
| ISPfu1 | PF0429 | Proline permease | 781 | Mem | |
| ISPfu1 | PF0823 | <Multiantimicrobial extrusion protein (MatE)> | 781 | 1 of 2 | Mem |
| ISPfu1 | PF1260 | Hypothetical protein | 781 | 1 of 2 | Cyt |
| ISPfu1 | PF1603 | Na antiporter | 781 | 1 of 3 | Mem |
| ISPfu1 | PF2059 | Aminopeptidase | 781 | 1 of 2 | Exc |
| Insertions in intergenic regionsg | |||||
| ISPfu1 | PF0189 | Dihydroorotase | 781 | 3 of 3 | Cyt |
| PF0190 | [Cold-induced protein A, CipA (57)] | Mem | |||
| ISPfu1 | PF0271 | Hypothetical protein | 781 | Exc | |
| PF0272 | [4-α-Glucanotransferase (35)] | Cyt | |||
| ISPfu1 | PF0401 | Methyltransferase | 781 | 4 of 4 | Exc |
| ISPfu1 | PF1738 | Sugar kinase | 781 | Cyt | |
| PF1739 | [Mal I transporter (31)] | 1 of 6 | Exc | ||
| ISPfu2 | PF_t006 | tRNA Glu anticodon TTC | 782 | ||
| ISPfu2 | PF0497 | <Winged helix-turn-helix transcription factor> | 782 | 1 of 9 | Cyt |
| ISPfu2 | PF1239 | Hypothetical protein | 782 | Mem | |
| PF1240 | Purine permease | 1 of 2 | Mem |
IS element nomenclature is based on the ISFinder database (52).
Annotations are NCBI gene names (no brackets) and literature cited (square brackets), except for hypothetical genes, for which the best InterPro (angle brackets) match is given if available.
Operon predictions are from reference 55 based on the NCBI reference sequence.
Predicted cellular location based on predicted transmembrane domains: proteins with ≥2 transmembrane domains are classified as membrane (Mem), proteins with <2 transmembrane domains and a predicted signal peptide using SignalP with the Gram-positive model of ≥0.6 are classified as extracellular (Exc), and proteins with <2 transmembrane domains and no predicted signal peptide are classified as cytoplasmic (Cyt).
Includes neighboring genes affected by IS excision, either full deletions (complete) or truncations (bp) at the chromosomal level.
Includes genes disrupted by IS insertion within their open reading frame.
Includes neighboring genes with transcriptional start sites downstream of IS insertion.
GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Transposases originally annotated in the NCBI reference sequence as PF0013 (ISPfu1), PF0408 (ISPfu1), PF2035 (ISPfu2), and PF0756 (ISPfu2) are deleted in COM1, and these include portions of or complete neighboring genes: PF0012 (39 bp, 3′ end), PF0407 (41 bp, 5′ end), PF2034 (41 bp, 5′ end), and a 1,714-bp region with PF0755 (300 bp deleted at 3′ end), PF0757, and PF0758 (deleted), respectively (Table 2). The affected neighboring genes are predicted to be nonfunctional since, in most cases, the deleted region includes the transcriptional start site. Additionally, IS-mediated gene disruptions include PF0061, PF0393, PF0429, PF0823, PF1260, PF1603, and PF2059, and these are also predicted to be nonfunctional. Nucleotide alignments of each gene are identical to the NCBI reference sequence minus the IS element and duplicated target repeat. However, at the protein level, the COM1 sequences terminate with premature stop codons ∼16 bp into the IS element. An example of IS element gene disruption is shown in Fig. 3, which compares the sequences of the genes encoding riboflavin synthase subunit alpha (PF0061). There are some other IS-disrupted genes that have a specific rather than a general predicted function; these include GAPN, aminopeptidase, and proline permease (Table 2), and they are considered further below. Similarly, several intergenic regions were also disrupted by IS elements and may potentially affect the regulation of genes with specific known or predicted functions, which include alpha-amylase, sugar kinase, trehalose/maltose binding protein, dihydroorotase, methyltransferase, purine permease, and cold-induced protein A (57), as shown in Fig. 3.
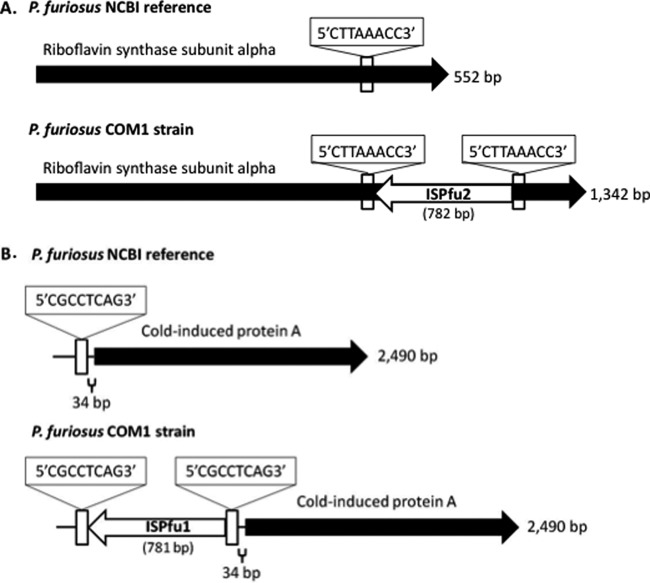
Fig 3.
