Abstract
Magnetotactic bacteria (MTB) synthesize unique organelles, the magnetosomes, which are intracellular nanometer-sized, membrane-enveloped magnetite. The biomineralization of magnetosomes involves the uptake of large amounts of iron. However, the iron metabolism of MTB is not well understood. The genome of the magnetotactic bacterium Magnetospirillum gryphiswaldense strain MSR-1 contains two ferrous iron transport genes, feoB1 and feoB2. The FeoB1 protein was reported to be responsible mainly for the transport of ferrous iron and to play an accessory role in magnetosome formation. To determine the role of feoB2, we constructed an feoB2 deletion mutant (MSR-1 ΔfeoB2) and an feoB1 feoB2 double deletion mutant (MSR-1 NfeoB). The single feoB2 mutation did not affect magnetite crystal biomineralization. MSR-1 NfeoB had a significantly lower average magnetosome number per cell (∼65%) than MSR-1 ΔfeoB1, indicating that FeoB2 plays a role in magnetosome formation when the feoB1 gene is deleted. Our findings showed that FeoB1 has a greater ferrous iron transport ability than FeoB2 and revealed the differential roles of FeoB1 and FeoB2 in MSR-1 iron metabolism. Interestingly, compared to the wild type, the feoB mutants showed increased sensitivity to oxidative stress and lower activities of the enzymes superoxide dismutase and catalase, indicating that the FeoB proteins help protect bacterial cells from oxidative stress.
INTRODUCTION
Iron is an essential component of almost all organisms and is required as a cofactor for many enzymes involved in key biological pathways and processes. At neutral pH, iron is often biologically unavailable because of the poor solubility of ferric iron (8). To obtain sufficient iron for optimal growth, bacteria have evolved a variety of specialized iron transport mechanisms. Many bacteria excrete ferric chelators, known as siderophores, to take up ferric iron (Fe3+). Soluble ferrous iron (Fe2+) can often be directly transported via the ferrous iron transport protein FeoB, sometimes accompanied by the reduction of Fe3+ to Fe2+ through ferric reductase (2, 21). The FeoB family has been identified in a number of bacterial species by genomic analysis (6). The predicted FeoB proteins are integral cytoplasmic membrane proteins, 700 to 800 amino acids (aa) in length, that have 7 to 12 transmembrane-spanning α-helices. The N-terminal region of FeoB proteins includes a G protein domain that is essential for ferrous iron uptake in bacteria (14).
Magnetotactic bacteria (MTB) are Gram-negative prokaryotes that synthesize unique intracellular magnetic nanoparticles composed of Fe3O4 or Fe3S4, termed magnetosomes, in order to navigate along geomagnetic field lines and search for microaerophilic environments (7, 11). Magnetosomes are membrane-bound crystals aligned in chain-like structures within the cell (12, 22). The biosynthesis of magnetosomes requires the acquisition of large amounts of iron from the environment (3). The iron content in MTB may represent >2% of the total dry weight; this value is 100-fold higher than that for Escherichia coli (4). The iron metabolism of MTB is not well understood. Magnetospirillum magnetotacticum strain MS-1 and M. magneticum strain AMB-1 secrete siderophores, and Taoka et al. also reported that ferric-siderophore receptor homologues are highly expressed in the M. magnetotacticum MS-1 cell under iron-rich conditions, indicating the ability to take up ferric iron (5, 16, 28). In studies of M. magneticum AMB-1, Yang et al. demonstrated that the supplementation of ferrous sulfate enhances magnetosome formation (31), and Suzuki et al. showed that cytoplasmic ATPase is involved in ferrous iron uptake and is essential for magnetosome formation (27). M. gryphiswaldense MSR-1 does not produce siderophores (23), but it has more than five isozymes of ferric reductase (30). The ferrous iron transporter FeoB1 was shown to play an important role in the iron uptake required for magnetosome formation in MSR-1, as an feoB1 deletion mutant took up less ferrous and ferric iron and displayed a smaller number and diameter of magnetosomes than the wild type (19). However, magnetosome formation was not completely abolished in the feoB1 mutant, indicating that another iron transport pathway(s) must be involved in iron uptake in MSR-1. Following the publication of the genomic sequence of MSR-1 in 2007 (18), a similarity search revealed an open reading frame (ORF) having 35% identity to FeoB1, termed FeoB2. We present the first demonstration of FeoB2 in magnetosome formation in MSR-1. Our findings indicate that FeoB2 plays a role in magnetosome formation when FeoB1 is deficient and is also required for the protection of the bacteria from oxidative stress.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. M. gryphiswaldense MSR-1 was cultured in sodium lactate medium (SLM) at 30°C as described previously (32). Escherichia coli strains were cultured in Luria broth (LB) at 37°C. Antibiotics and concentrations (in μg ml−1) used for E. coli were the following: ampicillin (Amp), 100; kanamycin (Km), 50; chloramphenicol (Cm), 34; and gentamicin (Gm), 5. Those used for MSR-1 were the following: nalidixic acid (Nx), 5; Km, 5, Cm, 5; and Gm, 5. Antibiotics were purchased from Amresco. Chemicals used for cultured bacteria were purchased from Sinopharm Chemical Reagent Co., Ltd. (China).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| M. gryphiswaldense MSR-1 | Wild type, Nxr | DSM 6361 |
| M. gryphiswaldense MSR-1 ΔfeoB1 | feoB1-deficient mutant, Nxr Gmr | 19 |
| M. gryphiswaldense MSR-1 ΔfeoB2 | feoB2-defective mutant, Nxr Kmr | Present study |
| M. gryphiswaldense MSR-1 NfeoB | feoB1 feoB2 double mutant, Nxr Gmr Kmr | Present study |
| E. coli DH5α | F′ f80dlacZΔM15 Δ(lacZYA-argF) U169 deoR recA1 endA1 | 9 |
| E. coli S17-1 | thi endA recA hsdR with RP4-2-Tc::Mu-Km::Tn7 integrated in chromosome, Smr | 25 |
| Plasmids | ||
| pMD19 T-simple | Cloning vector, Ampr | TaKaRa |
| pUC4K | Cloning vector with Km cassette, Kmr | 29 |
| pSUP202 | Suicide vector for M. gryphiswaldense MSR-1, Cmr Tcr Ampr | 13 |
| pSUPB2 | pSUP202 containing Km cassette, feoB2 upstream region and downstream region, Cmr Kmr Ampr | Present study |
Molecular techniques.
Unless noted otherwise, molecular techniques were performed by standard protocols (20). DNA was sequenced using BigDye Terminator (v3.1) chemistry on an ABI 3730 DNA analyzer (Applied Biosystems). Restriction endonuclease and DNA-modifying enzymes were purchased from TaKaRa (Japan). KOD DNA polymerase (Toyobo, Japan) was used for PCR amplification. Primers (see Table S1 in the supplemental material) were purchased from Invitrogen.
Construction of feoB2 deletion mutant and feoB1 feoB2 double deletion mutant.
A 1,196-bp upstream region and a 1,230-bp downstream region were amplified using primers B2uf/B2ur and B2df/B2dr, respectively. A Km cassette was digested by PstI from the pUC4K vector. These three fragments were fused by cloning into the BamHI and SphI sites of the pSUP202 vector, yielding pSUPB2. pSUPB2 was introduced into wild-type MSR-1 by biparental conjugation and screened for Kmr Cms colonies. The feoB2 single mutant was termed MSR-1 ΔfeoB2. pSUPB2 was introduced into MSR-1 ΔfeoB1 by biparental conjugation and screened for Kmr Gmr Cms colonies. The feoB1 feoB2 double deletion mutant was termed MSR-1 NfeoB. The mutants were confirmed by PCR using two pair primers (B2verif/B2verir and B2vf/B2vr).
Determination of ion content.
