Abstract
Prophages are encoded in most genomes of sequenced Clostridium difficile strains. They are key components of the mobile genetic elements and, as such, are likely to influence the biology of their host strains. The majority of these phages are not amenable to propagation, and therefore the development of a molecular marker is a useful tool with which to establish the extent and diversity of C. difficile prophage carriage within clinical strains. To design markers, several candidate genes were analyzed including structural and holin genes. The holin gene is the only gene present in all sequenced phage genomes, conserved at both terminals, with a variable mid-section. This allowed us to design two sets of degenerate PCR primers specific to C. difficile myoviruses and siphoviruses. Subsequent PCR analysis of 16 clinical C. difficile ribotypes showed that 15 of them are myovirus positive, and 2 of them are also siphovirus positive. Antibiotic induction and transmission electron microscope analysis confirmed the molecular prediction of myoviruses and/or siphovirus presence. Phylogenetic analysis of the holin sequences identified three groups of C. difficile phages, two within the myoviruses and a divergent siphovirus group. The marker also produced tight groups within temperate phages that infect other taxa, including Clostridium perfringens, Clostridium botulinum, and Bacillus spp., which suggests the potential application of the holin gene to study prophage carriage in other bacteria. This study reveals the high incidence of prophage carriage in clinically relevant strains of C. difficile and correlates the molecular data to the morphological observation.
INTRODUCTION
Clostridium difficile is a Gram-positive, spore-forming, toxin-producing anaerobic bacillus which is the commonest cause of infectious diarrhea in United Kingdom hospitals, with onset often following from broad-spectrum antibiotic treatment (5, 22, 23, 49, 53). Whole-genome sequencing has offered insights into the biology and evolution of C. difficile. The highly flexible genome is genetically diverse and encodes several mobile elements, including transposons and prophages (18, 43, 47, 48). Further evidence of C. difficile prophage presence comes from transmission electron microscopy (TEM) analysis of six distinct ribotypes and hybridization analysis of 37 clinical isolates, which both suggested phage carriage was common (8, 11).
At least one prophage has been identified in all of the five C. difficile ribotypes sequenced to date, with CD630 (ribotype 012) harboring two prophages (11, 43, 48). Temperate bacteriophages in C. difficile strains may contribute to the pathogenicity of their host either by encoding novel toxins or by differentially regulating the expression of bacterial toxins (10, 15, 44). No previous studies have described phage carriage within a large set of known clinically relevant ribotypes or have linked molecular data to TEM characterization and the ability of phages to be induced following antibiotic treatment. Before an understanding of phage contribution to virulence can be developed, a survey of phage carriage according to ribotype is necessary.
Five C. difficile temperate phages have been characterized and fully sequenced: three myoviruses (ϕC2, ϕCD119, and ϕCD27) and two siphoviruses (ϕCD6356 and ϕCD38-2) (11, 14, 19, 30, 44). They were all identified following the same procedures; phages were induced from a large number of strains and extensive screening identified a host strain for a small subset of phages. These phages therefore represent a very small fraction (∼3 to 6%) of the total phages present in clinical strains since most do not have a suitable alternative host strain to support virulent phage production (10, 19, 30). Therefore, new approaches have to be developed to study the vast majority of C. difficile phages, which are not amenable to propagation by plaque assays and which are uncharacterized.
Bacteriophage genes that have been used to examine phage diversity include gp23 and gp20, which encode for the major capsid protein and portal protein in myoviruses, respectively. These have been predominantly used to study diversity in T4-type phages (9, 28, 36, 46). Unfortunately, neither of these genes are conserved enough to be targeted in all known C. difficile bacteriophages (e.g., gp23 is not recognizable in ϕCD27, gp20 is too diverse to facilitate alignment). The DNA polymerase gene has also been used to assess both myovirus and podovirus diversity, but no polymerase genes have been identified in C. difficile phage genomes (3, 4, 9, 28, 46).
One gene that may be a suitable molecular marker to assess genetic diversity within C. difficile phages is the holin gene. This gene encodes for the protein that punctures the cytoplasmic membrane of the bacterial cell and is essential for phage release (54). Although this gene is often too diverse within some phage groups to be useful, it has been used successfully as a marker in studying molecular phylogeny of phi29-like phages, which infect Bacillus sp., and prophage carriage in Streptococcus pneumonia (35, 38, 51). This gene is present in all of the five sequenced C. difficile phages and is in the four fully annotated C. difficile genomes (CD630, CD196, R20291, and QCD-63q42) (11, 14, 18, 30, 43, 47, 48).
Degenerate PCR primers targeting C. difficile myoviruses and siphoviruses, respectively, were designed based on the conserved regions of 5′ and the 3′ ends of the alignment of the holin genes. We then sequenced this gene from the 15 distinct ribotypes from which a PCR product was obtained to allow us to estimate the diversity of C. difficile prophages. To put the molecular data into morphological context, the phages were characterized using TEM. We therefore present a plaque assay independent way to reveal the genetic diversity of C. difficile bacteriophages. We also show that the holin-based marker may also be a useful tool with which to assess the genetic diversity of bacteriophages in other bacterial genera where prophage carriage may be contributing to bacterial phenotype.
