Abstract
Acinetobacter species show high levels of intrinsic resistance to many antibiotics. The major protein species in the outer membrane of Acinetobacter baumannii does not belong to the high-permeability trimeric porin family, which includes Escherichia coli OmpF/OmpC, and instead is a close homolog of E. coli OmpA and Pseudomonas aeruginosa OprF. We characterized the pore-forming function of this OmpA homolog, OmpAAb, by a reconstitution assay. OmpAAb produced very low pore-forming activity, about 70-fold lower than that of OmpF and an activity similar to that of E. coli OmpA and P. aeruginosa OprF. The pore size of the OmpAAb channel was similar to that of OprF, i.e., about 2 nm in diameter. The low permeability of OmpAAb is not due to the inactivation of this protein during purification, because the permeability of the whole A. baumannii outer membrane was also very low. Furthermore, the outer membrane permeability to cephalothin and cephaloridine, measured in intact cells, was about 100-fold lower than that of E. coli K-12. The permeability of cephalothin and cephaloridine in A. baumannii was decreased 2- to 3-fold when the ompAAb gene was deleted. These results show that OmpAAb is the major nonspecific channel in A. baumannii. The low permeability of this porin, together with the presence of constitutive β-lactamases and multidrug efflux pumps, such as AdeABC and AdeIJK, appears to be essential for the high levels of intrinsic resistance to a number of antibiotics.
INTRODUCTION
Besides Pseudomonas aeruginosa, there are other genera of Gram-negative bacteria, such as the Acinetobacter species, that often produce multidrug-resistant and even pan-resistant strains (4, 32). Since P. aeruginosa produces a major porin of unusually low permeability, or a “slow porin” (40, 43), that plays a major role in its high levels of intrinsic resistance (3, 49), it is suspected that a similar situation may also exist in Acinetobacter species. It has been reported that the permeability coefficients of the Acinetobacter calcoaceticus outer membrane (OM) to zwitterionic cephalosporins were 2 to 7 times lower than the already very low values for the same β-lactams in the OM of P. aeruginosa (37). However, the identity of the major porin species in Acinetobacter has not been established so far.
The major protein of the Acinetobacter baumannii OM was named OmpAb (18), or HMP-AB (15), and belongs to the OmpA-like family, as sequence comparison revealed a clear homology with the monomeric OM protein A (OmpA) of Enterobacteriaceae and OM protein F (OprF) of Pseudomonas spp. OmpAb and HMP-AB were reported, however, to produce permeability comparable to or even higher than that of the classical trimeric porins of Escherichia coli, such as OmpF and OmpC (15, 18). If this is correct, the presence of such porins does not explain the low permeability of the Acinetobacter OM. In contrast, another study found that the OmpA homolog in A. baumannii had very low permeability (30). Furthermore, the high permeability of the OmpA homolog is not consistent with our earlier observation that E. coli OmpA (41) and P. aeruginosa OprF (25, 50) behave as slow porins (43) that produce very slow permeation of solutes yet have pore sizes that are either similar to, or larger than, that of OmpF. Thus, the existing literature is quite confusing, and indeed, one review (47) argues that “one of the limitations of our knowledge of A. baumannii is the lack of information concerning its OM proteins and the permeability.”
Because the properties of the porin channel in Acinetobacter species are crucial for the understanding of their resistance mechanism and also the development of antibiotics for the control of this important pathogen, we reexamined the properties of the OmpA family protein in A. baumannii (hereafter it is called OmpAAb to distinguish it from E. coli OmpA). The results show that it produces a low-permeability channel that is similar in both permeability and size to that of P. aeruginosa OprF. We then show that an A. baumannii mutant that lacks OmpAAb is defective in the uptake of a model antibiotic, hydrophilic cephalosporins, indicating that this protein is, indeed, the major nonspecific porin of this organism.
MATERIALS AND METHODS
Bacterial strains.
A. baumannii ATCC 17978 and its derivatives were used for the study of OM permeability. OmpAAb was purified either from this strain or after its expression in a hexahistidine-tagged form in porinless E. coli strain HN705 (ompC ompF::Tn5) (41). A. baumannii strains BM4651 (ΔadeABC), BM4679 (ΔadeIJK), and BM4652 (ΔadeABC ΔadeIJK) and its parent strain BM4454 (11, 21) were obtained from P. Courvalin.
Expression plasmid for hexahistidine-tagged OmpAAb.
The mature part of the ompAAb gene (coding for amino acids 23 to 346) was cloned by PCR amplification using the Expand Long Template PCR system (Roche) and inserted between the PstI and NotI sites of the vector plasmid, pKY9790 (40), which was previously modified by inserting the signal sequence of the E. coli OmpA protein followed by a hexahistidine tag just in front of the PstI site, generating the plasmid pKY-OmpAAb.
Purification of the hexahistidine-tagged OmpAAb.
Purification of hexahistidine-tagged OmpAAb from HN705 cells with freshly transformed pKY-OmpAAb was performed essentially as described earlier for His-tagged OprF (39). Briefly, a 1-liter LB culture was grown at 30°C overnight with shaking in the presence of 30 μg/ml chloramphenicol but without IPTG (isopropyl-β-d-thiogalactopyranoside) induction. The crude envelope fraction, prepared with a French pressure cell disruption followed by centrifugation, was extracted with 0.5% Sarkosyl to remove inner membrane proteins, and then the OM proteins were solubilized with a buffer containing 1.5% dodecyl-β-d-maltoside and 0.25 mg/ml lysozyme. Hexahistidine-tagged OmpAAb was isolated with a Ni-NTA Superflow column (Qiagen) as described for His-tagged OprF (39). Imidazole was then removed, and the protein sample was concentrated by using centrifugal filtration with Vivaspin20 (molecular weight cutoff, 10,000; Vivascience, Inc.) (39).
Purification of unmodified OmpAAb.