Maps of IS element insertions in both coding and potential regulatory regions of the COM1 genome compared to the NCBI reference sequence. The riboflavin synthase subunit alpha (PF0061) gene (A) and the region upstream of the cold-induced protein A (PF0190) (B) in COM1 are disrupted by IS elements. IS elements are represented by white arrows and are flanked by direct repeat sequences (boxed). A single copy of the direct repeat sequence is located at the same positions on the reference sequences, and the nucleotide sequence alignments are identical for the two genomic regions minus the IS element insertion and additional direct repeats. In the case of the riboflavin synthase gene, at the protein level the COM1 sequence has a premature stop codon (∼16 bp into the IS element).
In addition to the IS-mediated excisions observed in COM1, there are seven genes affected by large chromosomal deletions compared to the reference sequence that appear to be independent of these elements. However, given the obvious recent IS activity, it is impossible to rule out previous IS activity in these regions prior to excision. The coding sequences of the seven genes are partially or completely deleted (Table 3), and the encoded proteins are therefore predicted to be nonfunctional. These include CelB (PF0073), which has a 774-bp deletion at the 3′ end resulting in a protein length of 214 compared to 472 amino acid residues in the NCBI reference sequence. The other six genes are in a single large chromosomal region (6,238 bp) that includes the genes PF1249 to PF1253 (deleted) and PF1254 (904 bp deleted at the 5′ end). These encode an ABC transporter, a sodium-dependent transporter, aromatic aminotransferase II, and three hypothetical proteins.
Table 3.
Additional large chromosomal deletions in P. furiosus COM1 strain relative to the NCBI reference strain
| Gene | Gene annotationa | Deletion length (bp) | Operon positionb | Predicted locationc |
|---|---|---|---|---|
| PF0073 | Beta-glucosidase (CelB) | 774 of 1,419 (3′ end) | Cyt | |
| PF1249d | ABC transporter | Complete | 1 of 2 | Cyt |
| PF1250 | Hypothetical protein | Complete | 2 of 2 | Exc |
| PF1251 | <Alpha-galactosidase, NEW3 domain> | Complete | 1 of 2 | Cyt |
| PF1252 | Hypothetical protein | Complete | 2 of 2 | Mem |
| PF1253 | [Aromatic aminotransferase II (56)] | Complete | Cyt | |
| PF1254 | Sodium-dependent transporter | 904 of 1,737 (5′ end) | 1 of 2 | Mem |
Annotations are NCBI gene names (no brackets) and literature cited (square brackets), except for hypothetical genes, for which the best InterPro (angle brackets) match is given if available.
Operon predictions are from reference 55 based on the NCBI reference sequence.
Predicted cellular location; for an explanation, see footnote d of Table 2.
Adjacent to transposase (PF1248, ISPfu3) in both COM1 and reference sequences.
Protein-level genome differences.
In addition to the IS-mediated gene disruptions and large chromosomal deletions observed in COM1, the protein products of 102 of the total 2,134 genes in the NCBI reference sequence are disrupted in COM1 at the translational level through nonsynonymous mutations. These result in amino acid changes, insertions and deletions introducing premature stop codons, and frameshifts producing longer or shorter products with alternate coding sequences. These were subdivided into 36 major changes (Table 4) and 66 minor changes (see Table S1 in the supplemental material) based on the Needleman-Wunsch global alignment identity compared with the protein sequences in the NCBI reference sequence (49) using ≥90% identity cutoff as a major difference. Since the deletion of the pyrF gene (PF1114) is the auxotrophic selectable marker for uracil biosynthesis in the COM1 strain (37), it was omitted from the list of major differences in Table 4 but is included in the complete list of disrupted genes in COM1 relative to the NCBI reference (see Table S1 in the supplemental material).
Table 4.