MATERIALS AND METHODS
C. difficile isolates and phages.
Clinical fecal samples were collected from patients in Leicester Royal Infirmary who had previously tested positive for either or both toxin A and B using C. difficile Tox A/B II (Techlab, Blacksburg, VA). National standard methods were followed for C. difficile isolation and identification (http://www.hpa.org.uk/SMI/pdf). Bacterial strains were stored in cooked meat broth (Bioconnections, Leeds, United Kingdom) at room temperature and in Protect bacterial preservers (Technical Service Consultants, Ltd., Heywood, United Kingdom) at −80°C. C. difficile strain NC11204 (ribotype 001) was obtained from the National Collection of Type Cultures (NCTC; Central Public Health Laboratory, London, United Kingdom). C. difficile reference strains, including ribotype 027 and CD630 (ribotype 012) strains, were kindly provided by N. Fairweather (Imperial College, London, United Kingdom). Nine other 027 reference strains (12L, 14L, 17L, 25L, 28L, 87L, 91L, 96L, and 73L) with different multilocus variable-number tandem-repeat analysis (MLVA) types were obtained from M. Wilcox (University of Leeds, Leeds, United Kingdom). When required, C. difficile strains were subcultured on brain heart infusion (BHI; Oxoid, Ltd., United Kingdom) agar with 7% defibrinated horse blood (TCS Biosciences, Ltd., United Kingdom). Growth of C. difficile was carried out using BHI broth in a MiniMACS anaerobic chamber (Don Whitley Scientific, United Kingdom) at 37°C. C. difficile phage ϕCD27 was kindly provided by Melinda Mayer, Institute of Food Research, London, United Kingdom.
C. difficile ribotyping.
Ribotyping PCR primers sequence were 5′-GTG CGG CTG GAT CAC CTC CT-3′ (16S primer) and 5′-CCC TGC ACC CTT AAT AAC TTG ACC-3′ (23S primer) (34). The 16S primer was labeled with carboxyfluorescein (FAM) at the 5′ end (Applied Biosystems, United Kingdom). PCRs were carried out as described by O'Neill et al. (34). A total of 50 μl of PCR products were concentrated to final volumes of 15 μl by heating at 75°C for 1 h. The PCR products were analyzed using capillary gel electrophoresis by an ABI 3730 genetic analyzer with a 41-cm capillary loaded with a POP7 gel (Applied Biosystems). The samples were injected with 1.6 kV over 15 s with a total running time of 103 min at 8 kV run voltage. A 20- to 1,200-bp GeneScan ladder (Applied Biosystems) was added to each well as an individual marker. The sizes of the peaks were resolved using Peak Scanner v1.0 (Applied Biosystems). Peaks that were at least 3% of the highest peak of the run in question were counted as bands. Double bands were accepted if >1.5-bp separations were recorded. A matrix was constructed based on the presence or absence of bands of specific sizes in each C. difficile isolate (see Table S1 in the supplemental material). Sorensen's distance was calculated between each combination of isolates and clustered using MultiVariate statistical package (MVSP) (Kovach Computing Services, Anglesey, United Kingdom). All of the ribotypes obtained in the present study were confirmed by W. Fawley and M. Wilcox (University of Leeds) with reference to Clostridium difficile Ribotyping Network (CDRN) database for England and Northern Ireland. The official ribotype names were determined as 002, 005, 013, 014, 015, 020, 023, 027, 056, 076, 078, 081, 087, 107, and 220.
Phage induction, purification, and TEM.
Mitomycin C or norfloxacin (Sigma-Aldrich, United Kingdom) was added to a final concentration of 3 μg ml−1 to C. difficile culture with the optical density at 550 nm of 0.6 to 0.8. After 48 h, the bacterial cultures were centrifuged at 8,000 × g for 15 min. The resulting supernatants were filtered through 0.22-μm-pore-size and examined by TEM. The filtrates were treated with DNase I and RNase A (Sigma-Aldrich) at a final concentration of 1 μg ml−1 for 2 h at 37°C before polyethylene glycol (PEG) 8000 was used to concentrate the phage particles (42). TEM was carried out by The Electron Microscope Laboratory, University of Leicester. Briefly, samples were placed on Athene 3-mm copper grids (Agar Scientific, United Kingdom), prepared by coating them with 0.25% pioloform, and carbon coated for 1 to 3 s before using high-voltage glow discharged for 30 to 60 s. Staining was performed by transferring 8 μl of sample to the grids, incubation at room temperature for 5 min, two washes with distilled water, and staining with 5 μl of 1% uranyl acetate (wt/vol) for 5 to 10 s. TEM was performed with a JEOL 1220 (JEOL, United Kingdom) run at 80 kV, and images were captured using an SIS Megaview III camera with analySIS software (Olympus).
Degenerate PCR and sequencing.
Phenol-chloroform extraction and isopropanol precipitation were used for DNA extraction (42). Degenerate PCR primers were designed manually against conserved regions in C. difficile myoviruses and siphoviruses holin gene sequences (see Fig. S2 and S3 in the supplemental material). The myoviruses and siphoviruses primer pairs CDHmyoF (5′-TATACCAGAGCAGTTRCTRA-3′)/CDHmyoR (5′-CMTCCTTCAAYTGTTTGTAA-3′) and CDHsiphoF (5′-TTATGCGCTTTGCTRTTYAA-3′)/CDHsiphoR (5′-MGTTTTCATTGCTCCCATTT-3′) amplify 227- and 150-bp products, respectively. Both PCRs were carried out in a LabCycler (SensoQuest GmbH, Göttingen, Germany) in total volumes of 50 μl, containing 0.25 mM deoxynucleoside triphosphates, 3 mM MgCl2, 2 μM concentrations of the primers, 50 ng of template DNA, 0.5 U of Taq polymerase (Bioline, United Kingdom), and 5 μl of 10× Taq buffer (Bioline). The amplification conditions were 94°C for 2 min, followed by 35 cycles of 94°C for 45 s, 48°C for 45 s (46°C for 45 s for siphoviruses), and 72°C for 1 min, with a final extension of 10 min at 72°C. PCR products were gel purified using a Qiagen gel extraction kit and subjected to TOPO TA cloning (Invitrogen). Sequencing was carried out by GATC Biotech. Sequencing results were edited using Chromas 2.33. A BLASTn (National Center for Biotechnology Information [NCBI]) search on the sequenced PCR products confirmed that all of the sequences were encoded by C. difficile phages.