The OmpAAb protein without a hexahistidine tag was isolated from the outer membrane of A. baumannii ATCC 17978. Cells were disrupted with a French pressure cell treatment, and the envelope fraction was extracted with 0.5% Sarkosyl to remove inner membrane proteins. The outer membrane proteins were extracted overnight with 68 mM octyl β-d-glucoside in 20 mM HEPES buffer (pH 7.5) containing 5 mM EDTA and 0.1 mg/ml lysozyme at 0°C. The extract was then mixed with an equal volume of the sample buffer (10% glycerol, 1.5 M aminocaproic acid, 100 mM Bis Tris-HCl [pH 7.0], 0.5% Coomassie brilliant blue G250), and the proteins were separated by blue native gel electrophoresis (38) at 100 V for 4 h at 4°C in a gel system consisting of 4% stacking gel (pH 7.0) and 10% separating gel (pH 7.0). For visualization of protein bands, a narrow lane was cut out and stained with Coomassie brilliant blue R250 staining solution (0.25% Coomassie brilliant blue R250, 50% methanol, 5% acetic acid) for 5 min. The band containing OmpAAb was cut out from the unstained portion of the gel and was subjected to electroelution in a buffer containing 0.1% octylpolyoxyethylene (octyl-POE), 192 mM glycine, and 25 mM Tris base as described by Dé et al. (12). The sample consisted of an essentially pure preparation of OmpAAb, as seen in Fig. 1.
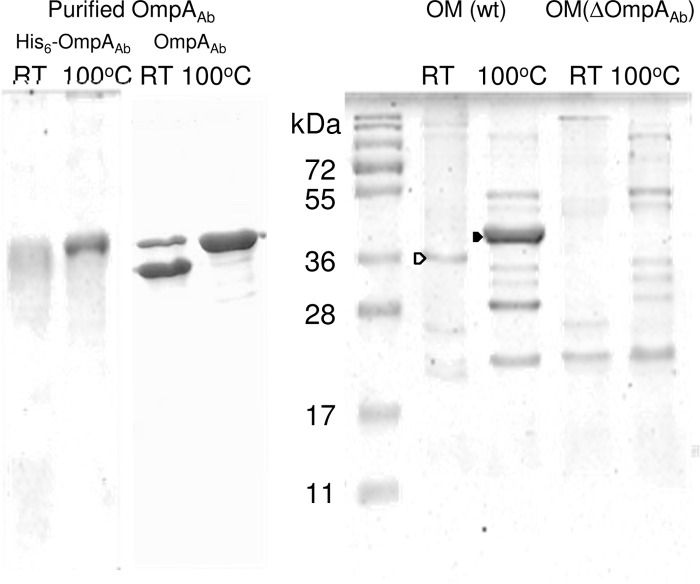
Fig 1.
SDS-PAGE of the purified OmpAAb preparations and the OM proteins from A. baumannii ATCC 17978 and its ΔompAAb mutant. Samples containing 5 μg of purified hexahistidine-tagged OmpAAb or untagged OmpAAb or 10 μg of total OM proteins from the parent or its ΔOmpAAb mutant strain were applied to SDS-PAGE either unheated (room temperature [RT]) or after heating at 100°C for 5 min in the sample buffer (62.5 mM Tris-HCl [pH 6.8], 10% glycerol, 5% β-mercaptoethanol, 2% SDS). The gels were analyzed by Coomassie blue staining. The bands containing the partially denatured (heat-modifiable) and completely denatured OmpAAb are indicated by open and filled arrowheads, respectively.
Determination of pore-forming activity.
Pore-forming activity was routinely assayed by determining the osmotic swelling rates of proteoliposomes containing different amounts of OmpAAb in an isotonic solution of sugars, usually l-arabinose (25). As a measure of pore-forming activity, rates of swelling were expressed in milliunits of optical density (OD)/min/μg protein (mOD/min/μg protein). The pore size was inferred from the dependence of the swelling rates on the sizes of the permeating solutes, as described earlier (25).
Construction of an ompAAb null mutant of A. baumannii.
In order to assess the contribution of OmpAAb in the nonspecific diffusion across the OM, the ompAAb gene was replaced by a kanamycin resistance gene (Kanr) by using a gene replacement plasmid, pEX18Tc (17). Two approximately 1,000-bp regions located upstream and downstream from the ompAAb gene were amplified by PCR using primers (UpFw, 5′-ACGCGTCGACCGAATGCTTCGTCAGTTTGAGGCAACATGGCAAAAACAGCTG; UpRv, 5′-CGGAATTCTTTGAGTTCTTGAACAGTAAAAAAGCGACTCGTTAGAGTCGC; DownFw, 5′-AACTGCAGGGATATCCTCCAGAGATAACAATTGTTGTTCAAGCTCAGCCT; and DownRv, 5′-CGGGATCCGCAGTATATTGCAAAATGAAGAACGGGTTAGCTTCTGCATTA). These PCR products, Upstream and Downstream, were digested and ligated between SalI and EcoRI and PstI and BamHI sites, respectively, of pEX18Tc. The kan gene (816 bp) of pACYC was amplified by PCR using primers containing PstI and EcoRI restriction sites. This PCR product was digested and ligated between PstI and EcoRI sites of pEX18Tc containing the Upstream and Downstream segments.
For gene replacement, the recombinant plasmid was electroporated into A. baumannii ATCC 17978. Integrants of the Kanr plasmid were selected on LB plates containing 30 μg/ml kanamycin, and these were then plated on LB plates containing 5% sucrose in order to select for strains that have lost the plasmid sequence. Sucrose-resistant (Sucr) colonies were ascertained to have the tetracycline-sensitive phenotype as the result of plasmid eviction. The absence of the ompAAb gene sequence was monitored by PCR and was further confirmed by the absence of the OmpAAb band in the SDS-PAGE of OMs.
MIC determination.
MIC values were determined with 96-well microtiter plates using a standard 2-fold broth microdilution method with LB broth containing 5 mM MgCl2.
Expression of the chromosomally encoded β-lactamases.
The strain of A. baumannii used is known to contain two β-lactamase genes coding for a class C AmpC enzyme and a class D oxacillinase of the OXA-51 family (1). The standard isoelectric focusing (IEF) procedure was used with a precast gel (Novex pH 3-10, Invitrogen), followed by nitrocefin staining to evaluate the expression of these genes essentially as described by Philippon et al. (34). We used pI 4.45 to 9.6 IEF standards from Bio-Rad as well as the TEM enzyme of pI 5.4 as standards.
Assay of EtBr influx in intact cells.
The rate of entry of ethidium bromide (EtBr) into intact cells was measured as an indicator of the permeability of the lipid bilayer domains of OM, because the diffusion across the inner membrane bilayer, composed of the conventional phospholipids, is expected to be orders of magnitude faster, and thus, the flux across the OM becomes the rate-limiting step. Overnight cultures were diluted 20-fold in fresh LB containing 5 mM MgCl2, and the cultures were grown with shaking at 37°C. Ten-milliliter portions were harvested by centrifugation at room temperature, and the cells were washed twice with 50 mM K-phosphate buffer, pH 7, by centrifugation at room temperature. The amount of cells corresponding to 0.4 optical density at 600 nm (OD600) units was added into 2 ml of the same K-phosphate buffer containing a 100 μM concentration of the proton conductor carbonyl cyanide 3-chlorophenylhydrazone (CCCP). After addition of EtBr (6 μM final concentration), the fluorescence of the EtBr-nucleic acid complex generated by the influx of EtBr into the cells was determined at room temperature by using a Shimadzu RF6301 spectrofluorometer with excitation and emission wavelengths of 545 nm and 600 nm, respectively.