Major protein-level genome differences in P. furiosus COM1 strain relative to the NCBI reference strain (<90% identity)
| Gene | Gene annotationa | No. of identical residuesd | Alignment length (amino acids)d | Identity (%)d | Operon positionb | Predicted locationc |
|---|---|---|---|---|---|---|
| PF0054 | AsnC family transcriptional regulator | 91 | 155 | 58.7 | 6 of 7 | Cyt |
| PF0067 | Cobalt ABC transporter | 157 | 243 | 64.6 | 1 of 2 | Mem |
| PF0147 | Potassium channel-like protein | 23 | 205 | 11.2 | 2 of 2 | Cyt |
| PF0225 | N-Acetyltransferase | 111 | 201 | 55.2 | 1 of 2 | Cyt |
| PF0228 | Hypothetical protein | 49 | 89 | 55.1 | 2 of 3 | Cyt |
| PF0244 | Hypothetical protein | 38 | 89 | 42.7 | Cyt | |
| PF0314 | Signal peptidase | 58 | 173 | 33.5 | 2 of 3 | Cyt |
| PF0334 | Flagellum-like protein | 24 | 194 | 12.4 | 2 of 6 | Exc |
| PF0351 | Hypothetical protein | 22 | 285 | 7.7 | Cyt | |
| PF0352 | [CRISPR-associated protein, Cmr1-2 (21)] | 189 | 451 | 41.9 | Cyt | |
| PF0363 | Beta-galactosidase | 11 | 775 | 1.4 | 7 of 7 | Cyt |
| PF0423 | Hypothetical protein | 45 | 117 | 38.5 | 1 of 3 | Cyt |
| PF0424 | Hypothetical protein | 34 | 172 | 19.8 | 2 of 3 | Cyt |
| PF0439 | Hypothetical protein | 297 | 401 | 74.1 | Cyt | |
| PF0472 | <Class III signal peptide motif> | 42 | 54 | 77.8 | 2 of 2 | Exc |
| PF0524 | <Ribbon-helix-helix (Met_repress_like)> | 57 | 72 | 79.2 | 1 of 4 | Cyt |
| PF0611 | Hypothetical protein | 295 | 382 | 77.2 | Cyt | |
| PF0622 | Hypothetical protein | 7 | 33 | 21.2 | Cyt | |
| PF0710 | <Transcription repressor, DNA binding> | 79 | 157 | 50.3 | 3 of 3 | Cyt |
| PF0764 | DEXX-box ATPase | 232 | 434 | 53.5 | Cyt | |
| PF0777 | Hypothetical protein | 191 | 215 | 88.8 | Mem | |
| PF0785.2n | Hypothetical protein | 148 | 192 | 77.1 | NAf | |
| PF0873 | Hypothetical protein | 36 | 196 | 18.4 | Cyt | |
| PF0874 | Membrane dipeptidase | 9 | 379 | 2.4 | 1 of 2 | Cyt |
| PF0901 | Hypothetical protein | 121 | 142 | 85.2 | Cyt | |
| PF0960 | Hypothetical protein | 15 | 112 | 13.4 | 2 of 2 | Exc |
| PF1075 | Hypothetical protein | 3 | 219 | 1.4 | Mem | |
| PF1109 | <Carbohydrate-binding domain (CARDB)> | 959 | 1,132 | 84.7 | 1 of 2 | Exc |
| PF1113 | <tRNA amidotransferase GAD domain> | 199 | 223 | 89.2 | 1 of 3 | Cyt |
| PF1182 | <Thioredoxin-like fold> | 7 | 126 | 5.6 | Cyt | |
| PF1182.1n | Hypothetical protein | 53 | 169 | 31.4 | NA | |
| PF1206 | <Nucleic acid binding, PIN, PH0500> | 15 | 156 | 9.6 | 1 of 3 | Cyt |
| PF1748 | Sulfate permease, ABC transporter | 321 | 543 | 59.1 | 4 of 6 | Mem |
| PF1749 | Sulfate transport integral membrane protein | 10 | 228 | 4.4 | 5 of 6 | Mem |
| PF1761 | <Putative cell wall binding repeat 2> | 204 | 389 | 52.4 | 1 of 2 | Exc |
| PF1935e | Amylopullulanase | 1,114 | 1,355 | 82.2 | 3 of 6 | Exc |
Annotations are NCBI gene names (no brackets) and literature cited (square brackets), except for hypothetical genes, for which the best IPR (angle brackets) match is given if available.
Operon predictions from reference 55 based on the NCBI reference sequence.
Predicted cellular location; for an explanation, see footnote d of Table 2.
Based on Needleman-Wunsch global alignment with the NCBI reference sequence.
A fusion of PF1934 and PF1935 was previously reported (35). A longer product, which is closer in length to those in other pyrococci, was found in COM1.
NA, not applicable.
In general, although the COM1 genes that fall into the major difference category almost certainly encode nonfunctional proteins, those in the minor difference category could also result in protein products with diminished or abolished functions. Experimental assessment of function would be required for proteins deemed to have minor changes since a single amino acid change could obviously lead to inactivation. Functional annotation of all 102 genes in the reference sequence revealed that 46 of them encode conserved hypothetical proteins (see Table S1 in the supplemental material). The remaining 56 are predicted to be involved in a wide variety of cellular functions involving carbohydrate and peptide metabolism, DNA repair, CRISPR-associated defense, transcriptional regulation, and membrane transport. However, as described above for the IS-mediated disruptions, there are only some affected genes where a specific rather than a general function can be assigned. For example, the major difference category includes two enzymes involved in carbohydrate degradation, β-galactosidase (PF0363) and amylopullulanase (PF1935*; Table 4). β-Galactosidase in COM1 is predicted to be nonfunctional as it contains only 32 of the 772 amino acid residues encoded in the NCBI reference sequence due to a single base deletion only 18 bp into the coding sequence creating a premature stop. On the other hand, the gene encoding amylopullulanase was previously reported to have a sequencing error that revealed a gene fusion of PF1934-PF1935 (35). In COM1, we found that the gene fusion results in an even longer protein than previously reported (1,355 compared to 1,114 amino acids [35]) and is more similar in length to those in other Pyrococcus and Thermococcus species. The amylopullulanase in COM1 is therefore expected to be fully functional. In fact, PCR confirmed the same length product for the PF1935* gene in all three strains (DSMZ, Parent, and COM1 [data not shown]). The COM1 genome sequence also contained the gene fusion between PF1191 and PF1192 previously demonstrated by the native purification of spherical virus-like particles from P. furiosus (44).