Bacterial strains were grown in SLM supplemented with 50 μM ferric citrate at 30°C for 24 h. Cells were washed with 20 mM HEPES–4 mM EDTA, pH 7.4, and harvested by centrifugation. Cell pellets were dried to constant weight at 60°C, resuspended in 1 ml nitric acid, and incubated at 100°C for 3 h. Iron content was assayed using an inductively coupled plasma optical emission spectrometer (ICP-OES; Optima 5300DV; Perkin Elmer). The experiments were performed in triplicate. For determination of intracellular metal content, the strains were cultured in SLM with 10 μM MnSO4, 10 μM CuSO4, and 5 μM ZnSO4. Cell lysates of strains for ICP-OES analysis were prepared in the same manner as that described above for iron content.
Analysis of bacterial H2O2 tolerance.
MSR-1 wild-type, ΔfeoB1, ΔfeoB2, and NfeoB strains were cultured in SLM to an optical density of 1.0 and inoculated into 50 ml SLM containing various concentrations of H2O2. After 24 h of incubation, the cell density was measured at 600 nm using a spectrophotometer. The experiments were performed in triplicate.
SOD and CAT assays in MSR-1.
MSR-1 wild-type, ΔfeoB1, ΔfeoB2, and NfeoB strains were cultured in SLM. Cells were harvested, resuspended in 50 mM Tris · HCl, pH 7.4, and lysed by sonication. Cell debris was removed by centrifugation (16,000 × g for 20 min at 4°C). The supernatant was obtained as crude enzyme extract. Superoxide dismutase (SOD) and catalase (CAT) assays were performed using the SOD and CAT assay kit (Nanjing Jiancheng Co., China).
TEM.
For transmission electron microscopy (TEM), bacterial strains were grown in SLM for 12 h at 30°C and then supplemented with 80 μM ferric citrate or ferrous citrate, and growth was continued until stationary phase. Cell suspensions were adsorbed onto copper grids and observed directly with a JEM 1230 (JEM, Japan) transmission electron microscope. Thirty to 35 randomly chosen cells from each sample were examined.
RESULTS
Analysis of feoB2 gene sequence.
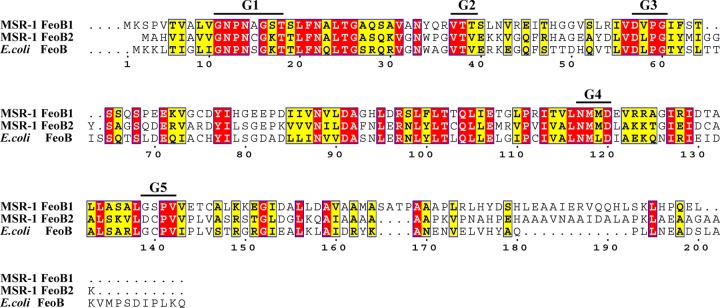
The feoB1 gene of M. gryphiswaldense MSR-1 encodes a 704-aa protein, and the feoB2 gene encodes a 786-aa protein. FeoB2 has 35 and 43% identity to MSR-1 FeoB1 and E. coli FeoB, respectively. The N termini of MSR-1 FeoB1 and FeoB2 have a G protein conserved motif (Fig. 1) which is essential for ferrous iron transport (14). The C terminus of MSR-1 FeoB is predicted to be located in the cytoplasmic membrane with nine membrane-spanning α-helices.
Fig 1.
Multiple alignments of the M. gryphiswaldense MSR-1 FeoB1, MSR-1 FeoB2, and E. coli FeoB N termini. FeoB contains a G protein conserved motif (G1 to G5). The multiple alignments were performed using the program ClustalW, and pictures were constructed using the program ESPript 2.2 (http://espript.ibcp.fr/ESPript/ESPript/). Red box, strict identity; yellow box, similarity in a group.
Magnetosome formation in feoB mutants.
