Abstract
Tailed double-stranded DNA (dsDNA) bacteriophages frequently harbor structural proteins displaying peptidoglycan hydrolytic activities. The tape measure protein from Staphylococcus aureus bacteriophage vB_SauS-phiIPLA35 has a lysozyme-like and a peptidase_M23 domain. This report shows that the lysozyme-like domain (TG1) has muramidase activity and exhibits in vitro lytic activity against live S. aureus cells, an activity that could eventually find use in the treatment of infections.
TEXT
The peptidoglycan layer is a barrier that bacteriophages must overcome to infect the bacterial host. In this regard, virions are provided with structural proteins containing catalytic domains that play roles in attachment to host cells through sugar binding or degradation of the polysaccharides on the cell surface (6). Murein hydrolases appear to be widespread in virions of bacteriophages infecting Gram-positive and Gram-negative bacteria, where the enzyme is either part of a large protein or found associated with other structural components of the virion (10). These peptidoglycan hydrolases have been related to modification in the peptidoglycan structure necessary for phage infection. Sequence analysis revealed that peptidoglycan-degrading domains are frequently located in tape measure proteins (TMPs) (6), although in most cases their activity has not been analyzed. The TMPs of several mycobacteriophages contain domains that mediate peptidoglycan degradation and play an essential role when these phages infect stationary-phase cells (12). In a similar way, phage T5 straight tail fiber is a multifunctional protein that not only acts as a tape measure protein but also carries fusogenic and muralytic activities (3). In other phages, virion-associated proteins with peptidoglycan hydrolytic activity might play a similar role in the phage infection process. Thus, the bacteriophage T7 gp16 with transglycosylase activity is not essential for phage growth but was shown to be beneficial during infection of Escherichia coli cells grown to high cell density or low temperatures, where the murein is more highly cross-linked (9). Furthermore, the amino-terminal end of the bacteriophage PRD1 structural protein P7 carries a conserved transglycosylase domain, which is located at the particle vertices and is involved in the early steps of the PRD1 life cycle (16). Additionally, the tail baseplate of bacteriophage T4 contains an essential protein, gp5, which possesses lysozyme activity and functions to locally dissolve the periplasmic cell wall (1).
Staphylococcus aureus bacteriophage vB_SauS-phiIPLA35 (in short, phiIPLA35) is a member of the Siphoviridae family, previously isolated from dairy samples (7). The complete genome sequence was determined (GenBank accession no. EU861005), and zymogram analysis of virions revealed the presence of a phiIPLA35 virion-associated muralytic enzyme (8). In this study, a muramidase domain has been described as part of the TMP, a structural component of phage phiIPLA35. The lytic activity of this protein against S. aureus cells has been determined, and its cleavage sites in the peptidoglycan have been identified.
The tape measure protein (TMP) of phage phiIPLA35 has two putative catalytic domains.
Computer-based similarity searches using BLASTp revealed that protein TMP (gp50; 2,066 amino acids [aa) (YP_002332413.1) showed 98% to 99% similarity to TMPs from the staphylococcal phages 42E, phi47, phi12, phiSLT, tp310-2, and phi3A. Conserved-domain analyses of TMP performed using InterProScan identified a structural conserved domain, tape_meas_TP901 (from aa 323 to 666), related to tail length in TP901-like phages (11). In addition, two typical catalytic domains were observed: a peptidase_M23 domain (from aa 1706 to 1796) and a lysozyme-like domain (from aa 1829 to 1920). Members of peptidase family M23 are zinc metallopeptidases with Gly-Gly endopeptidase activity, and many of them, such as lysostaphin, have specific hydrolytic activity with respect to peptidoglycan (17). Lysozyme activities catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-d-glucosamine (GlcNAc). Lysozyme catalytic domains are often found in cell wall hydrolases from S. aureus phages (14). However, only phages 42E, phi47, phi12, phiSLT, tp310-2, and phi3A revealed a TMP with a lysozyme-like domain homolog to the phiIPLA35 TMP.
The lysozyme-like domain (TG1) of TMP has peptidoglycan muramidase activity.
