Abstract
Following arrest by UV-induced DNA damage, replication is restored through a sequence of steps that involve partial resection of the nascent DNA by RecJ and RecQ, branch migration and processing of the fork DNA surrounding the lesion by RecA and RecF-O-R, and resumption of DNA synthesis once the blocking lesion has been repaired or bypassed. In vitro, the primosomal proteins (PriA, PriB, and PriC) and Rep are capable of initiating replication from synthetic DNA fork structures, and they have been proposed to catalyze these events when replication is disrupted by certain impediments in vivo. Here, we characterized the role that PriA, PriB, PriC, and Rep have in processing and restoring replication forks following arrest by UV-induced DNA damage. We show that the partial degradation and processing of the arrested replication fork occurs normally in both rep and primosome mutants. In each mutant, the nascent degradation ceases and DNA synthesis initially resumes in a timely manner, but the recovery then stalls in the absence of PriA, PriB, or Rep. The results demonstrate a role for the primosome and Rep helicase in overcoming replication forks arrested by UV-induced damage in vivo and suggest that these proteins are required for the stability and efficiency of the replisome when DNA synthesis resumes but not to initiate de novo replication downstream of the lesion.
INTRODUCTION
PriA, PriB, and PriC were originally identified as proteins required for replication of single-strand ϕX174 phage DNA in vitro and in vivo (70, 71). In vitro, the proteins function as a complex that is required for processive priming to occur behind the replicative helicase, DnaB (1, 2). PriA initially binds a hairpin structure on the ϕX174 chromosome, followed by PriB, DnaT, and PriC. The resulting complex then recruits DnaC, which loads the DnaB helicase onto the chromosome. The DnaG primase is then able to associate with DnaB to synthesize RNA primers. While DnaG and DnaB are sufficient for primer synthesis on ϕX174 DNA (1), specific and processive priming of single-stranded DNA binding protein-coated phage DNA requires PriA (2). In vivo, conversion of ϕX174 from its plus-strand form to its minus-strand replication intermediate requires PriA and other Escherichia coli host proteins (40). E. coli strains lacking PriA have reduced viability, growth rates, and culture densities relative to wild-type cells (36). priA mutants are also constitutively induced for the SOS response, and cells lacking PriA produce filaments extensively (49). Taken together, these observations led early researchers to propose that the primosomal proteins promote efficient priming for Okazaki fragments during lagging-strand replication (35, 38).
rep was originally identified as a mutant that was unable to support replication of DNA of several double-stranded phage, including ϕX174 (19, 20, 33, 56, 62). rep was subsequently shown to encode a DNA helicase that tracks on the leading-strand template and is essential to reconstitute replication of double-stranded phage in vitro (55). rep mutants are modestly hypersensitive to UV irradiation compared to wild-type cells (9) and were reported to have longer doubling times, abnormal nucleoids, and a reduced rate of replication fork movement (3, 33, 34). Recent studies have shown that Rep helicase is needed to facilitate replication fork progression along transcribed DNA or protein-bound DNA (4, 7, 24), suggesting the helicase plays a role in clearing impediments to leading-strand synthesis during replication.
Replication forks must deal with a variety of obstacles that may impede their progress, including DNA-bound proteins, secondary structures, strand breaks, and adducts or damage to the DNA bases themselves. With respect to DNA base damage, UV irradiation with 254-nm light has often served as a model to address the question of how replication recovers following encounters with this form of impediment. UV irradiation induces two primary photoproducts, cis, syn-cyclobutane pyrimidine dimers (CPDs) and 6,4 pyrimidine-pyrimidone photoproducts (6-4 PPs) (59, 67, 68). Although these lesions block DNA polymerases and arrest replication (28, 58), growing E. coli cultures survive doses that produce more than 2,000 lesions per genome (30), indicating that cells contain efficient mechanisms to process these lesions when they are encountered during replication.
The recovery of replication following arrest by UV-induced DNA damage occurs through a sequence of well-characterized steps. Following arrest, the nascent lagging strand is partially degraded by the combined action of the RecJ nuclease and RecQ helicase. This processing is thought to restore the lesion-containing region to a double-stranded form that can be accessed and repaired by the nucleotide excision repair complex (17). Consistent with this, in the absence of either repair or nascent DNA degradation, the recovery of replication is delayed, and both survival and recovery become dependent on translesion synthesis by DNA polymerase V (12, 13). RecF, RecO, and RecR limit the RecJ/RecQ-mediated degradation and enhance the formation of RecA filaments at the arrested region (11, 14, 60, 64). Biochemical characterizations suggest that the RecA filament formed in the presence of RecFOR is capable of promoting branch migration at the fork in a manner that could promote regression away from the lesion and subsequently reset the 3′ end of the fork once the impediment has been removed or overcome (47, 60, 64, 69). In vivo, cells lacking any one of these gene products fail to resume DNA synthesis, and the DNA at the replication fork is extensively degraded (11, 14, 15).
Several lines of evidence suggest that Rep and the primosome also participate in restoring replication following arrest at a UV-induced lesion, either through direct resumption of the arrested replisome or de novo initiation of a replisome downstream of the arrest site. Both priA and rep contribute to the DNA synthesis that occurs during recombinational processes (26, 32, 41, 52, 65). Although no single gene by itself is essential for viability, double mutants in priA and priC or priA and rep are lethal, and both priA and rep mutants are hypersensitive to DNA damage (53). It has also been widely postulated that frequent replication disruptions by endogenous DNA damage in vivo account for the poor growth and low viability of priA and rep mutants (8, 45, 57). In addition, one study has reported a delayed recovery of DNA synthesis in PriA mutants following low doses of UV light (51). In vitro, the addition of PriA and PriB, PriA and PriC, or PriC and Rep allows DNA synthesis to occur at synthetic DNA fork structures in the presence of the other core replication proteins (25, 26). However, the role of PriA, PriB, PriC, and Rep in the progressive steps of resection, processing, or resumption following replication arrest at UV-induced DNA damage has not been directly examined in vivo. Here, we characterize the molecular events that occur during the progressive steps associated with the recovery of replication in UV-irradiated cultures of mutants lacking each of these gene products.
MATERIALS AND METHODS
Bacterial strains.
The parent of all strains used in this study is SR108, a thyA36 deoC2 derivative of W3110 (44). The priB302 allele from SS138 (54) was linked to kanamycin resistance in two steps. First, the kanR gene was inserted 40 bp downstream of yjfC using PCR insertion with the primers priBpostyjfC-kanF (5′GAATGTTTTAGCAATCTCTTTCTGTCATGAATCCATGGCATATGGACAGCAAGCGAACCG) and priBpost-yjfC-kanR (5′GTCTCCCTCCATCAATGGCAGTCACCATTAGTATGGTCACATCAGAAGAACTCGTCAAGAAG), followed by recombineering into DY329 to generate CL1631 (75). yjfC::kan, from CL1631, next was transduced into SS138 using standard P1 transduction methods. The priB302 allele is 72% cotransducible with yjfC::kan. The priC gene was replaced from codons 3 to 175 with cat, conferring chloramphenicol resistance, using PCR replacement with the primers PriC-CatF (5′TAACAATTATCATTTCATTGAGGTCTTATCGTGAAAACCATGAGACGTTGATCGGCAC) and PriC-CatR (5′TTCCAGTGACATATTCTCTCCATTGCTAGCGGGTTAAACGCTTTCGAATTTCTGCCATTC), followed by recombineering into DY329. The rep gene was replaced from codons 3 to 673 with kanR, conferring kanamycin resistance, using PCR replacement with the primers Rep-KanF (5′TCCCCCCGTTCGAAGATTGAGCAATACACCTATGCGTCTATATGGACAGCAAGCGAACCG) and Rep-KanR (5′GCTGACGCATCTTTTCCGGCCTTGATTATTTCCCTCGTTTTCAGAAGAACTCGTCAAGAAG), followed by recombineering into DY329. Strains isogenic to SR108 and lacking priA2, priB302, priC, rep, recF, and recA were made using standard P1 transduction methods. Cells were transformed with plasmid pBR322 for experiments involving two-dimensional agarose gel electrophoresis. A list of the strains constructed and used in this study is shown in Table 1. Genotypes for all strains were confirmed by PCR and Southern blot analysis. UV sensitivity was assessed in every experiment to monitor priA2 strains for possible suppressors.
Table 1.