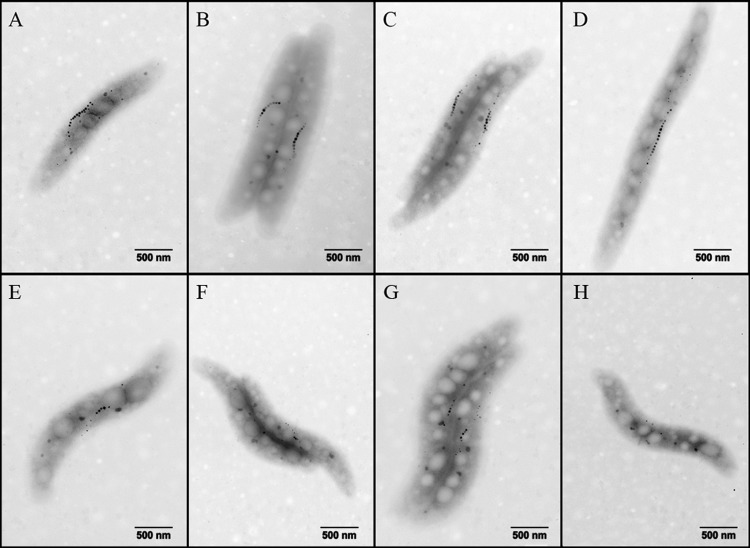
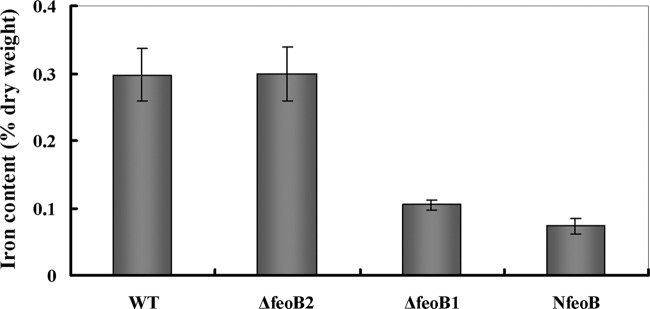
To determine the role of FeoB2 in magnetosome formation, an MSR-1 feoB2 deletion mutant (MSR-1 ΔfeoB2) and a mutant with two feoB genes deleted (MSR-1 NfeoB) were constructed. TEM observations indicated that the magnetosome number and magnetosome crystal size of MSR-1 ΔfeoB2 were similar to those of the wild type (Fig. 2 and Table 2). The distributions of these two parameters were also similar; i.e., in both the wild-type and MSR-1 ΔfeoB2 strains, crystals with diameters of 26 to 30 nm and magnetosome numbers of 11 to 15 per cell were most frequent. Magnetosome formation in MSR-1 ΔfeoB1 was significantly lower than in the wild type, as reported previously (19). Magnetosome formation in MSR-1 NfeoB was not completely abolished. MSR-1 NfeoB and MSR-1 ΔfeoB1 showed similar magnetosome crystal sizes (the most frequent diameter was 16 to 20 nm) but different magnetosome number distributions, i.e., the most frequent number was <5 for MSR-1 NfeoB and 6 to 10 for MSR-1 ΔfeoB1, and the mean number for MSR-1 NfeoB was ∼65% of that for MSR-1 ΔfeoB1 (P < 0.01) (Fig. 2 and Table 2). To confirm these findings, the four strains were cultured with 50 μM ferric citrate and the iron content was measured by ICP-OES. In both the wild type and the MSR-1 ΔfeoB2 mutant, the iron content was 0.30% of the dry weight (P > 0.05 by t test) (Fig. 3). The iron content of MSR-1 NfeoB (0.07% of dry weight) was significantly lower than that of MSR-1 ΔfeoB1 (0.10% of dry weight) (P < 0.05) (Fig. 3). These findings, taken together, indicate that FeoB2 played an important role in magnetosome formation when the feoB1 gene was deleted.
Fig 2.
TEM micrographs of wild-type MSR-1 and feoB mutants. (A) Wild type cultured in 80 μM Fe2+. (B) MSR-1 ΔfeoB2 cultured in 80 μM Fe2+. (C) Wild type cultured in 80 μM Fe3+. (D) MSR-1 ΔfeoB2 cultured in 80 μM Fe3+. (E) MSR-1 ΔfeoB1 cultured in 80 μM Fe2+. (F) MSR-1 NfeoB cultured in 80 μM Fe2+. (G) MSR-1 ΔfeoB1 cultured in 80 μM Fe3+. (H) MSR-1 NfeoB cultured in 80 μM Fe3+.