Determination of cephalosporin flux.
The diffusion rates of cephalosporins through the OM were determined by measuring the rates of their hydrolysis by periplasmic β-lactamase(s) in intact cells (28). Bacteria were grown in 5 ml LB overnight at 37°C, and 1.5 ml of the culture was diluted into 50 ml of LB containing 5 mM MgCl2. The cells were grown at 37°C for 3 h with aeration by shaking and were washed once with 50 mM potassium phosphate buffer, pH 7.0, containing 5 mM MgCl2. Washed cells were resuspended in the same buffer at an OD600 of 0.8 (corresponding to 0.24 mg [dry weight]/ml). One portion was sonicated to release all β-lactamase into the solution, and the other portion was used for the intact-cell assay. In order to correct for hydrolysis caused by the enzyme that leaked out into the suspension buffer, one portion of the cell suspension was centrifuged at 13,000 rpm in a microcentrifuge for 2 min and the supernatant was used for the assay. Hydrolysis was monitored with a Uvikon 860 spectrophotometer, custom designed to minimize the effect of light scattering, at 486 nm for nitrocefin (31). For cephalothin and cephaloridine, 260-nm absorption was used, with cells of 10- and 1-mm light paths, respectively (28). The permeability coefficient P was obtained from cells treated with 100 μM CCCP in order to inactivate the efflux pumps.
RESULTS
Characterization of the A. baumannii OmpAAb channel.
As described in the introduction, there is no consensus on the rate of solute influx through the OmpAAb channel. We therefore reexamined the properties of the OmpAAb channel by producing a recombinant OmpAAb containing the E. coli OmpA signal peptide and a hexahistidine tag at the N terminus. This protein was expressed in E. coli strain HN705, which lacks both OmpF and OmpC, so that the final product would not be contaminated by the high-permeability porins of E. coli. Under our condition of expression utilizing the basal transcription level of the pTac promoter, most of OmpAAb was inserted into the E. coli OM (not shown) and could be extracted under the conditions which led to extraction of OM proteins (39, 44).
Figure 1 shows the Coomassie-stained SDS-PAGE of the purified hexahistidine-tagged OmpAAb and the OM protein profile of A. baumannii grown at 37°C in L broth. OmpAAb is by far the most abundant protein in the OM of A. baumannii, as described earlier (18), and this protein showed a characteristic mobility shift, sometimes called “heat modifiability,” so that it travels more slowly after denaturation at 100°C, similar to E. coli OmpA and P. aeruginosa OprF (44). The purified hexahistidine-tagged OmpA protein (expected size, 38.8 kDa) appeared as a broad band with an apparent molecular mass of around 40 kDa, and this protein migrated more slowly after the heat denaturation. The SDS-PAGE profile in Fig. 1 suggests that the majority of OmpAAb is monomeric, especially after heating at 100°C, although it is possible that some of it is loosely associated in an oligomeric state before heating.
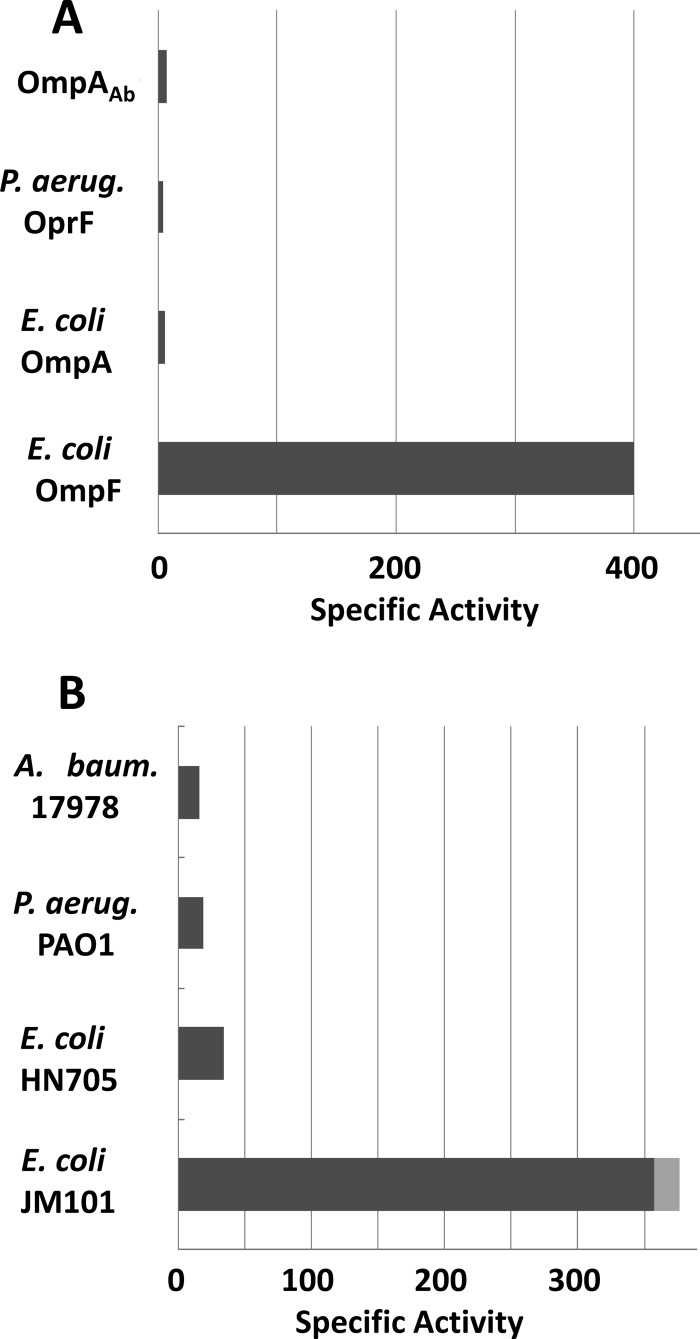
For the functional assay, the purified hexahistidine-tagged OmpAAb was reconstituted into proteoliposomes, and the rates of their osmotic swelling in isotonic l-arabinose were determined (Fig. 2A). The specific pore-forming activity of OmpAAb was 7.2 ± 0.9 mOD/min/μg, which was similar to the activities of E. coli OmpA (6 mOD/min/μg [41]) and P. aeruginosa OprF (4.5 mOD/min/μg [23]) and which was nearly 100-fold lower than the value obtained with E. coli OmpF (400 mOD/min/μg [27]). This low permeability is not caused by the hexahistidine tag sequence, because tag-free OmpAAb, purified from the outer membrane of A. baumannii ATCC 17978 as described in Materials and Methods, showed a similarly low permeability (7.9 ± 0.4 mOD/min/μg) (data not shown). Furthermore, it is not due to its inactivation during purification or to improper folding during expression in E. coli, as the proteoliposomes reconstituted with fragments of the intact OM of A. baumannii also showed about 200-fold lower permeability (that was similar to the results with the OM of P. aeruginosa) than the vesicles reconstituted with the OM of wild-type E. coli (Fig. 1B). The OM permeability of E. coli HN705 was quite low, as the high-permeability porins, OmpF and OmpC, were absent (Fig. 1B). These data strongly suggest that the OM of A. baumannii does not contain significant amounts of high-permeability porins, a situation similar to the OM of P. aeruginosa.
