Abstract
Plant- and animal-pathogenic bacteria utilize phylogenetically distinct type III secretion systems (T3SS) that produce needle-like injectisomes or pili for the delivery of effector proteins into host cells. Pantoea stewartii subsp. stewartii (herein referred to as P. stewartii), the causative agent of Stewart's bacterial wilt and leaf blight of maize, carries phylogenetically distinct T3SSs. In addition to an Hrc-Hrp T3SS, known to be essential for maize pathogenesis, P. stewartii has a second T3SS (Pantoea secretion island 2 [PSI-2]) that is required for persistence in its flea beetle vector, Chaetocnema pulicaria (Melsh). PSI-2 belongs to the Inv-Mxi-Spa T3SS family, typically found in animal pathogens. Mutagenesis of the PSI-2 psaN gene, which encodes an ATPase essential for secretion of T3SS effectors by the injectisome, greatly reduces both the persistence of P. stewartii in flea beetle guts and the beetle's ability to transmit P. stewartii to maize. Ectopic expression of the psaN gene complements these phenotypes. In addition, the PSI-2 psaN gene is not required for P. stewartii pathogenesis of maize and is transcriptionally upregulated in insects compared to maize tissues. Thus, the Hrp and PSI-2 T3SSs play different roles in the life cycle of P. stewartii as it alternates between its insect vector and plant host.
INTRODUCTION
The type III protein secretion systems (T3SS) of Gram-negative bacterial pathogens inject effector proteins into the cytosol of eukaryotic cells in order to modulate host cell defenses, enabling successful pathogen colonization and growth (45, 50). The host defense and surveillance systems impose intense selective pressures on T3SSs, resulting in a host-pathogen coevolutionary arms race with diversifying selection acting on the effectors (26, 50). The T3SS apparatus is composed of 20 to 25 structural proteins organized into three substructures: a transmembrane channel, a cytoplasmic domain, and an external injection needle, collectively known as an injectisome (4). Five T3SS families, adapted to colonize either animals or plants, can be distinguished (54). The T3SSs of animal pathogens are grouped into three families: the Inv-Mxi-Spa family, which enables bacterial cell invasion and survival; the Ssa-Esc family, which is required for intracellular replication of bacterial pathogens; and the Ysc family, which allows extracellular manipulation of animal cells by bacteria. The hrc-hrp-encoded T3SSs of plant pathogens cluster into two families, Hrc-Hrp1 and Hrc-Hrp2 (54), which generate thin pili designed to traverse the thick plant cell wall, enabling pathogen colonization of susceptible hosts (32). Interestingly, some groups of bacterial pathogens possess two T3SS clusters, each playing distinct roles in the same host at different phases of pathogenesis (13, 48). However, the roles of multiple T3SSs in bacterial pathogenesis and ecology, especially with regard to multiple hosts, are not well understood.
Pantoea stewartii subsp. stewartii Smith (referred to here as P. stewartii), the causative agent of Stewart's bacterial wilt and leaf blight of maize (Zea mays L.), requires a Hrc-Hrp1 family T3SS for plant pathogenesis (23, 27). Although this pathogen can be mechanically transmitted to maize under laboratory conditions by wounding, in nature it is largely dependent on its maize flea beetle vectors, predominantly Chaetocnema pulicaria (Melsh) and Chaetocnema denticulate (Illiger), for transmission, dissemination, and overwintering (15, 16). P. stewartii does not have a saprophytic phase in its life cycle, and it is thought to persist in the alimentary tracts of adult flea beetles that overwinter in the soil, thus enabling its transmission to new plants in the spring (16). P. stewartii is also suspected to overwinter as an endophyte within alternative grass hosts (16). Although several studies on P. stewartii transmission by its insect vector have been conducted (15, 16, 18), the genetic mechanisms that enable this bacterium-insect association are unclear.
In this study, we report that P. stewartii carries a second T3SS that is required for persistence in the gut of its flea beetle vector. Inactivation of this second T3SS reduced the bacterium's ability to persist within the insect vector so drastically that the beetles were subsequently unable to transmit an effective inoculum dose to maize plants. Complementation studies partially reversed these phenotypes. Our results demonstrate that this T3SS plays an important role in successful transmission of P. stewartii by its insect vector and suggest different roles for the two known P. stewartii T3SSs in the life cycle of this pathogen as it alternates between insect and plant hosts.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. P. stewartii strains were grown at 28°C in Luria-Bertani (LB) broth or on agar supplemented with appropriate antibiotics [nalidixic acid (Nal), 20 μg/ml; kanamycin (Km), 50 μg/ml; and tetracycline (Tc), 25 μg/ml]. Dickeya dadantii, Pantoea ananatis, and Escherichia coli were grown as previously described (10, 35).
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or phenotype | Source or reference |
|---|---|---|
| E. coli | ||
| DH10B | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 ΔlacX74 endA1 recA1 deoRΔ (ara,leu)7697 araD139 galU galK nupG rpsL λ− | Gibco BRL |
| HB101 | F− thi-1 hsd20 (rB− mB−) supE44 recA13 ara-14 leuB6 proA2 lacY1 rpsL20 (Smr) xyl-5 mtl-1 | 5 |
| S17-1 | pro thi, chromosomal tra operon and recA from plasmid RP4 | 49 |
| P. stewartii | ||
| DC283 | A Nalr derivative of wild-type strain SS104 | 8 |
| DC440 | Wild type, isolated in 1999 in Wooster, OH | 10 |
| DM223 | wceG cpsA glycosyl transferase mutant; EPS partial mutant | 9 |
| DM5044 | DC283 yhiN∷mini-Tn5-gus Nalr Kmr | This study |
| DM5106 | DC283 sapD∷mini-Tn5-gus Nalr Kmr | This study |
| DM5107 | DC283 mtlA∷mini-Tn5-gus | This study |
| DM5108 | DC283 sicA∷mini-Tn5-gus Nalr Kmr | This study |
| DM5115 | DC283 ysaG∷mini-Tn5-gus Nalr Kmr | This study |
| DM5121 | DC283 psaN1∷mini-Tn5-gus Nalr Kmr | This study |
| DM5123 | DC283 lepA∷mini-Tn5-gus Nalr Kmr | This study |
| DM5125 | DC283 pilD∷mini-Tn5-gus Nalr Kmr | This study |
| DM 5126 | DC283 ospC∷mini-Tn5-gus Nalr Kmr | This study |
| DM5127 | DC283 kuP∷mini-Tn5-gus Nalr Kmr | This study |
| DM5130 | DC283 ackA∷mini-Tn5-gus Nalr Kmr | This study |
| DM7003 | DC283 psaN2∷aphA3 Nalr Kmr | This study |
| ESΔR | Hypermucoid, esaR | 3 |
| P. ananatis DC131 | Wild type, isolated from maize in Missouri in 1976 | 10 |
| D. dadantii EC16 | Wild type | 35 |
| Plasmids | ||
| pBluescript SK(+) | ColE1 α-lacZ Apr | Stratagene |
| pDM3007 | 1.4-kb BamHI/KpnI fragment containing Plac-ysaN+ cloned into pRK415; Tcr | This study |
| pLD55 | Suicide vector with R6K ori, α-lacZ, tetAR, Apr | 42 |
| pMM6 | pRK415 containing Plac-hrpL+ | 41 |
| pRK415 | IncP a-lacZ, Tcr | 34 |
| pRK2013∷Tn7 | ColE1 mob+ traRK2 ΔrepRK2 repE kan∷Tn7 Tpr Smr Spr | 19 |
| pUC18K | pUC18 with promoterless, terminatorless aphA cassette; Kmr | 39 |
| pUTmini-Tn5-gus | Mini-Tn5-gus in suicide vector pGP704; Apr Kmr | 22 |
Virulence and hypersensitivity assays.
Maize seedlings (Zea mays var. rugosa, cv. ‘Seneca Horizon’) and tobacco (Nicotiana tabacum cv. ‘Bottom Special’) were grown and maintained in growth chambers at 30°C with an 18-h/6-h day/night cycle. Virulence assays for P. stewartii mutants in maize seedlings and hypersensitivity assays were performed as previously described (23, 27).
Collection of maize flea beetles.
Flea beetles (C. pulicaria) cannot be reared in the laboratory and were collected for each experiment in and around maize fields in Wooster, OH (40°46′3″N, 81°54′20″W), throughout the growing season. In each collection, subsamples of beetles were used for confirming species identity. Prior to P. stewartii colonization, persistence, and transmission experiments, beetles were held for at least 10 days on healthy maize at 27°C and 50% relative humidity with a 16-h photoperiod. Only beetles kept on plants that remained free from Stewart's wilt symptoms were used in subsequent experiments.
Detection of P. stewartii in field-collected flea beetles using viable cell counts and cytology.
