Abstract
Saccharomyces cerevisiae has evolved a highly efficient strategy for energy generation which maximizes ATP energy production from sugar. This adaptation enables efficient energy generation under anaerobic conditions and limits competition from other microorganisms by producing toxic metabolites, such as ethanol and CO2. Yeast fermentative and flavor capacity forms the biotechnological basis of a wide range of alcohol-containing beverages. Largely as a result of consumer demand for improved flavor, the alcohol content of some beverages like wine has increased. However, a global trend has recently emerged toward lowering the ethanol content of alcoholic beverages. One option for decreasing ethanol concentration is to use yeast strains able to divert some carbon away from ethanol production. In the case of wine, we have generated and evaluated a large number of gene modifications that were predicted, or known, to impact ethanol formation. Using the same yeast genetic background, 41 modifications were assessed. Enhancing glycerol production by increasing expression of the glyceraldehyde-3-phosphate dehydrogenase gene, GPD1, was the most efficient strategy to lower ethanol concentration. However, additional modifications were needed to avoid negatively affecting wine quality. Two strains carrying several stable, chromosomally integrated modifications showed significantly lower ethanol production in fermenting grape juice. Strain AWRI2531 was able to decrease ethanol concentrations from 15.6% (vol/vol) to 13.2% (vol/vol), whereas AWRI2532 lowered ethanol content from 15.6% (vol/vol) to 12% (vol/vol) in both Chardonnay and Cabernet Sauvignon juices. Both strains, however, produced high concentrations of acetaldehyde and acetoin, which negatively affect wine flavor. Further modifications of these strains allowed reduction of these metabolites.
INTRODUCTION
Saccharomyces sensu stricto species, in particular Saccharomyces cerevisiae, have evolved a strategy for sugar utilization that maximizes ethanol production (39). This adaptation permits energy extraction under fermentation and, perhaps more importantly, leads to the production of a toxic metabolite, ethanol, which inhibits the growth of competing microorganisms. Production of other metabolites by yeast is also important, particularly in the context of alcoholic beverage industries, as these molecules shape the organoleptic properties of beer and wines.
Largely driven by consumer demand for rich and ripe fruit flavor profiles, the alcohol content of some beverages has increased in recent years (29). High alcohol content in wine, for example, has several important consequences: it can compromise product quality, including increasing the perception of mouthfeel parameters such as hotness and viscosity, and to a lesser extent, sweetness, acidity, aroma, flavor intensity, and textural properties can be negatively impacted (20–22); costs to the consumer are higher in countries where taxes are levied according to ethanol content; and excessive alcohol consumption has negative health impacts.
The combination of quality, economic, and health issues associated with high-alcohol wines has led to significant interest in the development of technologies to produce wines with reduced ethanol concentrations that retain balance, flavor profile, and other sensory characteristics (29).
Several genetic modification (GM) strategies are available to divert yeast metabolism away from ethanol formation by redirecting carbon to other endpoints (29, 41). However, maintaining yeast fermentation performance and wine quality in low-ethanol GM yeast strains remains a major challenge.
One GM approach to decrease ethanol formation involves increasing glycerol production. Several gene modifications can achieve this end, for example overexpression of GPD1 and/or GPD2 genes, which encode glycerol 3-phosphate dehydrogenase (Gpd) isozymes (6, 10, 15, 35, 46); deleting PDC genes encoding pyruvate decarboxylase (35); impairing alcohol dehydrogenase (ADH/Adh) expression and activity (13, 27); deleting TPI1, which encodes triose phosphate isomerase (7, 8); and modifying the glycerol transporter encoded by FPS1 (56, 57).
Gpd catalyzes the conversion of dihydroxyacetone phosphate to glycerol 3-phosphate, which is subsequently dephosphorylated to glycerol by glycerol 3-phosphatase. Overexpression of GPD1 or GPD2 has been shown to increase glycerol yield by up to 548%, depending on the yeast strain, medium and fermentation conditions (29), and this is associated with lower ethanol production. However, production of other metabolites that might negatively impact wine quality is also observed; for instance, GPD overexpression leads to increased acetic acid production (10, 34, 46). At least some of these negative effects can be rectified by engineering further modifications. For example, the problem of increased acetic acid concentration can be ameliorated by deleting ALD6, which encodes aldehyde dehydrogenase (6, 15).
Yeasts engineered for GPD overexpression and ALD6 deletion have been reported to produce elevated concentrations of acetoin, which has the aroma of rancid butter and has a low sensory threshold. However, this compound can be converted to innocuous 2,3-butanediol by increasing expression of BDH1 which encodes 2,3-butanediol dehydrogenase (16).
Pyruvate decarboxylase catalyzes the decarboxylation of pyruvate to acetaldehyde and CO2. There are three pyruvate decarboxylase genes in Saccharomyces cerevisiae: PDC1, PDC5, and PDC6, which are upregulated by the transcription factor Pdc2p; only Pdc1p and Pdc5p are known to be active in yeast during fermentation (18, 19, 26). Yeast strains lacking all three PDC genes are unable to grow in medium containing glucose as a sole carbon source, with excess NADH production inhibiting glycolytic flux (43). However, PDC2 deletion reduced ethanol yield with associated increased glycerol production (35).