E. coli K-12 strains used
| Strain | Relevant genotype | Reference or construction |
|---|---|---|
| SR108 | λ− thyA36, deoC2 IN(rrnD-rrnE)1 rph | 44 |
| DY329 | ΔlacU169 nadA::Tn10 gal490 λcI857 Δ(cro-bioA) | 75 |
| JC19008 | priA2::kan dnaC809 (parent DM4000) | 52 |
| SS138 | priB302 | 54 |
| CL1070 | ΔlacU169 nadA::Tn10 gal490 λcI857 Δ(cro-bioA) priC::cat | DY329 × PCR fragment (priC-cat primers) |
| CL1073 | ΔlacU169 nadA::Tn10 gal490 λcI857 Δ(cro-bioA) rep::kan | DY329 × PCR fragment (rep-kan primers) |
| CL1631 | ΔlacU169 nadA::Tn10 gal490 λcI857 Δ(cro-bioA) yjfC::kan | DY329 × PCR fragment (priB-postyjfC-kan primers) |
| CL1633 | priB302 yjfC::kan | SS138 × P1 (CL1631) |
| Strains isogenic to SR108 | ||
| HL921 | recA::Tn10 | 14 |
| CL530 | pBR322 | 16 |
| CL579 | recF6206::tet | 16 |
| CL583 | recF6206::tet pBR322 | 16 |
| CL1102 | priC::cat | SR108 × P1 (CL1070) |
| CL1105 | rep::kan | SR108 × P1 (CL1073) |
| CL1122 | priA2::kan | SR108 × P1 (JC19008) |
| CL1638 | priB302 yjfC::kan | SR108 × P1 (CL1633) |
| CL1465 | priC::cat pBR322 | CL1102 transformed with pBR322 |
| CL1467 | rep::kan pBR322 | CL1105 transformed with pBR322 |
| CL2013 | priB302 yjfC::kan pBR322 | CL1638 transformed with pBR322 |
Degradation of nascent and genomic DNA.
UV irradiation used a 15-watt germicidal lamp (254 nm) at an incident dose of 0.9 J/m2/s (0.005 J/m2/s for doses below 5 J/m2). Overnight cultures were diluted 1:100 and grown in 10 ml Davis medium with 0.4% glucose, 0.2% Casamino Acids, and 10 μg/ml thymine (DGCthy medium) supplemented with 0.1 μCi/ml of [14C]thymine to an optical density at 600 nm (OD600) of 0.4 in a shaking incubator at 37°C. Cultures were pulse labeled with 1 μCi/ml [3H]thymidine for 5 s, filtered onto 0.45-μm Fisherbrand general filtration membranes, washed with NET buffer (100 mM NaCl, 10 mM EDTA, pH 8.0, 10 mM Tris, pH 8.0), and resuspended in 10 ml of prewarmed, nonradioactive DGCthy medium. Cultures were immediately UV irradiated at 27 J/m2. At the indicated time points, duplicate 0.2-ml aliquots (triplicates for the zero time point) were precipitated in cold 5% trichloroacetic acid (TCA; Fisherbrand). The precipitate was collected on Millipore glass fiber filters, and the amount of 3H and 14C on each filter was determined by scintillation counting.
Two-dimensional agarose gel electrophoresis.
Cultures containing the plasmid pBR322 were grown overnight in the presence of 100 μg/ml ampicillin. Aliquots (0.2 ml) of the overnight cultures were collected by centrifugation, resuspended in 20 ml of DGCthy medium, and grown without ampicillin to an OD600 of 0.5 in a shaking incubator at 37°C. At this time, cultures were irradiated at 50 J/m2 and transferred to a fresh, prewarmed flask. At the indicated times, a 0.75-ml aliquot of culture was transferred to an equal volume of NET (100 mM NaCl, 20 mM EDTA, pH 8.0, 10 mM Tris, pH 8.0), pelleted, resuspended in 0.15 ml of lysis solution (2 mg/ml lysozyme, 0.5 mg/ml RNAse A in 10 mM Tris [pH 8.0], 1 mM EDTA [pH 8.0]), and incubated for 30 min at 37°C. After this step, 0.01 ml each of 20% Sarkosyl and 10 mg/ml Proteinase K was added to the samples, and incubation continued for an additional 30 min at 37°C. Samples were then extracted once with four volumes of phenol-chloroform followed by one extraction with four volumes of chloroform, dialyzed against 200 ml of TE (2 mM Tris [pH 8.0], 1 mM EDTA [pH 8.0]) for 1 h on floating 47-mm Millipore 0.025-μm pore disks, and then digested with PvuII restriction endonuclease (Fermentas) overnight at 37°C. Samples were then extracted with two volumes of chloroform and loaded directly on a 0.4% agarose gel in 1× TBE (Tris-borate-EDTA, pH 8.0) buffer. Genomic DNA was initially separated in this first dimension at 1 V/cm for 15 h. For the second dimension, lanes were excised, rotated 90°, recast in a 1% agarose gel in 1× TBE, and electrophoresed at 6.5 V/cm for 7 h. DNA in the gels was transferred to a HybondN+ nylon membrane by standard Southern blotting, and the plasmid DNA was detected by probing with 32P-labeled pBR322 that was prepared by nick translation (Roche) using [α-32P]dCTP (MP Biomedicals) and visualized using a Storm PhosphorImager with its associated ImageQuant analysis software (GE LifeSciences).
DNA synthesis and accumulation.
Overnight cultures were diluted 1:100 and grown in DGCthy medium supplemented with 0.1 μCi/ml of [14C]thymine to an OD600 of precisely 0.3, at which point half of the culture was mock irradiated while the other half received an incident dose of 27 J/m2. At the times indicated, duplicate 0.5-ml aliquots of culture were pulse labeled with 1 μCi/ml [3H]thymidine for 2 min at 37°C. Cells were then lysed and the DNA precipitated in cold 5% TCA. The precipitate was collected on Millipore glass fiber filters, and the amount of 3H and 14C on each filter was determined by scintillation counting. The relative rate of synthesis was determined by normalizing the amount of 3H incorporated at each time point to the amount of 3H incorporated 10 min before UV irradiation (−10 min post-UV). The relative amount of total DNA was determined by normalizing the amount of 14C incorporated at each time point to the amount of 14C incorporated 10 min before UV irradiation.
UV survival assays.
Fresh overnight cultures were diluted 1:100 in DGCthy medium and grown to an OD600 between 0.4 and 0.5 (approximately 6 × 108 cells/ml). Ten-μl aliquots of serial 10-fold dilutions were spotted in triplicate on Luria-Bertani plates containing 10 μg/ml thymine and UV irradiated at the indicated doses. Viable colonies were counted following overnight incubation at 37°C.
Growth rates.
Fresh overnight cultures were10-fold serially diluted in DGCthy medium, and 0.2-ml aliquots then were plated in duplicate into the wells of a sterile 96-well microtiter dish. The microtiter cultures were then agitated at 37°C, and the OD600 for each culture was measured over time using a BIO-Whittaker ELx808 plate reader. The number of viable colonies per ml in each overnight culture was determined at the start of every experiment.
RESULTS
Mutants lacking PriA, PriB, PriC, or Rep partially degrade the nascent DNA following arrest by UV-induced DNA damage, similar to wild-type cells.
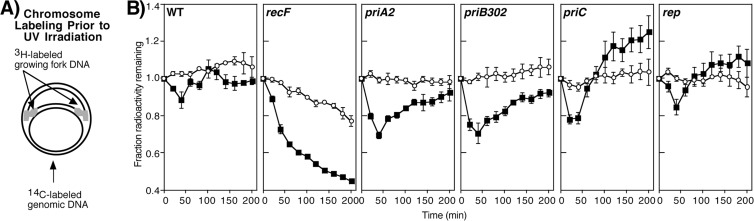
To determine what effect the absence of PriA, PriB, PriC, or Rep has on the DNA at UV-arrested replication forks, we monitored the amount of DNA degradation occurring at the arrested replication fork compared to that occurring on the overall genome for mutants in each of these genes. To this end, isogenic mutants of priA2, priB302, priC, and rep were constructed into the parental strain SR108. Cultures prelabeled with [14C]thymine were pulse labeled with [3H]thymidine for 5 s and then immediately transferred to nonradioactive medium and UV irradiated with 27 J/m2. The amounts of 3H and 14C remaining in the DNA were then monitored over time. This approach allowed us to measure the amount of cellular degradation occurring at the nascent DNA strands (Fig. 1A, 3H label) of the replication fork as well as in the total genomic DNA (14C label).
Fig 1.
Primosomal proteins and Rep do not act directly on UV damage-arrested replication forks. (A) [14C]thymine-labeled cultures were pulse labeled with [3H]thymidine for 5 s, filtered, rinsed, and resuspended in nonradioactive medium and immediately UV irradiated with 27 J/m2. (B) The fraction of total DNA, 14C (○), and nascent DNA, 3H (■), remaining in the culture at each indicated time point is plotted. Graphs represent averages from at least three independent experiments. Error bars represent one standard deviation. The initial values for 3H and 14C ranged from 1,300 to 4,000 cpm and 700 to 2,100 cpm, respectively, for all experiments.