In order to estimate the proportion of flea beetles naturally carrying P. stewartii, flea beetles were sampled (n = 25 to 60 for each date) (Table 2) from the fields described above in spring, summer, late summer, and fall of 2007 and 2008. Single insects were ground in 300 μl 0.01 M potassium phosphate buffer, pH 7, for 15 s. Serial dilutions of the homogenates were plated on Ivanoff's semiselective medium for P. stewartii (325 mM glycerol, 35 mM ferric ammonium citrate, 256 mM NaCl, 17.6 mM Na2SO4, 14.4 mM K2HPO4, 0.9 mM CaCl2, 0.4 mM MgSO4 · 7H2O, 5.57 mM sodium taurocholate, 1.5% agar) (31), and the number of CFU was determined. P. stewartii identity was verified for three randomly selected yellow colonies per insect by species-specific PCR (8). Using toothpicks, single colonies were dipped into 25-μl reaction mixtures containing 1× Green GoTaq reaction buffer (Promega, Madison, WI) (1.5 mM MgCl2, 2.5 mM deoxynucleoside triphosphates [dNTPs]), 20 pmol of each primer (ESR1-f, 5′ CGAAGCGAGGACACACG 3′, and ESIG2-r, 5′ GCGCTTGCGTGTTATGAG 3′), and 2.5 U Taq DNA polymerase. PCR conditions were: 3 min at 94°C followed by 35 cycles of 1 min at 94°C, 1 min at 55°C, and 3 min at 72°C. PCR products were separated in a 1% agarose gel. Additional insect samples (n = 139) were dissected and analyzed by immunofluorescence confocal microscopy (see below) (Table 2).
Table 2.
Proportion of field-collected flea beetles (FLB) naturally infected with Pantoea stewartii
| Collection date | No. ofP. stewartii-positive FLB/total (%) | Gut region | No. positive/total (%) |
|---|---|---|---|
| June 2007 | 5/30 (16.7)a | NDc | ND |
| August 2007 | 15/60 (25.0)a | ND | ND |
| Sept. 2007 | 1/30 (3.3)a | ND | ND |
| Sept./Oct. 2007 | 3/91 (3.3)b | Foregut | 0/23 (0) |
| Midgut | 0/24 (0) | ||
| Hindgut | 2/23 (8.7) | ||
| Mtd | 1/21 (4.8) | ||
| April 2008 | 2/25 (8.0)a | ND | ND |
| Sept./Oct. 2008 | 1/48 (2.0)b | Foregut | 0/12 (0) |
| Midgut | 1/14 (0.07) | ||
| Hindgut | 0/11(0) | ||
| Mt | 0/11 (0) |
P. stewartii was detected using semiselective medium and verified using PCR.
P. stewartii was detected using immunofluorescence confocal microscopy of beetle guts.
ND, not determined.
Mt, Malpighian tubules.
Transposon mutagenesis, high-throughput genetic screen, and sequencing.
Transposon mutagenesis was performed by filter mating of 2 × 109 S17-1 pir (pUT mini-Tn5-gus) cells with 4 ×109 DC283 cells for 18 h at 28°C on LB, followed by plating of the mating mixture onto 150-mm-diameter LB plates containing Nal, Km, and X-Gluc (50 μg/ml 5-bromo-4-chloro-3-indolyl-ß-d-glucuronic acid) to obtain ca. 500 to 1,000 colonies on each plate. After 4 days at 28°C, colonies were selected using a Genetix QPix robotic colony picker (Genetix USA, Boston, MA) and arrayed in 384-well microtiter plates containing 50 μl of buffered LB plus 4.4% glycerol per well. A charge-coupled device (CCD) camera (Hamamatsu Photonics, Hamamatsu City, Japan) was calibrated to preferentially pick blue colonies expressing uidA in order to eliminate from the screen most gene fusions that were not constitutively expressed in a rich medium. About 10,000 GUS+ DC283∷mini-Tn5-gus mutants were stored at −20°C after growth in the microtiter plates at 28°C for 48 h. To genetically screen DC283∷mini-Tn5-gus strains for repression of the gene fusion by HrpL, each microtiter plate was replicated onto an LB Nal Km X-Gluc plate (plate A) and onto an LB plate containing a lawn of E. coli DH10B (pMM6) and HB101 (pRK2013∷Tn7) mixed in a 1:1 ratio. This was done to introduce pMM6, which constitutively expresses hrpL+ in P. stewartii, into the mini-Tn5-gus mutants by triparental mating. pMM6 transconjugants were then selected by replica mating on LB Nal Kan Tc X-Gluc agar (plate B). Downregulated (GUS−) yellow mutants observed in plate B were selected from plate A after alignment and visual comparison. This resulted in the selection of 28 GUS− mutants that were checked again for downregulation of GUS on LB Nal Km Tc X-Gluc agar plates.
The transposon-genome junctions on each side of the mini-Tn5-gus insertion site for the 28 candidate HrpL-down-regulated mini-Tn5-gus insertion mutants were sequenced with the primers 5′GUS (5′CATTTCACGGGTTGGGGTTTCT3′) and O-END-mTn5 (5′CCGCACTTGTGTATAAGAGTCAG3′) using an ABI 3700 automated DNA sequencer at the Plant-Microbe Genomic Facility, The Ohio State University. Junction sequences were BLAST searched against the unannotated P. stewartii genome sequence (Human Genome Sequencing Center, Baylor College of Medicine [https://www.hgsc.bcm.edu/content/pantoea-stewartii]), and the full-length wild-type amino sequences from each corresponding gene found in the Baylor genome sequence were searched against the GenBank nonredundant (nr) database using BLASTp. (In 2012, 65 contigs from the P. stewartii genome project, which were obtained by whole-genome shotgun sequencing, were deposited in GenBank under accession numbers AHIE01000001 to AHIE01000065, along with most of the protein sequences resulting from this search.)
Cloning and site-directed mutagenesis of psaN.
The psaN gene was amplified by PCR using primers psaN-5′-Eco (GGAATTCCACGGCCACCTGGAGCTTCGC) and psaN-3′-Xho (CCGCTCGAGCGGCGATATCCTGCTGTAATACCTG). The 1,457-bp amplicon was verified by sequencing. It was then cut with EcoRI and XhoI and ligated into the corresponding sites of pBluescript SK. The BamHI/KpnI-cut insert from this plasmid was then recloned into pRK415 to make plasmid pDM3007, which was then used for complementation studies. A nonpolar psaN mutation was constructed by ligating the aphA3 cassette from pUC18K (39) into the HpaI site of pDM3004, which is located 335 bp from the start of psaN. The psaN∷aphA3 mutation was then subcloned into suicide plasmid pLD55 as a BamHI/KpnI fragment. The resulting plasmid was used to exchange the mutant allele into the P. stewartii DC283 chromosome, as previously described (41), to create strain DM7003. The replacement was confirmed by PCR using primers to amplify the psaN gene.
Antibody production.
Antigen was prepared as described by Tsuchiya et al. (55). Wild-type P. stewartii DC283 was grown in LB Nal broth to an optical density at 600 nm (OD600) of ∼1.0. Cells were washed three times in phosphate-buffered saline (PBS) [0.01 M potassium phosphate, 0.15 M NaCl (pH 7.4)] to remove exopolysaccharide slime and capsule and then resuspended in PBS at an OD600 of ∼0.6. Bacteria were fixed in 0.5% formaldehyde overnight at 4°C, washed once with PBS, then resuspended in sterile saline, and stored at −80°C. Antibodies were produced in two New Zealand White rabbits as described previously (28). Each rabbit was injected with a mixture of 500 μl of whole-cell suspension and 500 μl of Freund's incomplete adjuvant (Sigma-Aldrich, St. Louis, MO). Two booster infections were done at 2-week intervals, with 7 ml blood collected just prior to the booster injections for testing antibody titers by Western blotting. Animals were exsanguinated at 9 weeks after the first injection, and the resulting sera were centrifuged at 1,200 × g for 15 min at 4°C. The clarified antisera were cross-absorbed with a 1:1:1 mixture of Dickeya dadantii EC16, Pantoea ananatis DC131, and E. coli HB101 as described previously (47). Antisera were diluted in a 1:1 (vol/vol) with 100% glycerol, lyophilized, and stored at −20°C.
Beetle dissection and P. stewartii immunolocalization.