Alcohol dehydrogenases (encoded by ADH1, AHD3, ADH4, and ADH5) play an important role in yeast fermentation, catalyzing the reduction of acetaldehyde to ethanol (29). Yeast lacking the major fermentative isoform, ADH1, show decreased ethanol production and increased glycerol synthesis (27). In addition, deletion of ADH1 causes a considerable decrease in growth rate (9, 12, 27). Yeast strains lacking additional ADH genes (ADH3 and ADH4) exhibited a further decrease in ethanol production, greater glycerol formation, and significantly impaired growth (13).
Triose phosphate isomerase, encoded by TPI1, functions at the branch point of glycolysis, converting dihydroxyacetone phosphate to glyceraldehyde 3-phosphate. Yeast strains lacking TPI1 produced elevated glycerol concentration with a concomitant substantial decrease in ethanol production (8). Growth on glucose as a sole carbon source was not possible for TPI1 mutants; the addition of ethanol, however, restored growth, indicating an imbalance in NADH supply (7, 36).
Fps1p is a member of the major intrinsic protein (MIP) family of channel proteins and facilitates glycerol export and import in S. cerevisiae. Fps1p regulates intracellular glycerol concentrations, and its expression is controlled by the osmolarity of the surrounding medium (31, 57). Expression of a truncated form of Fps1p lacking the N-terminal domain in yeast resulted in continuous glycerol leakage from the cell, which was compensated for by increasing glycerol production (45). Yeast carrying the modified form of Fps1p, however, showed impaired growth on glucose and decreased biomass formation, suggesting that it may be of limited value in industrial yeast strains.
Another potential strategy to decrease ethanol production is to divert carbon toward the synthesis of organic acids, such as gluconic acid and acids involved in the tricarboxylic acid (TCA) cycle. Glucose oxidase catalyzes the conversion of glucose into gluconic acid and hydrogen peroxide. However, this enzyme is not encoded in the S. cerevisiae genome. Expression of the Aspergillus niger GOX gene in S. cerevisiae, and secretion of its product, led to reduced ethanol concentration during trial fermentations (33). However, the effect of hydrogen peroxide on the appearance of the resulting wine and the requirement for oxygen was not reported.
Deletions and overexpression of several genes involved in the TCA cycle have been shown to not only impact the formation of organic acids but also affect ethanol production (1, 37, 51). Although these modifications are promising, their real impact in a winemaking context has not been explored.
In this work, we evaluated several strategies aimed at decreasing ethanol production during wine fermentation. By performing stable chromosomal gene modifications in a wine yeast, we determined the most effective gene modifications for reducing ethanol concentration. Although the best low-ethanol strains carry no foreign DNA (i.e., they are self cloned), they are still considered genetically modified organisms (GMOs). The use of GMOs in food and beverages continues to be the subject of intense debate, particularly for winemakers and the wine sector (40, 42).
MATERIALS AND METHODS
Strains and molecular techniques.
The wine yeast AWRI1631 was used as the parental strain for all genetically modified constructs. AWRI1631 is a stable haploid generated by sporulation of a wine yeast and deletion of the HO locus (4).
Genetic modifications of AWRI1631 performed for work described in this paper were chromosomally integrated and included (i) gene deletions, where the open reading frame (ORF) of the target gene was deleted; (ii) promoter replacement, where the native promoter of the target gene was replaced with a strong constitutive yeast promoter; (iii) gene cassette insertion, where a gene under the control of a strong constitutive yeast promoter was inserted into the chromosome; and (iv) discrete modifications, including nucleotide substitutions and deletions. Strains generated for this study and their genetic modifications are listed in Table 1.
Table 1.
