Abstract
The Brucella BhuQ protein is a homolog of the Bradyrhizobium japonicum heme oxygenases HmuD and HmuQ. To determine if this protein plays a role in the ability of Brucella abortus 2308 to use heme as an iron source, an isogenic bhuQ mutant was constructed and its phenotype evaluated. Although the Brucella abortus bhuQ mutant DCO1 did not exhibit a defect in its capacity to use heme as an iron source or evidence of increased heme toxicity in vitro, this mutant produced increased levels of siderophore in response to iron deprivation compared to 2308. Introduction of a bhuQ mutation into the B. abortus dhbC mutant BHB2 (which cannot produce siderophores) resulted in a severe growth defect in the dhbC bhuQ double mutant JFO1 during cultivation under iron-restricted conditions, which could be rescued by the addition of FeCl3, but not heme, to the growth medium. The bhuQ gene is cotranscribed with the gene encoding the iron-responsive regulator RirA, and both of these genes are repressed by the other major iron-responsive regulator in the alphaproteobacteria, Irr. The results of these studies suggest that B. abortus 2308 has at least one other heme oxygenase that works in concert with BhuQ to allow this strain to efficiently use heme as an iron source. The genetic organization of the rirA-bhuQ operon also provides the basis for the proposition that BhuQ may perform a previously unrecognized function by allowing the transcriptional regulator RirA to recognize heme as an iron source.
INTRODUCTION
Iron represents an essential micronutrient for Brucella strains (27). Acquiring sufficient iron to meet their physiological needs is particularly challenging for the brucellae because these bacteria are found in nature almost exclusively in mammalian hosts, an environment where the iron restriction faced by pathogenic microbes is well documented (29). Brucella strains can use heme as an iron source in vitro, and studies with an isogenic mutant have shown that the presence of the TonB-dependent outer membrane heme transporter BhuA is required for the wild-type virulence of B. abortus 2308 in experimentally infected mice (21), suggesting that heme is a biologically relevant source of iron for the brucellae during infection.
Heme oxygenases catalyze the release of iron from heme, and these enzymes contribute to the ability of a variety of bacteria to utilize heme as an iron source (25, 30, 37). The product of the gene designated BMEII0706 in the Brucella melitensis 16M genome sequence shares 58 and 50% amino acid identity with the heme oxygenases HmuD and HmuQ, respectively, from Bradyrhizobium japonicum, and this Brucella protein exhibits heme oxygenase activity in an in vitro assay (23). Based on its documented biochemical activity and its homology to the IsdG/HmuQ family of heme oxygenases, we have given this protein the designation BhuQ (Brucella heme utilization oxygenase Q). The purpose of the experiments described in this report was to determine if the homologous protein in Brucella abortus 2308 (which is encoded by BAB2_0677) plays a role in the capacity of this strain to use heme as an iron source.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Brucella abortus strains (Table 1) were routinely grown in brucella broth at 37°C with aeration or on Schaedler agar supplemented with 5% bovine blood (SBA) at 37°C under 5% CO2. Kanamycin (45 μg/ml) (Sigma) and/or ampicillin (25 μg/ml) (Sigma) was added to these media as appropriate for the selection of strains with antibiotic resistance markers. Escherichia coli strain DH5α was used for the propagation of plasmids for procedures involving recombinant DNA, and this strain was cultivated at 37°C in LB broth or on LB agar plates containing either 100 μg/ml ampicillin or 45 μg/ml kanamycin when appropriate. Gerhardt's minimal medium (GMM) (11) and low-iron minimal medium (15) were prepared as previously described.
Table 1.
Bacterial strains used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia coli DH5α | F− ϕ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1 λ− | Invitrogen |
| Brucella abortus | ||
| 2308 | Virulent challenge strain | Laboratory stock |
| DCO1 | 70-bp gene deletion of bhuQ (BAB2_0677) with an ahpA3 kanamycin resistance gene insertion | This study |
| DCO1RC | DCO1 with bhuQ (BAB2_0677) reconstructed on the chromosome using pNPTS138 | This study |
| BHB2 | Nonpolar, in-frame deletion of dhbC | 5, this study |
| JFO1 | ΔdhbC ΔbhuQ | This study |
| JFO1RC | ΔdhbC ΔbhuQ with bhuQ (BAB2_0677) reconstructed on the chromosome using pNPTS138 | This study |
| BEA2 | Dirr | 1, this study |
| Plasmids | ||
| pGEM-T Easy | ColE1-based cloning vector; Apr | Promega |
| pKS + Kan | 794-bp aph3A gene from TnphoA cloned into SalI-HindIII-digested pBluescript II KS+ | 14 |
| pNPTS138 | sacB-containing counterselection vector; Ampr | 32 |
Construction of a B. abortus bhuQ mutant and a dhbC bhuQ double mutant.