Phenotypic properties.
The COM1, Parent, and DSMZ strains were screened for their ability to grow under various conditions to investigate the predictions of nonfunctional genes based on the COM1 sequence. For example, both IS and non-IS disruptions of gene products or gene regulatory regions (Tables 2 to 4) involve β-glucosidase (CelB; PF0073), 4-α-glucanotransferase (PF0272), sugar kinase (PF1738), trehalose/maltose binding protein (PF1739), and GAPN (PF0755). However, the growth rates and final cell densities of all three strains, COM1, Parent, and DSMZ, on cellobiose, maltose, and malto-oligosaccharides were comparable, with the proviso for COM1 that yeast extract (0.5 g/liter) was also added. This was due to the inability of COM1 to synthesize riboflavin due to a nonfunctional riboflavin synthase (PF0061) (Table 2). When riboflavin (0.1 μM) was added, the yeast extract was no longer required. COM1 also grew on a complex peptide-based medium (tryptone, 5 g/liter) in the presence of elemental sulfur (2 g/liter) to the same extent as the Parent and DSMZ strains, indicating that the disrupted genes in COM1 potentially related to peptide catabolism, including aromatic aminotransferase II (PF1253), proline permease (PF0429), and aminopeptidase (PF2059) (Tables 2 and 3), do not affect growth on peptides. In addition, all three strains grew well on the defined minimal medium (although COM1 required riboflavin), implying that the gene products involved in nucleotide metabolism, such as dihydroorotase (PF0189) and purine permease (PF1240) (Table 2), are still fully functional despite upstream IS insertion or not required for growth under these conditions.
A previous study (57) showed that a number of genes are upregulated when P. furiosus is grown at 72°C compared to the optimum near 100°C, the most highly expressed of which is a membrane glycoprotein termed cold-induced protein A (CipA [PF0190]). In COM1, cipA is not expressed due to the upstream insertion of an IS element (Fig. 3). However, no differences were observed in the abilities of the COM1, Parent, and DSMZ strains to grow at low temperature (72°C, data not shown). In addition, a number of genes involved in DNA repair, including an uncharacterized RadA domain protein (PF0872), DNA repair helicase Rad3 (PF0933), 5′-to-3′ exonuclease NurA (PF1168 [23]), and DNA repair helicase (PF1902), could potentially be inactive in COM1 as they fall into the minor protein-level differences category (see Table S1 in the supplemental material). However, COM1, Parent, and DSMZ showed no significant differences in their ability to recover from exposure to UV or gamma irradiation under conditions previously reported for P. furiosus (14). Consequently, in spite of the massive genome rearrangements in COM1 relative to the NCBI reference sequence, and the actual or potential inactivation of more than 120 genes, its ability to grow under various conditions previously used for the Parent strain in the laboratory is unaffected.
Enzyme assays were carried out to determine the effects of 5′-end deletion of the gene encoding nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase, GAPN (PF0755), and the 3′-end deletion of the gene encoding β-glucosidase (CelB; PF0073). The DSMZ strain was used as a control, as PCR and sequence analyses confirmed that it contained full-length versions of gapn and celB (data not shown). On the other hand, PCR analysis of the Parent strain revealed a full-length copy of gapn but its celB gene contained the same 5′-end deletion as that found in COM1. Surprisingly, all attempts to measure GAPN activity in the cell extracts of all three of the strains were unsuccessful, even in the presence of glucose-1-phosphate (1 mM), which is reportedly an activator of the enzyme in Thermococcus kodakarensis (41). As expected, the DSMZ control strain contained high CelB activity (1.68 μmol/min/mg at pH 6.0), and this was an order of magnitude greater than that measured in extracts of either the COM1 or the Parent strain (0.16 and 0.15 μmol/min/mg, respectively, at pH 6.0).
DISCUSSION
The availability of a highly efficient genetically tractable strain of P. furiosus (COM1) has dramatically expanded the potential to study this model hyperthermophilic species. Its sequence not only defines the genetic platform for these future studies but has also provided novel insights into the biology of this organism, including the dynamic nature of its genome and the utility of two classical carbohydrate metabolism genes, gapn and celB. The COM1 sequence assembly and scaffolding indicated the inversion of two large chromosomal segments relative to the published NCBI reference sequence (Fig. 1 and 2), and PCR confirmation revealed that both the reference order and inverted order of these segments were present in genomic DNA isolated from a small-scale culture originating from a single colony. This remarkable flexibility of chromosomal arrangement without apparent consequence for proper metabolic function has not previously been observed in this organism. The high rate of chromosomal breakage in hyperthermophilic environments and those where large doses of ionizing radiation are encountered require robust and accurate repair mechanisms (46). P. furiosus is able to completely reassemble its chromosome after gamma radiation-induced fragmentation into 30- to 500-kb fragments with up to 15 chromosomal copies per cell present in the exponential growth phase serving as the templates for reassembly (12). This ability, coupled with IS elements scattered throughout the genome that serve as the substrates for homologous recombination, is consistent with the observed alterations in genome structure in COM1. A comparative genomics study of three Pyrococcus species revealed that the genome of P. furiosus exhibits less bias in the preference of codirectionality of transcription with replication than do other sequenced organisms at the time (60). Given the ease with which the genome undergoes rearrangement, this is likely necessary to ensure proper transcription of essential genes. In addition, the high degree of operonization in the genome of P. furiosus places further constraints on the location of chromosomal shuffling hot spots (59).