To assess P. stewartii colonization of beetle intestinal tracts, beetles collected from the field were quarantined for at least 10 days on healthy maize (see above) and then were exposed to plants infected with wild-type P. stewartii or the P. stewartii psaN mutant for 2 days for acquisition and then transferred to healthy maize every 2 days. Individual beetles (n = 7 to 9) were harvested at 2 h and 4, 8, and 12 days after transfer to healthy plants for dissection of their intestinal tracts. Beetles were dissected under a stereomicroscope in 0.01 M potassium phosphate buffer, pH 7, using two fine-tip forceps. The head, with the foregut and part of the midgut attached to it, was pulled forward and separated from the thorax. Then, the rear end of the abdomen, with the hindgut and the rest of the midgut attached to it, was pulled backward and freed from the cuticle surrounding the abdomen. The dissected organs were fixed, washed, and permeabilized as described previously (1). For labeling with antibody, insect organs were immersed in blocking buffer [phosphate-buffered saline plus 0.1% Triton X-100 (PBS-T), containing 5% goat serum] for 30 min and then incubated with incubation buffer [1:500 (vol/vol) dilution of P. stewartii antiserum, containing 1% goat serum] for 3 h at room temperature. Tissues were washed three or four times with PBS-T, incubated for 1 h in a 1:600 dilution of goat anti-rabbit IgG conjugated with Alexa Fluor 488 (Molecular Probes, Eugene, OR), washed three or four times with PBS-T, and then stained for 5 min with 3 nM propidium iodide (Molecular Probes). Samples were examined using a confocal laser scanning microscope (Leica TCS SP), with parameters set as previously described (1). The percentage of infected insects was compared between the wild type and mutants. For a more quantitative estimate of bacterial infection, the number of fluorescent bacterial foci was estimated using a scale of 0 to 4, in which 0 indicates no fluorescent foci, 1 indicates few weak foci, 2 indicates bright independent foci, 3 indicates dense distribution of bright independent foci, and 4 indicates wider fluorescence areas. The experiment was conducted three times in total. Statistical analysis was carried out with MINITAB (Minitab Inc.). Differences among treatments were examined using one-way analysis of variance (ANOVA), and Fisher's protected least significant difference test (FLSD) was used for means separation.
Assessment of P. stewartii persistence in beetles using viable cell counts.
Beetles were collected and held on maize seedlings for 10 days. About 80 flea beetles were placed on 15 to 20 maize seedlings that had been infected with either mutant or parental strains of P. stewartii (8) for 2 days to allow bacterial acquisition. To prevent reacquisition of bacteria, beetles were moved to healthy seedlings every 2 days. At 0, 4, 8, and 10 days after acquisition, 5 to 10 insects per treatment were starved for 2 h and then were ground in 0.01 M potassium phosphate buffer, pH 7. Serial dilutions were plated on LB agar amended with appropriate antibiotics: nalidixic acid (20 μg/ml) for wild-type and bacterial mutants; a combination of nalidixic acid and kanamycin (50 μg/ml) for the mini-Tn5-gus and aphA3 insertion mutants (sapD, psaN, and ΔpsaN); and nalidixic acid plus kanamycin and tetracycline (5 μg/ml) for the psaN+ complemented strain. The number of CFU per insect was calculated after incubation of LB plates at 28°C for 2 days. Three independent repetitions of the experiment were done. The log10 CFU was used for statistical analysis (MINITAB), using one-way analysis of variance (ANOVA). Fisher's protected least significant difference test (FLSD) was used for means separation.
Semiquantitative RT-PCR.
Total RNA was isolated from P. stewartii cells cultured overnight on LB medium (OD600 ∼ 0.6), P. stewartii-infected maize, and insects fed on P. stewartii-infected plants at 4 days postinfection. A two-step RT-PCR was conducted with primer pairs specific for the Hrp ATPase (hrp-F, 5′GCCCTTATCACACCCCTTTATCTC3′, and hrp-R 5′TTTTGCCCTCAGCACGAAAC3′) and the Pantoea secretion island 2 (PSI-2) PsaN ATPase (psaN-F, 5′AATGTCTGGTTCATCTCGCACAC3′, and psaN-R, 5′GCTCCTCAACAAACTCCGTCAC3′). Two hundred nanograms of total RNA from P. stewartii cultures and from maize extracts and 400 ng of total RNA from beetles were used for first-strand cDNA synthesis. PCR was carried out for 20, 30, 35, and 40 cycles. 16S rRNA gene primers (ESR1-f, 5′ CGAAGCGAGGACACACG 3′, and ESIG2-r, 5′ GCGCTTGCGTGTTATGAG 3′) (10) were used as positive controls, and RT-PCRs without the addition of reverse transcriptase were included as negative controls to assess potential DNA contamination.
Transmission studies.
Subsamples of insects from the above colonization experiment were used to assess subsequent transmission of the pathogen to maize seedlings. Individual 10-day-old seedlings were exposed to five beetles each from day 0 or day 8 after acquisition of bacteria for a 2-day transmission period. A total of five seedlings per treatment were used, and the experiment was repeated three times. Plant growth conditions were as described above. P. stewartii infection of seedlings was assessed at 10 days postinoculation by estimating the number of CFU in 0.3 g of symptomatic tissue or tissues surrounding flea beetle feeding sites. Leaves were cut into ca. 2- to 3-mm strips and incubated in 2 ml of 0.01 M potassium phosphate buffer (pH 7) for 15 min, and then serial dilutions were plated onto LB agar amended with 20 μg ml nalidixic acid to determine the total number of CFU. Differences among treatments were examined using one-way analysis of variance (ANOVA). Fisher's protected least significant difference test (FLSD) was used for means separation with the MINITAB package (Minitab Inc.).
Phylogenetic analyses.
Protein sequences of the ATPases of the injectisomes and FliL, the ATPase of the E. coli flagellum, were aligned using Muscle (14), and a tree was generated using the neighbor-joining algorithm (gap excluded) of ClustalX with 1,000 bootstraps (53). The output file was viewed in Dendroscope (30), and names were added in Adobe Photoshop CS3 (Adobe Systems Inc.). FliL was selected as an outgroup. Accession numbers of T3SS islands of bacterial species (in alphabetical order) were as follows: Aeromonas salmonicida subsp. salmonicida AscN, CAD56760; Bradyrhizobium japonicum RhcN, AAG60799; Bordetella pertussis Tohama I BscN, CAC79575; Burkholderia pseudomallei K96243 SctN, YP_111406; B. pseudomallei K96243 SpaL, YP_111547; Chlamydia trachomatis A/HAR-13 SctN, AAX50947; Chlamydophila pneumoniae AR39 StcN, AAF37934; Chromobacterium violaceum ATCC 12472 CsaV, NP_902273; Desulfovibrio vulgaris DP4 BscN, YP_961187; Erwinia amylovora HrcN, AAB06001; Erwinia tasmaniensis Et1/99 SpaL, YP_001907837; Escherichia coli EivC, ACD01068; E. coli EscN, AAK26715; E. coli IAI39 FliL, CAR17249; Mesorhizobium loti MAFF303099 HrcN, BAB52652; Pantoea agglomerans HrcN, CAC43015; Pantoea stewartii HrcN, ABB77414; P. stewartii PsaN, GQ249669; Pectobacterium carotovorum subsp. carotovorum HrcN, ABZ05778; Photorhabdus luminescens LscN, AAO18044; Pseudomonas aeruginosa PscN, AAB86534; Pseudomonas cichorii HrcN, ABA47275; Pseudomonas syringae pv. tomato HrcN, AAG33879; Ralstonia solanacearum GMI1000 HrcN, CAD18021; Rhizobium sp. NGR234 HrcN, AAB91948; Salmonella enterica subsp. enterica serovar Choleraesuis str. SC-B67 SsaN, AAX65342; S. enterica subsp. enterica serovar Choleraesuis str. SC-B67 SpaL, YP_217813; Shigella flexneri SpaL, NP_085312; Sodalis glossinidius InvC, AAS66861; Vibrio parahaemolyticus RIMD 2210633 VpaN, NP_798047; V. parahaemolyticus RIMD 2210633 YscN, NP_800848; Xanthomonas campestris pv. vesicatoria str. 85-10 HrcN, CAJ22063; Xanthomonas oryzae pv. oryzae HrpB6, AAN46406; Yersinia enterocolitica YscN 1, AAK69223; Yersinia enterocolitica YsaN 2, AAB69192; Yersinia pestis CO92 YscN, CAB54917; and Yersinia pseudotuberculosis YscN, AAA20119.
Nucleotide sequence accession number.
Sequence information for PSI-2 has been deposited in GenBank under accession no. GQ249669.