PCR utilizing Taq polymerase (Invitrogen) with the oligonucleotide primers bhuQ-1F and bhuQ-1R (see Table S1 in the supplemental material) was used to amplify a 1,605-bp fragment encompassing the bhuQ gene (BAB2_0677) from B. abortus 2308 genomic DNA. This fragment was then cloned into pGEM-T Easy (Promega). Inverse PCR employing AccuPrime Pfx supermix (Invitrogen) with this plasmid as a template and the primers bhuQ-2F and bhuQ-2R (see Table S1) was then employed to generate a blunt-ended linear fragment from which 70 bp internal to the bhuQ coding region had been removed. This fragment was ligated with a 1,345-bp fragment containing the aph3a gene from pKS-Kn (14). The resulting construct, pGEMΔbhuQ, was introduced into B. abortus strain 2308 by electroporation and transformants were selected on SBA supplemented with 45 μg/ml kanamycin. Putative B. abortus bhuQ deletion mutants were identified based on their resistance to kanamycin and sensitivity to ampicillin, and their genotypes were confirmed by PCR analysis and DNA sequence analysis. Chromosomal DNA preparations from putative deletion mutants and strain 2308 were harvested, and oligonucleotides bhuQ F1 and Kan R (see Table S1) were used to determine the presence of the aph3a-based gene in the proper orientation, the absence of the ampicillin resistance gene from pGEM-T Easy (Promega) (amp F and amp R), and the absence of the 70 bp in the middle of bhuQ (bhuQ F1 and bhuQ R1). One confirmed B. abortus bhuQ mutant was selected for further phenotypic evaluation and given the designation DCO1.
The approach described in the previous paragraph was also used to introduce a bhuQ mutation into B. abortus BHB2 (6). BHB2 has an unmarked, in-frame deletion in its dhbC gene, which renders it unable to produce the two siderophores produced by Brucella strains, 2,3-dihydroxybenzoic acid (15) and brucebactin (12). The B. abortus dhbC bhuQ double mutant constructed in this fashion was given the designation JFO1.
Reconstruction of the bhuQ loci in the B. abortus bhuQ and dhbC bhuQ mutants.
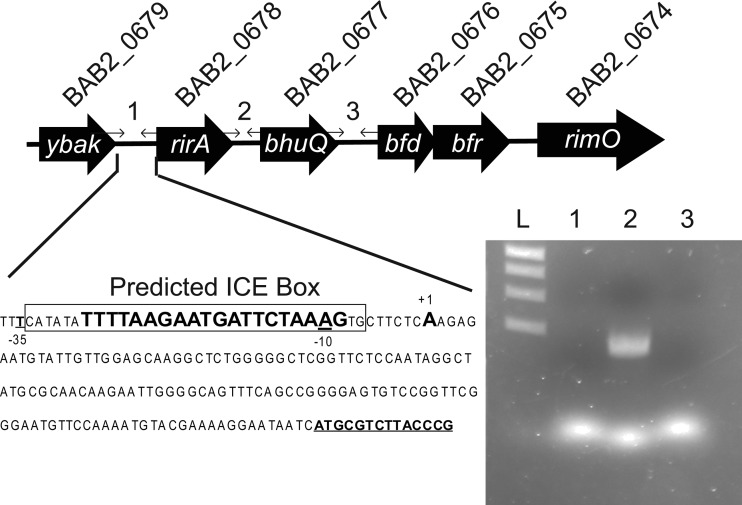
Because the bhuQ gene is the terminal gene in an operon and lies downstream of a transcriptional regulator (see Fig. 4), reconstruction of the mutated bhuQ genes in DCO1 and JFO1 was chosen as a strategy for verifying the link between genotype and the phenotypes exhibited by these strains, rather than genetic complementation with a plasmid-borne bhuQ gene. A 920-bp fragment encompassing the bhuQ gene from B. abortus strain 2308 genomic DNA was amplified by PCR using the primers bhuQ-3F and bhuQ-3F (see Table S1) and cloned into the BamHI and SalI sites of pNPTS138Ap (Table 1), an ampicillin-resistant derivative of the sacB-containing ColE1-based vector pNPTS138 (32). The resulting plasmid, designated pNPTS138bhuQ, was introduced into B. abortus DCO1 and JFO1 by electroporation, and a previously described sacB-based counterselection strategy (6) was used to select for derivatives of these mutants in which the mutated bhuQ genes had been replaced by the parental bhuQ gene. The genotypes of the resulting B. abortus strains, designated DCO1RC and JFO1RC were confirmed by PCR amplification and DNA sequence analysis.
Fig 4.
rirA and bhuQ are cotranscribed as an operon in B. abortus 2308. The sequence shows the start site (+1) for the rir-bhuQ transcript, located 134 bp upstream of the rirA ORF (bold underlined), and the location of an Irr binding motif predicted by Rodionov et al., labeled as an ICE box and shown in large boldface type (26), while the nucleotide sequence protected by Irr is boxed. At the top, primer sets 1, 2, and 3 denote the intragenic regions used to define the transcript, and the corresponding lanes are marked on the agarose gel below.
Measurement of siderophore production by Brucella strains.
Following growth of the B. abortus strains in low-iron minimal medium (15) at 37°C with shaking (165 rpm), bacterial cells from 1.5-ml portions of the cultures were pelleted by centrifugation (15,550 × g, 1 min, room temperature), 1 ml of the resulting supernatant was removed to a fresh tube (10 ml), and the level of catechol siderophore present was measured using the Arnow assay (4).
Determination of the growth characteristics of B. abortus strains in an iron-limited culture medium.
B. abortus strains were grown on SBA plates at 37°C with 5% CO2 for 48 h and harvested into phosphate-buffered saline (PBS). The resulting cell suspensions were used to inoculate 20 ml low-iron minimal medium in 125-ml Erlenmeyer flasks at a final concentration of approximately 106 CFU/ml. When applicable, the medium was supplemented with either 50 μM FeCl3 or 25 μM deferrated hemin (33). Cultures were incubated at 37°C with shaking at 165 rpm, and at 24-h time points postinoculation, these cultures were serially diluted in PBS and plated on SBA, followed by incubation at 37°C under 5% CO2.
Relative quantification of bhuQ transcript levels using real-time RT-PCR.