Following UV irradiation of wild-type cultures, very little degradation of the total genomic DNA is observed (Fig. 1B). However, in the pulse-labeled nascent DNA, some limited degradation occurs at times prior to when replication resumes, consistent with our previous observations (11, 14, 17). The amount of detectable degradation is restricted to 40 min post-UV irradiation, and an increase in 3H-labeled DNA is actually observed at later times after DNA synthesis resumes. Although in principle the amount of precipitable radioactivity should either remain constant or decrease over time, an increase in radioactivity is consistently observed in wild-type cells and other strains at times after replication resumes. The increase in 3H is likely due to the remaining intracellular pools of labeled nucleotides that we are unable to wash out (14, 17). Unlike wild-type cultures, in UV-irradiated recF cultures, which fail to resume DNA synthesis, the nascent DNA degradation is more extensive and continues, with approximately half of the 3H-labeled nascent DNA degrading over the time course (Fig. 1B).
When we examined UV-irradiated cultures of priA2, priB302, priC, or rep mutants, we observed that in each case, little degradation occurred in the overall genomic DNA, and the degradation of the nascent DNA ceased within 40 min after UV irradiation, similar to wild-type cells (Fig. 1B). Following this initial degradation, we observed that intracellular pools began to be reincorporated at the fork after the 40-min time point, similar to wild-type cells, in all mutant strains examined. We interpret these observations to suggest that PriA, PriB, PriC, and Rep are not required for the initial processing or degradation of the nascent DNA after the arrest of replication. The observed reincorporation of radionucleotides after 40 min in each strain also suggests that these mutants are able to synthesize some new DNA, and that the synthesis initially resumes at a time comparable to that observed in wild-type cultures.
Although degradation of 3H-labeled nascent DNA ceased and reincorporation of 3H-labeled nucleoside precursors resumed at times similar to those of wild-type cells, a modestly higher fraction of the nascent DNA was degraded in priA2 and priB302 mutants (approximately 25% in these strains compared to 13% in wild-type cells). This could be explained by basal replication in these mutants being modestly impaired and less DNA initially labeled in the 5-s pulse. Alternatively, it could suggest that the newly synthesized lagging-strand DNA in these mutants contains more fragments or DNA ends, which are susceptible to degradation by RecJ and RecQ.
Following arrest by UV-induced DNA damage, replication forks are processed normally in the absence of PriB, PriC, or Rep.
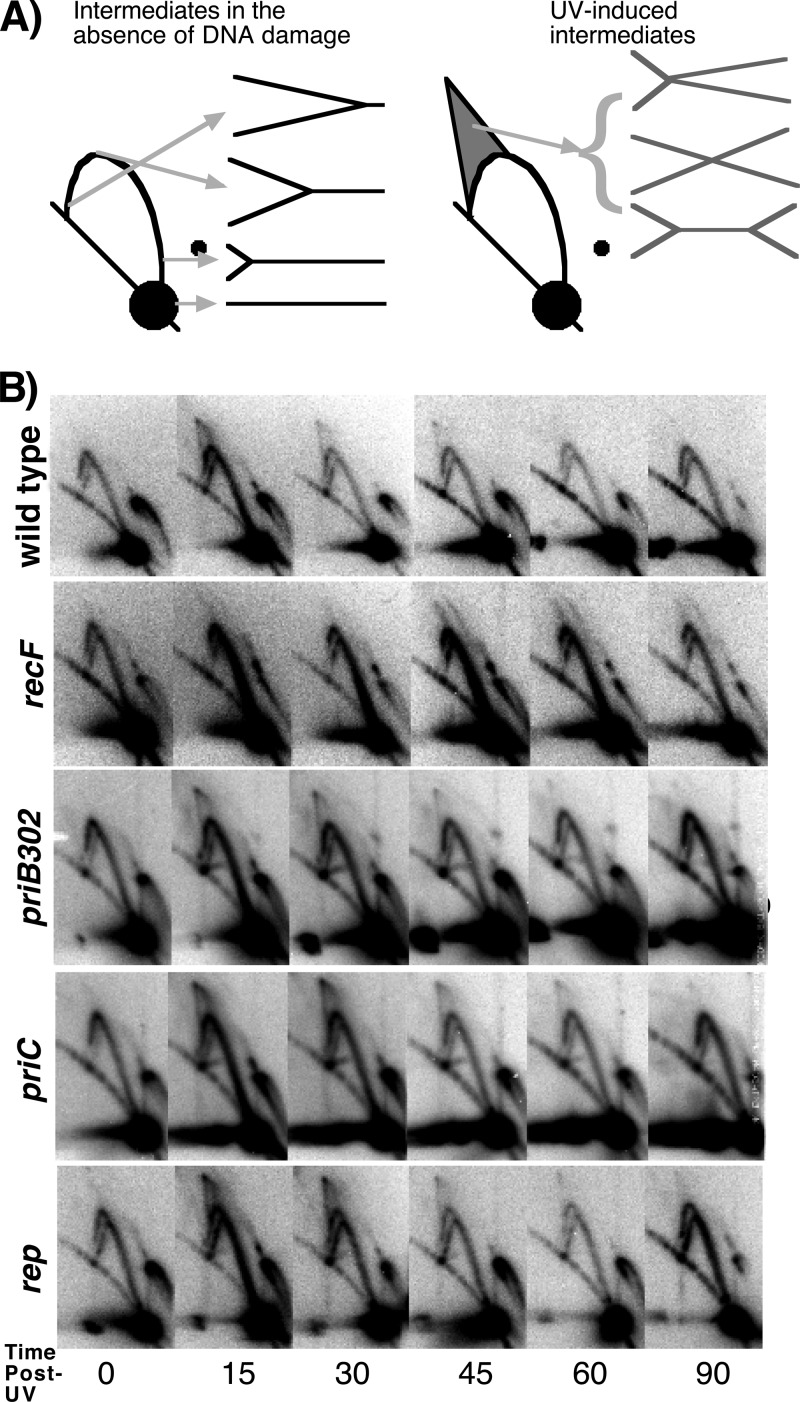
In vivo, the processing of replication forks following UV-induced arrest involves a transient regression of the fork structure that is catalyzed by RecFOR and RecA. The regressed structure persists until a time that correlates with when the lesions are repaired and replication resumes (11, 16). The structural intermediates that occur at the replication fork during the recovery process can be visualized on replicating plasmids such as pBR322 in vivo.
To determine whether the absence of the primosome or Rep helicase affects replication fork processing prior to the recovery of DNA synthesis, we examined the structural intermediates of replicating pBR322 plasmids in these mutants following UV irradiation using two-dimensional agarose gel analysis. To this end, strains containing the plasmid pBR322 were UV irradiated at 50 J/m2. This dose produces an average of one lesion per plasmid strand. At various times after irradiation, total genomic DNA was purified and digested with PvuII, which linearizes the plasmid just downstream from its origin of replication. The structural intermediates were then separated in two-dimensional agarose gels and visualized by Southern analysis. In the absence of UV damage, nonreplicating plasmids migrate through the gel as a linear 4.4-kb fragment, forming the large predominant spot observed in these gels (Fig. 2A). Replicating plasmids form simple Y-shaped structures that migrate more slowly through the gel due to their larger size and nonlinear shape. These structures form an arc that extends out from the linear fragments toward the origin of the gel. Following UV irradiation, the processing of replication forks by RecA and the RecF pathway proteins produces transient intermediates that are observed as double-Y- or X-shaped structures (11, 16). These complex, nonlinear shapes migrate through the gel at a lower rate and are observed in a cone region above the arc of replicating Y-shaped structures.
Fig 2.
UV-arrested replication forks are processed normally in cells lacking priB, priC, or rep. (A) The predicted migration pattern of PvuII-digested pBR322 plasmid observed by two-dimensional agarose gel analysis and subsequent visualization with 32P-labeled pBR322 is diagrammed. Nonreplicating plasmids run as a linear 4.4-kb fragment. Replicating plasmids form Y-shaped structures that migrate slower than nonreplicating DNA, forming an arc that extends above the linear region. Following UV irradiation, double-Y- or X-shaped intermediates are observed that migrate in the cone region behind the arc of Y-shaped molecules. (B) Two-dimensional agarose gels from wild-type, recF, priB302, priC, and rep cultures containing pBR322 at the indicated times following UV irradiation.
In wild-type cultures, only Y-shaped replication intermediates are observed at times prior to irradiation (Fig. 2B). Within 15 min after UV irradiation, a transient accumulation of Y-shaped and cone region intermediates are observed. These UV-induced intermediates begin to decrease after 30 min and return to preirradiation levels within 60 min of UV exposure. In recF mutants, the processing of the arrested replication forks does not occur and the cone region intermediates are not observed. Instead, the arrested forks remain and accumulate as simple Y-shaped structures (Fig. 2B).