Table 2.
Magnetosome numbers and diameters of the wild-type strain and three deletion mutants
| Strain and Fe type | Avg magnetosome no. | Avg magnetosome diam |
|---|---|---|
| Fe2 | ||
| WT | 12.89 ± 3.07 | 30.32 ± 8.71 |
| MSR-1 ΔfeoB1 | 6.31 ± 2.48 | 21.17 ± 7.60 |
| MSR-1 ΔfeoB2 | 11.18 ± 4.30 | 28.40 ± 11.07 |
| MSR-1 NfeoB | 4.13 ± 2.24 | 24.13 ± 11.77 |
| Fe3+ | ||
| WT | 13.47 ± 3.76 | 25.39 ± 8.93 |
| MSR-1 ΔfeoB1 | 8.50 ± 3.16 | 17.68 ± 6.02 |
| MSR-1 ΔfeoB2 | 13.81 ± 4.58 | 26.11 ± 8.86 |
| MSR-1 NfeoB | 5.47 ± 2.82 | 16.01 ± 5.64 |
Fig 3.
Intracellular iron contents (% dry weight) of the wild type and feoB mutants. Cells were grown in SLM supplemented with 50 μM ferric citrate and harvested at stationary phase. Iron content was determined by ICP-OES. Experiments were performed in triplicate.
Metal content in cells.
Intracellular metal content, including manganese, zinc, copper, and magnesium, were measured by ICP-OES. The ion contents of these four metals in MSR-1 ΔfeoB1, ΔfeoB2, and NfeoB were similar to those of the MSR-1 wild type (P > 0.05) (Table 3). The results showed that MSR-1 FeoB2 did not play a role in manganese, zinc, copper, and magnesium accumulation.
Table 3.
Metal ion content of M. gryphiswaldense MSR-1 wild type, ΔfeoB1, ΔfeoB2, and NfeoB
| Strain | Metal contenta |
|||
|---|---|---|---|---|
| Mn | Zn | Cu | Mg | |
| WT | 0.06 ± 0.01 | 0.66 ± 0.08 | 0.95 ± 0.10 | 24.99 ± 7.54 |
| MSR-1 ΔfeoB1 | 0.07 ± 0.01 | 0.50 ± 0.12 | 0.95 ± 0.14 | 21.38 ± 4.52 |
| MSR-1 ΔfeoB2 | 0.06 ± 0.01 | 0.61 ± 0.14 | 0.93 ± 0.04 | 24.96 ± 2.42 |
| MSR-1 NfeoB | 0.06 ± 0.01 | 0.54 ± 0.03 | 0.93 ± 0.09 | 22.71 ± 0.02 |
All values are presented as μmol/g cell dry weight and represent the means from three independent determinations.
Sensitivity to oxidative stress.
The growth of these four strains was not affected by culturing in 200 μM H2O2 but was inhibited to various degrees by culturing in 500 μM H2O2. The resistance to 500 μM H2O2 in the three mutants (MSR-1 ΔfeoB1, ΔfeoB2, and NfeoB) was 40, 71, and 77%, respectively, less than that of the wild type (Fig. 4), indicating greater sensitivity to oxidative stress.
Fig 4.
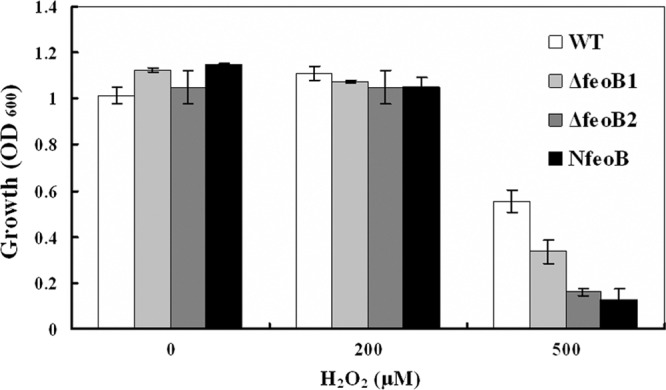
Effect of H2O2 on cell growth. Cells were grown in SLM supplemented with various concentrations of H2O2, and cell density was measured by spectrophotometry at 600 nm at stationary phase.