Genetic modifications of constructed strains and ethanol production compared to parental strain AWRI1631
| Strain | Modified gene(s) | Genetic modification | Ethanol (%)a | Significanceb |
|---|---|---|---|---|
| AWRI1631 | None | None (parental strain) | 100 ± 1.4 | NS |
| AWRI1631 ΔACO1 | ACO1 | ORF deletion | 99 ± 0.6 | NS |
| AWRI1631 ΔACO2 | ACO2 | ORF deletion | 99 ± 0.2 | NS |
| AWRI1631 ΔADH1 | ADH1 | ORF deletion | 100 ± 0.6 | NS |
| AWRI1631 ΔADH3 | ADH3 | ORF deletion | 99 ± 0.4 | NS |
| AWRI1631 ΔADH1ΔADH3 | ADH1, ADH3 | ORF deletion | 99 ± 0.6 | NS |
| AWRI1631 ΔFRDS1 | FRD1 | ORF deletion | 100 ± 0.9 | NS |
| AWRI1631 ΔGPH1 | GPH1 | ORF deletion | 99 ± 0.3 | NS |
| AWRI1631 ΔGRR1 | GRR1 | ORF deletion | 100 ± 1.5 | NS |
| AWRI1631 ΔHXK2 | HXK2 | ORF deletion | 97 ± 0.4 | S |
| AWRI1631 ΔIDH1 | IDH1 | ORF deletion | 99 ± 0.4 | NS |
| AWRI1631 ΔIDP2 | IDP2 | ORF deletion | 99 ± 0.6 | NS |
| AWRI1631 ΔKGD1 | KGD1 | ORF deletion | 100 ± 0.1 | NS |
| AWRI1631 ΔMDH1 | MDH1 | ORF deletion | 99 ± 1.4 | NS |
| AWRI1631 ΔMIG1 | MIG1 | ORF deletion | 97 ± 0.8 | S |
| AWRI1631 ΔMIG2 | MIG2 | ORF deletion | 99 ± 0.4 | NS |
| AWRI1631 ΔOSM1 | OSM1 | ORF deletion | 99 ± 0.4 | NS |
| AWRI1631 ΔPDC1 | PDC1 | ORF deletion | 100 ± 2.1 | NS |
| AWRI1631 ΔPDC5 | PDC5 | ORF deletion | 97 ± 0.3 | S |
| AWRI1631 ΔPYC1 | PYC1 | ORF deletion | 99 ± 0.8 | NS |
| AWRI1631 ΔPYC2 | PYC2 | ORF deletion | 99 ± 1.7 | NS |
| AWRI1631 ΔTPI1 | TPI1 | ORF deletion | Stuck fermentc | NA |
| AWRI1631 FPS1Δ11 | FPS1 | Truncated Fps1p | 92 ± 0.4 | S |
| AWRI1631 gcrTPI1 | TPI1 | Point mutation in TPI1 promoter at GCR1 binding site | 98 ± 0.9 | S |
| AWRI1631 rapTPI1 | TPI1 | Point mutation in TPI1 promoter at RAP1 binding site | 100 ± 0.7 | NS |
| AWRI1631 aspTPI1 | TPI1 | Point mutation in TPI1 to change Glu165 to Asp in Tpi1p active site | 94 ± 0.3 | S |
| AWRI1631 GOX | GOX | Expression of glucose oxidase from Aspergillus niger | 95 ± 2.1 | S |
| AWRI1631 PYC1 | PYC1 | Promoter replacement | 99 ± 0.4 | NS |
| AWRI1631 MDH2 | MDH2 | Promoter replacement | 98 ± 0.3 | S |
| AWRI1631 FUM1 | FUM1 | Promoter replacement | 100 ± 0.5 | NS |
| AWRI1631 FRDS1 | FRD1 | Promoter replacement | 98 ± 0.5 | NS |
| AWRI1631 ICL1 MLS1 | ICL1, MLS1 | Promoter replacement | 98 ± 0.7 | NS |
| AWRI1631 ADH2 | ADH2 | Promoter replacement | 100 ± 0.6 | NS |
| AWRI1631 ZWF1 | ZWF1 | Promoter replacement | 99 ± 0.8 | NS |
| AWRI1631 GND1 | GND1 | Promoter replacement | 99 ± 0.9 | NS |
| AWRI1631 GPD1 | GPD1 | Promoter replacement | 89 ± 0.3 | S |
| AWRI1631 2GPD1 | GPD1 | Two copies of the FBA1p-GPD1 cassette | 81 ± 0.4 | S |
| AWRI1631 3GPD1 | GPD1 | Three copies of the FBA1p-GPD1 cassette | 71 ± 0.3 | S |
| AWRI1631 GPD1 FPS1Δ11 | FPS1, GPD1 | GPD1 promoter replacement and truncated Fps1p | 87 ± 0.5 | S |
| AWRI1631 GPD1 ΔTPI1 | GPD1, TPI1 | GPD1 promoter replacement and TPI1 deletion | Stuck ferment | NA |
| AWRI1631 2GPD1 ACS1 | GPD1, ACS1 | Two copies of the FBA1p-GPD1 cassette, and ACS1 promoter replacement | 84 ± 0.9 | S |
| AWRI 2531 | GPD1, ALD6 | Two copies of the FBA1p-GPD1 cassette and ALD6 deletion | 71 ± 0.4 | S |
| AWRI 2532 | GPD1, ALD6 | Three copies of the FBA1p-GPD1 cassette and ALD6 deletion | 65 ± 1.2 | S |
Values are averages and standard deviations from nine replicates for the parental strain and three replicates for all other strains.
S, P < 0.05 (strain produced a significantly lower ethanol yield); NS, not significantly different; NA, not applicable.
Unfinished fermentation with high residual sugar.
Single gene deletions in AWRI1631 were obtained from the AWRI Wine Yeast Deletion Library (WYDL) collection. Deletions in this collection were carried out by replacing the ORFs with a KanMX cassette encoding G418 resistance. Cassettes (kindly provided by Charlie Boone, University of Toronto) were PCR amplified using primers containing 50-bp flanking regions corresponding to up- and downstream regions outside the ORF.