To confirm the predicted peptidoglycan hydrolytic activity of TG1, a truncated protein (from aa 1829 to 2066) containing the lysozyme-like domain was cloned and overexpressed. The phiIPLA35 tg1 fragment (717 bp) was codon optimized based on the E. coli codon usage and commercially synthesized (Biomedal, Sevilla, Spain). Then, tg1-opt was subcloned between the NdeI and XhoI sites in the multicloning site of the inducible expression vector pET21a (EMD Biosciences, San Diego, CA), which introduces a C-terminal 6×His tag. E. coli BL21(DE3)/pLysS was used in protein expression assays (19). Purification of TG1 (30.1 kDa) was performed by nickel-nitrilotriacetic acid (Ni-NTA) chromatography (15). Peptidoglycan cleavage site determination was carried out by reverse-phase high-performance liquid chromatography (HPLC) and mass spectrometry (MS) (2). S. aureus SA113 peptidoglycan (2 mg) was isolated by CeCoLabs UG, Tübingen, Germany, and incubated overnight with 27 μg of TG1 in a final volume of 250 μl at 37°C with shaking in MES-NaOH (morpholineethanesulfonic acid-NaOH) (50 mM, pH 5) buffer. Samples were boiled 3 min to stop the reaction and centrifuged at 14,000 × g for 5 min, and the soluble fraction was separated by reverse-phase HPLC using a Nucleosil 100 column (Maisch GmbH, Ammerbuch, Germany) (C18; 125 by 4.6 mm; 5 μm) and a water/0.1% trifluoroacetic acid (TFA):80% acetonitrile/0.1% TFA gradient for 150 min at a flow rate of 0.5 ml/min. The molecular masses of the muropeptides were determined by LC-MS. The primary product of TG1 digestion was the disaccharide pentapeptide GlcNAc-(β1–4)-MurNAc-(l-Ala-d-iGln-l-Lys-(Gly)-d-Ala-COOH) (Mr 951.7), which in MS yields peaks with m/z 952.6 (protonated) and m/z 950.5 (unprotonated) (Fig. 1). This main digestion product is also obtained with the mutanolysin digestion of the same peptidoglycan (data not shown). Therefore, TG1 is most likely a muramidase that can digest MurNAc-GlcNAc linkages.
Fig 1.
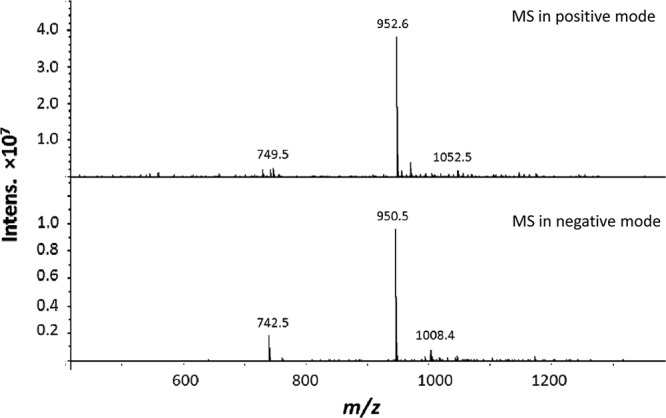
MS analysis of the overnight digestion of 2 mg of S. aureus SA113 peptidoglycan with 27 μg of TG1 in buffer (MES-NaOH [50 mM], pH 5). The primary product of the digestion was the disaccharide pentapeptide GlcNAc-(β1–4)-MurNAc-(l-Ala-d-iGln-l-Lys-(Gly)-d-Ala-COOH) (Mr, 951.7).
There are no previous reports about the involvement of virion-associated peptidoglycan hydrolases in S. aureus phage infection. However, the presence in phages phiIPLA88 and phiMR11 of proteins with muralytic activities might indicate its involvement in local cell wall degradation, allowing the subsequent introduction of DNA into the host cytoplasm (13, 14). In phiIPLA35, the presence of an active muramidase domain in the TMP could also indicate its contribution to the infection process, taking into account that no other virion-associated proteins with peptidoglycan hydrolase activity were identified in this phage (8).
The TG1 domain displays lytic activity against S. aureus cells.