Sequence and phylogenetic analysis.
The C. difficile holin sequences obtained from the present study have been submitted to the EMBL databases. A further 12 sequences, obtained from a protein-protein BLAST search from GenBank using holin protein from ϕCD27 as a query, were also used in the phylogenetic analysis. Accession numbers are listed in the Fig. S4 in the supplemental material.
Phylogenetic analyses were constructed using the program Molecular Evolutionary Genetics Analysis (MEGA) package, version 5.01 (24, 50). Alignment Explorer/CLUSTAL in MEGA 5.01 was used to align the DNA sequences. Neighbor-joining (NJ) and maximum-parsimony (MP) analyses were conducted on a nucleotide data set; for NJ a maximum composite likelihood model was used, and for MP a close-neighbor-interchange with a search level of 3 was used. Support for clades was estimated using a bootstrap analysis implemented in MEGA with 3,000 replicates. The trees were rooted with phage Lambda (NC_001416) as an outgroup.
Accession numbers.
The GenBank accession numbers of holin sequences obtained from the present study are as follows: C. difficile ribotypes 001, 002, 005, 013, 014, 015, 020, and 023, HE795757 to HE795764; C. difficile ribotype 027, FR666781; C. difficile ribotype 056, FR666782; C. difficile ribotype 076, FR666777; C. difficile ribotype 078, HE795765; C. difficile ribotype 087, FR666780; C. difficile ribotype 107, HE795766; C. difficile ribotype 220Sipho, HE795767; and C. difficile ribotype 220, FR666783.
The GenBank accession numbers of nine published C. difficile phage holin gene sequences are as follows: ϕCD27, YP_002290909.1; ϕC2, YP_001110752.1; ϕCD119, YP_529585.1; CD630, YP_001087452.1; QCD-63q42, ZP_05331963.1; R20291, FN545816; CD196, FN538970; ϕ38-2, YP_004508400; and ϕCD6356, ZP_05331929.
The GenBank accession numbers of holin homologous sequences (hypothetical proteins were indicated as HP) used in phylogenetic analyses are as follows: Clostridium perfringens B strain ATCC 3616, ZP_02636959.1; Clostridium perfringens E strain JGS1987, ZP_02633353.1; Clostridium perfringens CPE strain F4969, ZP_02640276.1; Bacillus cellulosilyticus DSM_2522 HP1, EFC16386.1; Bacillus sp. strain B14905_HP, ZP_01722281.1; Lysinibacillus sphaericus C3 41 HP, YP_001697516.1; Clostridium tetani E88 HP, NP_782152.1; Clostridium butyricum 5521, ZP_02948549.1; Clostridium botulinum E3 strain Alaska E43, YP_001921455.1; Clostridium beijerinckii NCIMB 8052 HP, YP_001309424.1; Bacillus cellulosilyticus DSM 2522 HP2, EFC16328.1; and Clostridium botulinum E1 strain “BoNT E Beluga,” ZP_04824030.1.
RESULTS AND DISCUSSION
C. difficile ribotype diversity.
To obtain a clear picture of C. difficile phage carriage with respect to ribotype, C. difficile isolated from clinical samples for the present study were PCR ribotyped and analyzed. Ribotyping is the standard method of genotyping C. difficile in the United Kingdom and is based on the number and size of the variable intergenic 16S-23S rRNA spacer regions of the strain (16, 34). Capillary gel electrophoresis revealed 4 to 11 distinct PCR products for each isolate ranging from 213 to 581 bp, which agrees well with previous reports (20, 41).
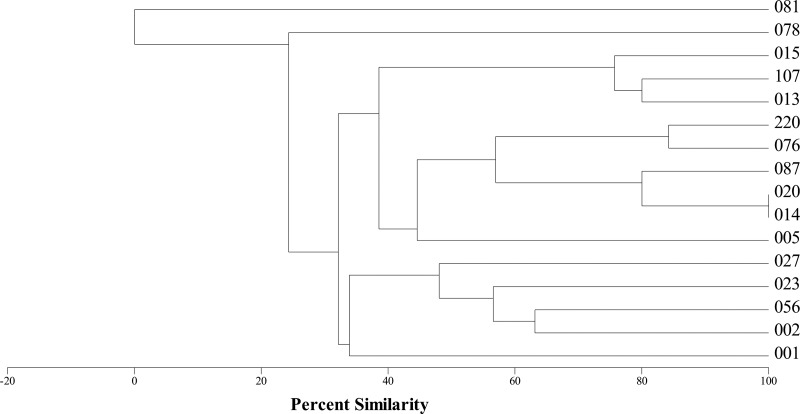
A total of 16 distinct ribotypes were selected for further analysis, and their similarity was assessed using a MVSP clustering analysis based on the presence or absence of bands of a particular size (Fig. 1). Of these, 14 ribotypes (all except 081 and 078) form a group with 32% similarity. Ribotype 081 has no similarity to other strains, and ribotype 078 only shares ∼25% similarity to the rest of the strains. As expected, the ribotype profiles from 014 and 020 are indistinguishable (27). Finally, the hypervirulent ribotype 027 has ∼50% similarity and appears therefore to be more closely related to ribotypes 023, 056, and 002 (1, 13, 23).