When we examined irradiated cultures of priB302, priC, and rep mutants, we observed that in each case, the UV-induced replication intermediates transiently appeared and were resolved in a manner that was similar to that observed in wild-type cells (Fig. 2B). These observations indicate that PriB, PriC, and Rep are not required for the replication fork processing that occurs following UV-induced replication arrest.
Consistent with previous reports (36, 49), we were unable to derive stable pBR322 transformants in priA2 mutants and therefore were unable to evaluate these mutants by two-dimensional agarose gel analysis. This is likely due to the requirement for PriA in the initiation of pBR322 lagging-strand synthesis (46). Some priB mutations have also been reported to affect the stable replication of plasmids (6), although this occurs to a more modest degree than with priA2. Irrespective of the basal plasmid replication capacity of these mutants, the priB302 mutation did not affect the processing that occurred after UV irradiation on replicating plasmids.
The absence of PriA, PriB, or Rep impairs the resumption of DNA synthesis after disruption by UV-induced damage.
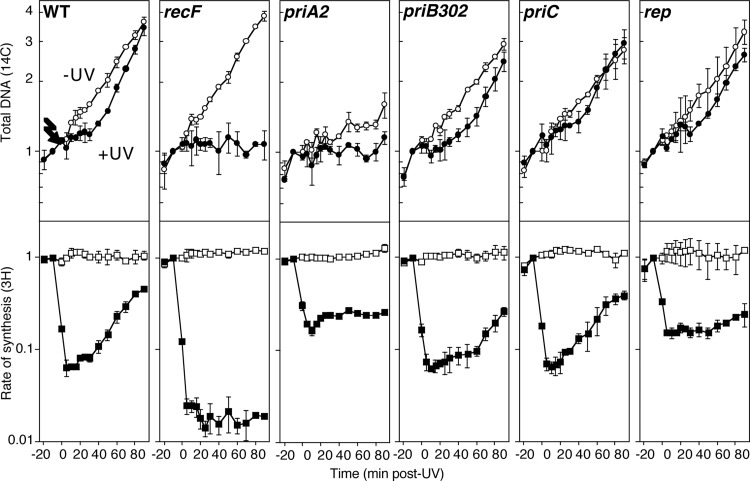
Although the precise composition of the arrested replisome remains uncharacterized, the lack of synthesis and accessibility of the nascent lagging-strand DNA to nucleolytic degradation suggests that some components of the replisome are partially disassembled following arrest. Reestablishing or reactivating the replisome depends on the initial processing and repair by RecA and the RecF pathway genes and is thought to require other gene products as well. To characterize the role that the primosome and Rep helicase have in restoring an active replisome, we monitored the rate at which replication recovered following UV-induced arrest in priA2, priB302, priC, and rep mutants. Cultures grown in the presence of [14C]thymine were split and then either UV irradiated at 27 J/m2 or mock irradiated. At various times after irradiation, duplicate aliquots of each culture were pulse labeled for 2 min with [3H]thymidine before the cells were lysed and the amount of radioactivity incorporated into the DNA was determined. In this way, both the total accumulation of genomic DNA (14C label) and rate of DNA synthesis (3H label) could be monitored in the cultures concurrently over time. To monitor the ability of each strain to replicate in the absence of damage and ensure that any observed effects were due specifically to UV, a mock-irradiated control was included in each case.
In wild-type cultures, the rate of DNA synthesis decreased immediately after UV irradiation by more than 90%, but it began to recover within 20 min and continued to increase until it approached unirradiated levels by the end of the time course (Fig. 3). At this time, the overall DNA accumulation in the irradiated wild-type cultures also approached that of the unirradiated cultures. In comparison, in UV-irradiated cultures of recF mutants, the rate of DNA synthesis remained low, and no further DNA was seen to accumulate following the inhibition of replication (Fig. 3), consistent with previous studies showing that the processing of the fork by RecF and RecA is required for the resumption of DNA synthesis following arrest (14, 15).
Fig 3.
Primosomal proteins PriA and PriB, but not PriC, and Rep are required for the recovery of replication following UV irradiation. [3H]thymidine was added to [14C]thymine-prelabeled cultures for 2 min at the indicated times following either 27 J/m2 UV irradiation (filled symbols) or mock irradiation (open symbols) at time zero. The amount of total DNA at each time point relative to −10 min post-UV treatment, 14C (○), and DNA synthesis/2 min, 3H (□), is plotted. Graphs represent averages from at least three independent experiments. Error bars represent one standard deviation.
When we examined UV-irradiated cultures of priA2, priB302, or priC mutants, we observed that following the initial inhibition, the rate of DNA synthesis began to recover within the first 20 min of irradiation in each strain (Fig. 3), similar to wild-type cultures. However, following an initial burst of DNA synthesis where the rate increased, the recovery appeared to stall in both priA2 and priB302 mutants. In priA2 mutants, the recovery of DNA synthesis remained stalled for the duration of the time course. In priB302 mutants, on the other hand, any further recovery of DNA synthesis was delayed for approximately 60 min, at which time a more robust recovery of synthesis was observed. In comparison, no defect in the recovery of DNA synthesis was detected in priC mutants, and DNA synthesis was restored with kinetics that were similar to those of wild-type cultures, suggesting that PriC is not necessary for the resumption of synthesis. We interpret the stalled recovery of synthesis in priA2 and priB302 mutants to indicate that cells are able to resume some DNA synthesis but fail to reestablish or reset an efficient replisome after the disruption or arresting event. Consistent with this interpretation, suppression of priA2 by dnaC809, which is thought to bypass the requirement for PriA in reestablishing a bona fide replisome from disrupted replication fork substrates (39), completely restores the ability of these cells to resume replication after UV-induced DNA damage (see Fig. S1A in the supplemental material).
The impaired replication in priA2 mutants explains the apparent difference between the extents of inhibition observed in the wild-type and priA2 cells. priA2 mutants are impaired in their ability to replicate in the absence of DNA damage, which, based on the accumulation of DNA (Fig. 3, 14C-label), occurs at 32% of the rate of wild-type cultures. Based on the raw counts of [3H]thymidine incorporated during the 2-min pulse, the rate of replication was inhibited to a similar extent in both wild-type cells and priA2 mutants after UV irradiation, 2,178 and 2,286 cpm, respectively. However, prior to irradiation, the incorporation of [3H]thymidine in wild-type cultures was much higher than in priA2 cultures, 33,902 versus 7,217 cpm. Thus, although the difference between the replication rate before and after inhibition by UV is less in priA2 than in wild-type cultures, the actual amount of synthesis remaining in both cultures after UV irradiation was similar. Notwithstanding the inherent basal replication defect in priA2 mutants, the apparent lack of a sustained recovery in the rate of DNA synthesis after UV irradiation suggests that PriA function is needed to reset or reestablish an active replisome following arrest at UV-induced damage.
In UV-irradiated Rep mutants, the recovery of DNA synthesis was also delayed until 60 min postirradiation (Fig. 3). The delayed recovery in rep cultures was more severe than would be expected based on the amount of DNA accumulating in these mutants, and it was not as robust as that observed in wild-type cultures. This result suggests that while replication can recover in cells lacking functional Rep protein, it proceeds at a much lower rate than in wild-type cells.
In the absence of exogenous DNA damage, the replication competence of strains other than priA2 was similar to that of wild-type cultures (Fig. 3, 14C-labeled DNA). Based on the total DNA accumulation, wild-type cultures doubled once every 45 min under our conditions. In comparison, in recF, priB302, priC, and rep cultures, DNA doubled at a rate that was 104, 88, 85, and 89% of that observed in wild-type cells, respectively.