IS elements also participate in genome evolution by disrupting gene coding sequences, influencing the expression of genes downstream of insertions (10). Sequencing of the COM1 genome revealed that two IS element types, ISPfu1 and ISPfu2 (both of the IS6 family), have been recently active under laboratory conditions and involved in four excisions and 14 insertions within genes and upstream of genes in potential regulatory regions (Table 2). LGT of a bacterial-type composite transposon was previously documented in P. furiosus and Thermococcus litoralis with the acquisition in the former organism of the Mal I ABC transport system for maltose and trehalose (13, 22, 45). Among the seven IS annotated archaeal genomes (30), P. furiosus harbors far more IS elements (29 full, 6 partial) than does any other member of the Thermococcales but less than a quarter of the IS elements identified in the Sulfolobus solfataricus genome (146 full, 297 partial). In fact, no full-length IS elements have been identified in the genomes of Pyrococcus abyssi or Pyrococcus horikoshii, and only one full-length IS element has been identified in T. kodakarensis (30).
In addition to IS-mediated disruptions, 102 differences in sequence were observed in the COM1 genome that would lead to gene products differing from those encoded by the published NCBI reference sequence. Some are likely to have been sequencing or assembly errors in the original published NCBI sequence, some could have arisen in the lab strain through random mutagenesis, and some may be unique to the COM1 strain. However, these categories are difficult to assess quantitatively. Although the NCBI reference sequence is labeled as the type strain (DSM 3638), the strain that provided the reference sequence (49) was not obtained directly from the DSMZ (R. Kelly and F. Robb, personal communication). Originally a gift from Karl Stetter (19), the strain was maintained at 4°C in a laboratory setting with periodic clonal selection for more than 10 years prior to producing DNA for genome sequencing in 1999 (49). Similarly, the Parent strain that gave rise to COM1 (37) was maintained in the laboratory for 2 years at ambient temperature with multiple transfers on various media after being obtained from DSMZ in 2007. Therefore, it is assumed that the DSMZ control strain (2010) used in this study is a good representative of the ancestor of Parent and COM1. That maintenance under laboratory conditions can lead to numerous mutations in strains originating from the same isolate is not surprising and is now well established in other organisms. For example, this was recently revealed by a large-scale resequencing effort of Bacillus subtilis strains from multiple laboratories (53), emphasizing the need to maintain a permanent storage capacity within the laboratory setting.
Despite extensive genomic changes in the COM1 strain, including multiple excisions, insertions, and protein-level changes, its major metabolic functions have not been disrupted. Select phenotypic properties were examined based on the changes in the COM1 strain, but no significant differences in growth under a variety of conditions were detected between it and the Parent and DSMZ strains. However, these analyses did reveal surprisingly nonessential roles for two carbohydrate metabolism enzymes, GAPN and CelB, whose functions should be abolished in COM1 due to large deletions in their coding regions. GAPN has been shown to be a key glycolytic enzyme in the related euryarchaeon T. kodakarensis, since a strain with a targeted gene knockout (ΔgapN) was unable to grow on malto-oligosaccharides (41). Similarly, GAPN plays a key glycolytic role in various crenarchaeota, including Thermoproteus (5), Aeropyrum (47) and Sulfolobus (1, 17) species. The oxidation of glyceraldehyde-3-phosphate during glycolysis in P. furiosus can be catalyzed by the tungsten-containing enzyme glyceraldehyde-3-phosphate ferredoxin oxidoreductase (GAPOR), which uses ferredoxin rather than NADP as the electron acceptor (43). In T. kodakarensis, both GAPOR and GAPN are required for growth on sugars (41), but clearly this is not the case in P. furiosus, as COM1 grew well on maltose, cellobiose, and malto-oligosaccharides in the absence of a functional GAPN. COM1 also lacks the gene encoding CelB, which has been shown to be the cellobiose-hydrolyzing enzyme in P. furiosus that constitutes up to 5% of the total cell protein during growth on this β-1,4-linked disaccharide (29). CelB activity measured in COM1 cell extracts was an order of magnitude less than that of the DSMZ control strain harboring a full-length celB gene. Therefore, growth of COM1 on cellobiose must be accomplished via other beta-specific glycosidases present in P. furiosus (15).