Fig 2.
Comparison of specific pore-forming activities of OmpA homolog porins from different origins, including A. baumannii (A. baum.), P. aeruginosa (P. aerug.), and E. coli. (A) Specific pore-forming activity for l-arabinose diffusion through various purified porins in mOD min−1 mg−1. (B) Specific pore-forming activity measured for l-arabinose diffusion through the proteoliposomes reconstituted from crude OM fragments of different strains (0.5 μg of OM from E. coli JM101, 5 μg of OM from E. coli HN705 [OmpF−, OmpC−], P. aeruginosa PAO1, and A. baumannii ATCC 17978). With JM101, the standard deviation is shown as a gray box. The unit is mOD min−1 μg−1.
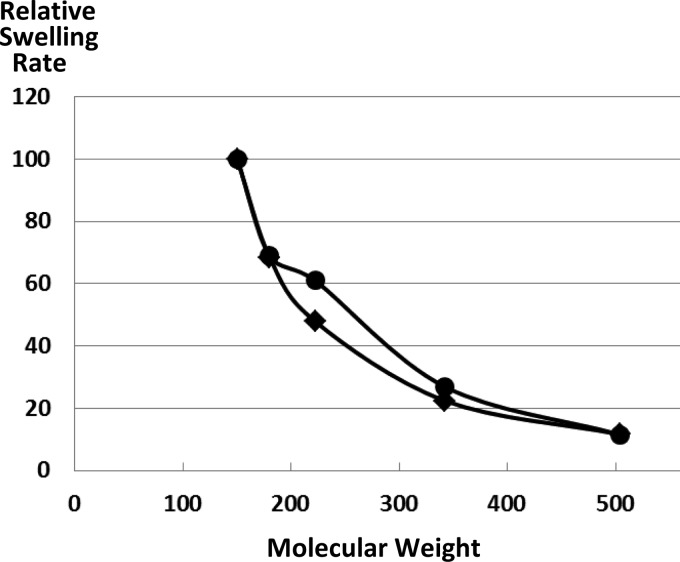
The pore size of OmpAAb was estimated by the osmotic swelling of proteoliposomes in sugars of different sizes (Fig. 3). The hexahistidine tag-free OmpAAb behaved in a nearly identical manner (data not shown). The behavior of OmpAAb appeared to be very similar to that of P. aeruginosa OprF (25), and thus, the size of the OmpAAb channel is larger than those of the classical high-permeability porins, such as E. coli OmpF. This combination of large channel size and a slow permeation rate, which may appear paradoxical, is common among OmpA family “slow porins” and is caused by the fact that only a minority of these protein molecules folds into an open-channel conformation (23, 39, 40, 43).
Fig 3.
Diffusion rates of solutes of various sizes through the A. baumannii OmpAAb porin. A total of 20 μg of hexahistidine-tagged OmpAAb was reconstituted into proteoliposomes as described in Materials and Methods. They were diluted in isosmotic solutions of l-arabinose (molecular weight, 150), d-glucose (molecular weight, 180), N-acetyl-d-glucosamine (molecular weight, 221), sucrose (molecular weight, 342), and raffinose (molecular weight, 504), and the initial rates of swelling of the liposomes are shown relative to that in l-arabinose. The data with hexahistidine-tagged OmpA (circles) were similar to previous data with OprF purified from P. aeruginosa PAO1 (squares) (26, 28).
Influx of antibiotics and dyes into an A. baumannii ΔompAAb mutant.
In order to confirm that OmpAAb plays a major role in the penetration of solutes across the OM, we made a mutant lacking OmpAAb by replacing the chromosomal ompAAb gene with the Kanr cassette. The OM from the ΔompAAb strain indeed showed a complete absence of OmpAAb (Fig. 1).
The ΔompAAb mutant grew relatively well in L broth with a doubling time of 29 min, only slightly slower than that of the parent strain (26 min). However, we noted that the ΔompAAb mutant had a significantly increased permeability for hydrophobic dyes, as seen by the uptake rates of EtBr in the presence of the proton conductor CCCP (Fig. 4). Since EtBr is large, lipophilic, and rather rigid, it is believed to diffuse mainly across the bilayer region of OM rather than through the narrow porin channel. Thus, it seems likely that the absence of this major protein, OmpAAb, produces empty spaces in the OM, which becomes filled by phospholipids, producing phospholipid bilayer domains in the OM, as seen in the “deep rough” mutants of enteric bacteria (2). A similar phenotypic change was observed in P. aeruginosa when the oprF gene was deleted, as shown by the severalfold increases in the entry of the hydrophobic fluorescent probe 1-N-phenylnaphthylamine (48).