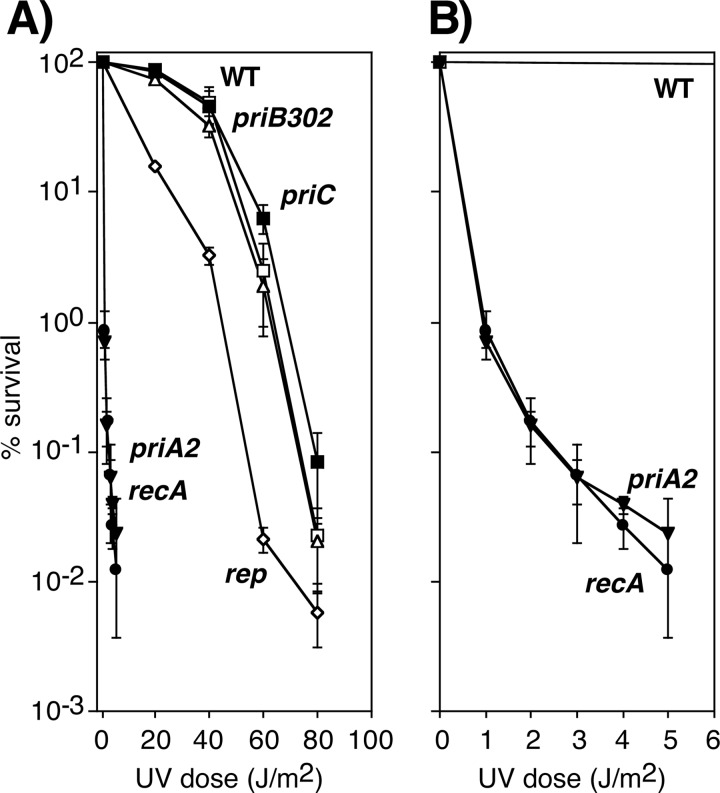
The UV hypersensitivity of priA2 and rep mutants mirrored their defective or delayed ability to resume DNA synthesis (Fig. 4). However, the delayed recovery of replication in priB302 mutants did not translate into a UV-hypersensitive phenotype. Both priB302 and priC mutants were as resistant to UV irradiation as wild-type cells, as was the dnaC809-suppressed priA2 mutant (Fig. 4; also see Fig. S1B and S2 in the supplemental material). These results are generally consistent with those previously reported for these mutations and confirm that PriA and Rep contribute to cell survival after UV irradiation (9, 22, 54). It is noteworthy that the hypersensitivity of priA2 was nearly identical to that of a recA mutant and had a mean (37%) lethal dose (LD37, or e−1 survival) occurring at 0.2 J/m2 or ∼6 lesions per genome (Fig. 4; also see Fig. S2). Previous studies have shown that recA is essential for survival when replication encounters a UV-induced lesion (14, 15, 29, 31). Thus, analogous to recA, the hypersensitivity of priA2 is consistent with the idea that PriA is nearly essential for survival when replication encounters a UV-induced DNA lesion.
Fig 4.
PriA and Rep are required for cell viability following UV irradiation. (A) The survival of wild-type (□), priA2 (▼), priB302 (△), priC (■), rep (♢), and recA (●) cultures after UV irradiation at the indicated doses. (B) The survival of wild-type (□), priA2 (▼), and recA (●) cultures replotted on a different scale. Graphs represent averages from at least three independent experiments. Error bars represent one standard deviation. A comparison of the viability of all strains across a range of UV doses used is shown in Fig. S2 in the supplemental material.
priA2 mutant cells are impaired for growth and viability in the absence of damage.
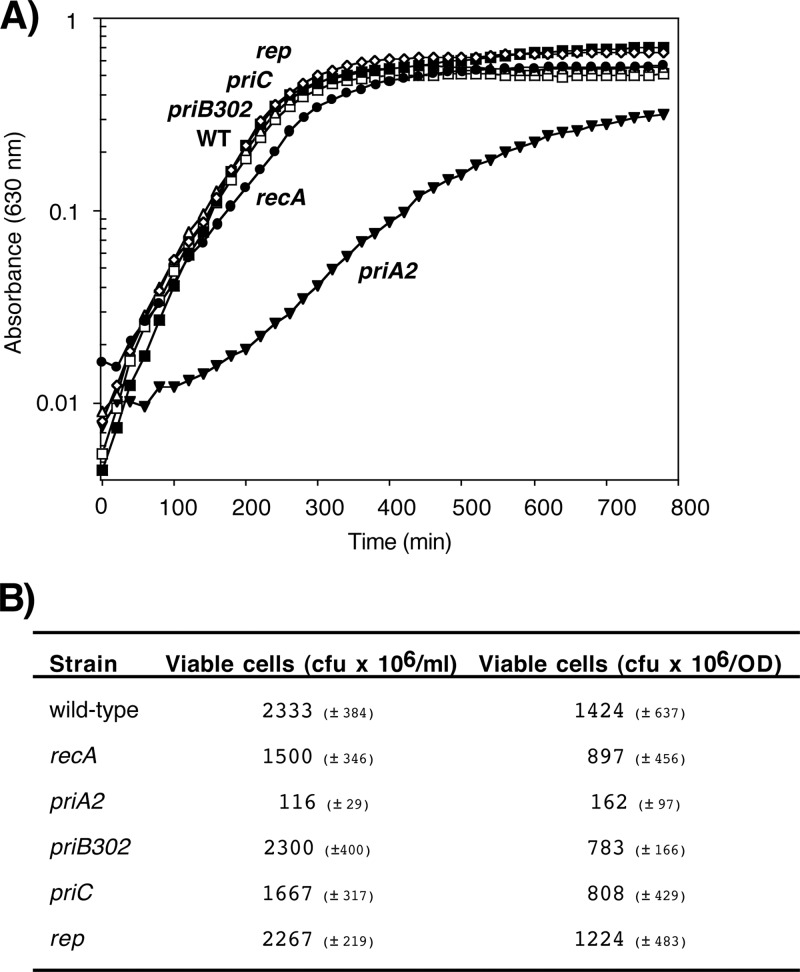
Of the primosomal mutants examined, only priA2 mutants reduced the quantity of DNA replicated by the cell in the absence of exogenous DNA damage. However, it is possible that the absence of the other primosomal proteins or Rep helicase impairs the overall integrity of the DNA or increases DNA strand exchanges or rearrangements that would compromise the cell's ability to grow or divide. To assess this, we compared the growth rate and viability of wild-type cells to those of isogenic priA2, priB302, priC, and rep mutants as well as recA mutants. Viable cells (105) were used to inoculate media, and the culture's growth at 37°C was monitored over time by examining the absorbance at 600 nm.
The maximum rate of growth for wild-type cells under our conditions was 0.0164 A600/min (Fig. 5A). In comparison, the maximal growth rates for priB302, priC, and rep were similar to those of wild-type cultures, with rates of 0.0161, 0.0191, and 0.0160 A600/min, respectively. recA mutants reached a maximal growth rate of 0.0119 A600/min (73% of wild-type cells) after normalization for cell viability, consistent with previous reports for recA mutants (10). In contrast, the absence of priA2 caused a severe growth impairment under normal conditions even after normalizing for the low cell viability in this strain, reaching a maximum of 0.0061 A600/min (37% of the rate of wild-type cells). This rate of growth closely mirrors the reduced rate of replication seen in this strain. Growth in the priA2 mutant was restored to wild-type rates by dnaC809 (see Fig. S1C in the supplemental material).
Fig 5.
PriA is required for robust growth in the absence of DNA damage. (A) The OD600 of wild-type (□), priA2 (▼), priB302 (△), priC (■), rep (♢), and recA (●) strains is plotted over time. (B) The number of viable cells in overnight cultures of wild-type, recA, priA2, priB302, priC, and rep strains is indicated in CFU × 106/ml and CFU × 106/OD.
In addition, the relative viability of priA2 mutants, as measured by CFU per OD600 and CFU per ml, was reduced by approximately 10- to 20-fold, similar to what was previously reported for a priA1 mutant (37), whereas the viability of recA and all other mutants was within 2-fold of that of wild-type cultures (Fig. 5B; also see Fig. S1D in the supplemental material). Thus, although both PriA and RecA are nearly essential for survival following arrest by UV-induced damage, only PriA affects the viability and growth in the absence of DNA damage, consistent with its proposed role in maintaining efficient lagging-strand synthesis during replication (35, 38).
DISCUSSION
In this study, we characterized the molecular events occurring at UV-arrested replication forks in priA2, priB302, priC, and rep mutants. Following arrest by UV-induced DNA damage, we observed that the arrested forks are initially processed normally in the absence of the primosome or Rep helicase. The nascent DNA is partially degraded, and a transient regression and resetting of the replication fork DNA occurs with normal kinetics. In addition, DNA synthesis initially begins to resume in each of the UV-irradiated primosomal protein and Rep mutants at times similar to those for wild-type cultures, as evidenced by the reincorporation of [3H]thymidine pools when the nascent DNA degradation ceases (Fig. 2) and an increase in the DNA synthesis rate at early times after fork arrest (Fig. 3). However, the recovery of synthesis stalls in the absence of PriA, PriB, or Rep, and it is less robust when it does resume. These observations indicate that the primosome and Rep helicase operate at a late step in the recovery or reestablishment of an active replisome after arrest by UV-induced DNA damage.
