Abstract
Seven Propionibacterium freudenreichii strains exhibited similar responses when placed at 4°C. They slowed down cell machinery, displayed cold stress responses, and rerouted their carbon metabolism toward trehalose and glycogen synthesis, both accumulated in cells. These results highlight the molecular basis of long-term survival of P. freudenreichii in the cold.
TEXT
Propionibacterium freudenreichii is a bacterium of food and probiotic interest, widely used as a ripening culture in the manufacture of Swiss cheese varieties (4, 16). It grows in cheese during ripening at warm temperatures (20 to 24°C) but remains metabolically active during the storage of cheese at low temperatures (10). We previously investigated the adaptation strategies of P. freudenreichii type strain CIRM-BIA1T by -omic approaches under conditions mimicking cheese ripening in the cold (6). Our previous results suggest in particular that CIRM-BIA1T reroutes its metabolism toward glycogen synthesis. In the present study, we confirmed the actual accumulation of glycogen in cells and investigated the response in the cold of six other P. freudenreichii strains.
Choice of strains and culture conditions.
The transcriptomic response of six P. freudenreichii subsp. shermanii strains (CIRM-BIA9, CIRM-BIA118, CIRM-BIA122, and CIRM-BIA123 from CIRM-BIA [Centre International de Ressources Microbiennes—Bactéries d'Intérêt Alimentaire, INRA, Rennes, France] and CIRM-BIA472 and CIRM-BIA482 from Valio Ltd., Helsinki, Finland) was studied during their transfer from 30°C to 4°C under conditions mimicking cheese ripening, previously applied to strain CIRM-BIA1T (6). All experiments were made in triplicate independent cultures. The six strains were chosen with different sequence types (7) and phenotypes. For example, they produce methylbutanoate and ethyl propionate, two cheese aroma compounds, at concentrations varying by factors of 6 and 12, respectively, depending on the strain (data not shown).
Growth and metabolite production in the cold.
All the strains stopped their growth when placed at 4°C, whereas in the control cultures maintained at 30°C, cells went on growing for about 20 h (Fig. 1A). They went on producing propionate and acetate, the two main products of lactate fermentation, but at a markedly lower production rate in the cold (Fig. 1C and D) (3.4 ± 0.6 [mean ± standard deviation] mM per day at 4°C versus 76 ± 15 mM per day at 30°C, i.e., a 23- ± 6-fold decrease for propionate). The rate of methylbutanoate production also decreased but at a markedly lower extent (from 69 ± 55 μM per day at 30°C to 12 ± 12 μM per day at 4°C, i.e., a mean fold decrease of 7 ± 4) (Fig. 1B).
Fig 1.
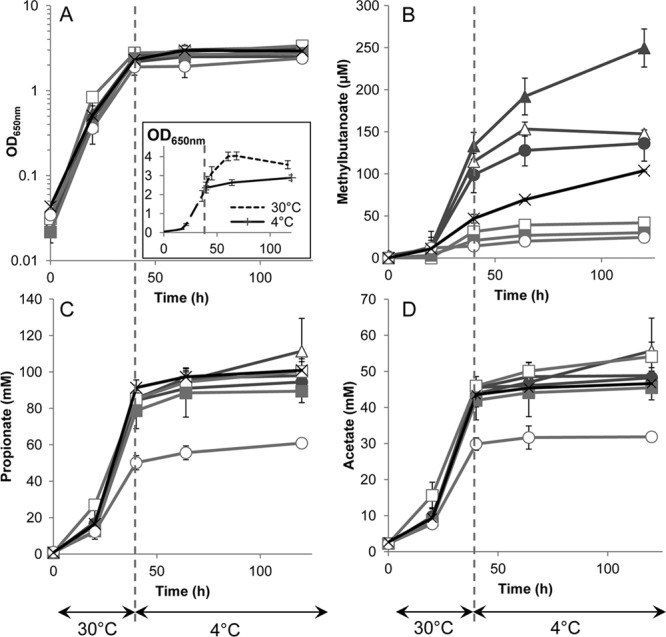
Time course of metabolic activity of seven P. freudenreichii strains over a 40-h incubation at 30°C followed by a further 80 h at 4°C. Growth (OD650nm, optical density at 650 nm) (A), concentrations of methylbutanoate (sum of 2-methylbutanoate and 3-methylbutanoate) (B), propionate (C), and acetate (D). Error bars show the standard deviations of the results of triplicate independent experiments. The inset in panel A shows the growth curves at 4°C and 30°C. Values are means for the 7 strains: CIRM-BIA1T (×), CIRM-BIA9 (△), CIRM-BIA118 (□), CIRM-BIA122 (▲), CIRM-BIA123 (○), CIRM-BIA472 (●), CIRM-BIA482 (■).
Transcriptomic approach applied to all strains.