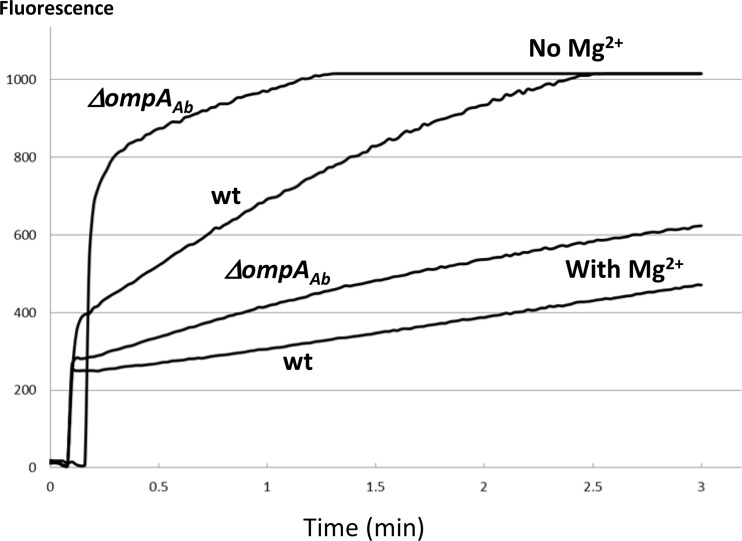
Fig 4.
Accumulation of ethidium bromide (EtBr) in intact cells of the ΔompAAb mutant and its parent strain. The amount of cells corresponding to 0.4 OD600 units was added into 2 ml of 50 mM K-phosphate buffer, pH 7.0, containing 100 μM CCCP with or without 5 mM MgCl2. Ten seconds later, EtBr (final concentration, 6 μM) was added to the mixture, and the fluorescence of the EtBr-nucleic acid complex generated by the influx of EtBr into cells was determined as described in Materials and Methods. Intensity of the fluorescence emission (ordinate) is in arbitrary units.
Some of this increased EtBr permeability was suppressed by the addition of 5 mM MgCl2 to the assay mixture (Fig. 4). Therefore, we used LB broth supplemented with 5 mM MgCl2 for the measurement of antibiotic susceptibility. The broth 2-fold dilution assay of MICs (Table 1) showed that instead of the expected increase in MICs, in the ΔompAAb mutant, there was a small decrease in the MICs of lipophilic compounds, such as benzylpenicillin, nitrocefin, and novobiocin, and a large decrease for the agent that attacks the lipopolysaccharide (LPS)-phospholipid asymmetric bilayer of the OM, polymyxin B. Although many common antibiotics are normally thought to traverse the OM barrier mainly through the water-filled channels of porins (24, 29), in the ΔompAAb mutant, the increased diffusion across the modified lipid bilayer domain (described above) now presumably plays a predominant role, causing this increased susceptibility to lipophilic agents.
Table 1.
Antibiotic susceptibilities of the wild type and the ΔompAAb mutant of A. baumannii
| Agent | MIC (μg/ml) |
|
|---|---|---|
| Wild type | ΔompAAb mutant | |
| Ampicillin | 250 | 125 |
| Benzylpenicillin | 125 | 62 |
| Carbenicillin | 31 | 31 |
| Nitrocefin | 500 | 125 |
| Cephaloridine | 250 | 250 |
| Cephalothin | 500 | 500 |
| Ceftriaxone | 31 | 31 |
| Cefotaxime | 31 | 31 |
| Aztreonam | 31 | 31 |
| Imipenem | 0.3 | 0.3 |
| Tetracycline | 6 | 6 |
| Chloramphenicol | 250 | 250 |
| Novobiocin | 10 | 5 |
| Gentamicin | 6 | 6 |
| Polymyxin B | 20 | 2.5 |
Expression of endogenous β-lactamases in A. baumannii ATCC 17978.
Since we used the endogenous β-lactamase(s) in the OM permeability assay described below, it was important to assess the expression levels of the two endogenous enzymes, AmpC (pI > 9.0) (33) and OXA-51 (pI = 7.0) (7). When about 0.1 mg of the extract of ATCC 17978, grown in LB, was analyzed by isoelectric focusing (see Materials and Methods), the only activity seen had a pI close to 9 and no activity was seen in the area close to a pI of 7 (results not shown). We therefore assume that under our growth conditions, only the AmpC enzyme is expressed to a significant degree.
When the kinetics of hydrolysis of cephalosporins were examined by using crude extracts of ATCC 17978 cells, Kan values estimated by curve fitting were 63, 72, and 550 μM for nitrocefin, cephalothin, and cephaloridine, respectively, quite similar to the values reported earlier for two clinical strains of A. baumannii (33) and for a strain of A. calcoaceticus (presumably A. baumannii) (37). The Vmax values were 0.40, 0.48, and 0.70 nmol/s/mg (dry weight) cells, respectively, and again the relative rates among the three agents were similar to those reported earlier (33).
Permeability of cephalosporins through the OM of A. baumannii in an intact-cell assay.
Because the OM integrity is compromised as described above in the ΔompAAb mutant, we turned to the direct quantitative determination of OM permeability by using cephalosporins as test solutes, partly because cephalosporins, owing to their strong acidic groups, are likely to traverse the OM mainly through aqueous porin channels (as shown experimentally in E. coli [28]). In this assay, we measured the periplasmic concentration of cephalosporins from the rates of hydrolysis of cephalosporins by the periplasmic β-lactamase(s) in intact cells and calculated the permeability coefficient P from the hydrolysis rate and the concentration gradient of the drugs across the OM (28, 51). We used the endogenous chromosomally coded AmpC enzyme, which apparently plays a predominant role in the hydrolysis of β-lactams in the uninduced cells of ATCC 17978, as described above.
The hydrolysis rates of three cephalosporins, including nitrocefin, cephalothin, and cephaloridine, were determined in intact cells treated with 100 μM CCCP in order to inactivate multidrug efflux pumps (Table 2). For nitrocefin and cephalothin, the rates were corrected for hydrolysis due to enzyme that had leaked out into the suspension buffer. Even after correction for the enzyme leakage, the apparent permeability coefficient for nitrocefin was surprisingly high (Table 2). This most likely means that most of this quite lipophilic compound is crossing the OM through the lipid bilayer region that has become more permeable, as described above (rather than through the porin channel).
Table 2.
OM permeability to cephalosporins
| Substrate (concn [mM]) | Strain | Hydrolysis rate (nmol/s/mg)b |
Permeability coefficient (cm/s) | |
|---|---|---|---|---|
| Intact cells | Extract | |||
| Nitrocefin (0.1) | Parent | 0.006 ± 0.002 | 0.253 ± 0.013 | 0.4 × 10−6 |
| ΔompAAb mutant | 0.018 ± 0.006 | 0.250 ± 0.013 | 1.4 × 10−6 | |
| Cephalothin (0.1) | Parent | 0.022 ± 0.007 | 0.295 ± 0.045 | 1.7 × 10−6 |
| ΔompAAb mutant | 0.011 ± 0.008 | 0.248 ± 0.020 | 0.9 × 10−6 | |
| Cephaloridine (1) | Parent | 0.071 ± 0.020 | 0.434 ± 0.048 | 0.57 × 10−6 |
| Parent (no CCCP) | 0.027 ± 0.013 | 0.434 ± 0.048 | 0.21 × 10−6 | |
| ΔompAAb mutanta | 0.033 ± 0.014 | 0.539 ± 0.066 | 0.26 × 10−6 | |
Although we also attempted to determine the cephaloridine permeability without CCCP in this mutant, the net influx was so low that it was impossible to obtain reproducible values.