RESULTS
P. stewartii resides in the midgut and hindgut of field-collected beetles.
Although it has been suggested that P. stewartii overwinters in flea beetles (16), the specific site(s) where P. stewartii is located within the beetles was unknown. To investigate this, beetles were collected in and around maize fields at various times from April to October during 2007 and 2008. P. stewartii was detected in field-collected flea beetles by culturing beetle extracts on a semiselective medium, and the identities of P. stewartii-like single colonies were confirmed by PCR using P. stewartii-specific primers. P. stewartii was detected in some beetles collected in April, prior to local maize planting, but P. stewartii-positive insects were most frequent in late August (Table 2), consistent with previous observations (18). To examine the location of P. stewartii inside the beetles, C. pulicaria individuals that were collected in September and October 2007 and 2008 were subjected to immunofluorescence microscopy analyses using P. stewartii-specific antibodies. Labeling was detected in the beetle's midgut and hindgut, but not in the foregut (Table 2). These results indicate that P. stewartii predominantly colonizes the middle and posterior parts of the gut in C. pulicaria.
Identification of P. stewartii genes that are downregulated by the HrpL alternative sigma factor.
To find genes that might be important for bacterium-host interactions, P. stewartii DC283 was randomly mutated with the mini-Tn5-gus transposon (22) and screened for genes that were both up- and downregulated by the alternative sigma factor HrpL, which activates the Hrp regulon in planta (40). The screen for HrpL-activated genes identified mostly known hrp-hrc and wts genes and several candidate effector genes that were subsequently shown to be defective (M. Merighi, D. R. Majerczak, and D. L. Coplin, unpublished data). We also hypothesized that HrpL might be indirectly involved in the repression of genes that were either nonessential for or detrimental to plant pathogenesis. Initially, we did not expect that this screen would also turn up the genes described in this paper that promote insect colonization. The mini-Tn5-gus transposon makes uidA transcriptional fusions and contains stop codons in all reading frames just upstream of the β-glucuronidase (GUS) reporter gene. To screen for HrpL-downregulated genes, ca. 10,000 mini-Tn5-gus insertion mutants that exhibited GUS activity in a rich medium (LB) were screened for decreased GUS activity following introduction of a plasmid that constitutively expressed the wild-type hrpL (hrpL+) gene. Downregulated mutants were selected by visualizing the difference in GUS activity between the mutants with and without the hrpL+ plasmid on plates containing X-Gluc, resulting in the selection of 28 potential mutants. Sequencing the DNA flanking the mini-Tn5-gus insertion sites indicated that we had 12 independent insertions and that 10 of them corresponded to open reading frames in the incomplete P. stewartii genome sequence at Baylor College of Medicine.
Database searches revealed that in 11 of the mutants, the transposon had disrupted genes encoding proteins with similarities to the T3SS components SycD, MxiH, SpaL, and OspC1, ABC transporter components SapD/DppD and MtlA, prepilin peptidase PulO, acetate kinase AckA, potassium uptake protein Kup, FAD/NAD(P)-binding oxidoreductase YhiN, and translation elongation factor LepA (Table 3). The 12 independent mutants were fully virulent on maize, produced a hypersensitive reaction on tobacco, did not express GUS activity in hrp-inducing minimal medium, and grew normally in a minimal salts-glucose medium (see Table S1 in the supplemental material) and in LB (data not shown), indicating that these genes are not required for virulence in plants and that the mutations did not cause any growth defects.
Table 3.
Homology searches of transposon junctions of 12 HrpL-downregulated mini-Tn5-gus insertion mutants
| Insertion mutant | Gene name in P. stewartiia | Putative gene function in P. stewartii | Closest homologue (GenBank accession no.)b | Protein sequence length (aa); identity (E-value)c |
|---|---|---|---|---|
| DM5106 | sapD | Peptide transport system ATP-binding protein | Pantoea ananatis SapD (YP_00519557) | 331; 99 (0.0) |
| DM5107 | mtlA | PTS system, mannitol-specifc EII ABC component | P. ananatis MtlA (AER34649) | 644; 98 (0.0) |
| DM5108 | pchA* | Type III secretion chaperone protein | Yokenella regensburgei lcrH-sycD (EHM45774) | 184; 86 (4e-92) |
| DM5115 | psaG* | Type III secretion apparatus protein | Y. regensburgei mxiH (EHM45790) | 86; 85 (1e-37) |
| DM5121 | psaN* | Type III secretion cytoplasmic ATP synthase | Y. regensburgei invC (EHM45782) | 431; 89 (0.0) |
| DM5123 | lepA | GTP-binding protein | P. ananatis lepA (ADD78062) | 599; 99 (0.0) |
| DM5125 | pilD | Leader peptidase (prepilin peptidase)/N-methyltransferase | Yersinia enterocolitica pulO (CAC83039) | 162; 98 (6e-40) |
| DM 5126 | ospC1 | Type III secretion effector | Shigella flexneri ospC1 (AAL72322) | 480; 54 (6e-161) |
| DM5127 | kup | Kup system potassium uptake protein | P. ananatis kup (ADD77115) | 622; 97 (0.0) |
| DM5130 | ackA | Acetate kinase A/propionate kinase 2 | P. ananatis ackA (YP_005194842) | 400; 99 (0.0) |
| DM5044 | yhiN | FAD/NAD(P)-binding oxidoreductase | P. ananatis yhiN (AER30810) | 394; 97 (0.0) |
GenBank accession numbers for P. stewartii DC283 query sequences: sapD EHU00723; mtlA, EHT98766; pchA, ACT68037; psaG, ACT68022; psaN, ACT68029; lepA, EHT99695; ospC1, EHT98016; kup, EHU00817; ackA, EHT99841; and yhiN, EHT99058. *, gene within PSI-2 (Fig. 1A).
Nucleotide sequences of genome regions flanking the mini-Tn5-gus were compared with protein-coding genes of the P. stewartii genome sequence in ASAP (24) using BLASTx. The full-length protein sequence of each P. stewartii gene was then BLAST searched against the GenBank nr protein database using BLASTP, and the closest homologue in another species is indicated.
aa, amino acids.
Some HrpL-downregulated genes are located in a T3SS pathogenicity island.
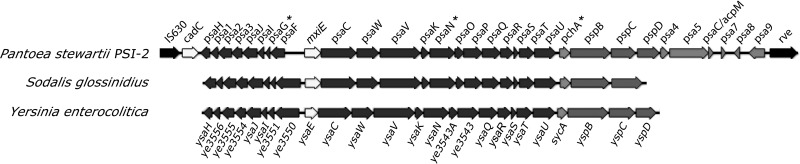
We searched the P. stewartii DC283 genome to determine the genomic context of the four T3SS genes identified in the transposon screen and found a gene cluster for a complete T3SS with gene order and sequence similarity to the Yersinia enterocolitica Ysa pathogenicity island (21), which is involved in the gastrointestinal phase of bacterial infection in mammals (56), and the Sodalis glossinidius SSR-1 pathogenicity island (11), which is required for bacterial invasion of insect gut cells (12) (Fig. 1). The transposon mutant screen identified three genes in the P. stewartii island with homology to S. glossinidius ysaG and Y. enterocolitica sycD and ysaN (Fig. 1; Table 3). We named this novel secretion system Pantoea secretion island 2 (PSI-2). The P. stewartii PSI-2 gene with similarity to Y. enterocolitica sycA and Yokenella regensburgei sycD was named pchA, the one with similarity to S. glossinidus ysaG was named psaG, and those similar to other ysa and ysp genes were named psa and psp, respectively (Fig. 1). P. stewartii PSI-2 pspB, pspC, and pspD are homologs of Shigella flexneri ipaB, ipaC, and ipaD, which are required for internalization and T3SS effector delivery (17, 39). PSI-2 is flanked by an IS630 insertion sequence and an rve integrase (Fig. 1), suggesting that PSI-2 integrated into the P. stewartii genome following horizontal gene exchange. The twelfth HrpL-downregulated transposon insertion in mutant DM5113 was located in the intergenic region between psa9 and rve.
Fig 1.
Organization of the Pantoea stewartii type III secretion system (T3SS), PSI-2. PSI-2 is aligned with T3SS pathogenicity islands from S. glossinidius (11, 12) and Y. enterocolitica (20, 21). PSI-2 genes that are downregulated by HrpL (Table 3) are indicated by asterisks. Putative gene functions include recombination and transposition (IS630 and rve), transcriptional regulators (cadC, mxiE, and ysaE), secretion apparatus structural (dark gray) and regulatory (light gray) functions (psa, ysa, and ye3543A), secreted effector chaperones (pchA and sycA), and secreted effectors (psp and ysp).