Gene expression after an 80-h period at 4°C (t = 120 h) was compared to that at 20 h during growth at 30°C for the 6 strains, using the methodology and microarrays previously described for strain CIRM-BIA1T (6) (NCBI GEO, http://www.ncbi.nlm.nih.gov/geo/, platform accession number GPL13959). The transcriptomic data for CIRM-BIA1T at sampling times 20 h and 3 days (accession number GSE30841) were added to the new data set (six strains, accession number GSE34227) to facilitate the comparison between the present and previous results. Microarray data were normalized and analyzed as previously described (6). An analysis of variance (ANOVA) was performed to evaluate the effects of time, strain, and their interactions on expression. Raw P values were adjusted for multiple comparisons by the Benjamini-Hochberg procedure. Since the microarray used was designed from the genome of strain CIRM-BIA1T, we first checked the quality of hybridization with DNA of all the strains used, to avoid any bias in the interpretation of results due to possible mismatches between the oligonucleotides and the DNA sequence of the 6 other strains. DNA was extracted from pure cultures as previously described (10). A signal intensity of >8 (expressed as log2) was obtained for all oligonucleotides using DNA from CIRM-BIA1T, whereas a low signal intensity (<6) was observed using DNA from the other strains for a small number of oligonucleotides. Therefore, we discarded from the data set the 281 genes for which 50% or more of the oligonucleotides targeting a gene showed a signal intensity of <6 for at least one strain. This resulted in a final data set consisting of 88% of the 2,300 genes targeted in the microarray. Significant (P < 0.01) changes in expression exceeding 2× (i.e., |fold change (log2)| > 1) for at least one strain were considered differentially expressed (DE), resulting in 1,079 DE genes.
A similar transcriptomic response for all strains.
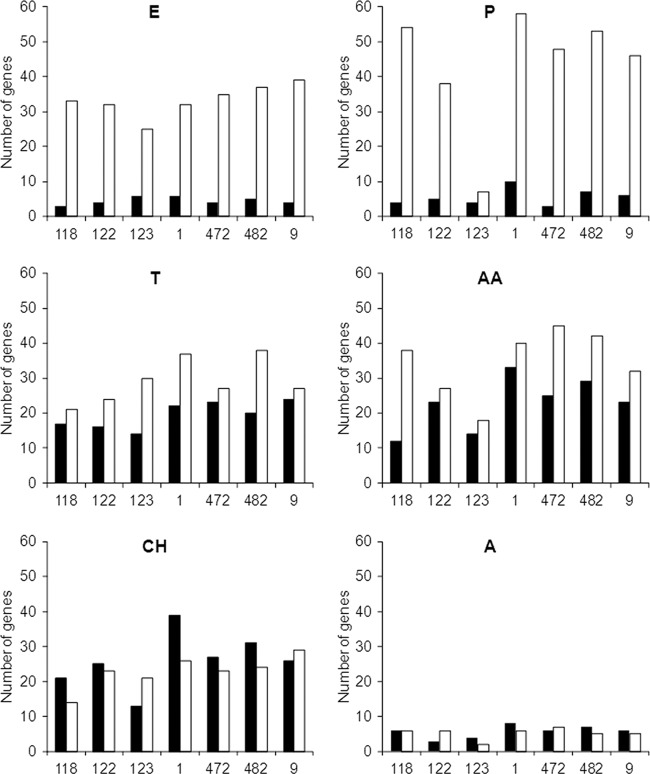
Like CIRM-BIA1T (6), the 6 strains downregulated most of the DE genes related to the general cell machinery, such as genes involved in energy production and protein synthesis, whereas both down- and upregulated genes were observed in some gene categories, like transport and metabolism of amino acids and carbohydrates (Fig. 2). The main features are briefly described below.
Fig 2.
Number of differentially expressed genes (|fold change| > 1) after 80 h at 4°C in comparison with gene expression at the reference time of 20 h for seven P. freudenreichii strains (CIRM-BIA118, -122, -123, -1, -472, -482, and -9). Downregulated (white bars) or upregulated (black bars) genes with known functions are presented according to their metabolic category: E, energy metabolism; P, protein synthesis; T, transport of peptides and inorganic ions; AA, transport and metabolism of amino acids; CH, transport and metabolism of carbohydrates; A, adaptation to atypical conditions.
General slowdown of cell machinery and cold stress response.
All strains slowed their metabolism, as indicated, for example, by the downregulation of ftsX, involved in cell division (fold changes ranging from −1.7 to −4.3) (Table 1), and of most of the genes involved in energetic metabolism (Table 1). Genes involved in the conversion of pyruvate into propionate (sdhABC and pccB) and into CO2 and acetate (aceE and lpd) were also downregulated at 4°C (Table 1). Many bacteria exhibit a general slowdown in the cold; for example, Lactococcus lactis in model cheeses when placed at 12°C (5). The 6 strains exhibited cold stress responses similar to those of CIRM-BIA1T (6) and other bacteria (2, 15). For example, the genes cspA and cspB, encoding cold shock proteins, were upregulated, as well as DEAD box RNA helicases, which facilitate translation and, thus, protein synthesis in the cold (Table 2). In contrast, several chaperone- and heat shock protein-coding genes (groSL and dnaKJ operons and hsp20, clpB2, and grpE) were downregulated at 4°C for most strains (Table 2).
Table 1.