Fig 1.
Dendrogram showing the relationships of C. difficile ribotypes based on the presence or absence of PCR products of a specific size as determined by capillary-based gel electrophoresis.
Use of the holin gene as a marker of C. difficile prophage carriage.
Unfortunately, there is no genetic marker that is common to all bacteriophage families, in the way that the 16S rRNA gene is found in all bacteria and archaea (40). However, two phage structural genes encoding the major capsid protein and the portal protein, the phage DNA polymerase, and the terminase genes are often used to study the diversity of myoviruses and siphoviruses (2–4, 9, 28, 46). Unfortunately, none of these genes are universal for the study of C. difficile bacteriophages. For example, no recognizable major capsid gene can be identified in the genome of ϕCD27 (30). As a result, a set of degenerate PCR primer based on the available capsid genes failed to generate PCR products on ribotypes 087 and 220 (32). However, both strains were demonstrated to carry inducible phages (Fig. 2 and Fig. 3). The portal protein and the terminase genes were also ruled out as a molecular marker as they were too variable to allow primer design (see Fig. S5 and S6 in the supplemental material). In addition, C. difficile phage genomes all lack DNA polymerase genes (11, 14, 19, 30, 44). We also investigated the possibility of using the endolysin gene as a marker, but it was less conserved than the holin gene and therefore universal C. difficile phage endolysin primers would have needed to be significantly more degenerate.
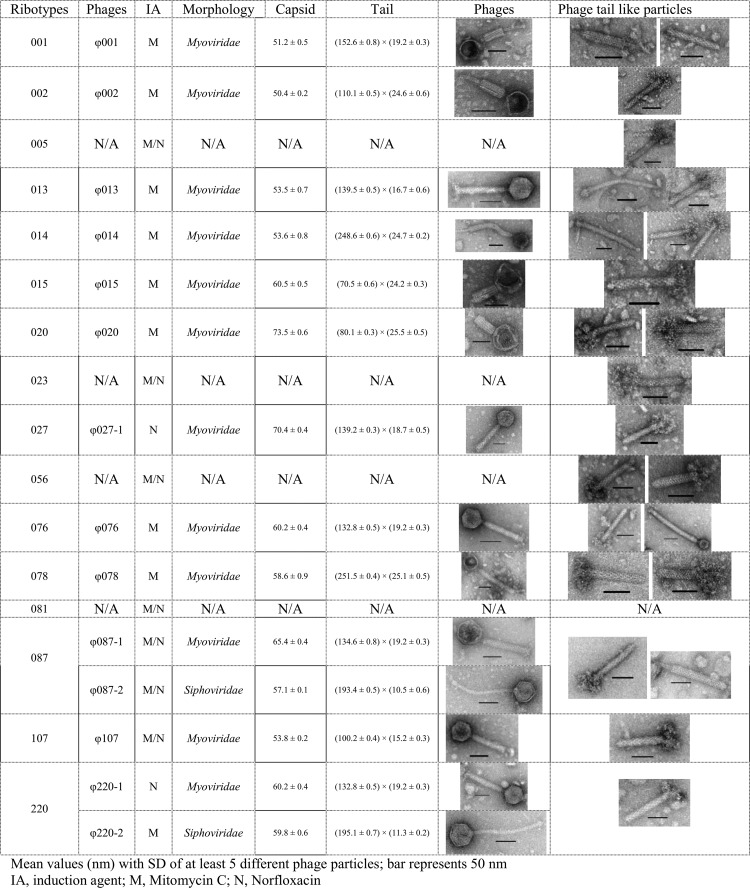
Fig 2.
Morphological diversity of phage particles induced from 16 different C. difficile ribotypes.
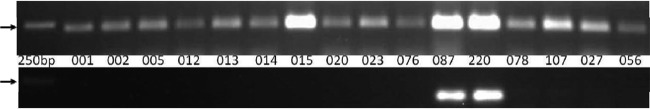
Fig 3.
One percent agarose gels with holin gene PCR products. The upper panel shows C. difficile myoviruses PCR products. The bottom panel shows C. difficile siphoviruses PCR products. DNA templates were extracted from 16 C. difficile strains with different PCR ribotypes. The arrow indicates a 250-bp DNA ladder.
The holin gene has been identified from all of the published C. difficile phage/bacterial genomes (11, 14, 18, 30, 43, 47, 48). To ensure that the holin sequence targeted was specific to C. difficile phages, a nucleotide blast search (NCBI) was carried out using the holin sequences from the myovirus ϕCD27 and the siphovirus ϕCD38-2 as queries against database nucleotide collection (nr/nt), respectively (conducted on 5 April 2012). The specificity of the holin gene to C. difficile phage was confirmed by the large e-value difference between C. difficile and the next closest match (8e-41 to 0.097 for the myovirus and from 2e-67 to 0.008 for the siphovirus).
Sequence alignments of holin genes from seven myoviruses and two siphoviruses, respectively, show conserved termini and a variable mid-section, which enables PCR primers to be placed. Two sets of degenerate primers targeting C. difficile myoviruses and siphoviruses, were designed (see Fig. S2 and S3 in the supplemental material). To establish the specificity of both primer pairs, in silico PCR (http://insilico.ehu.es/PCR/index.php) was conducted with Bacillus, Listeria, Staphylococcus, Enterococcus, Streptococcus, Lactobacillus, Lactococcus, Leuconostoc, Pediococcus, Clostridium, Eubacterium, and Heliobacterium genomes. Only C. difficile generated PCR products of the expected size.