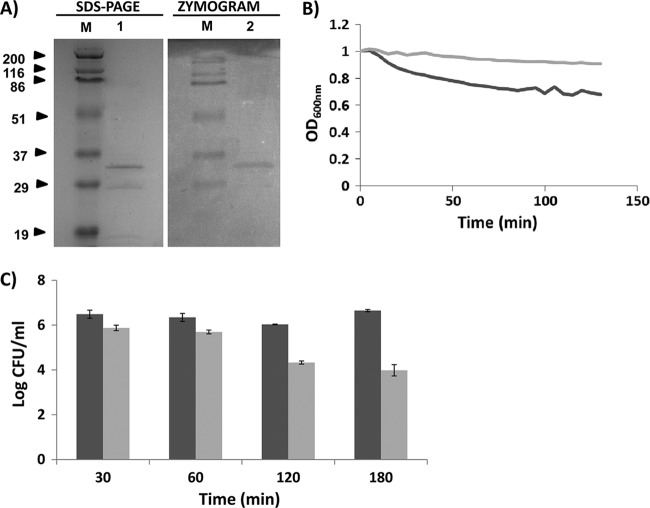
The peptidoglycan hydrolytic ability of TG1 was assayed by zymogram analysis using 10 ml of 15% (wt/vol) sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with or without S. aureus Sa9 cells from a 300-ml culture (A600 = 0.5) embedded in the gel. A clear band consistent with the predicted molecular mass (30.1 kDa) was observed after analysis of 5 μg of TG1 protein in the SDS-PAGE gel containing the staphylococcal cells. This clear band was the result of the lysis of the S. aureus cells embedded in the gel (Fig. 2A, zymogram). Moreover, in order to assess the lytic activity of TG1 against live S. aureus Sa9 cells, turbidity reduction assays were performed with S. aureus cells grown to the logarithmic phase (optical density at 600 nm [OD600] = 0.4 to 0.6), harvested, and resuspended in buffer A (MES-NaOH [50 mM], pH 5) to an OD600 of 1.0. As shown in Fig. 2B, A 32.7% decrease in OD600 was obtained after 120 min of incubation at 37°C with 5 μM TG1 (specific activity of 0.00138 expressed as ΔOD600 min−1 μM−1), which indicated the lytic ability of the TG1 protein. This result was confirmed by a cell viability assay using exponentially growing cells which were recovered by centrifugation, washed, and resuspended in buffer A to an OD600 of 0.1. Then, 5 μM TG1 was mixed with S. aureus Sa9 live cells to a final concentration of 4 × 106 CFU/ml and incubated for 30 min and 1, 2, and 3 h at 37°C; staphylococcal viable counts were reduced by 75.5%, 77.5%, 98%, and 99.7%, respectively, compared with the untreated control cultures (Fig. 2C). These results support the functionality of the putative lysozyme-like domain found by the bioinformatics analysis. Nevertheless, the activity seems to be somewhat weaker than that shown by other staphylococcal virion-associated peptidoglycan hydrolases such phiIPLA88 HydH5 (14) and phiMR11 gp61 (13). This weaker specific activity (about 23-fold lower than that of HydH5) could have been due to the presence of only one catalytic domain, the lysozyme-like domain, in contrast to HydH5, which showed two active catalytic domains (14). Nevertheless, the peptidoglycan hydrolytic activity of TMP as part of phiIPLA35 virions might also be enhanced by the peptidase_M23 domain. Therefore, although TG1 lytic activity was moderate against staphylococci, further investigation is worthwhile. We previously reported that the fusion of a staphylococcal SH3b cell wall-binding domain to the HydH5 CHAP domain resulted in a 4.8-fold increase of its lytic activity (15). Furthermore, it is plausible to swap different catalytic domains by protein engineering, generating proteins with improved lytic activity (4, 15). In this context, the search of catalytic domains with new biochemical properties is justified. Moreover, with the increasing prevalence of antibiotic-resistant pathogens, the possibility of using phage lytic proteins as novel therapeutic antibacterial agents has a renewed interest. This idea is supported not only by its ability to kill bacteria but also by the low probability of resistance development. In fact, some phage endolysins have already been successfully assayed in preclinical trials involving animal models of human diseases (5). Additionally, chimeric lysins consisting of an endopeptidase domain of the streptococcal phage lysin λSA2 and SH3b cell-binding domains from the staphylococcal phage lysin LysK and lysostaphin showed antimicrobial activity against S. aureus-induced mastitis in mouse models (18).
Fig 2.
Lytic activity of TG1 protein against S. aureus. (A) A 5-μg volume of nickel affinity-purified TG1 (30.1 kDa) was resolved on a 15% SDS-PAGE gel in the absence (SDS-PAGE, lane 1) or in the presence (zymogram, lane 2) of S. aureus Sa9 cells. Gels were either stained with Coomassie blue (SDS-PAGE) or incubated in water for 1 h at room temperature (zymogram). Lanes M, standard molecular mass marker in kilodaltons (Prestained SDS-PAGE Standards, broad range; Bio-Rad Laboratories). (B) Representative turbidity reduction assay performed by challenging S. aureus Sa9 cells with 5 μM TG1 for 120 min at 37°C. The light-gray line indicates the untreated cultures (S. aureus Sa9 cells plus MES-NaOH [50 mM], pH 5); the dark-gray line indicates treated cultures (S. aureus Sa9 cells plus 5 μM TG1). (C) Viability test performed by challenging S. aureus Sa9 cells with 5 μM TG1 for 30 min and 1, 2, and 3 h at 37°C (light-gray bars). The dark-gray bars indicate untreated cultures (S. aureus Sa9 plus MES-NaOH [50 mM], pH 5). Error bars represent the means ± standard deviations of the results of two independent assays.
In conclusion, the identification of muramidase activity associated with the phage phiIPLA35 tape measure protein showing lytic capabilities against live S. aureus cells is reported. This novel peptidoglycan lytic domain may be of help for designing potent antimicrobials against this pathogen.