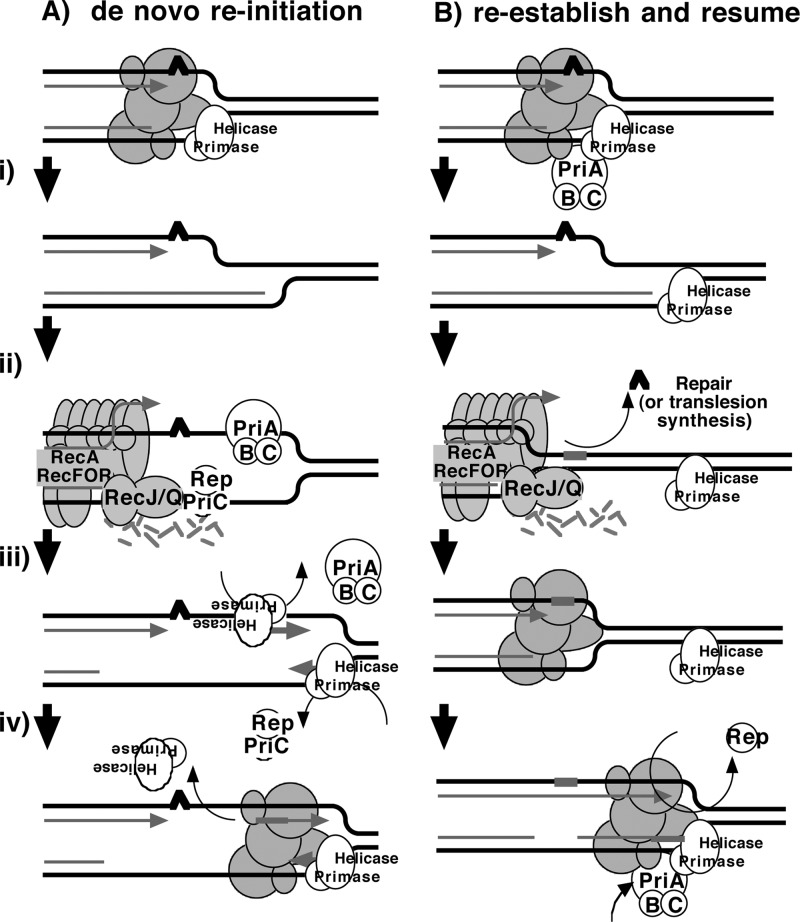
Two models that have been proposed for how the primosome and Rep helicase participate in restoring an active replisome following arrest by DNA damage are summarized in Fig. 6. Both models propose late functions for the primosome and Rep helicase but differ in the mechanism by which they promote replication recovery. The first model proposes that following arrest, the replisome and helicase are disrupted. Combinations of either PriA, PriB, or Rep with PriC participate in the displacement of the nascent lagging strand. These proteins then facilitate a transient loading of the helicase and primase complex on the leading-strand template, which serves as a primer, allowing a replisome to reinitiate downstream from the site of arrest (Fig. 6A). This model arose from the observation that, in vitro, the helicase activity of either PriA or Rep was capable of displacing the strands of a synthetic replication fork structure. In the presence of the helicase loader, DnaC, this is sufficient for the helicase and primase to prime the resulting single-stranded regions that are generated on the leading- and lagging-strand templates in vitro (22, 27).
Fig 6.
Two models for primosome and Rep function following disruption by DNA damage. (A) A model proposing that PriA and Rep function specifically to reinitiate DNA synthesis following disruption events. (i) Following the disruption of the replication machinery (grayed circles) by DNA damage (∧), (ii) PriA or Rep functions in a reaction to transiently load DnaB and DnaG to prime the leading strand and then (iii) stably load DnaB and DnaG on the lagging strand (22, 27). (iv) The leading-strand primer allows for the de novo formation of an active replisome downstream from the site of disruption. (B) A model in which PriA and Rep are required by the replisome to maintain efficient replication. (i) Following disruption by DNA damage, the recovery of DNA synthesis requires that the lesion is either repaired (ii) or bypassed (iii) by translesion synthesis (not shown), as found in previous studies (13). (iv) Since PriA and Rep are needed to maintain replication in the absence of damage, PriA and Rep would also be required for an active replisome to be maintained once the replisome is reestablished and DNA synthesis resumes.
The second model proposes that the primosome's primary contribution relates to enhancing the replisome's stability or priming efficiency during basal replication. Following arrest by UV-induced DNA damage, the helicase remains associated with the lagging strand, but other components of the holoenzyme may be displaced or disrupted. RecQ and RecJ contribute to the displacement and partial degradation of the nascent lagging strand, while the RecFOR proteins, together with RecA, process the fork DNA such that the lesion can either be repaired or bypassed. Once the block to replication has been overcome, the replisome can resume from the original arrest site. However, reestablishing an efficient replisome requires the primosome protein PriA and, to a lesser extent, PriB and PriC to coordinate the helicase/primase complex with the progressing replisome. The Rep helicase may also contribute to this reaction by helping to clear the region of other protein factors, such as recombination proteins, repair enzymes, or translesion polymerases, that may impair or compete with the replisome's ability to bind its forked substrate (Fig. 6B).
We interpret the in vivo observations presented here to be more consistent with the second model. A prediction of the first model is that the absence of PriA or Rep would result in attenuation of the nascent DNA unwinding and degradation at the arrested replication fork. However, the absence of the primosomal proteins or Rep does not impair the degradation of the nascent DNA, suggesting that these proteins do not contribute to the displacement of the nascent lagging strand in vivo (Fig. 3). The observation that DNA synthesis initially resumes and then stalls in the primosome and Rep mutants is also more consistent with a model in which the polymerase resumes from the existing 3′ end on the leading-strand template where replication was originally arrested. Reinitiation from a primer synthesized in a PriA- or Rep-dependent manner would predict that the initial resumption of DNA synthesis is severely delayed or fails to occur in the absence of these proteins.
The first model also speculates that the DnaB helicase is disrupted following encounters with UV-induced damage and suggests that new helicases are reloaded onto both the leading and lagging strands of DNA. This would necessitate that the helicase on the leading strand is also removed somehow, since its presence and polarity on this strand would lead to replication initiating in the wrong direction (Fig. 6A). However, other studies have observed that DnaB forms an unusually stable complex with its DNA substrate and remains associated with the DNA following encounters with UV damage in vitro (42, 43). Further, disruption of the DnaB helicase in vivo using a thermosensitive protein results in the degradation of the nascent DNA by exonuclease I and produces extensive single-stranded intermediates that are unlike any of the intermediates observed during the processing and recovery of replication at UV-induced lesions (5). These observations argue against the idea that the helicase dissociates or is able to transiently associate on the leading DNA strand following arrest by UV-induced damage.
Considering that PriA and Rep are both needed to maintain replication in the absence of DNA damage, we propose that their requirements after replication disruption relate more to their roles in maintaining efficient genome duplication. In this capacity, PriA reloads the primase and helicase on this template after disruption, while Rep helicase activity clears protein-bound replication fork DNA following repair (Fig. 6B). In effect, the loss of these activities would prevent or delay robust replication from resuming in their absence, as is observed experimentally.
Although these experiments do not exclude the possibility of de novo reinitiation, there is good evidence to suggest that in vivo, replication frequently resumes from the original site of disruption, as proposed in Fig. 6B. In the absence of nucleotide excision repair, the recovery of DNA synthesis in the presence of blocking lesions is severely impaired and results in high rates of DNA rearrangements and cell lethality in vivo, suggesting that lesions must be removed prior to the reestablishment of the replication fork (12–16, 21, 58). Further, a dramatic observation we consider important to this question is that in the absence of fork processing by RecJ or nucleotide excision repair, which allows the blocking lesion to be removed, the recovery of DNA synthesis becomes entirely dependent on translesion synthesis by DNA polymerase V (13). The dependence of recovery, in vivo, on a DNA polymerase that polymerizes through a blocking lesion strongly suggests that replication is frequently resuming from the site of the original blocking lesion rather than reinitiating de novo downstream of the lesion.
While both priA2 and recA mutants are extremely hypersensitive to DNA damage, only priA2 mutants exhibit a severe growth and replication impairment in the absence of damage. This observation indicates that PriA must have a more fundamental role in maintaining efficient chromosome replication, consistent with a number of previous studies. In vitro, PriA, together with PriB and PriC, enhances the processivity of priming by DnaB and DnaG and is required for minus-strand ϕX174 DNA synthesis in vivo and in vitro (2, 40, 61, 72). In addition, priA mutants are unable to maintain colE1-, R1-, or oriC-based plasmids (36, 49). The small size and high copy number of these minichromosomes make it unlikely that replication disruption events alone account for this phenotype and suggest a more basal function for PriA in their replication. Finally, a number of point mutants in other known subunits of the core replication complex, including the beta sliding clamp DnaN, the primase DnaG, and the proofreading exonuclease DnaQ, exhibit reduced viability, growth defects, and chronic SOS induction similar to priA mutants (23, 48–50, 63). In contrast, mutants such as recF, recO, recR, and recJ that are impaired in their ability to restore replication after disruption by DNA damage are not diminished for growth and are not chronically induced for the SOS response (14, 17). These observations argue that the chronic SOS induction in priA2 is the result of a fundamental deficiency in maintaining efficient chromosomal replication rather than frequent disruption by DNA damage.