One significant phenotypic difference between COM1 and the Parent and DSMZ strains is obviously its ability to be genetically manipulated. The high efficiency in transformation and recombination of COM1 is likely the result of changes in multiple genes, but genome sequence comparisons between the strains cannot define the responsible gene(s). One group potentially involved in conferring competence is the five of the CRISPR-associated (Cas) proteins that are disrupted (Tables 2 and 4; see also Table S1 in the supplemental material). This is the best-characterized defense system against invasion by foreign nucleic acids in both archaea and bacteria (20, 39). In P. furiosus, Cas6 has been shown to recognize and cleave foreign RNA that matches spacer sequences present in the host's CRISPR loci (6, 21). A possible role of these disrupted proteins in conferring competence is intriguing and an obvious target for future research. Other disrupted genes in COM1 that could also play a role in DNA uptake and recombination include 20 membrane transporters, one or more of which could affect membrane integrity, thereby facilitating DNA uptake. Similarly, numerous proteins involved in DNA replication, transcription, and repair are actually or potentially disrupted in COM1 (Tables 2 and 4). These include four DNA helicases (PF0085, PF0572, PF0933, and PF1902), two RNA helicases (PF0592 and PF1120), a DNA primase (PF1725), a cell division control protein (PF1882), an uncharacterized RadA domain protein (PF0872), five putative transcription factors (DNA-binding proteins PF0054, PF0524, PF0621, PF0710, and PF1206), an RNA-binding protein (PF1573), and 5′-to-3′ exonuclease NurA (PF1168 [23]). In addition, of the 122 genes disrupted by IS- and non-IS-mediated events in the COM1 strain, 56 of them encode conserved hypothetical proteins, some of which could be partially or solely responsible for the genetic properties of COM1. Clearly, determining how many of these genes directly or indirectly affect competence will not be an easy task.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy, through grant DE-FG05-95ER20175 for funding the phenotypic analyses and the ENIGMA project supported by the U.S. Department of Energy under contract no. DE-AC02-05CH11231 for funding the sequencing and data analysis.
We thank the UGA Sequencing Facility for 454 sequencing, K. Stirrett and J. Westpheling for providing a sample of COM1 DNA for 454 sequencing, Ryan Weil at the Emory GRA Genomics Core for assistance with Illumina sequencing, Mirko Basen for assistance with low-temperature cultures, and Gina Lipscomb for many helpful discussions.
Footnotes
Published ahead of print 25 May 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Albers SV, et al. 2009. SulfoSYS (Sulfolobus Systems Biology): towards a silicon cell model for the central carbohydrate metabolism of the archaeon Sulfolobus solfataricus under temperature variation. Biochem. Soc. Trans. 37:58–64 [DOI] [PubMed] [Google Scholar]
- 2. Atomi H. 2005. Recent progress towards the application of hyperthermophiles and their enzymes. Curr. Opin. Chem. Biol. 9:166–173 [DOI] [PubMed] [Google Scholar]
- 3. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 4. Bridger SL, et al. 2011. Deletion strains reveal metabolic roles for key elemental sulfur-responsive proteins in Pyrococcus furiosus. J. Bacteriol. 193:6498–6504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brunner NA, Brinkmann H, Siebers B, Hensel R. 1998. NAD+-dependent glyceraldehyde-3-phosphate dehydrogenase from Thermoproteus tenax. The first identified archaeal member of the aldehyde dehydrogenase superfamily is a glycolytic enzyme with unusual regulatory properties. J. Biol. Chem. 273:6149–6156 [DOI] [PubMed] [Google Scholar]
- 6. Carte J, Pfister NT, Compton MM, Terns RM, Terns MP. 2010. Binding and cleavage of CRISPR RNA by Cas6. RNA 16:2181–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chandrayan SK, et al. 2012. Engineering hyperthermophilic archaeon Pyrococcus furiosus to overproduce its cytoplasmic [NiFe]-hydrogenase. J. Biol. Chem. 287:3257–3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chinen A, Uchiyama I, Kobayashi I. 2000. Comparison between Pyrococcus horikoshii and Pyrococcus abyssi genome sequences reveals linkage of restriction-modification genes with large genome polymorphisms. Gene 259:109–121 [DOI] [PubMed] [Google Scholar]
- 9. Cohen GN, et al. 2003. An integrated analysis of the genome of the hyperthermophilic archaeon Pyrococcus abyssi. Mol. Microbiol. 47:1495–1512 [DOI] [PubMed] [Google Scholar]
- 10. Craig NL, Craigie R, Gellert M, Lambowitz AM. (ed). 2002. Mobile DNA II. American Society for Microbiology, Washington, DC [Google Scholar]
- 11. Cvetkovic A, et al. 2010. Microbial metalloproteomes are largely uncharacterized. Nature 466:779–782 [DOI] [PubMed] [Google Scholar]
- 12. DiRuggiero J, Brown JR, Bogert AP, Robb FT. 1999. DNA repair systems in archaea: mementos from the last universal common ancestor? J. Mol. Evol. 49:474–484 [DOI] [PubMed] [Google Scholar]
- 13. Diruggiero J, et al. 2000. Evidence of recent lateral gene transfer among hyperthermophilic archaea. Mol. Microbiol. 38:684–693 [DOI] [PubMed] [Google Scholar]