Values are means and standard deviations.
With the more-hydrophilic cephalothin, we observed a 2-fold decrease in permeability in the ΔompAAb mutant (Table 2). We expected a somewhat larger difference in the permeability coefficients of cephaloridine, since the diffusion rate of this zwitterionic compound across phospholipid bilayers is very low (19). The mutant lacking OmpAAb showed a significant decrease in the permeability coefficient of this compound, although the difference was only 2.2-fold. We note also that the permeability coefficient of cephaloridine in the A. baumannii wild-type OM was 0.57 × 10−6 cm/s: this value is about 100-fold lower than the value obtained for the E. coli OM (28) and similar to that for P. aeruginosa (around 1 × 10−6 cm/s) (49). Still, these values were probably overestimates as a measure of porin permeability because of the contributions from leaked-out enzymes and the penetration across the bilayer region of OM.
In this intact-cell assay, measurements were carried out with the active efflux inactivated by de-energization of the cytoplasmic membrane with a proton conductor, 100 μM CCCP. We examined, in two ways, if the efflux pumps in A. baumannii affected the measurement of OM permeability. First, carrying out assays in the absence of CCCP strongly decreased the apparent permeability coefficient of cephaloridine (Table 2). Second, in the mutant BM4652, lacking two known efflux pumps, AdeABC and AdeIJK, we found that there was a 2- to 4-fold decrease in MIC for nitrocefin, cephalothin, and cephaloridine compared to that of the parent strain BM4454 (data not shown), as was found earlier for more-recently introduced cephalosporins (10).
DISCUSSION
Clinical isolates of A. baumannii often show elevated resistance levels to a large number of antibiotics (4, 32). The strain we used (ATCC 17978) was isolated back in 1951, but it is already resistant to penicillins and cephalosporins introduced early, presumably owing to the intrinsic resistance of this species (Table 1). There are two β-lactamase genes in the A. baumannii ATCC 17978 genome (1) which code for class C AmpC and class D Blaoxa-51 enzymes. We found that the AmpC enzyme that hydrolyzes early cephalosporins rapidly was expressed at a fairly high level under our conditions of growth. However, β-lactamases alone cannot produce significant resistance levels, and we need the collaboration between the periplasmic β-lactamase and the low OM permeability (26). Indeed, the OM of a related species, A. calcoaceticus, was reported to show very low permeability (37).
In this work, we purified the major OM protein OmpAAb from A. baumannii and showed that it produced permeability channels that allowed only a very slow penetration of solutes (Fig. 2). Since the permeability of the unfractionated OM was similarly low (Fig. 2), it appears that OmpAAb is the major porin in this species. In order to confirm this conclusion, we measured the OM permeability for cephalosporins in intact cells with the classical Zimmermann-Rosselet method (51) using an ΔompAAb mutant. However, measurement of permeability in intact cells of A. baumannii was difficult because, owing to the extremely slow permeation through the porin channels, the spontaneous diffusion across the asymmetric lipid bilayer region became relatively significant, especially when the composition of the lipid domain became altered due to the absence of the major OM protein (2). Since β-lactams contain strongly acidic carboxylate groups, this may seem surprising. However, we have shown experimentally that lipophilic β-lactams, such as benzylpenicillin, can traverse the usual phospholipid bilayers with measurable rates most probably in their rare protonated forms (19). Thus, when the lipophilic nitrocefin was used as the probe, the permeation rate was unexpectedly high already in the wild type and became even higher in the ΔompAAb mutant (Table 2). The situation was somewhat better with the more hydrophilic cephalothin and cephaloridine, which do not easily diffuse across bilayers (19): here the ompAAb deletion decreased permeability to about one-half (Table 2). Nevertheless, these data do not necessarily mean that one-half of cephalothin and cephaloridine molecules permeate through a porin(s) other than OmpAAb, because they may be crossing the OM through the bilayer or the hydrolysis could have occurred by leaked-out enzyme that was not fully corrected for. It should also be noted that the permeability coefficient for cephaloridine in the wild-type strain (0.57 × 10−6 cm/s) (Table 2) was more than two orders of magnitude smaller than those found for OmpF- and OmpC-producing E. coli strains (5.3 × 10−4 and 0.45 × 10−4 cm/s, respectively [28]).
Our results thus show that OmpAAb is (a) a slow porin (defined in reference 43) and (b) the porin largely responsible for the nonspecific diffusion across the OM of A. baumannii. On the first point, we are confirming the results of Nitzan and coworkers (30) as well as those of Sato and Nakae (37). Jyothisri et al. (18) concluded that their OmpAAb preparation had a pore-forming specific activity about equal to that of E. coli OmpF; however, the data in their paper show a specific activity about 100-fold lower than that of OmpF, and their conclusion was apparently due to a mistake in calculation. Gribun et al. (15) concluded that their OmpAAb preparation had an activity three to four times lower than that of E. coli OmF. This is still quite high, but we note that the same investigator in this team, Y. Nitzan, had earlier published data showing that OmpAAb had specific activity 35- to 50-fold lower than that of E. coli OmpF (30).
Thus, the major porin of Acinetobacter is the OmpA homolog, OmpAAb, a situation similar to that found in P. aeruginosa, in which another OmpA homolog, OprF, is the major porin (25). This is not surprising, because these two species belong to the same order, Pseudomonadales (6). OmpA has long been believed to fold exclusively as a two-domain protein, with its N-terminal half inserted into the OM and the C-terminal half folded into a globular domain in the periplasm (36). However, this conformation belongs only to the majority conformer within the population of OmpA (and OprF) (42, 43). With OprF, we have shown that the low permeability is due to the fact that only a minority of the protein folds into a different, one-domain, open-channel conformation (40), and we found several factors that affect the divergent folding pathway of this protein (39). It is reasonable to assume that a similar mechanism applies to OmpAAb, although this has not yet been experimentally proven.