The enzymes SOD and CAT, which function to remove hydrogen peroxide and superoxide anion free radicals and thereby protect cells from injury, play important roles in the balance between oxidation and antioxidation (15, 24). To investigate why the three feoB mutants showed increased sensitivity to oxidative stress, we measured their SOD and CAT activities. All three mutants showed SOD and CAT activities that were significantly lower than those of the wild type (P < 0.05) (Table 4).
Table 4.
CAT and SOD enzyme activities of the wild-type strain and three deletion mutants
| Strain | Activity (U) of: |
|
|---|---|---|
| CATa | SODb | |
| WT | 5.48 ± 0.37 | 34.03 ± 0.79 |
| MSR-1 ΔfeoB1 | 4.58 ± 0.01 | 32.19 ± 0.51 |
| MSR-1 ΔfeoB2 | 4.65 ± 0.35 | 30.69 ± 0.35 |
| MSR-1 NfeoB | 3.83 ± 0.32 | 31.77 ± 0.76 |
One CAT activity unit is defined as 1 μmol hydrogen peroxide decomposed by CAT per mg protein in 1 min.
When the SOD inhibitory rate to superoxide anion free radicals reaches 50%, the quantity of SOD per milligram protein is 1 SOD activity unit.
DISCUSSION
We showed previously that the ferrous iron transport protein FeoB1 plays an important role in magnetosome formation in M. gryphiswaldense MSR-1 (19). The feoB1 deletion mutant MSR-1 ΔfeoB1 produced fewer and smaller magnetite crystals than the wild type but was still able to form magnetosomes, indicating that other proteins besides FeoB1 must be involved in magnetosome formation. We studied the role of another ferrous iron transport protein, FeoB2, that is also encoded by the MSR-1 genome. We constructed an feoB2 deletion mutant (MSR-1 ΔfeoB2) and an feoB1 feoB2 double deletion mutant (MSR-1 NfeoB). Magnetosome formation by MSR-1 ΔfeoB2 was similar to that of the wild type, i.e., the single feoB2 mutation did not affect magnetite crystal biomineralization. The average magnetosome number per cell of MSR-1 NfeoB was significantly less (∼65%) than that of MSR-1 ΔfeoB1, i.e., FeoB2 participated in magnetosome formation when FeoB1 was deleted. Our findings indicated that FeoB1 has a greater ferrous iron transport ability than FeoB2 and revealed the differential roles of FeoB1 and FeoB2 in MSR-1 iron metabolism. FeoB1 is responsible mainly for magnetosome formation in MSR-1, while FeoB2 is involved mainly in general iron metabolism under normal conditions. When the feoB1 gene was deleted, there was no other ferrous iron transporter responsible for magnetosome formation, and the ferrous iron that was transported by FeoB2 could be used in part for magnetite crystal biomineralization.