Differentially expressed genes involved in general cell machinery slowdown
| Name | Locus taga | Description | Categoryb |
P value |
Fold change (log2) for CIRM-BIA strainc: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time | Strain | Time × strain | 118 | 122 | 123 | 1 | 472 | 482 | 9 | ||||
| cstA | PFREUD_16500 | Carbon starvation protein | A | <0.01 | <0.01 | <0.01 | −1.0 | −1.2 | −3.4 | −1.5 | −2.0 | −3.7 | −2.9 |
| ftsX | PFREUD_09600 | Cell division protein | CD | <0.01 | <0.01 | <0.01 | −2.1 | −1.7 | −2.4 | −4.7 | −1.7 | −3.8 | −4.3 |
| icd | PFREUD_06870 | Putative isocitrate/isopropylmalate dehydrogenase | CH | <0.01 | <0.01 | 0.05 | −1.0 | −3.3 | −2.0 | −3.0 | −3.4 | −3.3 | −3.0 |
| pccB | PFREUD_07170 | Propionyl-coenzyme A carboxylase beta chain | CH | <0.01 | 0.01 | <0.01 | −1.2 | −1.6 | −0.4 | −1.8 | −1.8 | −1.5 | −1.2 |
| aceE | PFREUD_09470 | Pyruvate dehydrogenase E1 component | CH | <0.01 | <0.01 | 0.14 | −1.5 | −1.9 | −1.5 | −1.9 | −2.8 | −2.1 | −1.9 |
| lpd | PFREUD_10890 | Dihydrolipoyl dehydrogenase | CH | <0.01 | 0.28 | 0.44 | −0.5 | −0.9 | −0.7 | −1.5 | −2.2 | −0.8 | −1.1 |
| acn | PFREUD_12590 | Aconitase | CH | <0.01 | <0.01 | 0.47 | −0.7 | −0.8 | −1.0 | −1.0 | −1.5 | −0.8 | −1.0 |
| cydA | PFREUD_01720 | Cytochrome d ubiquinol oxidase, subunit I | E | <0.01 | <0.01 | 0.02 | −0.8 | −1.4 | −1.6 | −1.6 | −1.5 | −2.0 | −3.4 |
| cydB | PFREUD_01730 | Cytochrome d ubiquinol oxidase, subunit II | E | <0.01 | <0.01 | 0.03 | −1.0 | −1.3 | −1.3 | −1.1 | −0.7 | −0.9 | −2.2 |
| nuoA | PFREUD_05160 | NADH-quinone oxidoreductase chain A | E | <0.01 | <0.01 | 0.02 | −1.4 | −1.2 | −0.5 | −2.0 | −1.3 | −2.2 | −2.0 |
| nuoB | PFREUD_05170 | NADH-quinone oxidoreductase chain B | E | <0.01 | <0.01 | <0.01 | −1.9 | −1.8 | −1.0 | −2.3 | −1.4 | −3.3 | −2.3 |
| nuoC | PFREUD_05180 | NADH-quinone oxidoreductase chain C | E | <0.01 | <0.01 | <0.01 | −1.9 | −1.7 | −1.3 | −3.8 | −2.3 | −3.7 | −3.4 |
| nuoD | PFREUD_05190 | NADH-quinone oxidoreductase chain D | E | <0.01 | <0.01 | <0.01 | −2.0 | −1.9 | −1.6 | −3.8 | −2.7 | −4.9 | −5.2 |
| nuoE | PFREUD_05200 | NADH-quinone oxidoreductase chain E | E | <0.01 | <0.01 | <0.01 | −2.0 | −1.9 | −1.9 | −2.9 | −2.5 | −4.0 | −3.3 |
| nuoF | PFREUD_05210 | NADH-quinone oxidoreductase chain F | E | <0.01 | <0.01 | 0.08 | −2.2 | −2.0 | −1.9 | −2.7 | −2.4 | −3.4 | −2.9 |
| nuoG | PFREUD_05220 | NADH-quinone oxidoreductase chain G | E | <0.01 | <0.01 | 0.08 | −2.0 | −1.5 | −1.9 | −1.9 | −2.2 | −2.8 | −3.1 |
| nuoH | PFREUD_05230 | NADH-quinone oxidoreductase chain H | E | <0.01 | <0.01 | 0.01 | −1.8 | −1.0 | −1.6 | −0.9 | −1.6 | −2.4 | −1.7 |
| nuoI | PFREUD_05240 | NADH-quinone oxidoreductase chain I | E | <0.01 | <0.01 | 0.09 | −2.5 | −1.0 | −1.6 | −1.4 | −2.3 | −3.0 | −2.9 |
| nuoJ | PFREUD_05250 | NADH-quinone oxidoreductase chain J | E | <0.01 | <0.01 | 0.02 | −2.1 | −1.1 | −1.7 | −1.5 | −2.0 | −3.3 | −2.1 |
| nuoK | PFREUD_05260 | NADH dehydrogenase I chain K | E | <0.01 | <0.01 | 0.01 | −2.3 | −1.2 | −1.5 | −2.2 | −2.1 | −3.4 | −2.6 |
| nuoL | PFREUD_05270 | NADH dehydrogenase | E | <0.01 | <0.01 | 0.01 | −2.5 | −1.5 | −2.1 | −1.9 | −2.5 | −3.7 | −2.9 |