Gene deletions in strains intended for multiple modifications were conducted using a selection-counterselection system described previously (53), with some modifications. A counterselection (CORE) cassette was cloned in the plasmid pAG25 (EURSCARF collection). Briefly, the gene GIN11, which is lethal when expressed in S. cerevisiae, was cloned in pAG25 behind the galactose-inducible promoter GAL1, along with the gene natMX, which encodes resistance to the antibiotic ClonNAT. In a first step, the CORE cassette was amplified by PCR from plasmid pAG25-GIN11 using primers with 40-bp flanking regions complementary to up- and downstream sequences outside the targeted region. This PCR product was then transformed into yeast, where it was integrated into the targeted site.
In the counterselection step, the CORE cassette was replaced using oligonucleotides or PCR products depending on the gene modification. For gene deletions, 100-bp double-strand oligonucleotides, containing 50 bp complementary to the upstream and 50 bp complementary to the downstream regions, were used to remove the CORE cassette (53). For promoter replacement, a PCR product containing the strong constitutive yeast promoter FBA1 and 40 bp flanking each side of the CORE cassette was employed. For gene cassette insertions, a PCR product containing either the native GPD1 gene or the GOX gene from Aspergillus niger, both under the control of the FBA1 promoter and carrying 40-bp flanking sequences, complementary to regions to the CORE cassette was introduced in yeast. Nucleotide substitution and deletions were conducted similarly to gene deletions, except that the oligonucleotides carried base substitutions.
All yeast transformations were carried out using the lithium acetate-polyethylene glycol method (2). Transformed strains were selected on plates containing galactose as a sole carbon source and subsequently tested for ClonNAT sensitivity. ClonNAT-sensitive strains containing the desired genetic modifications were confirmed by sequencing.
Cloning of GOX gene.
The GOX gene, which encodes glucose oxidase, was amplified by PCR from genomic DNA of Aspergillus niger and cloned into plasmid pCV1-FBA1pMFα (AWRI plasmid collection). This plasmid encodes the secretion signal from the mating pheromone factor MFα immediately downstream of the FBA1 promoter. In the resulting plasmid (pCV1-MFαGOX), the GOX gene was cloned in frame with the MFα secretion signal to enable secretion of glucose oxidase to the extracellular medium.
Determination of glucose oxidase activity.
Yeast colonies transformed with the PCR product containing the MFα secretion signal-GOX gene construct were screened for their ability to secrete active glucose oxidase using a plate assay as described previously (33). Briefly, colonies on plates were overlaid with 10 ml of 0.1 M K2HPO4 buffer (pH 7.0) containing 10 g/liter glucose, 1% (wt/vol) agarose, 100 mg/liter o-dianisidine dihydrochloride (Sigma-Aldrich, Australia), and 15 U/ml peroxidase (Sigma-Aldrich, Australia). Plates were incubated at 37°C for 1 h once the overlay agar was set. Colonies secreting active glucose oxidase produced a brown halo.
Media and growth conditions.
All yeast strains were evaluated for decreased ethanol production in YPD10 medium (yeast extract, 10 g/liter; peptone, 20 g/liter; glucose, 100 g/liter) (30). Briefly, a yeast starter culture was made in 20 ml of YPD medium (yeast extract, 10 g/liter; peptone, 20 g/liter; glucose, 20 g/liter) and incubated at 28°C with shaking (180 rpm). The starter culture was then used to inoculate fermentation flasks at a cell density of 5 × 106 cells/ml. Fermentations were carried out in triplicate in 250-ml flasks equipped with fermentation locks and sampling ports closed with Suba-seals, and contained 100 ml of YPD10 medium. Aerobic conditions were attained by growing cultures in flasks covered with aluminum foil that allowed free gas exchange with the environment. Cultures were fermented at 28°C for 48 h. At the end of fermentation, samples were collected and centrifuged for 5 min at 15,000 × g, and the cell-free supernatants were kept at 4°C for high-performance liquid chromatography (HPLC) analysis.
Strains showing a considerable reduction in ethanol concentration after fermentation were assessed for their ability to ferment chemically defined grape juice medium (CDGJM) (48) and/or grape juice. Chardonnay juice was prepared commercially from grapes mechanically harvested from Adelaide Hills (South Australia), and Cabernet Sauvignon grapes were collected from Langhorne Creek (South Australia). CDGJM contained 200 g/liter of sugar and yeast assimilable nitrogen (YAN) at 300 mg N/liter. Sugar concentration in Chardonnay juice was increased from 210 g/liter and to 246 g/liter to match Cabernet Sauvignon. YAN concentrations in Chardonnay and Cabernet Sauvignon were 360 mg N/liter and 300 mg N/liter, respectively (Table 2). CDGJM and Chardonnay juice were filter sterilized (0.2 μm; Millipore) immediately after preparation. Cabernet Sauvignon grapes were aliquoted for individual fermentations and crushed prior to inoculation. CDGJM, Chardonnay, and Cabernet Sauvignon pH was adjusted to 3.5. CDGJM and Chardonnay fermentations were carried out in triplicate in 250-ml fermentation flasks (with fermentation locks and sampling ports), each containing 200 ml medium. Cabernet Sauvignon fermentations were conducted in 1-liter Schott bottles with fermentation locks and containing 400 ml juice and skins. CDGJM and grape juice fermentations were inoculated at a cell density of 5 × 106 cells/ml and incubated at 20°C with shaking, with progress monitored by measuring refractive index of culture supernatants. At the end of fermentation, cultures were cold settled and racked. Samples for HPLC analysis were centrifuged for 5 min at 15,000 × g, and the cell-free supernatants were stored at 4°C. Samples for volatile compound analysis were centrifuged in glass test tubes, poured into glass ampoules under nitrogen gas, and kept at 4°C. Glass was used to avoid stripping of volatile compounds.