The 16 clinical ribotypes were then used as a template for PCR for both the myovirus and the siphovirus primer sets. A myovirus PCR product of the expected size was obtained from all ribotypes except 081 and, in addition, a siphovirus PCR product was obtained from two ribotypes (Fig. 3). This suggests that ribotype 081 has no prophages, that 15 ribotypes carry myoviruses, and that 2 ribotypes (087 and 220) also carry siphoviruses.
To determine whether the PCR results were consistent with expected phage morphologies, TEM analysis after phage induction was performed. The results showed that 80% (12/15) of myovirus PCR-positive ribotypes produced intact myoviruses (Fig. 1). Despite being PCR positive, no phage particles could be induced from three ribotypes (005, 023, and 056) (Fig. 1). This could be due to the lack of the correct inducing agent to release the existing prophages, or the prophages in the genome are defective virions and therefore noninducible. Alternatively, the large amount of phage tail-like particles present in the induced culture could be induced defective C. difficile prophages that also encode for a holin protein (Fig. 2). Ribotypes 087 and 220 that generated a siphovirus PCR product also yielded siphoviruses under TEM analysis (Fig. 2). Reassuringly, despite several inductions and subsequent analysis, no siphoviruses were observed in any of the siphovirus PCR-negative strains.
Further confidence in our molecular markers being indicative of phage carriage can be seen from the observation that the PCR-negative ribotype 081 had no inducible particles (Fig. 2). Interestingly, only the non-phage-carrying ribotype 081 is also the only strain that is not related to the others in terms of ribotyping profile (Fig. 1). This implies that prophage carriage could contribute to, or at least is correlated with, C. difficile strain relatedness.
Molecular diversity of C. difficile phage holin gene.
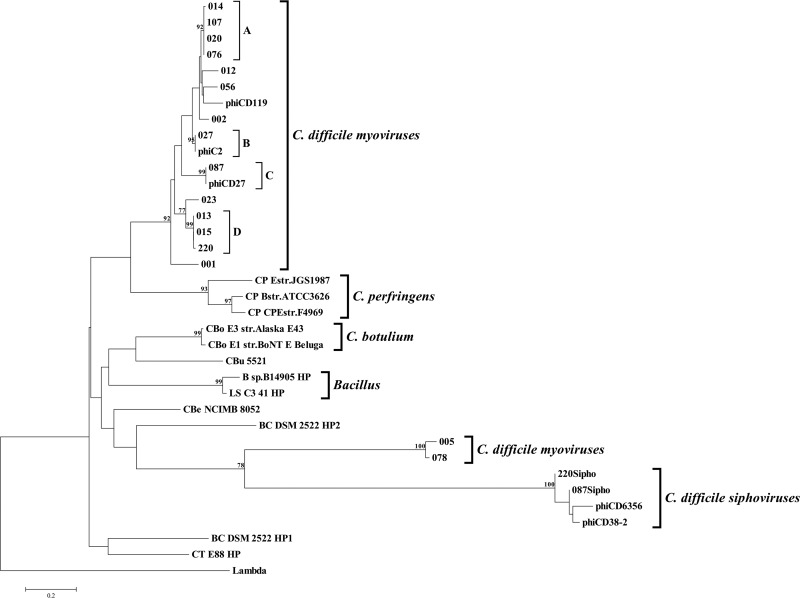
In addition to using the holin gene as a marker to detect prophage carriage, the holin genes were sequenced and phylogenetically analyzed to obtain a picture of phage relationship. This was determined from both NJ and MP analyses. Both algorithms gave a similar tree topology, and the tree from the NJ analysis is presented here. The tree shows two distinct clades of C. difficile myovirus holin sequences (supported by bootstrap values of 92 and 100%, respectively), which are clearly separated from the other clostridial species (Fig. 4). The four holin sequences from C. difficile siphoviruses fall within another independent and well-supported clade (Fig. 4). Interestingly, the siphovirus clade groups with the 005/078 myovirus clade (Fig. 4). These data demonstrate that the relationships between phages based on the holin sequences are consistent with those defined by morphology. It also suggests that there are at least three genetically distinct groups of C. difficile temperate phages: two within the myoviruses and a siphovirus group. Furthermore, the smaller myovirus group (carried by 005/078) may have a shared ancestry with the siphoviruses, presumably due to gene swapping during a mixed infection.
Fig 4.
Phylogenetic tree constructed using NJ method. The holin sequence from bacteriophage lambda was used as an outgroup to root the tree. Bootstrap values with levels of >75% are indicated at the nodes. The scale bar represents the proportion of nucleotides compared. Major clades and subclades are indicated. Abbreviations: CP, Clostridium perfringens; CT, Clostridium tetani; BC, Bacillus cellulosilyticus; B, Bacillus; LS, Lysinibacillus sphaericus; CBu, Clostridium butyricum; CBo, Clostridium botulinum; CBe, Clostridium beijerinckii; HP, hypothetical protein.
When the holin marker analysis is compared to the ribotyping analysis, we see that there are some relationships that are consistent with both analyses and others that are not. An example of the consistent relationships is the positioning of ribotype 078. It has a low similarity to the other ribotypes in the ribotyping dendrogram which mirror its holin gene position in the phylogenetic tree, where it falls in a discrete group away from the majority of the phages (Fig. 1 and 3). This divergent nature of 078 is consistent with the observation based on five fully sequenced C. difficile ribotypes (18, 47). The ribotype 078 relationship differs with respect to ribotype 005, where the holin gene is 95% identical, and yet the ribotype 005 is only 25% similar to the ribotype 078 in the ribotyping dendrogram, suggesting perhaps that there has been horizontal gene transfer between these strains. With reference to the TEM analysis, we see that the phage in ribotype 078 also has a distinct morphology, being the largest myovirus observed in the present study. Unfortunately, no inducible phage particles could be seen from the ribotype 005, which could be indicative of it carrying a defective prophage or simply a phage that was not inducible under the conditions tested (Fig. 2).