| nuoM | PFREUD_05280 | NADH dehydrogenase I chain M | E | <0.01 | <0.01 | 0.01 | −2.3 | −1.1 | −1.7 | −1.3 | −2.4 | −3.2 | −2.9 |
| nuoN | PFREUD_05290 | NADH dehydrogenase I chain N | E | <0.01 | <0.01 | 0.05 | −2.8 | −1.6 | −2.1 | −1.2 | −2.3 | −2.8 | −2.3 |
| sdhC1 | PFREUD_09240 | Succinate dehydrogenase, subunit C | E | <0.01 | <0.01 | <0.01 | −1.3 | −0.8 | 1.2 | −2.1 | −2.2 | −2.1 | −2.2 |
| sdhA | PFREUD_09250 | Succinate dehydrogenase, subunit A | E | <0.01 | <0.01 | <0.01 | −2.1 | −1.2 | 0.5 | −2.7 | −2.9 | −3.2 | −3.4 |
| sdhB | PFREUD_09260 | Succinate dehydrogenase, subunit B | E | <0.01 | <0.01 | 0.09 | −1.2 | −0.9 | 0.3 | −0.9 | −1.0 | −0.7 | −1.1 |
| atpB | PFREUD_10430 | ATP synthase A chain | E | <0.01 | <0.01 | 0.03 | −2.0 | −1.6 | −1.5 | −2.8 | −2.6 | −3.3 | −2.5 |
| atpE | PFREUD_10440 | ATP synthase C chain | E | <0.01 | <0.01 | 0.22 | −2.8 | −2.7 | −1.9 | −3.1 | −3.1 | −2.8 | −3.2 |
| atpF | PFREUD_10450 | ATP synthase B chain | E | <0.01 | <0.01 | 0.30 | −2.9 | −3.0 | −2.1 | −2.3 | −2.7 | −2.7 | −2.6 |
| atpH | PFREUD_10460 | ATP synthase delta chain | E | <0.01 | 0.25 | 0.06 | −2.8 | −2.8 | −2.4 | −3.4 | −3.5 | −3.0 | −3.6 |
| atpA | PFREUD_10470 | ATP synthase subunit alpha | E | <0.01 | 0.34 | 0.21 | −3.0 | −2.6 | −2.5 | −3.6 | −3.6 | −3.7 | −3.5 |
| atpG | PFREUD_10480 | ATP synthase gamma chain | E | <0.01 | <0.01 | 0.20 | −3.1 | −2.6 | −2.4 | −3.7 | −4.2 | −3.9 | −3.5 |
| atpD | PFREUD_10490 | ATP synthase subunit beta | E | <0.01 | 0.05 | 0.38 | −2.9 | −2.3 | −2.2 | −3.0 | −3.2 | −3.1 | −3.2 |
| atpC | PFREUD_10500 | ATP synthase epsilon chain | E | <0.01 | 0.12 | 0.23 | −3.8 | −2.5 | −2.4 | −3.0 | −3.6 | −3.2 | −3.8 |
| sdhB3 | PFREUD_14300 | Succinate dehydrogenase | E | <0.01 | <0.01 | 0.02 | −2.8 | −2.6 | −0.6 | −2.4 | −2.4 | −2.7 | −4.2 |
| sdhA3 | PFREUD_14310 | Succinate dehydrogenase flavoprotein subunit | E | <0.01 | <0.01 | <0.01 | −2.5 | −2.5 | −0.3 | −3.0 | −2.2 | −2.6 | −4.3 |
| sdhC2 | PFREUD_14320 | Succinate dehydrogenase cytochrome B-558 subunit | E | <0.01 | <0.01 | <0.01 | −2.4 | −2.2 | 0.3 | −2.3 | −1.5 | −2.3 | −2.8 |
Locus tag for CIRM-BIA1T.
A, adaptation to atypical conditions; CD, cell division; CH, transport and metabolism of carbohydrates; E, energy metabolism.
Values of |fold change (log2)| >1 are in boldface.
Table 2.
Differentially expressed genes involved in cold stress response
| Name | Locus taga | Description | Categoryb |
P value |
Fold change (log2) for CIRM-BIA strainc: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time | Strain | Time × strain | 118 | 122 | 123 | 1 | 472 | 482 | 9 | ||||
| pspC | PFREUD_06710 | Possible stress response transcriptional regulator protein | A | <0.01 | <0.01 | <0.01 | 3.8 | 2.0 | 2.9 | 3.5 | 2.6 | 4.3 | 4.0 |
| pspC | PFREUD_06710 | Possible stress response transcriptional regulator protein | A | <0.01 | <0.01 | <0.01 | 3.8 | 2.0 | 2.9 | 3.5 | 2.6 | 4.3 | 4.0 |
| cspA | PFREUD_09800 | Cold shock-like protein | A | <0.01 | <0.01 | 0.01 | −0.3 | 0.1 | 1.6 | 2.1 | 1.3 | 1.7 | 1.9 |
| cspB | PFREUD_18210 | Cold shock protein | A | <0.01 | <0.01 | 0.01 | 0.8 | 0.6 | 1.1 | 2.3 | 0.8 | 1.8 | 0.9 |
| PFREUD_04260 | DeaD/DeaH box helicase | DNA | <0.01 | <0.01 | 0.21 | 0.3 | 0.7 | 0.7 | 1.2 | 0.9 | 1.0 | 0.8 | |