Reactive oxygen species (ROS), which include the superoxide radical (O2−), hydrogen peroxide (H2O2), and hydroxyl radical (OH·), are generated by the incomplete reduction of oxygen (10). ROS can cause damage to proteins, nucleic acids, and membranes (26). During the course of evolution, bacteria have developed a variety of protective enzymatic systems to prevent ROS-mediated damage. The most common oxidative stress-protective enzymes in aerobic bacteria are SOD and CAT (24). He et al. showed that the feoB2 gene in Porphyromonas gingivalis encodes a major manganese transporter that is required for protection of the bacterium from oxidative stress generated by atmospheric oxygen and H2O2 (10). We observed that MSR-1 produces numerous hydroxyl radicals during the process of magnetosome formation (unpublished data); however, little is known regarding oxidative stress protection in M. gryphiswaldense. We also examined the role of FeoB2 in the uptake of other metal ions and in oxidative stress. The feoB2-deficient mutant (MSR-1 ΔfeoB2) did not display significantly reduced manganese, zinc, copper, or magnesium uptake ability compared to the wild type (P > 0.05) (Table 3). When the three mutant strains (MSR-1 ΔfeoB1, ΔfeoB2, and NfeoB) were cultured in normal SLM, their growth was similar to that of the wild type. Under oxidative stress (culturing in 500 μM H2O2), the growth of the three mutants was significantly inhibited relative to that of the wild type. In particular, the optical density at 600 nm of MSR-1 ΔfeoB2 and MSR-1 NfeoB was ∼70% less than that of the wild type. The MSR-1 genome contains genes that encode SOD and CAT. We found that the SOD and CAT activities of the mutants were significantly lower than those of the wild type (P < 0.05). Qi et al. reported that when the concentration of intracellular free iron was higher, the mRNA levels of sodB (which encodes SOD) and katG (which encodes CAT) were also higher in MSR-1 cells (17), so the concentration of free iron ions may be lower in MSR-1 ΔfeoB2 than in the wild type. Iron is an essential cofactor of SOD and CAT, accounting for the observation that the feoB mutants were more sensitive to oxidative stress.
In summary, our findings show that the ferrous iron transport protein FeoB2 of M. gryphiswaldense strain MSR-1 participates in magnetosome formation when the protein FeoB1 is deleted, and that the FeoB proteins indirectly protect the bacterial cells from oxidative stress.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by the Chinese National Natural Science Foundation (grant no. 30970041) and by the Innovation Program for Undergraduates from China Agricultural University.
We thank S. Anderson for his advice and correction of English grammar errors and Qing Peng for drawing high-resolution pictures.
Footnotes
Published ahead of print 25 May 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Anaya-Bergman C, et al. 2010. Porphyromonas gingivalis ferrous iron transporter FeoB1 influences sensitivity to oxidative stress. Infect. Immun. 78:688–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andrews SC, Robinson AK, Rodriguez-Quinones F. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215–237 [DOI] [PubMed] [Google Scholar]
- 3. Bazylinski DA, Frankel RB. 2004. Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2:217–230 [DOI] [PubMed] [Google Scholar]
- 4. Blakemore RP, Maratea D, Wolfe RS. 1979. Isolation and pure culture of a freshwater magnetic spirillum in chemically defined medium. J. Bacteriol. 140:720–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Calugay RJ, Miyashita H, Okamura Y, Matsunaga T. 2003. Siderophore production by the magnetic bacterium Magnetospirillum magneticum AMB-1. FEMS Microbiol. Lett. 218:371–375 [DOI] [PubMed] [Google Scholar]
- 6. Cartron ML, Maddocks S, Gillingham P, Craven CJ, Andrews SC. 2006. Feo–transport of ferrous iron into bacteria. Biometals 19:143–157 [DOI] [PubMed] [Google Scholar]
- 7. Frankel RB, Blakemore RP, Wolfe RS. 1979. Magnetite in freshwater magnetotactic bacteria. Science 203:1355–1356 [DOI] [PubMed] [Google Scholar]
- 8. Guerinot ML. 1994. Microbial iron transport. Annu. Rev. Microbiol. 48:743–772 [DOI] [PubMed] [Google Scholar]