B. abortus 2308 and BEA2 (2308 irr) (1) were grown in low-iron minimal medium and low-iron minimal medium supplemented with 50 μM FeCl3 or in Gerhardt's minimal medium with or without 25 μM deferrated hemin. Total cellular RNA was isolated from these cultures using a previously described procedure (7). The RNA was treated with RQ1 DNase (Ambion) following the manufacturer's instructions to remove residual contaminating DNA. The absence of DNA from the RNA preparations was confirmed via PCR analysis and lack of an amplified product as visualized on an agarose gel confirmed that the RNA sample was free of DNA contamination. Concentrations of RNA in the samples were determined by measuring the absorbance at 260 nm using a NanoDrop ND-1000 spectrophotometer.
A SuperScript III first-strand synthesis system for RT-PCR kit (Invitrogen) was used to convert 1 μg of RNA from these preparations into cDNA for each preparation following the manufacturer's instructions and using the random hexamers supplied with the kit. The cDNA preparations were then used as the templates in real-time RT-PCR analysis along with FastStart SYBR green master mix 2× (Roche) to evaluate the relative levels of gene-specific mRNA transcripts in the total cellular RNA preparations. Gene-specific oligonucleotide primers were utilized for the 16S gene, dhbC, bhuA, and bhuQ (see Table S1 in the supplemental material). The differences in the levels of the bhuQ-, bhuA-, and dhbC-specific transcripts present were calculated using methods described by Pfaffl (22), using the 16S gene as an internal standard. This gene encodes the ribosomal 16S protein, and its expression is constitutive in B. abortus 2308 under the experimental conditions used here.
Determination of the operonic organization of the ybaK, rirA, bhuQ, and bfr genes in B. abortus 2308.
Reverse transcriptase PCR was performed using cDNA prepared from clean RNA harvested from B. abortus 2308 grown for 72 h in low-iron minimal medium. Primers that span the intragenic regions between ybaK (BAB2_0679), rirA (BAB2_0678), bhuQ (BAB2_0677), and bfr (BAB2_0676) (see Table S1 in the supplemental material) were used in order to verify the presence or absence of a continuous transcript containing these genes. The resulting PCR products were separated by electrophoresis in a 0.7% agarose gel and visualized by staining the gel with ethidium bromide.
Determination of the transcriptional start site for the rirA-bhuQ operon.
The bhuQ gene is cotranscribed with the upstream gene, rirA. In order to determine a transcriptional start site for this operon, 5′ RNA ligase-mediated rapid amplification of the cDNA end (5′ RLM-RACE) was performed using a primer (Rev [see Table S1 in the supplemental material]) anchored in the reverse orientation within the rirA ORF following the manufacturer's instructions (FirstChoice RLM-RACE kit, no. AM1700; Ambion). The PCR product generated from this reaction was cloned into pCR2.1 (Invitrogen), and the authenticity of the PCR fragment verified by DNA sequence analysis.
Identification of the Irr binding site in the rirA promoter region.
A recombinant version of the Brucella Irr was purified and used in a DNase I footprint analysis with the rirA promoter region from B. abortus 2308 using previously described methods (1, 17). Briefly, the oligonucleotide primers rirA F and rirA R (see Table S1 in the supplemental material) were individually labeled with [γ-32P]ATP (Perkin Elmer) using the T4 polynucleotide kinase reaction (Promega, Madison, WI) prior to their use in PCRs with Pfx polymerase to generate 300-bp DNA fragments representing the rirA promoter and transcriptional start site. The resulting PCR products were subjected to agarose gel electrophoresis and purified by gel extraction (Fermentas, Glen Burnie, MD). DNA probes corresponding to 8,000 cpm of the forward-labeled and reverse-labeled templates were incubated separately in EMSA binding buffer (10 mM Tris-HCl [pH 8], 40 mM KCl, 1 mM MgCl2, 1 mM dithiothreitol [DTT], 5% glycerol) supplemented with 100 ng/ml bovine serum albumin (BSA) and 50 ng/ml salmon sperm DNA (nonspecific competitor) in the presence of 100 μM MnCl2 and increasing concentrations of the recombinant Brucella Irr protein. The reaction mixtures were incubated at room temperature for 30 min prior to treatment with 0.08 U of DNase I freshly diluted in 10× DNase I buffer (400 mM Tris-HCl [pH 8.0], 100 mM MgSO4, 10 mM CaCl2) for 4 min. Reactions were stopped by the addition of 5 mM EDTA and heating at 65°C for 10 min. Reaction mixtures were ethanol precipitated and resuspended in 4 μl of formamide loading buffer (98% formamide, 10 mM EDTA [pH 8.0], 1 mg/ml xylene cyanol FF, 1 mg/ml bromophenol blue). Digested DNA fragments were separated on a denaturing 6% (wt/vol) acrylamide and 7 M urea sequencing gel in glycerol-tolerant buffer (17). Gels were dried under vacuum and subjected to autoradiography. The sequence protected by Irr was determined by comparing the nucleotide sequences generated for a 100-bp region of the rirA promoter region using the SequiTherm Excel II DNA sequencing kit (Epicentre, Madison, WI) and B. abortus 2308 DNA preparations exposed to DNase I treatment with and without recombinant Irr as templates.
Statistical analysis.
All statistical analysis was performed using the Student two-tailed t test. P values of less than 0.05 were considered significant (28).