| PFREUD_13460 | Superfamily II RNA helicase, DeaD/DeaH box helicase | DNA | <0.01 | <0.01 | 0.04 | 1.3 | 1.2 | 1.5 | 2.0 | 0.9 | 1.9 | 1.8 | |
| dnaK2 | PFREUD_04630 | Chaperone protein | PM | <0.01 | <0.01 | <0.01 | −0.5 | −1.8 | −2.6 | −2.5 | −2.5 | −0.5 | 0.5 |
| grpE2 | PFREUD_04640 | Co-chaperone protein | PM | <0.01 | 0.04 | <0.01 | 0.9 | −0.9 | −1.5 | −2.4 | −1.5 | 0.0 | 0.9 |
| dnaJ2 | PFREUD_04650 | Chaperone protein DnaJ2 | PM | <0.01 | <0.01 | <0.01 | −0.3 | −0.9 | −1.4 | −1.1 | −0.7 | 0.4 | 0.6 |
| groS1 | PFREUD_06460 | 10-kDa chaperonin 1 | PM | <0.01 | 0.02 | 0.01 | −2.4 | −4.1 | −3.9 | −5.5 | −5.4 | −5.9 | −5.2 |
| groL1 | PFREUD_06470 | 60-kDa chaperonin 1 | PM | <0.01 | 0.24 | 0.22 | −3.0 | −3.9 | −3.8 | −4.0 | −4.6 | −4.9 | −4.7 |
| groS2 | PFREUD_07810 | 10-kDa chaperonin 2 | PM | <0.01 | <0.01 | 0.15 | −0.8 | −0.6 | 0.3 | −0.7 | −1.1 | −0.2 | −0.8 |
| dnaJ3 | PFREUD_08760 | Chaperone protein DnaJ3 | PM | <0.01 | <0.01 | <0.01 | −0.4 | 0.0 | −0.6 | −1.6 | −1.3 | −0.3 | −0.8 |
| hsp20 | PFREUD_09500 | Heat shock protein 20 2 | PM | <0.01 | 0.03 | 0.03 | −1.3 | −0.6 | −0.3 | −0.1 | −0.1 | −0.6 | 1.2 |
| dnaJ1 | PFREUD_17820 | Chaperone protein DnaJ1 | PM | <0.01 | <0.01 | <0.01 | −2.8 | 0.0 | −2.2 | −2.7 | −1.6 | −2.3 | −2.2 |
| grpE1 | PFREUD_17830 | Co-chaperone protein | PM | <0.01 | <0.01 | <0.01 | −3.0 | 0.1 | −2.8 | −3.2 | −2.0 | −3.0 | −2.3 |
| dnaK1 | PFREUD_17840 | Chaperone protein | PM | <0.01 | <0.01 | <0.01 | −2.9 | 0.2 | −3.2 | −2.0 | −2.5 | −4.5 | −1.7 |
| clpB 2 | PFREUD_17920 | Chaperone protein | PM | <0.01 | <0.01 | <0.01 | −2.9 | −0.4 | −3.5 | −3.3 | −2.1 | −1.9 | −4.1 |
| groL2 | PFREUD_18470 | 60-kDa chaperonin 2 | PM | <0.01 | 0.37 | 0.65 | −2.9 | −3.7 | −3.2 | −3.4 | −3.3 | −4.1 | −3.6 |
| clpB 1 | PFREUD_19250 | Chaperone protein | PM | <0.01 | <0.01 | <0.01 | 1.9 | 0.7 | −1.3 | 0.8 | 0.2 | 2.7 | 1.2 |
Locus tag for CIRM-BIA1T.
A, adaptation to atypical conditions; DNA, DNA metabolism; PM, protein modification and folding.
Values of |fold change (log2)| >1 are in boldface.
Production of aroma compounds in the cold.
P. freudenreichii contributes to the development of Swiss cheese flavor via different pathways involving esterases and branched-chain amino acid-converting enzymes (16). The 12 esterase-encoding genes identified in the P. freudenreichii genome (8), in particular pf279, encoding a lipolytic secreted esterase probably involved in lipolysis (9), kept the same level of expression at 4°C in the 6 strains tested, as previously observed for CIRM-BIA1T (Table 3). These results are in agreement with the observation that P. freudenreichii still contributes to the formation of free fatty acids in cheese at low temperatures (16). Most genes encoding branched-chain amino acid transporters and converting enzymes were downregulated (for example, fold changes ranging from −2.0 to −3.9 for bkdA2) (Table 3), whereas methylbutanoate was still produced at 4°C (Fig. 1B). It suggests that this pathway is posttranscriptionally regulated and/or that branched-chain amino acid-converting enzymes were accumulated in cells and remained active at 4°C.
Table 3.