The phylogenetic tree shows that multiple ribotypes can carry identical holin sequences. Nine different ribotypes can be clustered into four subclades (A to D) within the main C. difficile myoviruses clade. Each subclade encodes identical holin sequences within their group (Fig. 4). TEM analysis of these ribotypes revealed that they are carrying inducible myoviruses with different sizes (Fig. 2). The fact that prophages with different morphological sizes carry the same holin sequences suggests either that there has been horizontal gene transfer between clinical strains or that the phage holin gene is fairly conserved within C. difficile, possibly as a result of the constraints on the function of the holin gene (33). Despite this disadvantage of being “conserved,” the holin marker provides a level of resolution that can reveal genetic diversity within the same phage morphological type.
The discovery of two very similar ribotypes 014 and 020 carrying identical holin sequences prompted a further analysis of nine clinical 027strains with different MLVA types to determine their holin sequences. Analyses showed that all 027 strains tested have an identical sequence that separates them from all other tested ribotypes (subclade B in Fig. 4). Again, this shows a tight correlation between holin sequences and bacterial strain identity and suggests a possible application of using holin as a diagnostic marker to target the hypervirulent 027 strains, which could potentially aid in diagnosis and patient management.
In addition to different ribotypes encoding the same holin sequences, two ribotypes examined in the present study, 220 and 087, carried distinct holins for myoviruses and siphoviruses, respectively. This was confirmed by TEM analysis (Fig. 2). This multiple prophage carriage in C. difficile agrees well with patterns of phage carriage seen in other pathogenic bacteria (7, 31, 37). One possible selective advantage for C. difficile encoding multiple prophages lies in the fact that the presence of a prophage in a bacterial strain may preclude the invasion of a related phage by homoimmunity (25). This may explain why C. difficile phages normally have a narrow host range (11, 14, 19, 30, 44). In addition, multiple phage carriage may influence the phenotype of the host bacteria more than single phage carriage, such as providing beneficial effects for the host surviving in adverse environment and increasing host toxin production (7, 39, 52). It has been reported that single prophage carriage in the ribotype 027 strain increased the toxin production (44). Whether multiple prophage carriage would further increase the toxin production in C. difficile remains unknown.
Two other clostridial clades (C. perfringens and C. botulinum) and one Bacillus clade (Lysinibacillus sphaericus and Bacillus sp. strain B14905) can also be observed from an analysis of the holin gene (Fig. 4). This suggests a potential use of holin gene as a molecular marker to study the diversity and distribution of phages within other bacterial species. For example, holin sequences from three fully sequenced C. perfringens strains shared 191 of 252 bp (76%) (see Fig. S7 in the supplemental material). This enables the use of a pair of PCR primers to specifically amplify this holin gene region, therefore allows a large-scale investigation of the prevalence of prophage carriage in C. perfringens, which currently remains unknown.
TEM observation.
All isolates were subjected to phage induction and TEM to establish the morphological diversity of their prophages. In brief, the 12 induced myoviruses showed a rich morphological diversity with capsids ranging from 50 to 73.5 nm and tail lengths ranging from 70.5 to 251.5 nm. The two siphoviruses induced here had similar sizes, with capsids from 51.7 to 59.8 nm and tail lengths from 193.4 to 195.1 nm (Fig. 2). Both siphoviruses have much shorter tails than previously reported C. difficile siphoviruses (8, 12, 19). Apart from intact phage particles, a large number of phage tail-like particles were observed from all of the induced samples except ribotype 081 that was holin PCR negative (Fig. 2).
We used both mitomycin C and norfloxacin as inducing agents to characterize phage morphology since both antibiotics do not always induce the same bacteriophages (32). Mitomycin C has been routinely used to induce temperate phages from C. difficile (6, 17, 45). Norfloxacin in comparison has initially been used to induce phages from Escherichia coli, and recently been used in C. difficile phage induction (26, 29, 32). Although both antibiotics function by interfering with DNA synthesis, they use different mechanisms (21, 29). Two phages (ϕ027-1 and ϕ220-1) were only induced following norfloxacin induction, a further nine (ϕ001, ϕ002, ϕ013, ϕ014, ϕ015, ϕ020, ϕ076, ϕ078, and ϕ220-2) were only induced following mitomycin C induction (Fig. 2). The other three phages (ϕ087-1, ϕ087-2, and ϕ107) were induced by both antibiotics (Fig. 2). A clear example of the impact of the use of both antibiotics can be seen from the observation that ribotype 220 yielded a myovirus when induced with norfloxacin and a siphovirus when induced with mitomycin C (Fig. 2), which agrees well with the PCR predictions.
In conclusion, there are several potential applications of the phage holin gene. It can be used as a diagnostic marker because of its sequence specificity for C. difficile. This sequence-based marker will enable identification of C. difficile and providing the phage carriage information at the same time. Potentially, the holin marker can also be used to investigate phage presence/absence in the environment without phage isolation and purification, which is currently the bottleneck for characterization of C. difficile phages. By using two sets of PCR primers, the holin marker could also provide researchers with “morphological data” directly from environmental samples.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by an MRC New Investigator Research Grant (G0700855) to M.R.J.C.