RESULTS AND DISCUSSION
Pfam analysis of the Brucella BhuQ protein indicates that it belongs to the antibiotic monooxygenase (ABM) family of heme oxygenases along with the HmuD and HmuQ proteins from B. japonicum (23) and the IsdI and IsdG proteins from Staphylococcus aureus (31). Moreover, BhuQ contains the conserved Asn 7, Trp 67, and His 77 residues, which have been shown experimentally to be important for the heme oxygenase activity of IsdG (36) (Fig. 1), and BhuQ has been shown to bind and degrade heme in vitro (23). Despite the documented heme oxygenase activity of BhuQ, however, an isogenic bhuQ mutant (DCO1) constructed from B. abortus 2308 exhibited a comparable growth pattern in low-iron minimal medium and an equivalent resistance to the iron-specific chelator ethylenediamine-N,N′-bis(2-hydroxyphenylacetic acid) (EDDHA) to that displayed by the parental strain. More importantly, both strains were also able to use heme as an iron source with equivalent efficiency in a chelator-based disk assay on a solid growth medium (data not shown).
Fig 1.
The Brucella abortus (Ba) BhuQ protein shares amino acid homology with the heme oxygenases HmuD and HmuQ from Bradyrhizobium japonicum (Bj) and IsdG and IsdI from Staphylococcus aureus (Sa) and contains the conserved Asn 7, Trp 67 and His 77 residues (large bold font), which have been shown experimentally to be important for the heme oxygenase activity of IsdG (31).
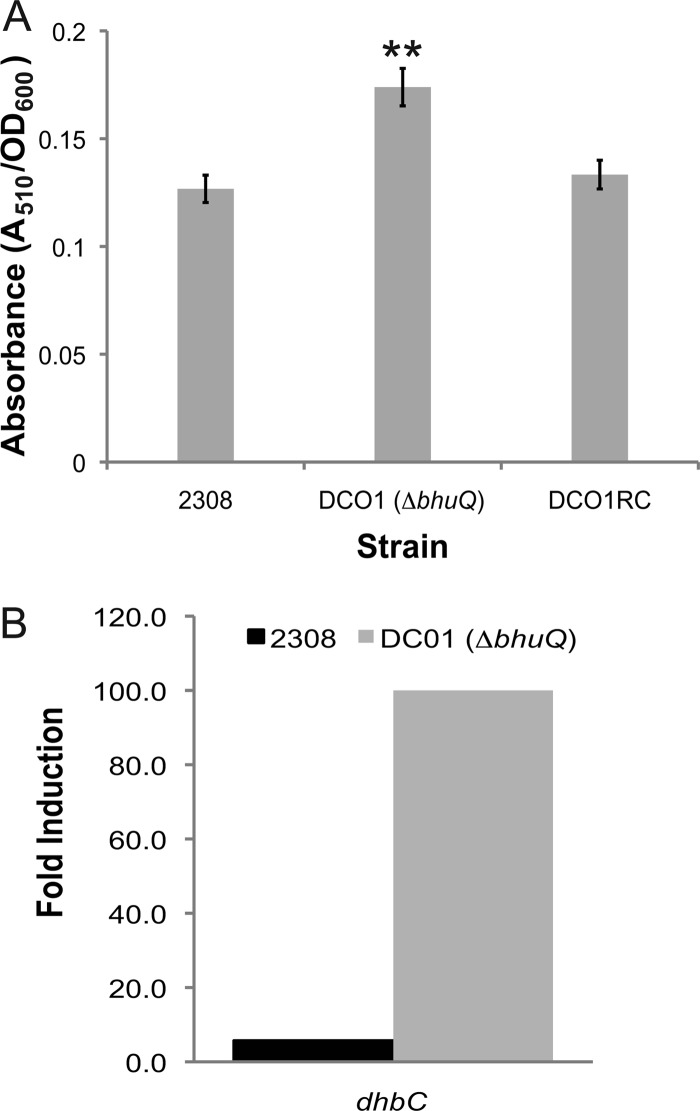
A distinctive characteristic of the B. abortus bhuQ mutant that was noticed during its phenotypic characterization, however, was that this mutant consistently and reproducibly produces more siderophore during growth in low-iron minimal medium than the parental 2308 strain (Fig. 2A). This increase in siderophore production is accompanied by a corresponding increase in transcription of dhbC in the bhuQ mutant in comparison to the parental strain (2308) following growth under iron-limiting conditions (Fig. 2B). This phenotype suggests that the B. abortus bhuQ mutant is experiencing a greater degree of iron deprivation in the low-iron minimal medium than 2308 and in turn increasing its siderophore production to compensate. Enhanced siderophore production could explain why the B. abortus bhuQ mutant does not exhibit a readily detectable iron acquisition defect in in vitro assays such as growth in low-iron minimal medium or sensitivity to EDDHA. The ability of this mutant to use heme as an iron source in the disk diffusion assays, however, indicates that B. abortus 2308 possesses one or more additional heme oxygenases that can compensate for the loss of BhuQ and allow the bhuQ mutant to use heme as an iron source. The identity of these other heme oxygenases is currently unknown.
Fig 2.
(A) Siderophore production by B. abortus 2308, DCO1 (ΔbhuQ), and DCO1RC (DCO1 bhuQ+) following 72 h growth in low-iron minimal medium. The values on the y axis are the levels of catechol siderophore detected by the Arnow assay (4). **, P < 0.01 for comparisons of the data obtained for these strains in the Student two-tailed t test (28). (B) Iron-responsive expression of dhbC in B. abortus 2308 and DCO1 (2308 bhuQ). Values on the y axis are the difference between the levels of dhbC transcripts detected in RNA preparations from B. abortus 2308 and DCO1 cultures after 72 h of growth in low-iron minimal medium compared to RNA preparations from these cultures after growth in low-iron minimal medium supplemented with 50 μM FeCl3.