- 9. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 10. He J, et al. 2006. Role of Porphyromonas gingivalis FeoB2 in metal uptake and oxidative stress protection. Infect. Immun. 74:4214–4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jogler C, Schuler D. 2009. Genomics, genetics, and cell biology of magnetosome formation. Annu. Rev. Microbiol. 63:501–521 [DOI] [PubMed] [Google Scholar]
- 12. Komeili A. 2007. Molecular mechanisms of magnetosome formation. Annu. Rev. Biochem. 76:351–366 [DOI] [PubMed] [Google Scholar]
- 13. Li L, Rock JL, Nelson DR. 2008. Identification and characterization of a repeat-in-toxin gene cluster in Vibrio anguillarum. Infect. Immun. 76:2620–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marlovits TC, Haase W, Herrmann C, Aller SG, Unger VM. 2002. The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria. Proc. Natl. Acad. Sci. U. S. A. 99:16243–16248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McCord JM, Fridovich I. 1969. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 244:6049–6055 [PubMed] [Google Scholar]
- 16. Paoletti LC, Blakemore RP. 1986. Hydroxamate production by Aquaspirillum magnetotacticum. J. Bacteriol. 167:73–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qi L, et al. 2012. Fur in Magnetospirillum gryphiswaldense influences magnetosomes formation and directly regulates the genes involved in iron and oxygen metabolism. PLoS One 7:e29572 doi:10.1371/journal.pone.0029572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Richter M, et al. 2007. Comparative genome analysis of four magnetotactic bacteria reveals a complex set of group-specific genes implicated in magnetosome biomineralization and function. J. Bacteriol. 189:4899–4910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rong C, et al. 2008. Ferrous iron transport protein B gene (feoB1) plays an accessory role in magnetosome formation in Magnetospirillum gryphiswaldense strain MSR-1. Res. Microbiol. 159:530–536 [DOI] [PubMed] [Google Scholar]
- 20. Sambrook J, Russel DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 21. Schroder I, Johnson E, de Vries S. 2003. Microbial ferric iron reductases. FEMS Microbiol. Rev. 27:427–447 [DOI] [PubMed] [Google Scholar]
- 22. Schuler D. 2004. Molecular analysis of a subcellular compartment: the magnetosome membrane in Magnetospirillum gryphiswaldense. Arch. Microbiol. 181:1–7 [DOI] [PubMed] [Google Scholar]
- 23. Schuler D, Baeuerlein E. 1996. Iron-limited growth and kinetics of iron uptake in Magnetospirillum gryphiswaldense. Arch. Microbiol. 166:301–307 [DOI] [PubMed] [Google Scholar]
- 24. Schwartz CE, et al. 1983. Catalase and superoxide dismutase in Escherichia coli. J. Biol. Chem. 258:6277–6281 [PubMed] [Google Scholar]
- 25. Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat. Biotechnol. 1:784–791 [Google Scholar]
- 26. Storz G, Tartaglia LA, Farr SB, Ames BN. 1990. Bacterial defenses against oxidative stress. Trends Genet. 6:363–368 [DOI] [PubMed] [Google Scholar]
- 27. Suzuki T, Okamura Y, Arakaki A, Takeyama H, Matsunaga T. 2007. Cytoplasmic ATPase involved in ferrous ion uptake from magnetotactic bacterium Magnetospirillum magneticum AMB-1. FEBS Lett. 581:3443–3448 [DOI] [PubMed] [Google Scholar]
- 28. Taoka A, Umeyama C, Fukumori Y. 2009. Identification of iron transporters expressed in the magnetotactic bacterium Magnetospirillum magnetotacticum. Curr. Microbiol. 58:177–181 [DOI] [PubMed] [Google Scholar]
- 29. Taylor LA, Rose RE. 1988. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 16:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xia M, Wei J, Lei Y, Ying L. 2007. A novel ferric reductase purified from Magnetospirillum gryphiswaldense MSR-1. Curr. Microbiol. 55:71–75 [DOI] [PubMed] [Google Scholar]
- 31. Yang C, Takeyama H, Tanaka T, Matsunaga T. 2001. Effects of growth medium composition, iron sources and atmospheric oxygen concentrations on production of luciferase-bacterial magnetic particle complex by a recombinant Magnetospirillum magneticum AMB-1. Enzyme Microb. Technol. 29:13–19 [DOI] [PubMed] [Google Scholar]
- 32. Yijun H, Weijia Z, Wei J, Chengbo R, Ying L. 2007. Disruption of a fur-like gene inhibits magnetosome formation in Magnetospirillum gryphiswaldense MSR-1. Biochemistry 72:1247–1253 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.