Table 2.
Chemical parameters for juices fermented with modified strainsa
| Juice | Sugar concn (g/liter) | YAN (mg N/liter) |
|---|---|---|
| CDGJM | 200 | 300 |
| Chardonnay | 246b | 360 |
| Cabernet Sauvignon | 246 | 300 |
The pH was adjusted to 3.5 for each juice.
Increased from 210 g/liter to match Cabernet Sauvignon composition.
Analytical methods.
Yeast growth was monitored spectrophotometrically by measuring absorbance at 600 nm. Concentrations of residual sugar, ethanol, glycerol, malic acid, succinic acid, gluconic acid, and acetic acid were determined by HPLC using a Bio-Rad HPX-87H column as described previously (58). Ammonia concentration was determined using the glutamate dehydrogenase enzymatic bioanalysis UV method test (Roche, Mannheim, Germany). Free α-amino acid nitrogen (FAN) was determined using the o-phthaldehyde–N-acetyl-l-cysteine spectrophotometric assay (NOPA) (14). YAN was calculated by adding the nitrogen present in ammonium to the FAN concentration.
Analysis of volatile compounds.
Concentrations of acetaldehyde, acetoin, and 2,3-butanediol in wine were determined using headspace solid-phase microextraction coupled with gas chromatography-mass spectrometry (HS-SPME/GCMS), with polydeuterated internal standards for stable isotope dilution analysis (SIDA) as follows. Briefly, an Agilent 6890 gas chromatograph was equipped with a Gerstel MPS2 multipurpose sampler and coupled to an Agilent 5973N mass selective detector. The instrument was controlled with Agilent GC ChemStation software (rev. D.02.00.275) and Maestro software (integrated version 1.3.3.51/3.3), and the data were analyzed with Agilent MassHunter quantitative analysis software, version B.04.00. The gas chromatograph was fitted with a 60-m by 0.250-mm J&W DB-wax fused silica capillary column, with a 0.25-μm film thickness and a 0.5-m by 0.250-mm Restek Siltek-deactivated retention gap. Helium (BOC ultrahigh purity) with a linear velocity of 30 cm/s and a flow rate of 1.4 ml/min was used as carrier gas in constant flow mode. The oven temperature was started at 40°C, held for 4 min, increased to 160°C at 5°C/min, increased to 240°C at 40°C/min, and held for 5 min. The inlet was fitted with a borosilicate glass SPME inlet liner (0.75-mm inside diameter; Supelco) and was held at 220°C. Samples were diluted 1:10 or 1:5 with MilliQ water in a 20-ml headspace vial. Vials were immediately sealed (magnetic, polytetrafluoroethylene [PTFE] septum). Subsequently, 100 μl of combined deuterated internal standards solution was injected through the septum and the vial was thoroughly shaken. The concentrations of each deuterated internal standard in the vial were 29.75 mg/liter for d4-acetaldehyde, 36.33 mg/liter for d7-acetoin, and 29.92 mg/liter for d8-2,3-butanediol. A Supelco polydimethylsiloxane-divinylbenzene (PDMS/DVB, blue) 65-μm fiber was exposed to the headspace of the sample vials for 10 min at 35°C and desorbed in the GC inlet in splitless mode for 10 min.
Polydeuterated standards.
d4-Acetaldehyde was purchased from Sigma-Aldrich (Castle Hill, Australia). d7-Acetoin and d8-2,3-butanediol were synthesized from d6-2,3-butanedione, which was prepared from 2,3-butanedione (Alfa Aesar, Australia), deuterium sulfate (D2SO4) and D2O (Sigma-Aldrich, Australia) as previously described (59). d7-Acetoin was prepared from d6-2,3-butanedione as described previously (59). Briefly, d6-2,3-butanedione (1.0 g, 11 mmol) and Zn powder (1.5 g, 22 mmol) were stirred under an N2 atmosphere in 12.8 g D2SO4 (20% in D2O) for 15 min; the mixture was then filtered, saturated with NaCl, and extracted with CH2Cl2 (seven times, 30 ml each time). The organic layer was dried, concentrated and purified by Kugelrohr distillation to give the product as a colorless liquid (341 mg, 32%, 91.2% purity by GC-MS). d8-2,3-Butanediol was prepared by reducing d6-2,3-butanedione using a variation of the method described by Maier et al. (32). A solution of d6-2,3-butanedione (310 mg in 5 ml diethyl ether) was slowly added to a cooled suspension of LiAlD4 (Sigma-Aldrich, Australia) (110 mg in 5 ml diethyl ether) under an N2 atmosphere. After stirring for 2.5 h, the suspension was hydrolyzed with KOH solution (50% wt/vol, 5 ml). The product was extracted with diethyl ether (10 times, 5 ml each time), dried with MgSO4, and concentrated to give a pale yellow oil. The aqueous layer was then taken up with Na2SO4, washed with diethyl ether (five times, 5 ml each time), dried, and concentrated in vacuo. The resultant oil was purified by column chromatography (2:3 ethyl acetate-petroleum ether) to give deuterated 2,3-butanediol (270 mg, 82%, 98.9% purity by GC-MS).