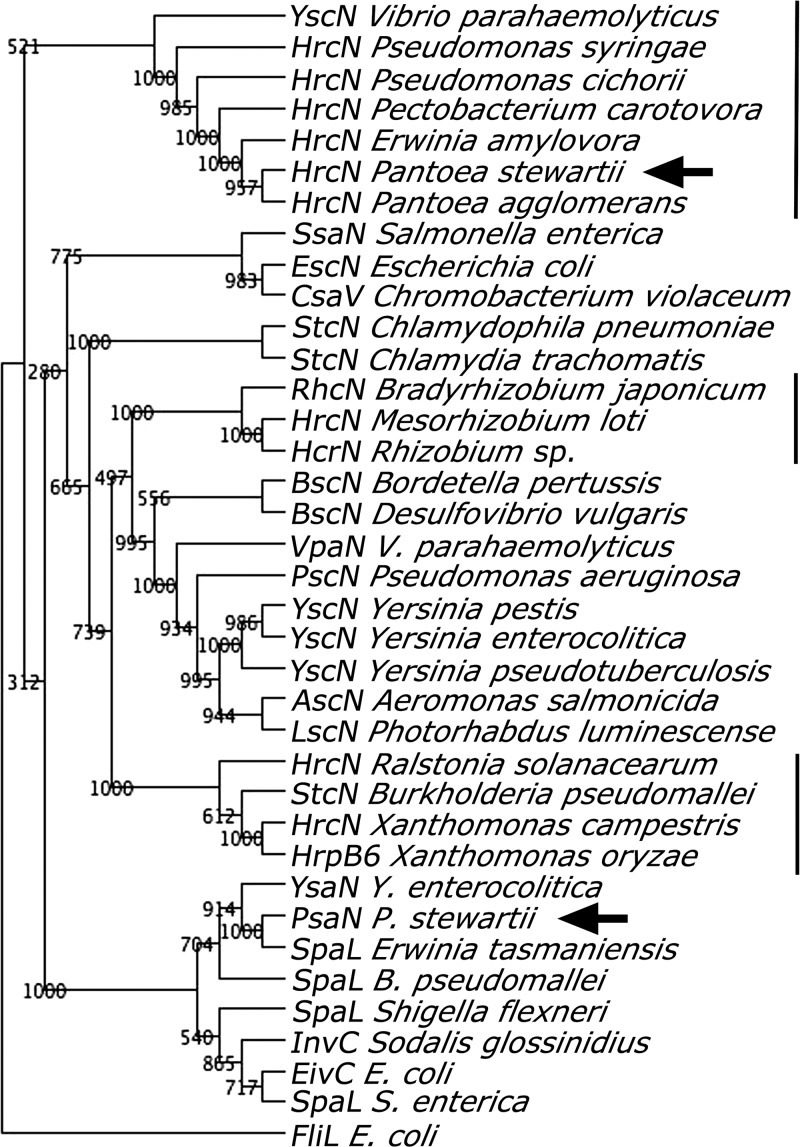
The genetic similarity of PSI-2 with T3SSs of other bacteria was examined using phylogenetic analysis of P. stewartii PsaN (GenBank accession no. ACT68029.1), which belongs to a conserved family of T3SS ATPases located in the bacterial inner membrane that are essential for building the pilus and for secretion of effectors (38, 44, 54). PsaN grouped phylogenetically with YsaN of Y. enterocolitica, SpaL of S. flexneri and InvC of the insect symbiont S. glossinidius (Fig. 2). In contrast, the HrcN ATPase homolog of the P. stewartii Hrc-Hrp system clustered with the HrcN homologs of other plant pathogens (Fig. 2). These findings indicate that P. stewartii PSI-2 belongs to the Inv-Mxi-Spa group of bacterial T3SSs required for bacterial invasion of animal cells (54).
Fig 2.
Phylogenetic analysis of type three secretion system (T3SS) ATPases from animal- and plant-associated microbes. Accession numbers for the gene and species names shown are given in Materials and Methods. Numbers to the left of the branches are bootstrap values for 1,000 replications, as outlined in Materials and Methods. Plant-associated microbes are designated by a solid line. The positions of the Pantoea stewartii hrcN from the hrc-hrp T3SS and psaN from PSI-2 are indicated by arrows.
In addition to the three PSI-2 genes, the mini-Tn5-gus screen identified a homolog of the S. flexneri ospC1 type III effector gene (Table 3) that is involved in postinvasion pathogenesis (60). A homolog of MxiE/YsaE, which activates ospC1 and other T3SS genes in Salmonella and Yersinia (33), was also present in P. stewartii PSI-2 (Fig. 1). OspC1 is involved in postinvasion pathogenesis of S. flexneri (60). In the P. stewartii genome, ospC1 is flanked by a gene similar to the integrase gene rve and by genes encoding hypothetical proteins that do not have similarities to protein sequences in the GenBank nr database. This suggests that, similar to PSI-2, OspC1 lies on a genomic segment that may have been introduced into the P. stewartii genome by horizontal gene exchange. The mini-Tn5-gus screen also identified a homolog of Salmonella enterica serovar Typhimurium ackA (Table 3), which encodes an acetate kinase that is required for producing the formate necessary for inducing the T3SS pathogenicity island 1 (29). Together these data suggest that P. stewartii has a second functional T3SS, as well as several unlinked genes that that are similarly downregulated by HrpL. We hypothesized that some of these genes might play a role in colonization of flea beetles by P. stewartii.
Expression of the PSI-2 psaN gene is increased in insects compared to maize tissues.
Our finding that P. stewartii psaN∷mini-Tn5-gus has decreased GUS expression in a rich medium (LB broth) upon introduction of wild-type hrpL or during growth in a hrp-inducing minimal medium suggests that psaN is highly expressed in a nutrient-rich environment, such as the insect gut, but not under conditions similar to those in the plant apoplast. Indeed, psaN transcripts were readily detected in wild-type DC283 grown in LB broth (see Fig. S1 in the supplemental material). In addition, investigation of the relative expression levels of PSI-2 psaN and Hrc-Hrp hrcN in P. stewartii-infected flea beetles and plants showed relatively higher psaN than hrcN expression in flea beetles, whereas the hrcN expression level was higher than that of psaN in maize (see Fig. S1). It was known that the Hrp-Hrc T3SS was expressed in plants but not in rich media. Therefore, we expected that it would not be highly expressed in the insect intestine. These results suggest that PSI-2 is less active in plants and is expressed in the insect and that it may have an important role in the insect host.
The PSI-2 T3SS is required for P. stewartii persistence in flea beetle guts.
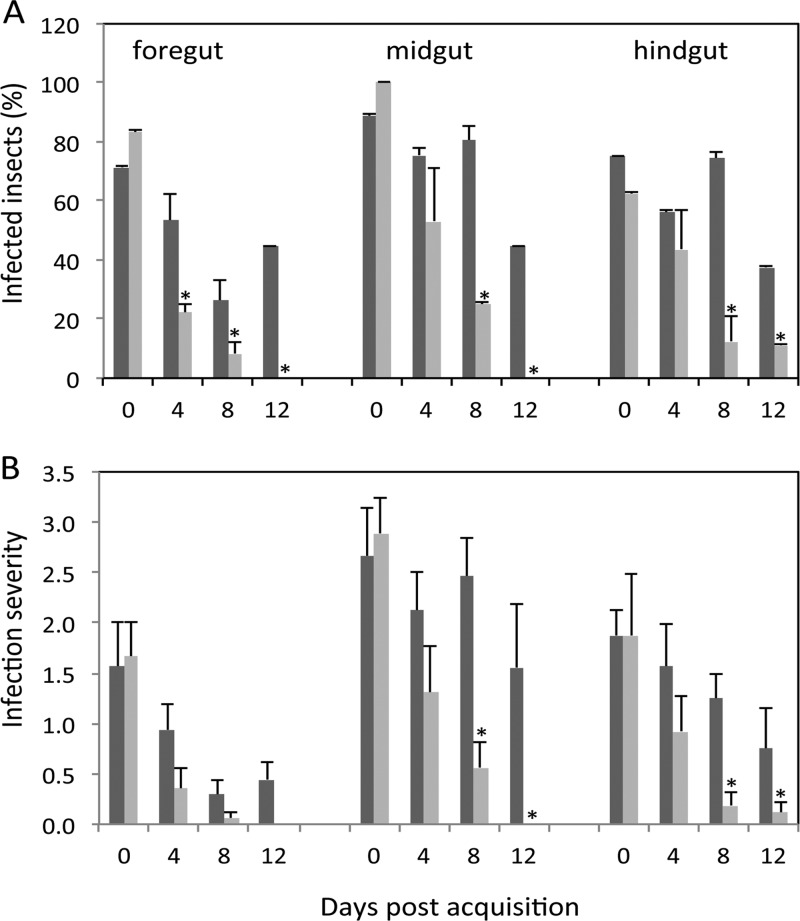
To further investigate PSI-2 involvement in P. stewartii flea beetle colonization, flea beetles were allowed to feed on maize infected with the psaN∷mini-Tn5-gus mutant (DM5121) and DC283 (wild-type). Immediately after feeding on infected maize plants (day 0), both mutant and wild-type bacteria were detected in the lumen of all gut regions by immunofluorescence confocal microscopy, including the foregut, midgut and hindgut of 71 to 90% of beetles (Fig. 3). However, 8 days later, the psaN mutant was detected in significantly fewer beetles than was the wild-type strain, and infested beetles had lower levels of mutant bacteria in the midgut and hindgut than wild-type bacteria (Fig. 4).