Genes encoding esterases and branched-chain amino acid-converting enzymes
| Type of enzyme and name | Locus taga | Descriptionb | Categoryc |
P value |
Fold change (log2) for each CIRM-BIA straind: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time | Strain | Time × strain | 118 | 122 | 123 | 1 | 472 | 482 | 9 | ||||
| Esterases | |||||||||||||
| pf1861 | PFREUD_03560 | Putative carboxylic ester hydrolase | L | 0.18 | <0.01 | 0.06 | −0.2 | −0.6 | −0.4 | 0.2 | 0.4 | 0.2 | −0.6 |
| pf774 | PFREUD_04240 | Putative carboxylic ester hydrolase | L | 0.60 | <0.01 | 0.11 | 0.4 | −0.7 | −0.4 | 0.3 | 0.2 | −0.3 | 0.6 |
| pf279 | PFREUD_04340 | Carboxylic ester hydrolase | L | 0.32 | 0.01 | 0.11 | 0.3 | −0.6 | −0.6 | 0.6 | 0 | 0.1 | 0.4 |
| pf962 | PFREUD_04810 | Carboxylic ester hydrolase | L | <0.01 | 0.05 | 0.61 | 0.5 | 0.4 | 0.5 | 0.7 | 0.2 | 0.5 | 0.2 |
| pf1509 | PFREUD_10540 | Putative carboxylic ester hydrolase | L | 0.50 | <0.01 | 0.03 | 0.0 | 0.5 | 0.2 | 0.3 | −0.7 | 0.9 | −0.5 |
| pf1758-2887 | PFREUD_10790-PFREUD_10800 | Putative carboxylic ester hydrolasese | L | 0.57 | 0.02 | 0.34 | −0.3 | −0.2 | −0.3 | —c | 0.2 | 0.5 | —c |
| pf1637 | PFREUD_12910 | Putative carboxylic ester hydrolase | L | <0.01 | <0.01 | 0.07 | 0.0 | −0.4 | 0.3 | 0.0 | −0.8 | −0.3 | −0.4 |
| pf379 | PFREUD_13000 | Putative carboxylic ester hydrolase | L | <0.01 | <0.01 | <0.01 | 0.8 | 0.7 | 0.6 | 1.2 | 0.6 | 1.6 | 0.6 |
| pf169 | PFREUD_14330 | Carboxylic ester hydrolase | L | 0.13 | 0.19 | 0.13 | −0.1 | −0.2 | −0.1 | 0.3 | 0.1 | 0.5 | 0.1 |
| pf1655 | PFREUD_18110 | Carboxylic ester hydrolase | L | 0.59 | <0.01 | 0.55 | 0.3 | 0.7 | −0.4 | −0.1 | 0 | 0 | −0.3 |
| pf667 | PFREUD_23150 | Carboxylic ester hydrolase | L | <0.01 | <0.01 | <0.01 | 0.1 | −0.1 | 0.2 | 0.9 | 0.3 | 0.5 | −0.2 |
| pf2042 | PFREUD_23770 | Putative carboxylic ester hydrolase | L | <0.01 | <0.01 | 0.04 | −0.3 | −0.6 | −0.6 | −0.2 | 0.1 | −0.6 | −0.4 |
| Branched-chain amino acid transport and conversion | |||||||||||||
| livG | PFREUD_10850 | ABC protein of branched-chain amino acid ABC transporter | AA | <0.01 | <0.01 | 0.02 | −1.4 | −0.8 | −2.3 | −3.6 | −2.4 | −3 | −2.1 |
| braE | PFREUD_10860 | IM protein of branched-chain amino acid ABC transporter | AA | <0.01 | <0.01 | <0.01 | −0.5 | −0.3 | −0.8 | −1.5 | −1.1 | −1.5 | −0.6 |
| braD | PFREUD_10870 | IM protein of branched-chain amino acid ABC transporter | AA | <0.01 | <0.01 | <0.01 | −1.0 | −0.4 | −1.3 | −1.9 | −1 | −2.4 | −0.9 |
| braC | PFREUD_10880 | BP of branched-chain amino acid ABC transporter | AA | <0.01 | <0.01 | 0.01 | −1.0 | −0.5 | −1.2 | −2.9 | −1.6 | −2.2 | −0.6 |
| ydaO | PFREUD_12690 | IM protein of branched-chain amino acid ABC transporter | AA | <0.01 | <0.01 | 0.06 | −2.1 | −2 | −1.6 | −2.4 | −1.9 | −2.4 | −2.2 |
| ilvE | PFREUD_13350 | Branched-chain amino acid aminotransferase | AA | 0.45 | 0.01 | 0.10 | −0.2 | 0.2 | −0.4 | 0.3 | −1.5 | 0.4 | 0.4 |
| bkdA2 | PFREUD_02200 | 2-Oxoisovalerate dehydrogenase subunit beta | AA | <0.01 | <0.01 | 0.03 | −2.1 | −3.7 | −2.0 | −3.3 | −3.3 | −3.5 | −3.9 |
| bkdB | PFREUD_02210 | Dihydrolipoyllysine residue (2-methylpropanoyl) transferase | AA | <0.01 | <0.01 | 0.03 | −0.8 | −1.7 | −0.7 | −1.8 | −1.4 | −0.7 | −1.5 |
Locus tag for CIRM-BIA1T.
IM, integral membrane; BP, binding protein.
L, lipid metabolism; AA, transport and metabolism of amino acids.
Values of |fold change (log2)| >1 are in boldface.
This gene presents a frameshift in strains CIRM-BIA1T and CIRM-BIA9.
Rerouting of carbon metabolism toward glycogen synthesis.
The phosphoenolpyruvate-pyruvate-oxaloacetate node interconnects the major pathways of carbon metabolism in bacteria (13). The main changes in the expression of pyruvate-related genes in P. freudenreichii in the cold are shown in Fig. 3. Three genes involved in generating pyruvate from alanine, serine, and lactate were upregulated, as previously observed in CIRM-BIA1T (6), as well as genes involved in gluconeogenesis (ppdK, eno1, eno2, fba2, and pgi) (Table 4). PpdK is a pyrophosphate-dependent enzyme, and accordingly, an inorganic pyrophosphatase-coding gene was found to be overexpressed in the cold in all strains (ppa) (Table 4). Genes coding for enzymes of glycogen synthesis by the classical pgmA-glgC-glgA pathway were overexpressed (Fig. 4). Moreover, we showed that glgE, encoding a maltosyltransferase, present in the newly described treS-pep2-glgE pathway of glycan synthesis from trehalose in mycobacteria (3), was also upregulated in all of the strains (fold changes ranging from 0.9 to 1.4) (Fig. 4). To confirm that glycogen was effectively synthesized by P. freudenreichii, it was quantified in cells over the incubation time (incubation at 4°C extended for 250 h), along with trehalose, since the syntheses of these two compounds are interconnected (3). Both compounds were analyzed in CIRM-BIA1T cells by enzymatic methods as previously described (12). Our results showed that the concentrations of glycogen and trehalose increased by factors of 3 and 18, respectively, between the end of growth at 30°C (40 h) and 120 h of incubation at 4°C (Fig. 5). The present study provides the first quantification of glycogen accumulation in propionibacteria, confirms the results of an in vivo 13C nuclear magnetic resonance (NMR) study showing the ability of the same strain to synthesize glycogen (11), and shows that low temperature and not only nutrient starvation can induce the synthesis of glycogen in bacteria. The synthesis of trehalose by propionibacteria was early reported (14), with O2, NaCl, and pH stresses known to induce its synthesis in propionibacteria (1).