RESULTS
Screening gene modifications that impact ethanol production.
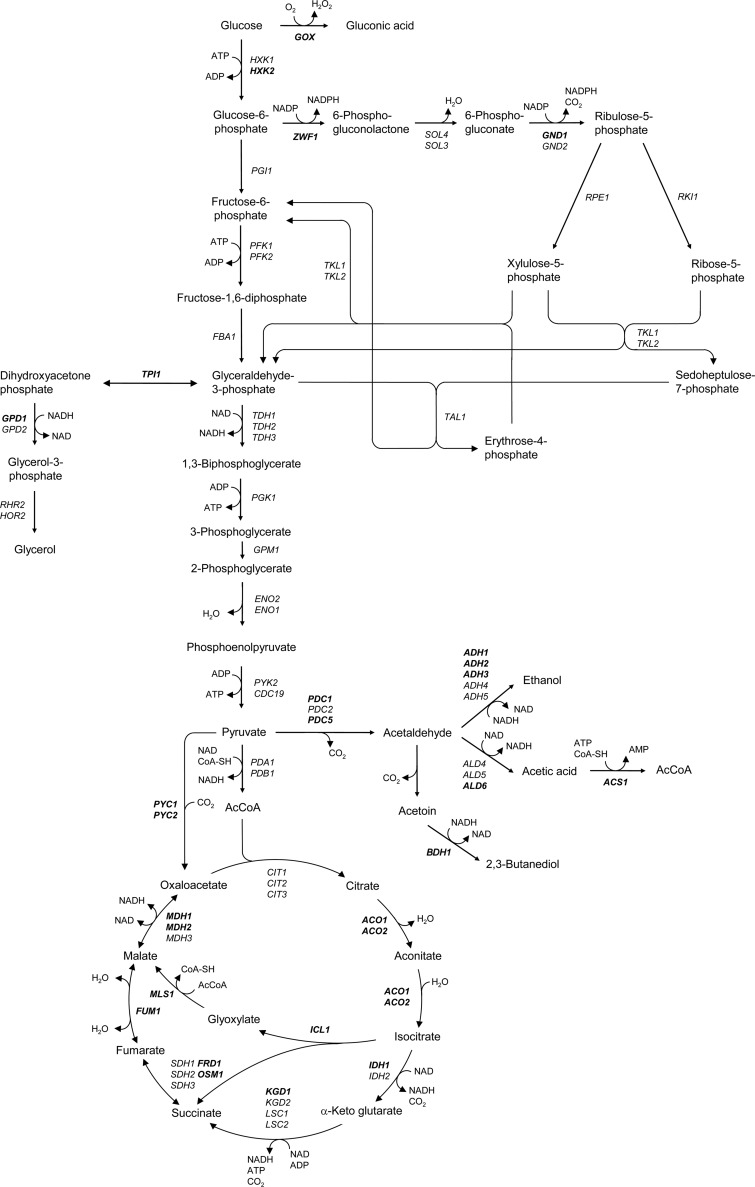
A collection of mutant S. cerevisiae wine yeast strains was generated from the same parent, AWRI1631, to identify the best candidate genes for constructing low-ethanol strains. Expression of genes encoding enzymes that divert carbon away from ethanol production was increased, whereas genes that contribute to ethanol formation were deleted or downregulated. Target genes included some involved in glycolysis, pentose phosphate pathway, and TCA cycle; some regulating glucose repression; and some with the potential to catabolize glucose in the medium (Fig. 1).
Fig 1.