ACKNOWLEDGMENTS
This research study was supported by grants AGL2009-13144-C02-01 (Ministry of Science and Innovation, Spain), IB08-052 (Science, Technology and Innovation Programme, Principado de Asturias, Spain), and PIE200970I090 (CSIC, Spain). L.R.-R. is a fellow of the Science, Technology and Innovation Programme (Principado de Asturias, Spain).
Footnotes
Published ahead of print on 22 June 2012.
REFERENCES
- 1. Arisaka F, Kanamaru S, Leiman P, Rossmann MG. 2003. The tail lysozyme complex of bacteriophage T4. Int. J. Biochem. Cell Biol. 35: 16–21 [DOI] [PubMed] [Google Scholar]
- 2. Biswas R, et al. 2006. Activity of the major staphylococcal autolysin Atl. FEMS Microbiol. Lett. 259: 260–268 [DOI] [PubMed] [Google Scholar]
- 3. Boulanger P, et al. 2008. Phage T5 straight tail fiber is a multifunctional protein acting as a tape measure and carrying fusogenic and muralytic activities. J. Biol. Chem. 283: 13556–13564 [DOI] [PubMed] [Google Scholar]
- 4. Donovan DM, et al. 2006. Peptidoglycan hydrolase fusions maintain their parental specificities. Appl. Environ. Microbiol. 72: 2988–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fischetti VA. 2010. Bacteriophage endolysins: a novel anti-infective to control Gram-positive pathogens. Int. J. Med. Microbiol. 300: 357–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fraser JS, Maxwell KL, Davidson AR. 2007. Immunoglobulin-like domains on bacteriophage: weapons of modest damage? Curr. Opin. Microbiol. 10: 382–387 [DOI] [PubMed] [Google Scholar]
- 7. GarcíA P, Madera C, Martínez B, Rodríguez A. 2007. Biocontrol of Staphylococcus aureus in curd manufacturing processes using bacteriophages. Int. Dairy J. 17: 1232–1239 [Google Scholar]
- 8. García P, et al. 2009. Functional genomic analysis of two Staphylococcus aureus phages isolated from the dairy environment. Appl. Environ. Microbiol. 75: 7663–7673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moak M, Molineux IJ. 2000. The role of the lytic transglycosylase motif of bacteriophage T7 in the initiation of infection. Mol. Microbiol. 37: 345–355 [DOI] [PubMed] [Google Scholar]
- 10. Moak M, Molineux IJ. 2004. Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol. Microbiol. 51: 1169–1183 [DOI] [PubMed] [Google Scholar]
- 11. Pedersen M, Ostergaard S, Bresciani J, Vogensen FK. 2000. Mutational analysis of two structural genes of the temperate lactococcal bacteriophage TP901-1 involved in tail length determination and baseplate assembly. Virology 276: 315–328 [DOI] [PubMed] [Google Scholar]
- 12. Piuri M, Hatfull GF. 2006. A peptidoglycan hydrolase motif within the mycobacteriophage TM4 tape measure protein promotes efficient infection of stationary phase cells. Mol. Microbiol. 62: 1569–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rashel M, et al. 2008. Tail-associated structural protein gp61 of Staphylococcus aureus phage ΦMR11 has bifunctional lytic activity. FEMS Microbiol. Lett. 284: 9–16 [DOI] [PubMed] [Google Scholar]
- 14. Rodríguez L, et al. 2011. Lytic activity of the virion-associated peptidoglycan hydrolase HydH5 of Staphylococcus aureus bacteriophage vB_SauS-phiIPLA88. BMC Microbiol. 11: 138 doi:10.1186/1471-2180-11-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rodríguez-Rubio L, Martínez B, Rodríguez A, Donovan DM, GarcíA P. 2012. Enhanced staphylolitic activity of the Staphylococcus aureus bacteriophage vB_SauS-phiIPLA88 HydH5 virion-associated peptidoglycan hydrolase: fusions, deletions, and synergy with LysH5. Appl. Environ. Microbiol. 78: 2241–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rydman PS, Bamford DH. 2000. Bacteriophage PRD1 DNA entry uses a viral membrane-associated transglycosylase activity. Mol. Microbiol. 37: 356–363 [DOI] [PubMed] [Google Scholar]
- 17. Schindler CA, Schuhardt VT. 1965. Purification and properties of lysostaphin—a lytic agent of Staphylococcus aureus. Biochim. Biophys. Acta 97: 242–250 [DOI] [PubMed] [Google Scholar]
- 18. Schmelcher M, Powell AM, Becker SC, Camp MJ, Donovan DM. 2012. Chimeric phage lysins act synergistically with lysostaphin to kill mastitis-causing Staphylococcus aureus in murine mammary glands. Appl. Environ. Microbiol. 78: 2297–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high level expression of cloned genes. J. Mol. Biol. 189: 113–130 [DOI] [PubMed] [Google Scholar]