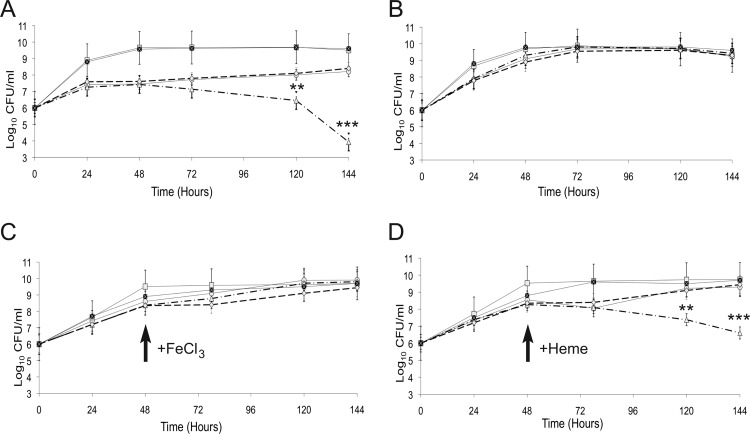
Brucella strains have the capacity to use multiple iron sources during in vitro growth (27), and it is not usual for bacterial strains with mutations affecting single iron transporters to exhibit little or no defect in iron utilization assays due to compensation by other iron transport systems. Indeed, the fact that the B. abortus bhuQ mutant exhibits increased siderophore production in response to iron deprivation compared to the parental strain provided an experimental avenue for assessing the role of BhuQ in iron metabolism. Specifically, if the loss of BhuQ from B. abortus DCO1 is leading to an increased demand for iron, and siderophore production is being increased in this mutant to meet this demand, then a derivative of this strain that cannot produce siderophore (e.g., a B. abortus dhbC bhuQ double mutant) would be expected to show an enhanced iron deprivation phenotype compared to B. abortus 2308 or the bhuQ mutant when grown under iron-limiting conditions. This is in fact the relationship that was observed. As shown in Fig. 3A, the dhbC bhuQ double mutant JFO1 showed a greatly enhanced growth defect compared to the parental BHB2 (dhbC mutant) strain when they were cultivated in low-iron minimal medium, a phenotype that was not observed when these strains were grown in iron-replete medium (Fig. 3B). More importantly, the enhanced growth defect exhibited by the B. abortus dhbC bhuQ double mutant during cultivation in low-iron minimal medium could be rescued by the addition of FeCl3 (Fig. 3C), but not heme (Fig. 3D), to the growth medium 48 h after inoculation of the bacterial cultures. In contrast, either FeCl3 (Fig. 3C) or heme (Fig. 3D) was able to rescue the growth defect exhibited by the dhbC mutant BHB2 during growth under low-iron conditions, although in both cases the alleviation of the growth restriction occurred more rapidly when FeCl3 was added. To verify the link between the bhuQ mutations in B. abortus DCO1 and JFO1 and the phenotypes exhibited by these strains, a sacB-based counterselection strategy (6) was used to reconstruct the bhuQ genes in these mutants. The resulting strains, designated DCO1RC and JFO1RC, exhibited the expected parental phenotype with regard to their production of siderophore in response to iron deprivation (Fig. 2), their sensitivities to iron deprivation (Fig. 3A and B), and their abilities to use FeCl3 and heme as iron sources (Fig. 3C and D). Although these experimental findings establish a role for BhuQ in the capacity of the B. abortus dhbC mutant to use heme as an iron source, this activity appears to be masked in B. abortus 2308 by the activity of other heme oxygenases when alternative iron sources are readily available in the growth medium. The capacity of the B. abortus bhuQ mutant to compensate for the loss of one heme oxygenase was also observed in experimental models of infection, as this strain exhibited wild-type virulence in cultured murine macrophages and experimentally infected BALB/c mice (data not shown).
Fig 3.
Growth of B. abortus 2308 (filled squares), B. abortus DCO1 (2308 ΔbhuQ) (squares), BHB2 (2308 ΔdhbC) (circles), JFO1 (2308 ΔdhbC ΔbhuQ) (triangles), and JFO1RC (JFO1 bhuQ+) (diamonds) in (A) low-iron minimal medium, (B) low-iron minimal medium containing 50 μM FeCl3, (C) low-iron minimal medium supplemented with 50 μM FeCl3 at 48 h postinoculation, and (D) low-iron minimal medium supplemented with deferrated 25 μM hemin at 48 h postinoculation. The data presented are from one experiment but representative of multiple experiments (>3) in which similar trends were observed. The data were compared with the Student two-tailed t test (28). **, P < 0.01; ***, P < 0.001.