- 14. DiRuggiero J, Santangelo N, Nackerdien Z, Ravel J, Robb FT. 1997. Repair of extensive ionizing-radiation DNA damage at 95 degrees C in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 179:4643–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Driskill LE, Bauer MW, Kelly RM. 1999. Synergistic interactions among beta-laminarinase, beta-1,4-glucanase, and beta-glucosidase from the hyperthermophilic archaeon Pyrococcus furiosus during hydrolysis of beta-1,4-, beta-1,3-, and mixed-linked polysaccharides. Biotechnol. Bioeng. 66:51–60 [PubMed] [Google Scholar]
- 16. Escobar-Paramo P, Ghosh S, DiRuggiero J. 2005. Evidence for genetic drift in the diversification of a geographically isolated population of the hyperthermophilic archaeon Pyrococcus. Mol. Biol. Evol. 22:2297–2303 [DOI] [PubMed] [Google Scholar]
- 17. Ettema TJ, Ahmed H, Geerling AC, van der Oost J, Siebers B. 2008. The non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) of Sulfolobus solfataricus: a key-enzyme of the semi-phosphorylative branch of the Entner-Doudoroff pathway. Extremophiles 12:75–88 [DOI] [PubMed] [Google Scholar]
- 18. Farkas J, Chung D, DeBarry M, Adams MW, Westpheling J. 2011. Defining components of the chromosomal origin of replication of the hyperthermophilic archaeon Pyrococcus furiosus needed for construction of a stable replicating shuttle vector. Appl. Environ. Microbiol. 77:6343–6349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fiala G, Stetter K. 1986. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch. Microbiol. 145:56–61 [Google Scholar]
- 20. Garrett RA, Vestergaard G, Shah SA. 2011. Archaeal CRISPR-based immune systems: exchangeable functional modules. Trends Microbiol. 19:549–556 [DOI] [PubMed] [Google Scholar]
- 21. Hale CR, et al. 2009. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139:945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hamilton-Brehm SD, Schut GJ, Adams MW. 2005. Metabolic and evolutionary relationships among Pyrococcus species: genetic exchange within a hydrothermal vent environment. J. Bacteriol. 187:7492–7499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hopkins BB, Paull TT. 2008. The P. furiosus mre11/rad50 complex promotes 5′ strand resection at a DNA double-strand break. Cell 135:250–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hopkins RC, et al. 2011. Homologous expression of a subcomplex of Pyrococcus furiosus hydrogenase that interacts with pyruvate ferredoxin oxidoreductase. PLoS One 6:e26569 doi:10.1371/journal.pone.0026569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jenney FE, Jr, Adams MW. 2008. The impact of extremophiles on structural genomics (and vice versa). Extremophiles 12:39–50 [DOI] [PubMed] [Google Scholar]
- 26. Jun X, et al. 2011. Complete genome sequence of the obligate piezophilic hyperthermophilic archaeon Pyrococcus yayanosii CH1. J. Bacteriol. 193:4297–4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kanoksilapatham W, Gonzalez JM, Maeder DL, DiRuggiero J, Robb FT. 2004. A proposal to rename the hyperthermophile Pyrococcus woesei as Pyrococcus furiosus subsp. woesei. Archaea 1:277–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kawarabayasi Y, et al. 1998. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 5:55–76 [DOI] [PubMed] [Google Scholar]
- 29. Kengen SW, Luesink EJ, Stams AJ, Zehnder AJ. 1993. Purification and characterization of an extremely thermostable beta-glucosidase from the hyperthermophilic archaeon Pyrococcus furiosus. Eur. J. Biochem. 213:305–312 [DOI] [PubMed] [Google Scholar]
- 30. Kichenaradja P, Siguier P, Perochon J, Chandler M. 2010. ISbrowser: an extension of ISfinder for visualizing insertion sequences in prokaryotic genomes. Nucleic Acids Res. 38:D62–D68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Koning SM, Konings WN, Driessen AJ. 2002. Biochemical evidence for the presence of two alpha-glucoside ABC-transport systems in the hyperthermophilic archaeon Pyrococcus furiosus. Archaea 1:19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lancaster WA, et al. 2011. A computational framework for proteome-wide pursuit and prediction of metalloproteins using ICP-MS and MS/MS data. BMC Bioinformatics 12:64 doi:10.1186/1471-2105-12-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lecompte O, et al. 2001. Genome evolution at the genus level: comparison of three complete genomes of hyperthermophilic archaea. Genome Res. 11:981–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee HS, et al. 2011. Complete genome sequence of hyperthermophilic Pyrococcus sp. strain NA2, isolated from a deep-sea hydrothermal vent area. J. Bacteriol. 193:3666–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee HS, et al. 2006. Transcriptional and biochemical analysis of starch metabolism in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 188:2115–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Leigh JA, Albers SV, Atomi H, Allers T. 2011. Model organisms for genetics in the domain Archaea: methanogens, halophiles, Thermococcales and Sulfolobales. FEMS Microbiol. Rev. 35:577–608 [DOI] [PubMed] [Google Scholar]