Fig 3.

The PSI-2 psaN is required for P. stewartii persistence in the flea beetle gut. (A) Diagram of flea beetle foregut (fg), midgut (mg), hindgut (hg), and Malpighian tubules (mt). (B to G) Immunochemical localization of P. stewartii (green fluorescence) in propidium iodide-stained guts (red fluorescence). Beetles fed on healthy control plants for 2 h (B) and 12 days (C), beetles fed on wild-type (DC283) P. stewartii-infected plants for 2 h (day 0) (D) and 12 h (E), and beetles fed on psaN-deficient (DM5121) P. stewartii for 2 h (day 0) (F) and 12 h (G) are shown. Bars, 100 µm.
Fig 4.
Persistence of wild-type and psaN-deficient P. stewartii in the flea beetle gut. Beetles were allowed to feed on maize infected with wild-type (DC283; dark gray) or psaN-deficient (DM5121; light gray) strains of P. stewartii for 2 days and then moved to healthy maize, as outlined in Materials and Methods. (A) Percentage of P. stewartii-infected beetles; (B) relative abundance of bacterial foci in beetle guts (rated on a scale of 0 to 4). Data are mean ± standard errors (SE) for 7 to 9 individuals from three biological replicates. *, significantly different (P < 0.05).
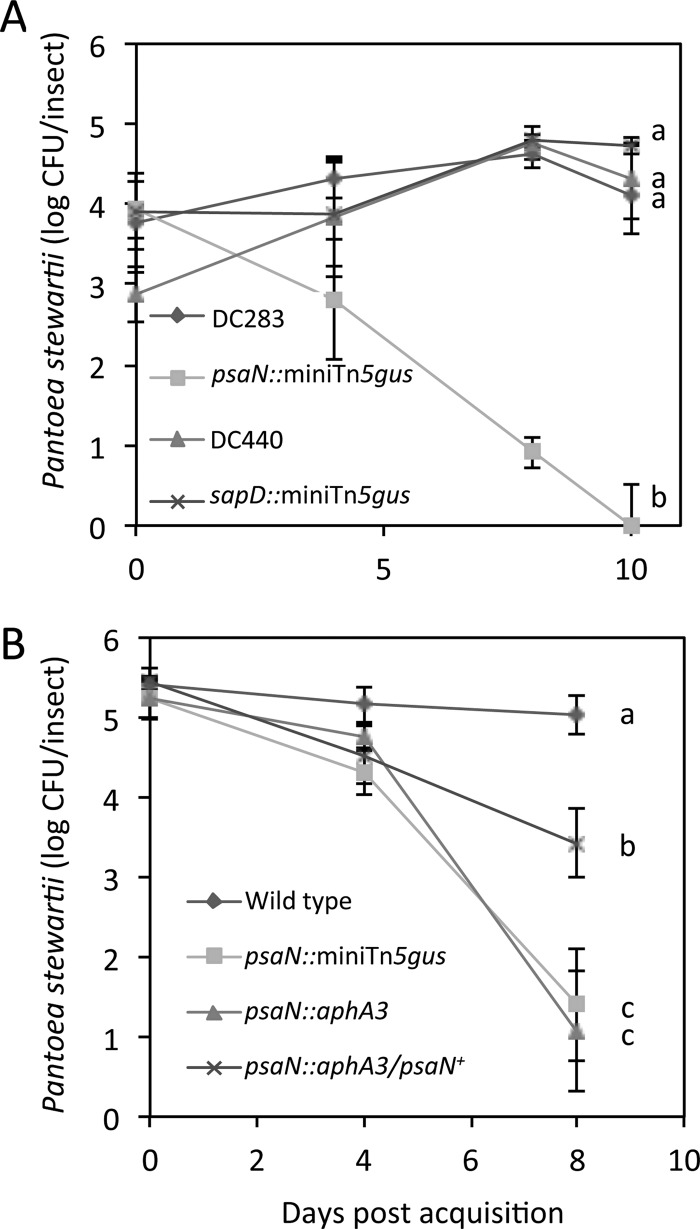
To obtain independent confirmation of these findings, we measured viable cell populations of the T3SS psaN mutant and the wild-type parent strain (DC283) during colonization of flea beetles. For comparison, we included a second, more recently isolated P. stewartii wild-type strain (DC440), since DC283 could have lost some of its ability to colonize insects after more than 40 years in storage. Since resistance to antimicrobial peptides might be a colonization factor, we also included the P. stewartii sapD∷mini-Tn5-gus (DM5106), which has a mini-Tn5-gus insertion in the Sap (sensitivity to antimicrobial peptides) ABC transporter gene sapD. The initial infection levels of the two mutants and the two wild-type strains were similar in beetles immediately after acquisition and increased by an average of 25% over a period of 10 days for DC283, DC440 and the sapD mutant (Fig. 5A). In contrast, the initial population level of the psaN mutant decreased to 72% on day 4, to 32% on day 8, and no viable cells were detected in the beetles on day 10. Thus, PSI-2 psaN appears to be required for P. stewartii persistence but not initial colonization of flea beetles, whereas sapD appears to have no apparent involvement in P. stewartii colonization and persistence.
Fig 5.
Mutation of psaN reduces persistence of colonization of P. stewartii in flea beetles. Beetles were allowed to feed on maize infected with two wild-type strains of P. stewartii (DC283 and DC440) or a psaN (DM5121) or sapD (DM5107) mutant strain. (B) Genetic complementation of the P. stewartii nonpolar psaN∷aphA3 mutant (DM7003) with a plasmid carrying wild-type PsaN (pDM3007) increases P. stewartii persistence in flea beetles. The number of viable P. stewartii cells was estimated as CFU from two or three beetle samples in each of three independent experiments. Data are mean log CFU per insect ± SE. Letters (a, b, and c) indicate significant differences in the number of CFU on day 10 (A) or 8 (B) (P < 0.05).
The psaN gene is located in the middle of an operon that contains a total of 13 genes. Thus, the mini-Tn5-gus insertion in the T3SS psaN∷mini-Tn5-gus mutant could affect expression of seven genes downstream of psaN. To investigate whether the observed mutant phenotype was associated with disruption of the psaN gene itself, a nonpolar aphA3 cassette was inserted into psaN after the codon for amino acid 111, then recombined into the DC283 chromosome to generate the nonpolar psaN mutant DM7003 psaN∷aphA3. After initial colonization of beetles, populations of both the T3SS psaN polar (psaN∷mini-Tn5-gus) and nonpolar (psaN∷aphA3) mutants started to decline at day 4 and were reduced to 27% and 20%, respectively, on day 8, whereas the infection levels of wild-type DC283 remained the same over 8 days (Fig. 5B). In contrast, a P. stewartii mutant in which the nonpolar psaN mutation in psaN∷aphA3 was genetically complemented with plasmid pDM3007 that constitutively expresses wild-type psaN (generating P. stewartii psaN∷aphA3/psaN+) had a population level of 63% that of the wild-type on day 8 (Fig. 5B), indicating that complementation with psaN+ alone can restore P. stewartii persistence in the flea beetles. However, persistence was not restored to full wild-type levels (Fig. 5B). Possible explanations for this may be that constitutive overexpression of psaN from a plasmid turns down PSI-2 expression by feedback repression, high levels of PsaN negatively affect growth, or pDM3007 may be slightly unstable in the insect gut. It was previously reported that the PsaN homologs from Yersinia (20) and Xanthomonas (38), YsaN and HrcN, respectively, are required for a functional T3SS. Thus, loss of PsaN probably disables the PSI-2 T3SS.
Reduced vector transmission of psaN mutants.
We also investigated if PSI-2 is involved in P. stewartii transmission by flea beetles to maize. Shortly after acquisition on day 0, the psaN∷mini-Tn5 polar mutant, the psaN∷aphA3 nonpolar mutant, the psaN+-complemented nonpolar mutant, and wild-type strain DC283 were transmitted equally well by the beetles to maize plants at 90 to 100% efficiency (Fig. 6). However, at 8 days after acquisition, the T3SS psaN polar and nonpolar mutants were transmitted at 10% and 12.5% efficiency, respectively, and wild-type P. stewartii and P. stewartii psaN∷aphA3/psaN+ were transmitted at 80% and 60% efficiency, respectively (Fig. 6). Thus, vector transmission of P. stewartii can also depend on a functional PSI-2.
Fig 6.