Central carbon metabolism in S. cerevisiae, including glycolysis, the pentose phosphate pathway, and the TCA cycle. Genes in bold were modified in this work. ACO1 and -2, aconitase; ACS1, acetyl coenzyme A (CoA) synthetase; ADH1 to -5, alcohol dehydrogenase; ALD4 to -6, aldehyde dehydrogenase; BDH1, butanediol dehydrogenase; CDC19, pyruvate kinase; CIT1 to -3, citrate synthase; ENO1 and -2, enolase; FBA1, fructose-bisphosphate aldolase; FRD1, fumarate reductase; FUM1, fumarase; GND1 and -2, 6-phophogluconate dehydrogenase; GOX, glucose oxidase; GPD1 and -2, glycerol-3-phosphate dehydrogenase; GPM1, phosphoglycerate mutase; HOR2, glycerol-3-phosphatase; HXK1 and -2, hexokinase; ICL1, isocitrate lyase; IDH1 and -2, isocitrate dehydrogenase; KGD1 and -2, α-ketoglutarate dehydrogenase; LSC1 and -2, succinyl-CoA ligase; MDH1 to -3, malate dehydrogenase; MLS1, malate synthase; OSM1, fumarate reductase; PDA1, pyruvate dehydrogenase alpha unit; PDB1, pyruvate dehydrogenase beta unit; PDC1, -2, and -5, pyruvate decarboxylase; PFK1 and -2, phosphofructokinase; PGK1, phosphoglycerate kinase; PGI1, phosphoglucose isomerase; PYC1 and -2, pyruvate carboxylase; PYK2, pyruvate kinase; RKI1, ribose-5-phosphate ketol-isomerase; RHR2, glycerol-3-phosphatase; RPE1, ribulose-5-phosphate epimerase; SDH1 to -3, succinate dehydrogenase; SOL3 and -4, 6-phosphogluconolactonase; TAL1, transaldolase; TDH1 to -3, glyceraldehyde-3-phosphate dehydrogenase; TKL1 and -2, transketolase; TPI1, triosephosphate isomerase; ZWF1, glucose-6-phosphate dehydrogenase.
Only 15 of the 41 strains that were constructed showed significantly lower ethanol production than the parental strain AWRI1631 when grown in YPD10 medium (Table 1). Increased expression of genes involved in the reductive branch of the TCA cycle (PYC1, MDH2, FUM1, and FRD1) affected the formation of organic acids (data not shown), but only MDH2 and FRD1 showed a marginal (2%) decrease in ethanol production. Deletion of PDC5, HXK2, and MIG1, the last two involved in glucose repression, produced a minor (3%) decrease in ethanol concentration. Under aerobic conditions, deletion of genes involved in glucose repression did not show any further decrease in ethanol formation (data not shown). Expression of the A. niger GOX gene showed a 5% decrease in ethanol. When AWRI1631 GOX was grown under aerobic conditions in the same medium, a decrease of 6% in ethanol production was achieved (data not shown), consistent with the requirement of glucose oxidase for oxygen, as noted previously by others (3). Interestingly, AWRI1631 GOX darkened the color of the medium relative to the parental strain, most likely due to hydrogen peroxide production during the formation of gluconic acid.
Both TPI1 deletion strains (AWRI1631 ΔTPI1 and AWRI1631 GPD1 ΔTPI1) were unable to complete fermentation; however, they showed the greatest glycerol-to-glucose yields (data not shown). For this reason, four additional modifications aimed at decreasing TPI1 expression or Tpi1p activity were tested, but only one showed a significant effect on ethanol production. Substituting glutamic acid-165 with aspartic acid in the active site of Tpi1p was previously shown to lower the activity of this enzyme (54); this mutation resulted in 6% decreased ethanol production compared to AWRI1631.
Fps1p, the glycerol transporter, has been shown to regulate the intracellular concentration of glycerol by closing the pore channel (57). Removal of the channel gate allows glycerol to leak out of the cell and increases glycerol production (56, 57). AWRI1631 FPS1Δ11, carrying the “open” transporter, diverted carbon toward glycerol generation and showed an 8% decrease in ethanol production.
Seven of the yeast constructs delivering reduced amounts of ethanol involved increased expression of GPD1. AWRI1631 GPD1 carried the single gene modification most effective at reducing ethanol production (10% decrease, compared to AWRI1631). Introduction of two (AWRI1631 2GPD1) and three copies (AWRI1631 3GPD1) of the FBA1p-GPD1 cassette enabled further reductions in ethanol production, of 19% and 29%, respectively. AWRI1631 GPD1 FPS1Δ11, carrying a high-GPD1-expression construct and the open glycerol transporter, exhibited a 13% decrease in ethanol concentration.
As described previously, increased glycerol production leads to a higher concentration of acetic acid (10, 46), which is detrimental to wine quality. Two strategies were attempted to reduce acetic acid production: increased expression of ACS1 and deletion of ALD6 (6, 15). AWRI1631 2GPD1 ACS1 produced amounts of ethanol similar to those produced by AWRI1631 2GPD1 (Table 1) with no effect on elevated levels of acetic acid (data not shown), while AWRI2531 (with two copies of the FBA1p-GPD1 cassette and an ALD6 deletion) showed a further 10% decrease in ethanol concentration compared to AWRI1631 2GPD1. The same behavior was observed when ALD6 was deleted in the strain carrying three copies of the FBA1p-GPD1 cassette. Thus, AWRI2532 produced the fermentation product with the lowest ethanol concentration, 35% lower than that of AWRI1631.
Characterization of low-ethanol strains under winemaking conditions in CDGJM.