We can examine if the measured low permeability and the properties of the β-lactamase can explain the high MIC values for the cephalosporins (Table 1). We can calculate, from the permeability coefficient and the kinetic properties of the β-lactamase, the periplasmic concentration (Cp) of the β-lactam by solving equation 3 of reference 26. Thus, at the MIC of cephaloridine (250 μg/ml or 0.6 mM), its periplasmic concentration is predicted to be 18 μg/ml. This is somewhat on the high side, as the minimal periplasmic concentration inhibiting cell growth for these β-lactams is usually expected to be in the range of 1 to 5 μg/ml (26). The most likely explanation of this discrepancy is the significant contribution from the constitutive multidrug efflux pumps (10) that would lower the periplasmic concentration of the drugs. Indeed, if we use the apparent permeability coefficient obtained without the inactivation of pumps by CCCP (Table 2), the predicted periplasmic concentration becomes 10 μg/ml, close to the expected range. On the other hand, if the A. baumannii OM had the high permeability of the E. coli OM for cephaloridine, we can calculate that the MIC will be essentially identical to its periplasmic concentration, and thus, the constitutive AmpC enzyme alone cannot produce any resistance, underscoring the importance of the OM permeation barrier.
P. aeruginosa also shows high intrinsic resistance levels to earlier cephalosporins, but it used to be quite susceptible to imipenem. It was discovered that imipenem bypasses the inefficient nonspecific porin OprF and uses a basic amino acid channel, OprD (14, 45, 46). In the organisms with slow major porins, we expect to see numerous specific channels for the uptake of various nutrients (16), and imipenem “hijacks” one of them. Interestingly, A. baumannii appears to use a similar strategy, as numerous studies implicated minor proteins other than OmpAAb for the influx of carbapenems. Thus, a decreased expression of 22- and 33-kDa proteins was reported in an imipenem-resistant isolate (5). A 29-kDa protein called CarO is apparently important in the permeation of carbapenems (8, 20, 22). Imipenem-resistant Acinetobacter isolates were also reported to have diminished levels of 47-, 44-, and 37-kDa OM proteins (35). Yet another investigator reported that an imipenem-resistant A. baumannii isolate had decreased levels of a 33- to 36-kDa protein (9). Yet another candidate for a carbapenem channel is the homolog of P. aeruginosa OprD, identified through proteomics (13). Possibly there is more than one channel that can serve for the penetration of carbapenems in Acinetobacter spp.
ACKNOWLEDGMENTS
We thank Patrice Courvalin for the generous gift of strains.
H.N. acknowledges support from NIH grant 1 R01AI009644-41.
Footnotes
Published ahead of print 25 May 2012
REFERENCES
- 1. Adams MD, et al. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 190:8053–8064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ames GF, Spudich EN, Nikaido H. 1974. Protein composition of the outer membrane of Salmonella typhimurium: effect of lipopolysaccharide mutations. J. Bacteriol. 117:406–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Angus BL, Carey AM, Caron DA, Kropinski AM, Hancock RE. 1982. Outer membrane permeability in Pseudomonas aeruginosa: comparison of a wild-type with an antibiotic-supersusceptible mutant. Antimicrob. Agents Chemother. 21:299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bonomo RA, Szabo D. 2006. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infect. Dis. 43(Suppl 2):S49–S56 [DOI] [PubMed] [Google Scholar]
- 5. Bou G, Cervero G, Dominguez MA, Quereda C, Martinez-Beltran J. 2000. Characterization of a nosocomial outbreak caused by a multiresistant Acinetobacter baumannii strain with a carbapenem-hydrolyzing enzyme: high-level carbapenem resistance in A. baumannii is not due solely to the presence of β-lactamases. J. Clin. Microbiol. 38:3299–3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brenner DJ, Krieg NR, Staley JT. 2005. Bergey's manual of systematic bacteriology, 2nd ed, vol 2, part A Springer, New York, NY [Google Scholar]
- 7. Brown S, Young HK, Amyes SG. 2005. Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin. Microbiol. Infect. 11:15–23 [DOI] [PubMed] [Google Scholar]
- 8. Catel-Ferreira M, et al. 2011. Structure-function relationships of CarO, the carbapenem resistance-associated outer membrane protein of Acinetobacter baumannii. J. Antimicrob. Chemother. 66:2053–2056 [DOI] [PubMed] [Google Scholar]
- 9. Clark RB. 1996. Imipenem resistance among Acinetobacter baumannii: association with reduced expression of a 33-36 kDa outer membrane protein. J. Antimicrob. Chemother. 38:245–251 [DOI] [PubMed] [Google Scholar]
- 10. Coyne S, Courvalin P, Perichon B. 2011. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob. Agents Chemother. 55:947–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Damier-Piolle L, Magnet S, Bremont S, Lambert T, Courvalin P. 2008. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob. Agents Chemother. 52:557–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. DÉ E, De Mot R, Orange N, Saint N, Molle G. 1995. Channel-forming properties and structural homology of major outer membrane proteins from Pseudomonas fluorescens MFO and OE 28.3. FEMS Microbiol. Lett. 127:267–272 [DOI] [PubMed] [Google Scholar]
- 13. Dupont M, Pages JM, Lafitte D, Siroy A, Bollet C. 2005. Identification of an OprD homologue in Acinetobacter baumannii. J. Proteome Res. 4:2386–2390 [DOI] [PubMed] [Google Scholar]
- 14. Fukuoka T, et al. 1993. Activity of the carbapenem panipenem and role of the OprD (D2) protein in its diffusion through the Pseudomonas aeruginosa outer membrane. Antimicrob. Agents Chemother. 37:322–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gribun A, Nitzan Y, Pechatnikov I, Hershkovits G, Katcoff DJ. 2003. Molecular and structural characterization of the HMP-AB gene encoding a pore-forming protein from a clinical isolate of Acinetobacter baumannii. Curr. Microbiol. 47:434–443 [DOI] [PubMed] [Google Scholar]
- 16. Hancock RE, Brinkman FS. 2002. Function of Pseudomonas porins in uptake and efflux. Annu. Rev. Microbiol. 56:17–38 [DOI] [PubMed] [Google Scholar]
- 17. Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86 [DOI] [PubMed] [Google Scholar]
- 18. Jyothisri K, Deepak V, Rajeswari MR. 1999. Purification and characterization of a major 40 kDa outer membrane protein of Acinetobacter baumannii. FEBS Lett. 443:57–60 [DOI] [PubMed] [Google Scholar]