Recent studies have shown that Rep functions as a motor to remove protein bound to DNA during replication (4, 7, 24, 26). Considering that the absence of Rep does not alter the initial processing or resetting of the disrupted fork, we hypothesize that the observed delay and slow kinetics of resumption are the result of a requirement to clear bound proteins from the template DNA. Candidate proteins that might be bound to the template DNA following lesion removal include DNA repair proteins such as UvrC, which has been shown to have a slow turnover rate following incisions at the 5′ and 3′ ends of DNA lesions (18, 66). This interpretation may also partially explain the inviability of uvrD rep cells, since the absence of both of these proteins would be expected to inhibit the dissociation of repair enzymes from damaged sites on the DNA template and the subsequent reassembly of the replisome (7, 24). Such a role would also be consistent with recent studies demonstrating that the impaired replication phenotypes of rep mutants relate to their inability to resolve conflicts and remove proteins ahead of the fork during replication and is unrelated to PriC-directed replisome assembly (3, 24, 73).
Based on models similar to those shown in Fig. 6, recent biochemical approaches focusing on the ability of PriA, PriB, PriC, and Rep to prime and initiate replication de novo have observed that priming promoted by these factors occurs relatively nonspecifically, with partial redundancy, or even in the complete absence of the primosome or Rep proteins (3, 24, 73, 74). The observation that nascent DNA processing ceases and that DNA synthesis resumes in vivo, albeit inefficiently, in cells lacking PriA, PriB, PriC, or Rep argues that de novo priming either is not required or can occur in their absence. However, the impaired ability of rep, priA2, and priB302 mutants to reestablish a replisome that is capable of efficient, sustained DNA replication suggests that it is of interest to examine whether the presence of these proteins contributes to the overall stability of the replisome, its processivity, or its priming efficiency in vitro.
Supplementary Material
ACKNOWLEDGMENTS
We thank N. P. Higgins for providing strain DY329 and S. Sandler for providing strains SS138 and JC19008.
This work was supported by CAREER award MCB0551798 from the National Science Foundation and AREA grant R15 ES021594 from the NIEHS at the National Institutes of Health.
Footnotes
Published ahead of print 25 May 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Arai K, Kornberg A. 1981. Unique primed start of phage phi X174 DNA replication and mobility of the primosome in a direction opposite chain synthesis. Proc. Natl. Acad. Sci. U. S. A. 78:69–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arai K, Low RL, Kornberg A. 1981. Movement and site selection for priming by the primosome in phage phi X174 DNA replication. Proc. Natl. Acad. Sci. U. S. A. 78:707–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Atkinson J, et al. 2011. Localization of an accessory helicase at the replisome is critical in sustaining efficient genome duplication. Nucleic Acids Res. 39:949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baharoglu Z, Lestini R, Duigou S, Michel B. 2010. RNA polymerase mutations that facilitate replication progression in the rep uvrD recF mutant lacking two accessory replicative helicases. Mol. Microbiol. 77:324–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Belle JJ, Casey A, Courcelle CT, Courcelle J. 2007. Inactivation of the DnaB helicase leads to the collapse and degradation of the replication fork: a comparison to UV-induced arrest. J. Bacteriol. 189:5452–5462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berges H, Oreglia J, Joseph-Liauzun E, Fayet O. 1997. Isolation and characterization of a priB mutant of Escherichia coli influencing plasmid copy number of delta rop ColE1-type plasmids. J. Bacteriol. 179:956–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boubakri H, de Septenville AL, Viguera E, Michel B. 2010. The helicases DinG, Rep. and UvrD cooperate to promote replication across transcription units in vivo. EMBO J. 29:145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cadman CJ, Lopper M, Moon PB, Keck JL, McGlynn P. 2005. PriB stimulates PriA helicase via an interaction with single-stranded DNA. J. Biol. Chem. 280:39693–39700 [DOI] [PubMed] [Google Scholar]
- 9. Calendar R, Lindqvist B, Sironi G, Clark AJ. 1970. Characterization of REP− mutants and their interaction with P2 phage. Virology 40:72–83 [DOI] [PubMed] [Google Scholar]
- 10. Capaldo FN, Ramsey G, Barbour SD. 1974. Analysis of the growth of recombination-deficient strains of Escherichia coli K-12. J. Bacteriol. 118:242–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chow KH, Courcelle J. 2004. RecO acts with RecF and RecR to protect and maintain replication forks blocked by UV-induced DNA damage in Escherichia coli. J. Biol. Chem. 279:3492–3496 [DOI] [PubMed] [Google Scholar]
- 12. Courcelle CT, Belle JJ, Courcelle J. 2005. Nucleotide excision repair or polymerase V-mediated lesion bypass can act to restore UV-arrested replication forks in Escherichia coli. J. Bacteriol. 187:6953–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Courcelle CT, Chow KH, Casey A, Courcelle J. 2006. Nascent DNA processing by RecJ favors lesion repair over translesion synthesis at arrested replication forks in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 103:9154–9159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Courcelle J, Carswell-Crumpton C, Hanawalt PC. 1997. recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 94:3714–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Courcelle J, Crowley DJ, Hanawalt PC. 1999. Recovery of DNA replication in UV-irradiated Escherichia coli requires both excision repair and recF protein function. J. Bacteriol. 181:916–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Courcelle J, Donaldson JR, Chow KH, Courcelle CT. 2003. DNA damage-induced replication fork regression and processing in Escherichia coli. Science 299:1064–1067 [DOI] [PubMed] [Google Scholar]
- 17. Courcelle J, Hanawalt PC. 1999. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol. Gen. Genet. 262:543–551 [DOI] [PubMed] [Google Scholar]
- 18. Crowley DJ, Hanawalt PC. 2001. The SOS-dependent upregulation of uvrD is not required for efficient nucleotide excision repair of ultraviolet light induced DNA photoproducts in Escherichia coli. Mutat. Res. 485:319–329 [DOI] [PubMed] [Google Scholar]
- 19. Denhardt DT, Dressler DH, Hathaway A. 1967. The abortive replication of PhiX174 DNA in a recombination-deficient mutant of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 57:813–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eisenberg S, Scott JF, Kornberg A. 1976. An enzyme system for replication of duplex circular DNA: the replicative form of phage phi X174. Proc. Natl. Acad. Sci. U. S. A. 73:1594–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Friedberg EC, Wagner R, Radman M. 2002. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science 296:1627–1630 [DOI] [PubMed] [Google Scholar]
- 22. Gregg AV, McGlynn P, Jaktaji RP, Lloyd RG. 2002. Direct rescue of stalled DNA replication forks via the combined action of PriA and RecG helicase activities. Mol. Cell 9:241–251 [DOI] [PubMed] [Google Scholar]
- 23. Grompe M, Versalovic J, Koeuth T, Lupski JR. 1991. Mutations in the Escherichia coli dnaG gene suggest coupling between DNA replication and chromosome partitioning. J. Bacteriol. 173:1268–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guy CP, et al. 2009. Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Mol. Cell 36:654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heller RC, Marians KJ. 2005. The disposition of nascent strands at stalled replication forks dictates the pathway of replisome loading during restart. Mol. Cell 17:733–743 [DOI] [PubMed] [Google Scholar]
- 26. Heller RC, Marians KJ. 2005. Unwinding of the nascent lagging strand by Rep and PriA enables the direct restart of stalled replication forks. J. Biol. Chem. 280:34143–34151 [DOI] [PubMed] [Google Scholar]
- 27. Heller RC, Marians KJ. 2006. Replication fork reactivation downstream of a blocked nascent leading strand. Nature 439:557–562 [DOI] [PubMed] [Google Scholar]
- 28. Howard-Flanders P, Rupp WD, Wilkins BM, Cole RS. 1968. DNA replication and recombination after UV irradiation. Cold Spring Harb. Symp. Quant. Biol. 33:195–207 [DOI] [PubMed] [Google Scholar]
- 29. Howard-Flanders P, Theriot L, Stedeford JB. 1969. Some properties of excision-defective recombination-deficient mutants of Escherichia coli K-12. J. Bacteriol. 97:1134–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Howard-Flanders P, Boyce RP. 1966. DNA repair and genetic recombination: studies on mutants of Escherichia coli defective in these processes. Radiat. Res. Suppl. 6:156–184 [PubMed] [Google Scholar]