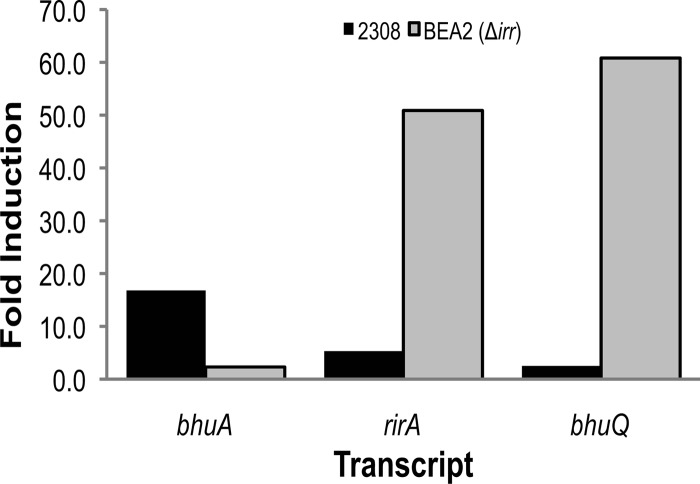
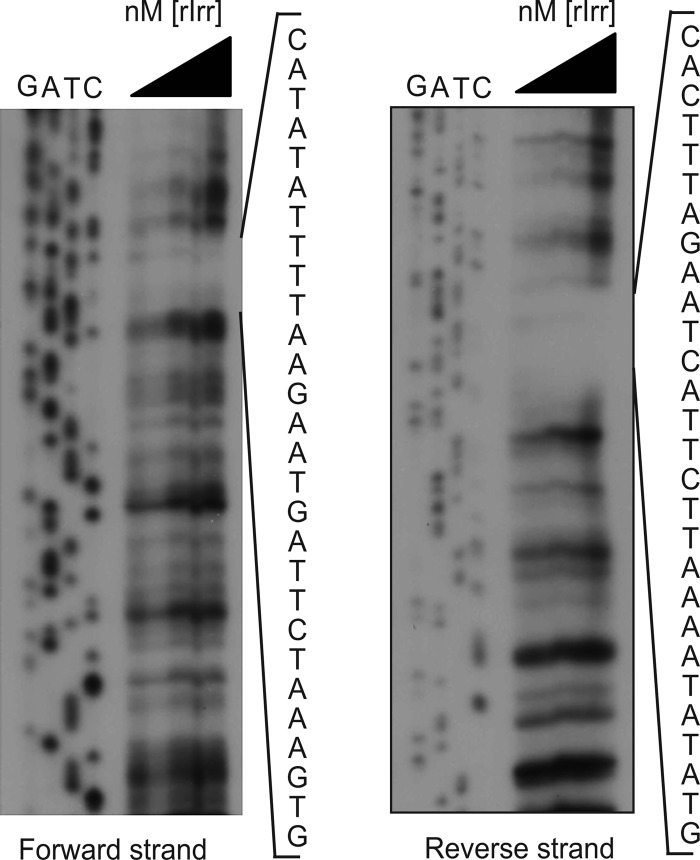
RT-PCR analysis indicates that bhuQ is the second gene in an operon transcribed as rirA (BAB2_0678)-bhuQ (BAB2_0677) in B. abortus 2308 (Fig. 4). RirA is a well-characterized regulator of iron metabolism genes in several other alphaproteobacteria (8, 13, 19, 35). A predicted iron control element (ICE) motif is located in the −10 region of the promoter of the rirA-bhuQ operon (Fig. 4), suggesting that the iron response regulator Irr regulates the expression of these genes in response to cellular iron levels (16). In fact, when the expression patterns of the rirA and bhuQ genes in B. abortus 2308 were independently evaluated by real-time PCR, both genes exhibited a modest induction in response to iron deprivation (Fig. 5). In contrast, the expression of both of these genes was elevated >50-fold in the B. abortus irr mutant BEA2 when this strain was grown under iron-limiting conditions. Thus, it appears that Irr represses the expression of rirA in Brucella strains during periods of iron deprivation in much the same manner as it does in the related alphaproteobacterium Agrobacterium tumefaciens (13). DNase I footprint analysis indicated that Irr directly binds to the rirA promoter (Fig. 6), protecting a 28-nucleotide sequence, 5′-CATATATTTTAAGAATGATTCTAAAGTG-3′. Notably, this sequence includes the conserved ICE motif in the promoter region of rirA (underlined) predicted by Rodionov et al. (26).
Fig 5.
rirA and bhuQ expression in response to iron deprivation in B. abortus 2308 and the isogenic irr mutant BEA2. Values on the y axis are the difference between the levels of rirA, bhuQ, and bhuA transcripts detected in RNA preparations from B. abortus 2308 and BEA2 after 72 h of growth in low-iron minimal medium compared to RNA preparations from these cultures after growth in low-iron minimal medium supplemented with 50 μM FeCl3. The pattern of bhuA transcription was included because this gene exhibits elevated expression in response to iron deprivation in B. abortus 2308, and this low-iron-responsive induction is dependent upon the presence of Irr (1).
Fig 6.
Irr binds directly to the rirA promoter in B. abortus 2308 and protects a 28-nucleotide sequence in a DNase I footprint analysis. The triangle above the lanes indicates that the corresponding reaction mixtures contain increasing concentrations (700 ng, 1.4 μg, 2.1 μg, and 3.5 μg) of recombinant Brucella Irr, and the nucleotide sequences shown to the right of the gels show the nucleotides protected from DNase I digestion in the forward and reverse strands of the rirA promoter sequence.