- 37. Lipscomb GL, et al. 2011. Natural competence in the hyperthermophilic archaeon Pyrococcus furiosus facilitates genetic manipulation: construction of markerless deletions of genes encoding the two cytoplasmic hydrogenases. Appl. Environ. Microbiol. 77:2232–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Makarova KS, et al. 2011. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9:467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martusewitsch E, Sensen CW, Schleper C. 2000. High spontaneous mutation rate in the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by transposable elements. J. Bacteriol. 182:2574–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Matsubara K, Yokooji Y, Atomi H, Imanaka T. 2011. Biochemical and genetic characterization of the three metabolic routes in Thermococcus kodakarensis linking glyceraldehyde 3-phosphate and 3-phosphoglycerate. Mol. Microbiol. 81:1300–1312 [DOI] [PubMed] [Google Scholar]
- 42. Menon AL, et al. 2009. Novel multiprotein complexes identified in the hyperthermophilic archaeon Pyrococcus furiosus by non-denaturing fractionation of the native proteome. Mol. Cell. Proteomics 8:735–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mukund S, Adams MW. 1995. Glyceraldehyde-3-phosphate ferredoxin oxidoreductase, a novel tungsten-containing enzyme with a potential glycolytic role in the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 270:8389–8392 [DOI] [PubMed] [Google Scholar]
- 44. Namba K, et al. 2005. Expression and molecular characterization of spherical particles derived from the genome of the hyperthermophilic euryarchaeote Pyrococcus furiosus. J. Biochem. 138:193–199 [DOI] [PubMed] [Google Scholar]
- 45. Noll KM, Lapierre P, Gogarten JP, Nanavati DM. 2008. Evolution of mal ABC transporter operons in the Thermococcales and Thermotogales. BMC Evol. Biol. 8:7 doi:10.1186/1471-2148-8-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Peak MJ, Robb FT, Peak JG. 1995. Extreme resistance to thermally induced DNA backbone breaks in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 177:6316–6318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reher M, Gebhard S, Schonheit P. 2007. Glyceraldehyde-3-phosphate ferredoxin oxidoreductase (GAPOR) and nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN), key enzymes of the respective modified Embden-Meyerhof pathways in the hyperthermophilic crenarchaeota Pyrobaculum aerophilum and Aeropyrum pernix. FEMS Microbiol. Lett. 273:196–205 [DOI] [PubMed] [Google Scholar]
- 48. Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276–277 [DOI] [PubMed] [Google Scholar]
- 49. Robb FT, et al. 2001. Genomic sequence of hyperthermophile, Pyrococcus furiosus: implications for physiology and enzymology. Methods Enzymol. 330:134–157 [DOI] [PubMed] [Google Scholar]
- 50. Santangelo TJ, Cubonova L, Reeve JN. 2010. Thermococcus kodakarensis genetics: TK1827-encoded beta-glycosidase, new positive-selection protocol, and targeted and repetitive deletion technology. Appl. Environ. Microbiol. 76:1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schut GJ, Brehm SD, Datta S, Adams MW. 2003. Whole-genome DNA microarray analysis of a hyperthermophile and an archaeon: Pyrococcus furiosus grown on carbohydrates or peptides. J. Bacteriol. 185:3935–3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34:D32–D36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Srivatsan A, et al. 2008. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet. 4:e1000139 doi:10.1371/journal.pgen.1000139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Strand KR, et al. 2010. Oxidative stress protection and the repair response to hydrogen peroxide in the hyperthermophilic archaeon Pyrococcus furiosus and in related species. Arch. Microbiol. 192:447–459 [DOI] [PubMed] [Google Scholar]
- 55. Tran TT, et al. 2007. Operon prediction in Pyrococcus furiosus. Nucleic Acids Res. 35:11–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ward DE, DE Vos WM, van der Oost J. 2002. Molecular analysis of the role of two aromatic aminotransferases and a broad-specificity aspartate aminotransferase in the aromatic amino acid metabolism of Pyrococcus furiosus. Archaea 1:133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Weinberg MV, Schut GJ, Brehm S, Datta S, Adams MW. 2005. Cold shock of a hyperthermophilic archaeon: Pyrococcus furiosus exhibits multiple responses to a suboptimal growth temperature with a key role for membrane-bound glycoproteins. J. Bacteriol. 187:336–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. White JR, Escobar-Paramo P, Mongodin EF, Nelson KE, DiRuggiero J. 2008. Extensive genome rearrangements and multiple horizontal gene transfers in a population of Pyrococcus isolates from Vulcano Island, Italy. Appl. Environ. Microbiol. 74:6447–6451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yoon SH, et al. 2011. Parallel evolution of transcriptome architecture during genome reorganization. Genome Res. 21:1892–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zivanovic Y, Lopez P, Philippe H, Forterre P. 2002. Pyrococcus genome comparison evidences chromosome shuffling-driven evolution. Nucleic Acids Res. 30:1902–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.