Fig 3.

Main routes of pyruvate formation and conversion in P. freudenreichii during storage at 4°C and relevant for this study. Genes upregulated at 4°C are shown in black, and downregulated genes in gray. Thick black arrows emphasize the metabolic pathways that are favored at 4°C, and thin gray arrows the pathways that are downregulated at 4°C. ace, pyruvate dehydrogenase, E1 component; ald, alanine dehydrogenase; ldh, l-lactate dehydrogenase; ppdk, pyruvate phosphate dikinase; sdaA, l-serine dehydratase; sdh, succinate dehydrogenase; CoA, coenzyme A. Values of fold changes are shown in Table 1, Table 4, and Fig. 5.
Table 4.
Differentially expressed genes involved in pyruvate generation and rerouting toward trehalose and glycogen synthesis
| Function and name | Locus taga | Description | Categoryb |
P value |
Fold change (log2) for each CIRM-BIA strainc: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time | Strain | Time × strain | 118 | 122 | 123 | 1 | 472 | 482 | 9 | ||||
| Generation of energy | |||||||||||||
| ppa | PFREUD_23500 | Inorganic pyrophosphatase | Ph | <0.01 | <0.01 | <0.01 | 2.5 | 1.8 | 1.7 | 3.3 | 2.7 | 2.8 | 2.5 |
| Generation of pyruvate | |||||||||||||
| ald | PFREUD_00370 | Alanine dehydrogenase | AA | <0.01 | <0.01 | <0.01 | 3.7 | 4.5 | 1.1 | 7.5 | 3.2 | 2.0 | 4.3 |
| sdaA | PFREUD_18570 | l-Serine dehydratase | AA | <0.01 | <0.01 | <0.01 | 1.6 | 1.3 | 0.5 | 2.0 | 1.0 | 1.9 | 0.9 |
| ldh2 | PFREUD_12840 | l-Lactate dehydrogenase | CH | <0.01 | 0.02 | 0.01 | 1.5 | 2.0 | 1.3 | 1.3 | 1.7 | 1.8 | 0.4 |
| Gluconeogenesis | |||||||||||||
| ppdk | PFREUD_03230 | Pyruvate phosphate dikinase | CH | <0.01 | <0.01 | <0.01 | 1.3 | 1.2 | −0.6 | 1.9 | 0.9 | 0.3 | 0.6 |
| eno1 | PFREUD_17320 | Enolase 1 | CH | <0.01 | <0.01 | 0.01 | 1.1 | 1 | 0.6 | 2.6 | 1.9 | 1.7 | 1.1 |
| eno2 | PFREUD_17250 | Enolase 2 | CH | <0.01 | <0.01 | 0.02 | 1.2 | 1.2 | 0.7 | 0.6 | 1.3 | 1.1 | 0.7 |
| fba1 | PFREUD_19150 | Fructose-bisphosphate aldolase class II | CH | <0.01 | <0.01 | 0.33 | −0.5 | −1.2 | −0.3 | −1.0 | −0.7 | −0.6 | −1.0 |
| fba2 | PFREUD_23890 | Fructose-bisphosphate aldolase class I | CH | <0.01 | <0.01 | <0.01 | 2.5 | 2.4 | 0.5 | 3.9 | 2.3 | 1.2 | 2.2 |
| pgi | PFREUD_04290 | Glucose-6-phosphate isomerase | CH | <0.01 | <0.01 | <0.01 | 1.0 | 0.4 | 0.5 | 1.6 | 0.8 | 1.2 | 1.5 |
Locus tag in CIRM-BIA1T.
Ph, metabolism of phosphate; AA, transport and metabolism of amino acids; CH, transport and metabolism of carbohydrates.
Values of |fold change (log2)| >1 are in boldface.
Fig 4.
Changes in expression of genes involved in trehalose and glycogen synthesis in P. freudenreichii (pathways adapted from Chandra et al. [3]). Each box shows the fold change, expressed as log2, for each gene in all seven strains (CIRM-BIA118, -122, -123, -1, -472, -482, and -9) after 80 h at 4°C in comparison with gene expression at the reference time (20 h). Values of |fold change (log2)| >1 and <−1 are shown in dark and light gray, respectively.
Fig 5.