The PSI-2 psaN is required for persistent transmission of P. stewartii by flea beetles. Beetles were allowed to feed on maize plants infected with wild-type (DC283), psaN-defective strains (DM5121 and DM7003), or a rescued psaN2∷aphA3/psaN+ strain [(DM7003(pDM3007)] for 2 days. Individual maize seedlings were then exposed to five beetles each from day 0 or day 8 after acquisition of bacteria for a 2-day transmission period. Data are means ± SE for five seedlings per treatment in three independent experiments. Asterisks indicate means significantly lower than unmarked bars (P < 0.05).
DISCUSSION
In this study, we demonstrated that, in addition to the Hrc-Hrp1 family T3SS necessary for plant infection (23, 27), P. stewartii contains an Inv-Mxi-Spa type T3SS that is required for bacterial persistence in its insect vector and subsequent transmission by the vector to host plants. Other bacteria, including Yersinia, Salmonella, and Sodalis spp., contain multiple T3SSs in their genomes, but these T3SSs additively contribute to successful invasion of a single host (12, 13, 48, 57, 58). In contrast, the two T3SSs of P. stewartii enable it to colonize multiple hosts belonging to different kingdoms—i.e., plants and insects. The PSI-2 cluster is not needed for plant pathogenicity, and its expression in plants may be detrimental to pathogenesis (6). Thus, strict regulation of the two T3SS clusters is probably necessary, and both may involve HrpL. The HrpL sigma factor is a positive regulator of the plant pathogenicity hrc-hrp T3SS that is controlled by a regulatory cascade that senses plant and environmental signals (40). The mini-Tn5-gus mutant screen provided evidence that HrpL also downregulates several PSI-2 genes (pchA, psaG, and the ospC1 homologue ackA) and several other genes (Table 3). Since HrpL activates transcription as an alternate sigma factor, it is most likely that it acts indirectly on PSI-2 and the other genes by activating expression of one or more negative transcriptional regulators, global regulators, or feedback mechanisms.
In nature, P. stewartii is not transmitted by wind, rain, or cultural practices, cannot survive as an epiphyte or in plant debris, and is only rarely seed-borne. Consequently, it must persist in the vector gut until it can be successfully transmitted to maize. Since P. stewartii resides primarily in the flea beetle midgut and hindgut lumen, the most likely mode of transmission involves P. stewartii passage through the hindgut into feces, followed by mechanical introduction into plants via feeding wounds (7, 43). This is similar to the transmission paths of other insect-transmitted plant pathogens, e.g., Xanthomonas axonopodis and Erwinia tracheiphila (43, 59). Since this mode of transmission is primarily mechanical, it is unlikely that PSI-2 is required for the actual transfer of bacteria from feces to the interior tissues of the plant. Therefore, the inability of flea beetles to transmit the psaN mutants probably indicates that high gut populations are required for the insect to deliver an effective inoculum.
Bacterial genes that are involved in interactions with insect vectors (2, 36, 51) and insect hosts (25, 52) have been identified and recently reviewed by Nadarasah and Stavrinides (46). However, the involvement of a separate T3SS required for persistence in an insect vector and subsequent transmission has not previously been reported for plant or vertebrate pathogens. Genome sequencing has revealed multiple T3SSs in Gram-negative bacteria associated with (in)vertebrate animals and plants (12, 37, 48, 54). The nonpathogenic epiphytic bacterium Erwinia tasmaniensis Et1/99, which is closely related to P. stewartii, is found on apple and pear flower surfaces. It has a bipartite Inv-Mxi-Spa-like T3SS, in addition to an Hrc-Hrp T3SS (37), suggesting that it may have an (in)vertebrate animal host. However, this Inv-Mxi-Spa gene cluster is missing a structural gene (37) and hence may be nonfunctional. Some plant-pathogenic Xanthomonas spp. carry animal-type T3SSs and persist in insect vectors, but it is not known whether these T3SSs enable these bacteria to invade (in)vertebrate animals (59). Conversely, the human pathogens Vibrio parahaemolyticus and Burkholderia spp. harbor plant-type T3SS Hrc-Hrp genes, suggesting that these bacteria may use plants as alternate hosts (54). Our results suggest that the life cycles of these and other bacterial pathogens may include currently unknown alternate host species that are relevant to food security.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kristen Willie, Angela Strock, Jane Todd, Mark Zimmerman, and Cameron Exner for excellent technical support, David Mackey, J. C. Jang, Allan Downie, Eric Stockinger, Xiaodong Bai, and Sophien Kamoun for their helpful comments and suggestions on earlier versions of the manuscript, and Eric Cabot and Nicole Perna for submission of the PSI-2 sequence to GenBank.
This work was supported by USDA, CSREES, National Research Initiative grants 2002-35319-11562 to D.L.C., 2005-35319-15328 to D.L.C. and S.A.H., and 2008-35319-04506 to D.L.C. and D. Mackey and by state and federal funds appropriated to The Ohio State University (OSU), Ohio Agricultural Research and Development Center (OARDC).
Preliminary genomic sequence data were obtained from Baylor College of Medicine Human Genome Sequencing Center website at http://www.hgsc.bcm.tmc.edu. The genomic DNA sequencing project of Pantoea stewartii DC283 was supported by National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, grant number 0198694 to Nicole T. Perna, at the University of Wisconsin—Madison.
The mention of trade names does not imply that they are endorsed or recommended by OSU or USDA, ARS over similar products not mentioned.
Footnotes
Published ahead of print on 6 July 2012.
This publication was assigned no. HCS09-10 by the OARDC.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Ammar ED, Hogenhout SA. 2008. A neurotropic route for Maize mosaic virus (Rhabdoviridae) in its planthopper vector Peregrinus maidis. Virus Res. 131: 77–85 [DOI] [PubMed] [Google Scholar]
- 2. Basset A, Tzou P, Lemaitre B, Boccard F. 2003. A single gene that promotes interaction of a phytopathogenic bacterium with its insect vector, Drosophila melanogaster. EMBO Rep. 4: 205–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beck von Bodman S, Farrand SK. 1995. Capsular polysaccharide biosynthesis and pathogenicity in Erwinia stewartii require induction by an N-acylhomoserine lactone autoinducer. J. Bacteriol. 177: 5000–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blocker A, et al. 2001. Structure and composition of the Shigella flexneri “needle complex”, a part of its type III secreton. Mol. Microbiol. 39: 652–663 [DOI] [PubMed] [Google Scholar]
- 5. Boyer HW, Roulland-Dussoix D. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41: 459–472 [DOI] [PubMed] [Google Scholar]
- 6. Cesbron S, Paulin JP, Tharaud M, Barny MA, Brisset MN. 2006. The alternative sigma factor HrpL negatively modulates the flagellar system in the phytopathogenic bacterium Erwinia amylovora under hrp-inducing conditions. FEMS Microbiol. Lett. 257: 221–227 [DOI] [PubMed] [Google Scholar]
- 7. Claflin LE. 1999. Stewart's bacterial wilt, 3rd ed. American Phytopathological Society, St. Paul, MN. [Google Scholar]
- 8. Coplin DL, Frederick RD, Majerczak DR, Haas ES. 1986. Molecular cloning of virulence genes from Erwinia stewartii. J. Bacteriol. 168: 619–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coplin DL, Majerczak DR. 1990. Extracellular polysaccharide genes in Erwinia stewartii: directed mutagenesis and complementation analysis. Mol. Plant Microbe Interact. 3: 286–292 [Google Scholar]
- 10. Coplin DL, et al. 2002. Identification of Pantoea stewartii subsp. stewartii by PCR and strain differentiation by PFGE. Plant Dis. 86: 304–311 [DOI] [PubMed] [Google Scholar]
- 11. Dale C, Young SA, Haydon DT, Welburn SC. 2001. The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc. Natl. Acad. Sci. U. S. A. 98: 1883–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dale C, Jones T, Pontes M. 2005. Degenerative evolution and functional diversification of type-III secretion systems in the insect endosymbiont Sodalis glossinidius. Mol. Biol. Evol. 22: 758–766 [DOI] [PubMed] [Google Scholar]
- 13. Deiwick J, et al. 1998. Mutations in Salmonella pathogenicity island 2 (SPI2) genes affecting transcription of SPI1 genes and resistance to antimicrobial agents. J. Bacteriol. 180: 4775–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dereeper A, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36: W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elliot C, Poos FW. 1934. Overwintering of Aplanobacter stewartii. Science 80: 289–290 [DOI] [PubMed] [Google Scholar]