- 31. Khidhir MA, Casaregola S, Holland IB. 1985. Mechanism of transient inhibition of DNA synthesis in ultraviolet-irradiated E. coli: inhibition is independent of recA whilst recovery requires RecA protein itself and an additional, inducible SOS function. Mol. Gen. Genet. 199:133–140 [DOI] [PubMed] [Google Scholar]
- 32. Kogoma T, Cadwell GW, Barnard KG, Asai T. 1996. The DNA replication priming protein, PriA, is required for homologous recombination and double-strand break repair. J. Bacteriol. 178:1258–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lane HE, Denhardt DT. 1974. The rep mutation. III. Altered structure of the replicating Escherichia coli chromosome. J. Bacteriol. 120:805–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lane HE, Denhardt DT. 1975. The rep mutation. IV. Slower movement of replication forks in Escherichia coli rep strains. J. Mol. Biol. 97:99–112 [DOI] [PubMed] [Google Scholar]
- 35. Lasken RS, Kornberg A. 1988. The primosomal protein N′ of Escherichia coli is a DNA helicase. J. Biol. Chem. 263:5512–5518 [PubMed] [Google Scholar]
- 36. Lee EH, Kornberg A. 1991. Replication deficiencies in priA mutants of Escherichia coli lacking the primosomal replication N′ protein. Proc. Natl. Acad. Sci. U. S. A. 88:3029–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee EH, Masai H, Allen GCJ, Kornberg A. 1990. The priA gene encoding the primosomal replicative N′ protein of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 87:4620–4624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee MS, Marians KJ. 1989. The Escherichia coli primosome can translocate actively in either direction along a DNA strand. J. Biol. Chem. 264:14531–14542 [PubMed] [Google Scholar]
- 39. Liu J, Xu L, Sandler SJ, Marians KJ. 1999. Replication fork assembly at recombination intermediates is required for bacterial growth. Proc. Natl. Acad. Sci. U. S. A. 96:3552–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Low RL, Arai K, Kornberg A. 1981. Conservation of the primosome in successive stages of phi X174 DNA replication. Proc. Natl. Acad. Sci. U. S. A. 78:1436–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mahdi AA, Buckman C, Harris L, Lloyd RG. 2006. Rep and PriA helicase activities prevent RecA from provoking unnecessary recombination during replication fork repair. Genes Dev. 20:2135–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McInerney P, O'Donnell M. 2007. Replisome fate upon encountering a leading strand block and clearance from DNA by recombination proteins. J. Biol. Chem. 282:25903–25916 [DOI] [PubMed] [Google Scholar]
- 43. McInerney P, O'Donnell M. 2004. Functional uncoupling of twin polymerases: mechanism of polymerase dissociation from a lagging-strand block. J. Biol. Chem. 279:21543–21551 [DOI] [PubMed] [Google Scholar]
- 44. Mellon I, Hanawalt PC. 1989. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature 342:95–98 [DOI] [PubMed] [Google Scholar]
- 45. Michel B, Ehrlich SD, Uzest M. 1997. DNA double-strand breaks caused by replication arrest. EMBO J. 16:430–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Minden JS, Marians KJ. 1985. Replication of pBR322 DNA in vitro with purified proteins. Requirement for topoisomerase I in the maintenance of template specificity. J. Biol. Chem. 260:9316–9325 [PubMed] [Google Scholar]
- 47. Morimatsu K, Kowalczykowski SC. 2003. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol. Cell 11:1337–1347 [DOI] [PubMed] [Google Scholar]
- 48. Nowosielska A, Wrzesinski M, Nieminuszczy J, Janion C, Grzesiuk E. 2005. Mutator activity and specificity of Escherichia coli dnaQ49 allele–effect of umuDC products. Mutat. Res. 572:113–122 [DOI] [PubMed] [Google Scholar]
- 49. Nurse P, Zavitz KH, Marians KJ. 1991. Inactivation of the Escherichia coli priA DNA replication protein induces the SOS response. J. Bacteriol. 173:6686–6693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. O'Reilly EK, Kreuzer KN. 2004. Isolation of SOS constitutive mutants of Escherichia coli. J. Bacteriol. 186:7149–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rangarajan S, Woodgate R, Goodman MF. 2002. Replication restart in UV-irradiated Escherichia coli involving pols II, III, V, PriA, RecA and RecFOR proteins. Mol. Microbiol. 43:617–628 [DOI] [PubMed] [Google Scholar]
- 52. Sandler SJ, Samra HS, Clark AJ. 1996. Differential suppression of priA2::kan phenotypes in Escherichia coli K-12 by mutations in priA, lexA, and dnaC. Genetics 143:5–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sandler SJ. 2000. Multiple genetic pathways for restarting DNA replication forks in Escherichia coli K-12. Genetics 155:487–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sandler SJ, et al. 1999. dnaC mutations suppress defects in DNA replication- and recombination-associated functions in priB and priC double mutants in Escherichia coli K-12. Mol. Microbiol. 34:91–101 [DOI] [PubMed] [Google Scholar]
- 55. Scott JF, Eisenberg S, Bertsch LL, Kornberg A. 1977. A mechanism of duplex DNA replication revealed by enzymatic studies of phage phi X174: catalytic strand separation in advance of replication. Proc. Natl. Acad. Sci. U. S. A. 74:193–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Scott JF, Kornberg A. 1978. Purification of the rep protein of Escherichia coli. An ATPase which separates duplex DNA strands in advance of replication. J. Biol. Chem. 253:3292–3297 [PubMed] [Google Scholar]
- 57. Seigneur M, Bidnenko V, Ehrlich SD, Michel B. 1998. RuvAB acts at arrested replication forks. Cell 95:419–430 [DOI] [PubMed] [Google Scholar]
- 58. Setlow RB, Swenson PA, Carrier WL. 1963. Thymine dimers and inhibition of DNA synthesis by ultraviolet irradiation of cells. Science 142:1464–1466 [DOI] [PubMed] [Google Scholar]
- 59. Setlow RB. 1966. Cyclobutane-type pyrimidine dimers in polynucleotides. Science 153:379–386 [DOI] [PubMed] [Google Scholar]
- 60. Shan Q, Bork JM, Webb BL, Inman RB, Cox MM. 1997. RecA protein filaments: end-dependent dissociation from ssDNA stabilization by RecO and RecR proteins. J. Mol. Biol. 265:519–540 [DOI] [PubMed] [Google Scholar]
- 61. Shlomai J, Kornberg A. 1980. An Escherichia coli replication protein that recognizes a unique sequence within a hairpin region in phi X174 DNA. Proc. Natl. Acad. Sci. U. S. A. 77:799–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sumida-Yasumoto C, Yudelevich A, Hurwitz J. 1976. DNA synthesis in vitro dependent upon phiX174 replicative form I DNA. Proc. Natl. Acad. Sci. U. S. A. 73:1887–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sutton MD. 2004. The Escherichia coli dnaN159 mutant displays altered DNA polymerase usage and chronic SOS induction. J. Bacteriol. 186:6738–6748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Umezu K, Kolodner RD. 1994. Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. J. Biol. Chem. 269:30005–30013 [PubMed] [Google Scholar]
- 65. Uzest M, Ehrlich SD, Michel B. 1995. Lethality of rep recB and rep recC double mutants of Escherichia coli. Mol. Microbiol. 17:1177–1188 [DOI] [PubMed] [Google Scholar]
- 66. Van Sluis CA, Mattern IE, Paterson MC. 1974. Properties of uvrE mutants of Escherichia coli K12. I. Effects of UV irradiation on DNA metabolism. Mutat. Res. 25:273–279 [DOI] [PubMed] [Google Scholar]
- 67. Varghese AJ, Wang SY. 1967. Ultraviolet irradiation of DNA in vitro and in vivo produces a 3D thymine-derived product. Science 156:955–957 [DOI] [PubMed] [Google Scholar]
- 68. Wang SY, Varghese AJ. 1967. Cytosine-thymine addition product from DNA irradiated with ultraviolet light. Biochem. Biophys. Res. Commun. 29:543–549 [DOI] [PubMed] [Google Scholar]
- 69. Webb BL, Cox MM, Inman RB. 1997. Recombinational DNA repair: the RecF and RecR proteins limit the extension of RecA filaments beyond single-strand DNA gaps. Cell 91:347–356 [DOI] [PubMed] [Google Scholar]
- 70. Weiner JH, McMacken R, Kornberg A. 1976. Isolation of an intermediate which precedes dnaG RNA polymerase participation in enzymatic replication of bacteriophage phi X174 DNA. Proc. Natl. Acad. Sci. U. S. A. 73:752–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wickner S, Hurwitz J. 1974. Conversion of phiX174 viral DNA to double-stranded form by purified Escherichia coli proteins. Proc. Natl. Acad. Sci. U. S. A. 71:4120–4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wickner S, Hurwitz J. 1975. Association of phiX174 DNA-dependent ATPase activity with an Escherichia coli protein, replication factor Y, required for in vitro synthesis of phiX174 DNA. Proc. Natl. Acad. Sci. U. S. A. 72:3342–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yancey-Wrona JE, Matson SW. 1992. Bound Lac repressor protein differentially inhibits the unwinding reactions catalyzed by DNA helicases. Nucleic Acids Res. 20:6713–6721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yeeles JT, Marians KJ. 2011. The Escherichia coli replisome is inherently DNA damage tolerant. Science 334:235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yu D, et al. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5978–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.