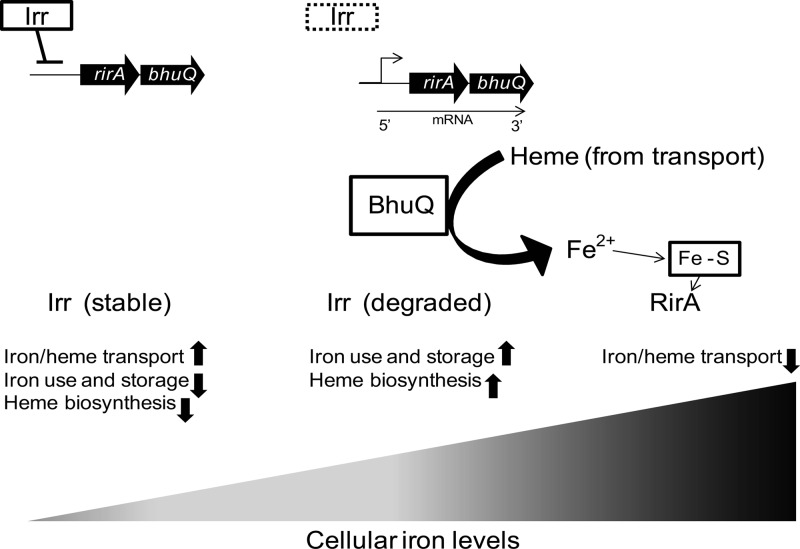
Genetic studies suggest that RirA functions as an iron-responsive repressor of iron acquisition genes in the alphaproteobacteria (35), in much the same fashion as Fur does in other bacteria. The potential benefit of Irr's repressing the expression of an iron-responsive repressor when the bacterial cell is experiencing iron deprivation is not difficult to envision. But such a regulatory link would appear to be counterproductive for the bhuQ gene if its product is solely dedicated to the utilization of heme as an iron source. One scenario that might explain a possible benefit of coregulation of bhuQ and rirA in B. abortus 2308 is that BhuQ production leads to an increased level of heme oxygenase activity which enhances the capacity of RirA to recognize exogenous heme being utilized as an iron source and represses the cell's iron acquisition systems accordingly (Fig. 7).
Fig 7.
Model showing the proposed role of BhuQ in allowing the transcriptional regulator RirA to recognize heme as an iron source in Brucella. Under iron-deprived conditions, Irr is stable and drives the expression of genes involved in iron and heme transport. As intracellular iron levels increase, Irr is degraded, which allows expression of the rirA-bhuQ operon. BhuQ degrades the heme being transported into the cell, releasing Fe2+. When cellular iron levels reach a threshold, RirA is postulated to repress the expression of the iron and heme acquisition genes.
In addition to their ability to provide iron from heme, some bacterial heme oxygenases also function to protect the cell against heme toxicity (2). Heme has a high redox potential, and too much heme inside a bacterial cell can be toxic (20). Neither B. abortus 2308 nor the isogenic bhuQ mutant display a growth defect in low-iron medium supplemented with up to 200 μM deferrated hemin. It is possible, however, that any role that BhuQ might be playing in the detoxification of heme is masked by the activity of another heme oxygenase. Another factor that may affect this observed lack of heme toxicity is that Brucella strains possess multiple homologs of the outer membrane heme-binding proteins (Hbps) that have been proposed to play a role in capturing heme and preventing its toxicity in Bartonella (18). In Brucella, these proteins are known as the Omp25/31 family of proteins (9). Heme export systems have also been proposed as a means by which bacteria protect themselves from heme toxicity (2). Although a heme exporter has not been identified in Brucella strains, genes that potentially encode orthologs of proteins linked to porphyrin (34) and heme (24) export in other bacteria can be found in the genome sequence of B. abortus 2308.
In order to fully understand the role of BhuQ in iron and heme metabolism in Brucella strains, it will be imperative to identify the other heme oxygenase(s) present in these bacteria. Phenotypic evaluation of mutants lacking combinations of these enzymes can then be used to assess the relative contributions of the heme oxygenases to iron and heme metabolism, as well as their potential role in modulating the regulatory capacity of RirA. Such studies may also provide an added practical benefit, as prokaryotic heme oxygenases have been proposed to be targets for the development of antimicrobial agents (10). Brucellosis in humans is notoriously difficult to treat, requiring a combination of antibiotics for a prolonged period (3). Hence, the development of improved chemotherapeutic regimens for treating this disease would be of great benefit to the medical community.
Supplementary Material
ACKNOWLEDGMENT
This work was funded by a grant (AI 63516) from the National Institute of Allergy and Infectious Diseases to R.M.R.
Footnotes
Published ahead of print 25 May 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Anderson ES, et al. 2011. The iron-responsive regulator Irr is required for wild-type expression of the gene encoding the heme transporter bhuA in Brucella abortus 2308. J. Bacteriol. 193:5359–5364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anzaldi LL, Skaar EP. 2010. Overcoming the heme paradox: heme toxicity and tolerance in bacterial pathogens. Infect. Immun. 78:4977–4989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ariza J, et al. 2007. Perspectives for the treatment of brucellosis in the 21st century: the Ioannina recommendations. PLoS Med. 4:e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arnow LE. 1937. Colorimetric determination of the components of 3,4-dihydroxyphenyalanin-tyrosine mixtures. J. Biol. Chem. 118:531–537 [Google Scholar]
- 5. Bellaire BH, Elzer PH, Baldwin CL, Roop RM., II 1999. The siderophore 2,3-dihydroxybenzoic acid is not required for virulence of Brucella abortus in BALB/c mice. Infect. Immun. 67:2615–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bellaire BH, Elzer PH, Baldwin CL, Roop RM., II 2003. Production of the siderophore 2,3-dihydroxybenzoic acid is required for wild-type growth of Brucella abortus in the presence of erythritol under low-iron conditions in vitro. Infect. Immun. 71:2927–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Caswell CC, Gaines JM, Roop RM., II 2012. The RNA chaperone Hfq independently coordinates expression of the VirB type IV secretion system and the LuxR-type regulator BabR in Brucella abortus 2308. J. Bacteriol. 194:3–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chao TC, Buhrmester J, Hansmeier N, Pühler A, Weidner S. 2005. Role of the regulatory gene rirA in the transcriptional response of Sinorhizobium meliloti to iron limitation. Appl. Environ. Microbiol. 71:5969–5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cloeckaert A, Verger JM, Grayon M, Vizcaino N. 1996. Molecular and immunological characterization of the major outer membrane proteins of Brucella. FEMS Microbiol. Lett. 145:1–8 [DOI] [PubMed] [Google Scholar]