- 16. Elliott C, Poos FW. 1940. Seasonal development, insect vectors, and host range of bacterial wilt of sweet corn. J. Agric. Res. 60: 645–686 [Google Scholar]
- 17. Epler CR, Dickenson NE, Olive AJ, Picking WL, Picking WD. 2009. Liposomes recruit IpaC to the Shigella type III secretion apparatus needle as a final step in secretion induction. Infect. Immun. 77: 2754–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Esker PD, Nutter FW. 2003. Temporal dynamics of corn flea beetle populations infested with Pantoea stewartii, causal agent of Stewart's disease of corn. Phytopathology 93: 210–218 [DOI] [PubMed] [Google Scholar]
- 19. Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76: 1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Foultier B, Troisfontaines P, Muller S, Opperdoes FR, Cornelis GR. 2002. Characterization of the ysa pathogenicity locus in the chromosome of Yersinia enterocolitica and phylogeny analysis of type III secretion systems. J. Mol. Evol. 55: 37–51 [DOI] [PubMed] [Google Scholar]
- 21. Foultier B, et al. 2003. Identification of substrates and chaperone from the Yersinia enterocolitica 1B Ysa type III secretion system. Infect. Immun. 71: 242–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fouts DE, et al. 2002. Genomewide identification of Pseudomonas syringae pv. tomato DC3000 promoters controlled by the HrpL alternative sigma factor. Proc. Natl. Acad. Sci. U. S. A. 99: 2275–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Frederick RD, et al. 2001. Genetic organization of the Pantoea stewartii subsp. stewartii hrp gene cluster and sequence analysis of the hrpA, hrpC, hrpN, and wtsE operons. Mol. Plant Microbe Interact. 14: 1213–1222 [DOI] [PubMed] [Google Scholar]
- 24. Glasner JD, et al. 2003. ASAP, a systematic annotation package for community analysis of genomes. Nucleic Acids Res. 31: 147–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grenier AM, Duport G, Pages S, Condemine G, Rahbe Y. 2006. The phytopathogen Dickeya dadantii (Erwinia chrysanthemi 3937) is a pathogen of the pea aphid. Appl. Environ. Microbiol. 72: 1956–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guttman DS, Gropp SJ, Morgan RL, Wang PW. 2006. Diversifying selection drives the evolution of the type III secretion system pilus of Pseudomonas syringae. Mol. Biol. Evol. 23: 2342–2354 [DOI] [PubMed] [Google Scholar]
- 27. Ham JH, Majerczak DR, Arroyo-Rodriguez AL, Mackey DM, Coplin DL. 2006. WtsE, an AvrE-family effector protein from Pantoea stewartii subsp. stewartii, causes disease-associated cell death in maize and requires a chaperone protein for stability. Mol. Plant Microbe Interact. 19: 1092–1102 [DOI] [PubMed] [Google Scholar]
- 28. Harlow E, Lane D. 1998. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 29. Huang Y, Suyemoto M, Garner CD, Cicconi KM, Altier C. 2008. Formate acts as a diffusible signal to induce Salmonella invasion. J. Bacteriol. 190: 4233–4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huson DH, et al. 2007. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics 8: 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ivanoff SS. 1933. Stewart's wilt disease of corn, with emphasis on the life history of Phytomonas stewartii in relation to pathogenesis. J. Agr. Res. 47: 749–770 [Google Scholar]
- 32. Jin Q, He SY. 2001. Role of the Hrp pilus in type III protein secretion in Pseudomonas syringae. Science 294: 2556–2558 [DOI] [PubMed] [Google Scholar]
- 33. Kane CD, Schuch R, Day WA, Jr, Maurelli AT. 2002. MxiE regulates intracellular expression of factors secreted by the Shigella flexneri 2a type III secretion system. J. Bacteriol. 184: 4409–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Keen NT, Tamaki S, Kobayashi D, Trollinger D. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70: 191–197 [DOI] [PubMed] [Google Scholar]
- 35. Keen NT, Ridgway D, Boyd C. 1992. Cloning and characterization of a phospholipase gene from Erwinia chrysanthemi EC16. Mol. Microbiol. 6: 179–187 [DOI] [PubMed] [Google Scholar]
- 36. Killiny N, Almeida RP. 2009. Xylella fastidiosa afimbrial adhesins mediate cell transmission to plants by leafhopper vectors. Appl. Environ. Microbiol. 75: 521–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kube M, et al. 2008. The genome of Erwinia tasmaniensis strain Et1/99, a non-pathogenic bacterium in the genus Erwinia. Environ. Microbiol. 10: 2211–2222 [DOI] [PubMed] [Google Scholar]
- 38. Lorenz C, Buttner D. 2009. Functional characterization of the type III secretion ATPase HrcN from the plant pathogen Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 191: 1414–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Menard R, Sansonetti PJ, Parsot C. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175: 5899–5906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Merighi M, Majerczak DR, Stover EH, Coplin DL. 2003. The HrpX/HrpY two-component system activates hrpS expression, the first step in the regulatory cascade controlling the Hrp regulon in Pantoea stewartii subsp. stewartii. Mol. Plant Microbe Interact. 16: 238–248 [DOI] [PubMed] [Google Scholar]
- 41. Merighi M, Majerczak DR, Zianni M, Tessanne K, Coplin DL. 2006. Molecular characterization of Pantoea stewartii subsp. stewartii HrpY, a conserved response regulator of the Hrp type III secretion system, and its interaction with the hrpS promoter. J. Bacteriol. 188: 5089–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Metcalf WW, et al. 1996. Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35: 1–13 [DOI] [PubMed] [Google Scholar]
- 43. Mitchell RF, Hanks LM. 2009. Insect frass as a pathway for transmission of bacterial wilt of cucurbits. Environ. Entomol. 38: 395–403 [DOI] [PubMed] [Google Scholar]
- 44. Moraes TF, Spreter T, Strynadka NC. 2008. Piecing together the type III injectisome of bacterial pathogens. Curr. Opin. Struct. Biol. 18: 258–266 [DOI] [PubMed] [Google Scholar]
- 45. Mueller CA, Broz P, Cornelis GR. 2008. The type III secretion system tip complex and translocon. Mol. Microbiol. 68: 1085–1095 [DOI] [PubMed] [Google Scholar]
- 46. Nadarasah G, Stavrinides J. 2011. Insects as alternative hosts for phytopathogenic bacteria. FEMS Microbiol. Rev. 35: 555–575 [DOI] [PubMed] [Google Scholar]
- 47. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 48. Shea JE, Hensel M, Gleeson C, Holden DW. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. U. S. A. 93: 2593–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology 1: 784–794 [Google Scholar]
- 50. Stavrinides J, McCann HC, Guttman DS. 2008. Host-pathogen interplay and the evolution of bacterial effectors. Cell Microbiol. 10: 285–292 [DOI] [PubMed] [Google Scholar]
- 51. Stavrinides J, McCloskey JK, Ochman H. 2009. Pea aphid as both host and vector for the phytopathogenic bacterium Pseudomonas syringae. Appl. Environ. Microbiol. 75: 2230–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stavrinides J, No A, Ochman H. 2010. A single genetic locus in the phytopathogen Pantoea stewartii enables gut colonization and pathogenicity in an insect host. Environ. Microbiol. 12: 147–155 [DOI] [PubMed] [Google Scholar]
- 53. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Troisfontaines P, Cornelis GR. 2005. Type III secretion: more systems than you think. Physiology (Bethesda) 20: 326–339 [DOI] [PubMed] [Google Scholar]
- 55. Tsuchiya K, d'Ursel CCM, Nozu YY. 2004. Production and preliminary characterization of monoclonal antibodies raised against Xanthomonas campestris pv. mangiferaeindicae. J. Gen. Plant Pathol. 70: 27–33 [Google Scholar]
- 56. Venecia K, Young GM. 2005. Environmental regulation and virulence attributes of the Ysa type III secretion system of Yersinia enterocolitica biovar 1B. Infect. Immun. 73: 5961–5977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Young BM, Young GM. 2002. Evidence for targeting of Yop effectors by the chromosomally encoded Ysa type III secretion system of Yersinia enterocolitica. J. Bacteriol. 184: 5563–5571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Young BM, Young GM. 2002. YplA is exported by the Ysc, Ysa, and flagellar type III secretion systems of Yersinia enterocolitica. J. Bacteriol. 184: 1324–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zandjanaku-Tachin M, Fanou A, Gall PL, Wydra K. 2007. Detection, survival and transmission of Xanthomonas axonopodis pv. manihotis and X. axonopodis pv. vignicola, causal agents of cassava and cowpea bacterial blight, respectively, in/by insect vectors. J. Phytopathol. 155: 159–169 [Google Scholar]
- 60. Zurawski DV, Mitsuhata C, Mumy KL, McCormick BA, Maurelli AT. 2006. OspF and OspC1 are Shigella flexneri type III secretion system effectors that are required for postinvasion aspects of virulence. Infect. Immun. 74: 5964–5976 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.