We thank Hemu Patel and David Jenkins (Department of Clinical Microbiology, University Hospitals of Leicester NHS Trust) for supplying fecal samples, Melinda Mayer (Institute of Food Research, Norwich, United Kingdom) for sending us ϕCD27, Mark Wilcox and Warren Fawley (Department of Microbiology, University of Leeds, Leeds, United Kingdom) for sending us the 027 ribotypes with different MLVA types. We also thank Kumar Rajakumar (Department of Infection, Immunity and Inflammation, University of Leicester, Leicester, United Kingdom) for useful discussions.
Footnotes
Published ahead of print 15 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Arvand M, Hauri AM, Zaiss NH, Witte W, Bettge-Weller G. 2009. Clostridium difficile ribotypes 001, 017, and 027 are associated with lethal C. difficile infection in Hesse, Germany. Euro Surveill. 14: 19403. [DOI] [PubMed] [Google Scholar]
- 2. Casjens SR, et al. 2005. The generalized transducing Salmonella bacteriophage ES18: complete genome sequence and DNA packaging strategy. J. Bacteriol. 187: 1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen F, Suttle CA. 1996. Evolutionary relationships among large double-stranded DNA viruses that infect microalgae and other organisms as inferred from DNA polymerase genes. Virology 219: 170–178 [DOI] [PubMed] [Google Scholar]
- 4. Chen F, Suttle CA, Short SM. 1996. Genetic diversity in marine algal virus communities as revealed by sequence analysis of DNA polymerase genes. Appl. Environ. Microbiol. 62: 2869–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cloud J, Kelly CP. 2007. Update on Clostridium difficile associated disease. Curr. Opin. Gastroenterol. 23: 4–9 [DOI] [PubMed] [Google Scholar]
- 6. Dei R. 1989. Observations on phage-typing of Clostridium difficile: preliminary evaluation of a phage panel. Eur. J. Epidemiol. 5: 351–354 [DOI] [PubMed] [Google Scholar]
- 7. Fogg PCM, Saunders JR, McCarthy AJ, Allison HE. 2012. Cumulative effect of prophage burden on Shiga toxin production in Escherichia coli. Microbiology 158: 488–497 [DOI] [PubMed] [Google Scholar]
- 8. Fortier LC, Moineau S. 2007. Morphological and genetic diversity of temperate phages in Clostridium difficile. Appl. Environ. Microbiol. 73: 7358–7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fuller NJ, Wilson WH, Joint IR, Mann NH. 1998. Occurrence of a sequence in marine cyanophages similar to that of T4 g20 and its application to PCR-based detection and quantification techniques. Appl. Environ. Microbiol. 64: 2051–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goh S, Chang BJ, Riley TV. 2005. Effect of phage infection on toxin production by Clostridium difficile. J. Med. Microbiol. 54: 129–135 [DOI] [PubMed] [Google Scholar]
- 11. Goh S, Ong PF, Song KP, Riley TV, Chang BJ. 2007. The complete genome sequence of Clostridium difficile phage phiC2 and comparisons to phiCD119 and inducible prophages of CD630. Microbiology 153: 676–685 [DOI] [PubMed] [Google Scholar]
- 12. Goh S, Riley TV, Chang BJ. 2005. Isolation and characterization of temperate bacteriophages of Clostridium difficile. Appl. Environ. Microbiol. 71: 1079–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goorhuis A, et al. 2008. Clostridium difficile PCR ribotype 078: an emerging strain in humans and in pigs? J. Clin. Microbiol. 46: 1157–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Govind R, Fralick JA, Rolfe RD. 2006. Genomic organization and molecular characterization of Clostridium difficile bacteriophage phiCD119. J. Bacteriol. 188: 2568–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Govind R, Vediyappan G, Rolfe RD, Dupuy B, Fralick JA. 2009. Bacteriophage-mediated toxin gene regulation in Clostridium difficile. J. Virol. 83: 12037–12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gurtler V. 1993. Typing of Clostridium difficile strains by PCR-amplification of variable length 16S–23S rDNA spacer regions. J. Gen. Microbiol. 139: 3089–3097 [DOI] [PubMed] [Google Scholar]
- 17. Hawkins CC, Buggy BP, Fekety R, Schaberg DR. 1984. Epidemiology of colitis induced by Clostridium difficile in hamsters: application of a bacteriophage and bacteriocin typing system. J. Infect. Dis. 149: 775–780 [DOI] [PubMed] [Google Scholar]
- 18. He M, et al. 2010. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc. Natl. Acad. Sci. U. S. A. 107: 7527–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Horgan M, et al. 2010. Genome analysis of the Clostridium difficile phage phiCD6356, a temperate phage of the Siphoviridae family. Gene doi:10.1016/j.gene.2010.04.010 [DOI] [PubMed] [Google Scholar]
- 20. Indra A, et al. 2008. Characterization of Clostridium difficile isolates using capillary gel electrophoresis-based PCR ribotyping. J. Med. Microbiol. 57: 1377–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iyer VN, Szybalski W. 1963. A molecular mechanism of mitomycin action: linking of complementary DNA strands. Proc. Natl. Acad. Sci. U. S. A. 50: 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kelly CP, Pothoulakis C, Lamont JT. 1994. Clostridium difficile colitis. N. Engl. J. Med. 330: 257–262 [DOI] [PubMed] [Google Scholar]
- 23. Kuijper EJ, Coignard B, Tüll P. 2006. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin. Microbiol. Infect. 12: 2–18 [DOI] [PubMed] [Google Scholar]
- 24. Kumar S, Nei M, Dudley J, Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 9: 299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Levin BR, Lenski RE. 1983. Coevolution in bacteria and their viruses and plasmids, p 99–127 In Futuyma DJ, Slatkin M. (ed), Coevolution. Sinauer, Sunderland, MA [Google Scholar]