- 10. Furci LM, et al. 2007. Inhibition of the bacterial heme oxygenases from Pseudomonas aeruginosa and Neisseria meningitidis: novel antimicrobial targets. J. Med. Chem. 50:3804–3813 [DOI] [PubMed] [Google Scholar]
- 11. Gerhardt P, Tucker LA, Wilson JB. 1950. The nutrition of brucellae: utilization of single amino acids for growth. J. Bacteriol. 59:777–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. González Carreró MI, Sangari FJ, Agüero J, García Lobo JM. 2002. Brucella abortus strain 2308 produces brucebactin, a highly efficient catecholic siderophore. Microbiology 148:353–360 [DOI] [PubMed] [Google Scholar]
- 13. Hibbing ME, Fuqua C. 2011. Antiparallel and interlinked control of cellular iron levels by the Irr and RirA regulators of Agrobacterium tumefaciens. J. Bacteriol. 193:3461–3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kovach ME, Elzer PH, Hill DS, Roop RM, II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 15. López-Goñi I, Moriyón I, Neilands JB. 1992. Identification of 2,3-dihydroxybenzoic acid as a Brucella abortus siderophore. Infect. Immun. 60:4496–4503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Martínez M, Ugalde RA, Almirón M. 2006. Irr regulates brucebactin and 2,3-dihydroxybenzoic acid biosynthesis, and is implicated in the oxidative stress resistance and intracellular survival of Brucella abortus. Microbiology 152:2591–2598 [DOI] [PubMed] [Google Scholar]
- 17. Menscher EA, Caswell CC, Anderson ES, Roop RM., II 2012. Mur regulates the gene encoding the manganese transporter MntH in Brucella abortus 2308. J. Bacteriol. 194:561–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Minnick MF, Battisti JM. 2009. Pestilence, persistence and pathogenicity: infection strategies of Bartonella. Future Microbiol. 4:743–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ngok-Ngam P, et al. 2009. Roles of Agrobacterium tumefaciens RirA in iron regulation, oxidative stress response, and virulence. J. Bacteriol. 191:2083–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nir U, Ladan H, Malik Z, Nitzan Y. 1991. In vivo effects of porphyrins on bacterial DNA. J. Photochem. Photobiol. B 11:295–306 [DOI] [PubMed] [Google Scholar]
- 21. Paulley JT, Anderson ES, Roop RM., II 2007. Brucella abortus requires the heme transporter BhuA for maintenance of chronic infection in BALB/c mice. Infect. Immun. 75:5248–5254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:2002–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Puri S, O'Brian MR. 2006. The hmuQ and hmuD genes from Bradyrhizobium japonicum encode heme-degrading enzymes. J. Bacteriol. 188:6476–6482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rasmussen AW, Alexander HL, Perkins-Balding D, Shafer WM, Stojiljkovic I. 2005. Resistance of Neisseria meningitidis to the toxic effects of heme iron and other hydrophobic reagents requires expression of ght. J. Bacteriol. 187:5214–5223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ratliff M, Zhu W, Deshmukh R, Wilks A, Stojiljkovic I. 2001. Homologues of neisserial heme oxygenase in Gram-negative bacteria: degradation of heme by the product of the pigA gene of Pseudomonas aeruginosa. J. Bacteriol. 183:6394–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rodionov DA, Gelfand MS, Todd JD, Curson AR, Johnston AW. 2006. Computational reconstruction of iron- and manganese-responsive transcriptional networks in alpha-proteobacteria. PLoS Comput. Biol. 2:e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roop RM, II, et al. 2011. Metal acquisition by Brucella strains, p 179–199 In López-Goñi I, O'Callaghan D. (ed), Brucella: molecular microbiology and genetics. Horizon Scientific Press, Norfolk, United Kingdom [Google Scholar]
- 28. Rosner B. 2000. Fundamentals of biostatistics, 5th ed Duxbury, Pacific Grove, CA [Google Scholar]
- 29. Schaible UE, Kaufmann SHE. 2004. Iron and microbial infection. Nat. Rev. Microbiol. 2:946–953 [DOI] [PubMed] [Google Scholar]
- 30. Schmitt MP. 1997. Utilization of host iron sources by Corynebacterium diphtheriae: identification of a gene whose product is homologous to eukaryotic heme oxygenases and is required for acquisition of iron from heme and hemoglobin. J. Bacteriol. 179:838–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Skaar EP, Gaspar AH, Schneewind O. 2004. IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J. Biol. Chem. 279:436–443 [DOI] [PubMed] [Google Scholar]
- 32. Spratt BG, Hedge PJ, te Heesen S, Edelman A, Broome-Smith JK. 1986. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene 41:337–342 [DOI] [PubMed] [Google Scholar]
- 33. Staggs TM, Perry RD. 1991. Identification and cloning of a fur regulatory gene in Yersinia pestis. J. Bacteriol. 173:417–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tatsumi R, Wachi M. 2008. TolC-dependent exclusion of porphyrins in Escherichia coli. J. Bacteriol. 190:6228–6233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Todd JD, et al. 2002. RirA, an iron-responsive regulator in the symbiotic bacterium Rhizobium leguminosarum. Microbiology 148:4059–4071 [DOI] [PubMed] [Google Scholar]
- 36. Wu R, et al. 2005. Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases. J. Biol. Chem. 280:2840–2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhu W, Hunt JD, Richardson AR, Stojiljkovic I. 2000. Use of heme compounds as iron sources by pathogenic Neisseriae requires the product of the hemO gene. J. Bacteriol. 182:439–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.