- 19. Li XZ, Ma D, Livermore DM, Nikaido H. 1994. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: active efflux as a contributing factor to β-lactam resistance. Antimicrob. Agents Chemother. 38:1742–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Limansky AS, Mussi MA, Viale AM. 2002. Loss of a 29-kilodalton outer membrane protein in Acinetobacter baumannii is associated with imipenem resistance. J. Clin. Microbiol. 40:4776–4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Magnet S, Courvalin P, Lambert T. 2001. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 45:3375–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mussi MA, Limansky AS, Viale AM. 2005. Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: natural insertional inactivation of a gene encoding a member of a novel family of β-barrel outer membrane proteins. Antimicrob. Agents Chemother. 49:1432–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nestorovich EM, Sugawara E, Nikaido H, Bezrukov SM. 2006. Pseudomonas aeruginosa porin OprF: properties of the channel. J. Biol. Chem. 281:16230–16237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nikaido H. 1989. Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob. Agents Chemother. 33:1831–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nikaido H, Nikaido K, Harayama S. 1991. Identification and characterization of porins in Pseudomonas aeruginosa. J. Biol. Chem. 266:770–779 [PubMed] [Google Scholar]
- 26. Nikaido H, Normark S. 1987. Sensitivity of Escherichia coli to various β-lactams is determined by the interplay of outer membrane permeability and degradation by periplasmic β-lactamases: a quantitative predictive treatment. Mol. Microbiol. 1:29–36 [DOI] [PubMed] [Google Scholar]
- 27. Nikaido H, Rosenberg EY. 1981. Effect on solute size on diffusion rates through the transmembrane pores of the outer membrane of Escherichia coli. J. Gen. Physiol. 77:121–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nikaido H, Rosenberg EY, Foulds J. 1983. Porin channels in Escherichia coli: studies with β-lactams in intact cells. J. Bacteriol. 153:232–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nikaido H, Vaara M. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49:1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nitzan Y, Pechatnikov I, Bar-El D, Wexler H. 1999. Isolation and characterization of heat-modifiable proteins from the outer membrane of Porphyromonas asaccharolytica and Acinetobacter baumannii. Anaerobe 5:43–50 [DOI] [PubMed] [Google Scholar]
- 31. O'Callaghan CH, Morris A, Kirby SM, Shingler AH. 1972. Novel method for detection of β-lactamases by using a chromogenic cephalosporin substrate. Antimicrob. Agents Chemother. 1:283–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Perilli M, et al. 1996. Characterization of the chromosomal cephalosporinases produced by Acinetobacter lwoffii and Acinetobacter baumannii clinical isolates. Antimicrob. Agents Chemother. 40:715–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Philippon LN, Naas T, Bouthors AT, Barakett V, Nordmann P. 1997. OXA-18, a class D clavulanic acid-inhibited extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 41:2188–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Quale J, Bratu S, Landman D, Heddurshetti R. 2003. Molecular epidemiology and mechanisms of carbapenem resistance in Acinetobacter baumannii endemic in New York City. Clin. Infect. Dis. 37:214–220 [DOI] [PubMed] [Google Scholar]
- 36. Ried G, Koebnik R, Hindennach I, Mutschler B, Henning U. 1994. Membrane topology and assembly of the outer membrane protein OmpA of Escherichia coli K12. Mol. Gen. Genet. 243:127–135 [DOI] [PubMed] [Google Scholar]
- 37. Sato K, Nakae T. 1991. Outer membrane permeability of Acinetobacter calcoaceticus and its implication in antibiotic resistance. J. Antimicrob. Chemother. 28:35–45 [DOI] [PubMed] [Google Scholar]
- 38. Schagger H, von Jagow G. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199:223–231 [DOI] [PubMed] [Google Scholar]
- 39. Sugawara E, Nagano K, Nikaido H. 2010. Factors affecting the folding of Pseudomonas aeruginosa OprF porin into the one-domain open conformer. mBio 1(4):e00228–10 doi:10.1128/mBio.00228-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sugawara E, Nestorovich EM, Bezrukov SM, Nikaido H. 2006. Pseudomonas aeruginosa porin OprF exists in two different conformations. J. Biol. Chem. 281:16220–16229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sugawara E, Nikaido H. 1992. Pore-forming activity of OmpA protein of Escherichia coli. J. Biol. Chem. 267:2507–2511 [PubMed] [Google Scholar]
- 42. Sugawara E, Nikaido H. 1994. OmpA protein of Escherichia coli outer membrane occurs in open and closed channel forms. J. Biol. Chem. 269:17981–17987 [PubMed] [Google Scholar]
- 43. Sugawara E, Nikaido H. 2004. OmpA/OprF: slow porins or channels produced by alternative folding of outer membrane proteins, p 119–138 In Benz R. (ed), Bacterial and eukaryotic porins: structure, function, mechanism. Wiley-VCH, Weinheim, Germany [Google Scholar]
- 44. Sugawara E, Steiert M, Rouhani S, Nikaido H. 1996. Secondary structure of the outer membrane proteins OmpA of Escherichia coli and OprF of Pseudomonas aeruginosa. J. Bacteriol. 178:6067–6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Trias J, Nikaido H. 1990. Outer membrane protein D2 catalyzes facilitated diffusion of carbapenems and penems through the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 34:52–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Trias J, Nikaido H. 1990. Protein D2 channel of the Pseudomonas aeruginosa outer membrane has a binding site for basic amino acids and peptides. J. Biol. Chem. 265:15680–15684 [PubMed] [Google Scholar]
- 47. Vila J, Marti S, Sanchez-Cespedes J. 2007. Porins, efflux pumps and multidrug resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 59:1210–1215 [DOI] [PubMed] [Google Scholar]
- 48. Woodruff WA, Hancock RE. 1988. Construction and characterization of Pseudomonas aeruginosa protein F-deficient mutants after in vitro and in vivo insertion mutagenesis of the cloned gene. J. Bacteriol. 170:2592–2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yoshimura F, Nikaido H. 1982. Permeability of Pseudomonas aeruginosa outer membrane to hydrophilic solutes. J. Bacteriol. 152:636–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yoshimura F, Zalman LS, Nikaido H. 1983. Purification and properties of Pseudomonas aeruginosa porin. J. Biol. Chem. 258:2308–2314 [PubMed] [Google Scholar]
- 51. Zimmermann W, Rosselet A. 1977. Function of the outer membrane of Escherichia coli as a permeability barrier to β-lactam antibiotics. Antimicrob. Agents Chemother. 12:368–372 [DOI] [PMC free article] [PubMed] [Google Scholar]