- 26. Los JM, Los M, Wegrzyn G, Wegrzyn A. 2009. Differential efficiency of induction of various lambdoid prophages responsible for production of Shiga toxins in response to different induction agents. Microb. Pathog. 47: 289–298 [DOI] [PubMed] [Google Scholar]
- 27. Marsden G, et al. 2010. Array comparative hybridisation reveals a high degree of similarity between UK and European clinical isolates of hypervirulent Clostridium difficile. BMC Genomics 11: 389 doi:10.1186/1471-2164-11-389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Marston MF, Sallee JL. 2003. Genetic diversity and temporal variation in the cyanophage community infecting marine Synechococcus species in Rhode Island's coastal waters. Appl. Environ. Microbiol. 69: 4639–4647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Matsushiro A, Sato K, Miyamoto H, Yamamura T, Honda T. 1999. Induction of prophages of enterohemorrhagic Escherichia coli O157:H7 with norfloxacin. J. Bacteriol. 181: 2257–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mayer MJ, Narbad A, Gasson MJ. 2008. Molecular characterization of a Clostridium difficile bacteriophage and its cloned biologically active endolysin. J. Bacteriol. 190: 6734–6740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McCarthy AJ, Witney AA, Lindsay JA. 2012. Staphylococcus aureus lysogenic bacteriophage: carriage and horizontal gene transfer (HGT) is lineage associated. Front. Cell Infect. Microbiol. 2: 6 doi:10.3389/fcimb.2012.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nale JY, et al. 2012. Diverse temperate bacteriophage carriage in Clostridium difficile 027 strains. PLoS One 7: e37263 doi:10.1371/journal.pone.0037263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oakley BB, et al. 2011. Comparative genomics of four closely related Clostridium perfringens bacteriophages reveals variable evolution among core genes with therapeutic potential. BMC Genomics 12: 282 doi:10.1186/1471-2164-12-282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. O'Neill GL, Ogunsola FT, Brazier JS, Duerden BI. 1996. Modification of a PCR ribotyping method for application as a routine typing scheme for Clostridium difficile. Anaerobe 2: 205–209 [Google Scholar]
- 35. Pecenkova T, Paces V. 1999. Molecular phylogeny of phi 29-like phages and their evolutionary relatedness to other protein-primed replicating phages and other phages hosted by Gram-positive bacteria. J. Mol. Evol. 48: 197–208 [DOI] [PubMed] [Google Scholar]
- 36. Petrov VM, et al. 2006. Plasticity of the gene functions for DNA replication in the T4-like phages. J. Mol. Biol. 361: 46–68 [DOI] [PubMed] [Google Scholar]
- 37. Ramirez M, Severina E, Tomasz A. 1999. A high incidence of prophage carriage among natural isolates of Streptococcus pneumoniae. J. Bacteriol. 181: 3618–3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Romero P, Garcia E, Mitchell TJ. 2009. Development of a prophage typing system and analysis of prophage carriage in Streptococcus pneumoniae. Appl. Environ. Microbiol. 75: 1642–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ronning CM, et al. 2010. Genetic and phenotypic diversity in Burkholderia: contributions by prophage and phage-like elements. BMC Microbiol. 10: 202 doi:10.1186/1471-2180-10-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roux S, Enault F, Bronner G, Debroas D. 2011. Comparison of 16S rRNA and protein-coding genes as molecular markers for assessing microbial diversity (Bacteria and Archaea) in ecosystems. FEMS Microbiol. Lett. 78: 617–628 [DOI] [PubMed] [Google Scholar]
- 41. Sadeghifard N, Gurtler V, Beer M, Seviour RJ. 2006. The mosaic nature of intergenic 16S-23S rRNA spacer regions suggests rRNA operon copy number variation in Clostridium difficile strains. Appl. Environ. Microbiol. 72: 7311–7323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, vol 1 Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 43. Sebaihia M, et al. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38: 779–786 [DOI] [PubMed] [Google Scholar]
- 44. Sekulovic O, Meessen-Pinard M, Fortier L-C. 2011. Prophage-stimulated toxin production in Clostridium difficile NAP1/027 lysogens. J. Bacteriol. 193: 2726–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sell TL, Schaberg DR, Fekety FR. 1983. Bacteriophage and bacteriocin typing scheme for Clostridium difficile. J. Clin. Microbiol. 17: 1148–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Short CM, Suttle CA. 2005. Nearly identical bacteriophage structural gene sequences are widely distributed in both marine and freshwater environments. Appl. Environ. Microbiol. 71: 480–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stabler RA, et al. 2006. Comparative phylogenomics of Clostridium difficile reveals clade specificity and microevolution of hypervirulent strains. J. Bacteriol. 188: 7297–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stabler RA, et al. 2009. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 10: R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Starr J. 2005. Clostridium difficile associated diarrhoea: diagnosis and treatment. BMJ 331: 498–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- 51. Wang IN, Smith DL, Young R. 2000. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54: 799–825 [DOI] [PubMed] [Google Scholar]
- 52. Wang X, et al. 2010. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 1: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Warny M, et al. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366: 1079–1084 [DOI] [PubMed] [Google Scholar]
- 54. Young RY. 1992. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56: 430–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.