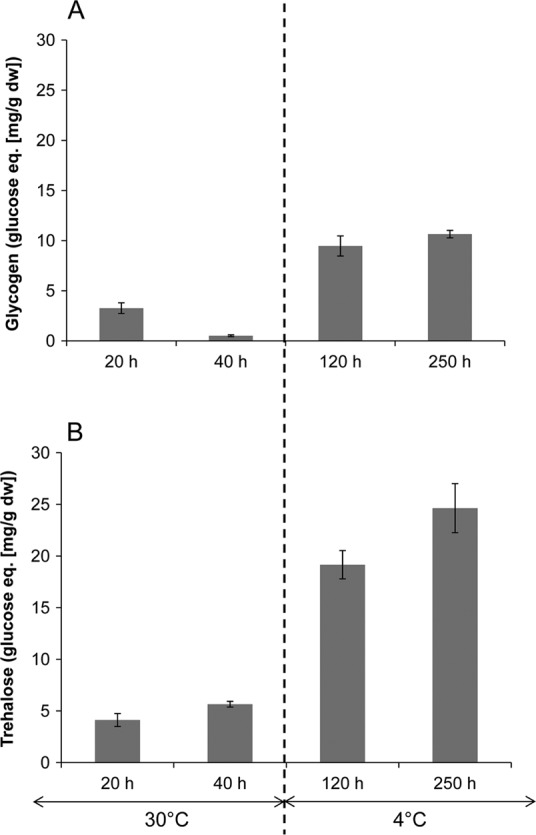
Accumulation of intracellular glycogen and trehalose in P. freudenreichii CIRM-BIA1T during growth at 30°C (up to 40 h of incubation) and further incubation for 250 h at 4°C. Values are means of the results of triplicate independent experiments; error bars show standard deviations. eq, equivalent.
Conclusions.
This study shows that adaptation strategies in the cold described for the type strain are general within P. freudenreichii species and gives clues on the molecular basis of the long-term survival and activity of this bacterium during prolonged incubation at low temperatures.
ACKNOWLEDGMENTS
We thank Pascal Pachot, Stat-Plan, for his support concerning statistical analysis and Victoria Chuat (CIRM-BIA, INRA, Rennes, France) for strain preparations.
INRA, Valio Ltd. (Helsinki, Finland), and Tekes (the Finnish Funding Agency for Technology and Innovation) supported this study.
Footnotes
Published ahead of print 22 June 2012
REFERENCES
- 1. Cardoso FS, Gaspar P, Hugenholtz J, Ramos A, Santos H. 2004. Enhancement of trehalose production in dairy propionibacteria through manipulation of environmental conditions. Int. J. Food Microbiol. 91: 195–204 [DOI] [PubMed] [Google Scholar]
- 2. Chan YC, Wiedmann M. 2009. Physiology and genetics of Listeria monocytogenes survival and growth at cold temperatures. Crit. Rev. Food Sci. Nutr. 49: 237–253 [DOI] [PubMed] [Google Scholar]
- 3. Chandra G, Chater KF, Bornemann S. 2011. Unexpected and widespread connections between bacterial glycogen and trehalose metabolism. Microbiol. 157: 1565–1572 [DOI] [PubMed] [Google Scholar]
- 4. Cousin F, Mater DDG, Foligné B, Jan G. 2 August 2010. Dairy propionibacteria as human probiotics: a review of recent evidence. Dairy Sci. Technol. 10.1051/dst/2010032 [Google Scholar]
- 5. Cretenet M, et al. 2011. Dynamic analysis of the Lactococcus lactis transcriptome in cheeses made from milk concentrated by ultrafiltration reveals multiple strategies of adaptation to stresses. Appl. Environ. Microbiol. 77: 247–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dalmasso M, et al. 2012. A temporal -omic study of Propionibacterium freudenreichii CIRM-BIA1T adaptation strategies in conditions mimicking cheese ripening in the cold. PLoS One 7: e29083 doi:10.1371/journal.pone.0029083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dalmasso M, et al. 2011. Multilocus sequence typing of Propionibacterium freudenreichii. Int. J. Food Microbiol. 145: 113–120 [DOI] [PubMed] [Google Scholar]
- 8. Dherbécourt J, Falentin H, Canaan S, Thierry A. 2008. A genomic search approach to identify esterases in Propionibacterium freudenreichii involved in the formation of flavour in Emmental cheese. Microb. Cell Fact. 7: 16 doi:10.1186/1475-2859-7-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dherbécourt J, et al. 2010. Identification of a secreted lipolytic esterase in Propionibacterium freudenreichii, a ripening process bacterium involved in Emmental cheese lipolysis. Appl. Environ. Microbiol. 76: 1181–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Falentin H, et al. 2010. Specific metabolic activity of ripening bacteria quantified by real-time reverse transcription PCR throughout Emmental cheese manufacture. Int. J. Food Microbiol. 144: 10–19 [DOI] [PubMed] [Google Scholar]
- 11. Meurice G. 2004. Biochimie, biologie cellulaire et moléculaire. Ph.D. thesis. Ecole Nationale Supérieure Agronomique de Rennes, Rennes, France [Google Scholar]
- 12. Parrou JL, Francois J. 1997. A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Anal. Biochem. 248: 186–188 [DOI] [PubMed] [Google Scholar]
- 13. Sauer U, Eikmanns BJ. 2005. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 29: 765–794 [DOI] [PubMed] [Google Scholar]
- 14. Stjernholm R. 1958. Formation of trehalose during dissimilation of glucose by Propionibacterium. Acta Chem. Scand. 12: 646–649 [Google Scholar]
- 15. Thieringer HA, Jones PG, Inouye M. 1998. Cold shock and adaptation. Bioessays 20: 49–57 [DOI] [PubMed] [Google Scholar]
- 16. Thierry A, et al. 2011. New insights into physiology and metabolism of Propionibacterium freudenreichii. Int. J. Food Microbiol. 149: 18–27 [DOI] [PubMed] [Google Scholar]