Four strains selected from the above screening were assessed for their ability to ferment CDGJM in winemaking conditions (Table 3). Only AWRI2532 was not able to complete fermentation, as was evident from the high concentration of residual sugar in the medium. All four strains produced fermentation products with higher glycerol and lower ethanol concentrations than AWRI1631. AWRI1631 aspTPI1 produced more malic acid, acetic acid, and glycerol with less succinic acid and a 2% lower ethanol concentration than the parental strain. Compared to AWRI1631, AWRI1631 3GPD1 produced slightly less malic acid and succinic acid, 6 times more glycerol, 20 times more acetic acid, and 20% less ethanol. AWRI2531 produced less malic acid, slightly more succinic acid, 6.1 times more glycerol, 3 times more acetic acid, and 22% less ethanol.
Table 3.
Chemical composition of wines obtained by fermentation of CDGJM
| Strain | Concn (g/liter) ofa: |
|||||
|---|---|---|---|---|---|---|
| Residual sugar | Malic acid | Succinic acid | Glycerol | Acetic acid | Ethanol | |
| AWRI1631 | 0.0 ± 0.0 | 2.6 ± 0.0 | 0.7 ± 0.0 | 5.7 ± 0.1 | 0.2 ± 0.0 | 102 ± 0.3 |
| AWRI1631 aspTPI1 | 0.0 ± 0.0 | 4.2 ± 0.1 | 0.0 ± 0.0 | 7.3 ± 0.2 | 0.3 ± 0.0 | 100 ± 0.4 |
| AWRI1631 3GPD1 | 0.0 ± 0.0 | 2.4 ± 0.1 | 0.5 ± 0.0 | 34.3 ± 0.1 | 4.1 ± 0.1 | 82 ± 0.4 |
| AWRI 2531 | 0.0 ± 0.0 | 2.1 ± 0.0 | 0.8 ± 0.0 | 35.0 ± 0.1 | 0.6 ± 0.0 | 81 ± 0.2 |
| AWRI 2532 | 118 ± 5.2 | 4.7 ± 0.3 | 0.0 ± 0.0 | 14.7 ± 0.3 | 0.3 ± 0.0 | 24 ± 0.5 |
Values are averages and standard deviations from three replicates.
Characterization of low-ethanol strains in grape juice.
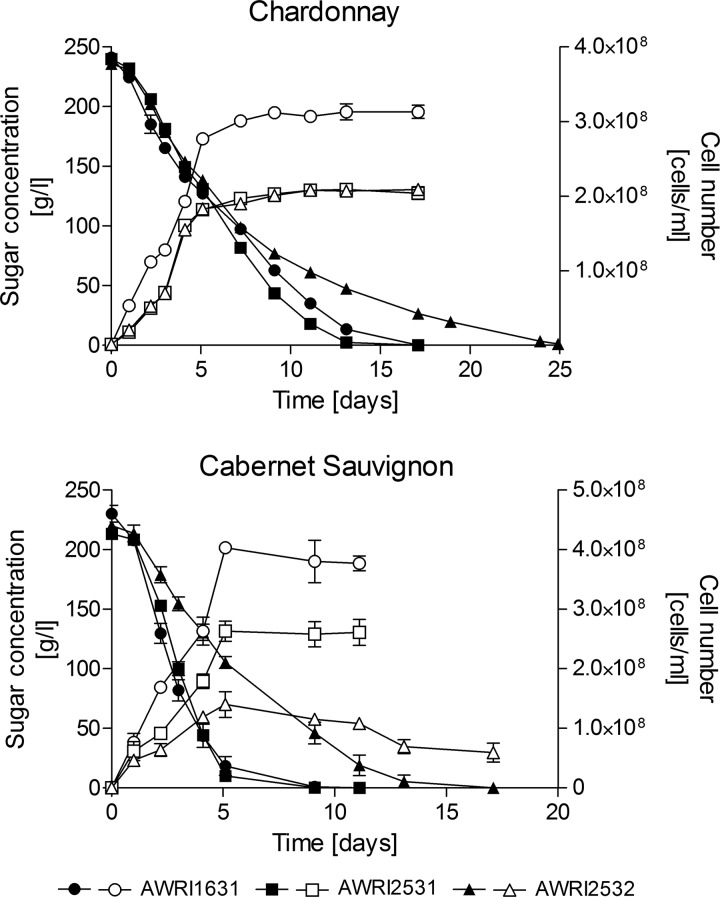
Only two strains, AWRI2531 and AWRI2532, were tested for their ability to ferment Chardonnay and Cabernet Sauvignon grape juice. Both strains fermented these juices to dryness, although AWRI 2532 took longer to consume all sugar (Fig. 2). Cell numbers were lower for the modified strains than for the parental strain in both Chardonnay and Cabernet Sauvignon (Fig. 2). AWRI 2531 and AWRI 2532 produced higher concentrations of glycerol, acetic acid, acetaldehyde, acetoin, and 2,3-butanediol and less ethanol than the parental strain (Tables 4 and 5).
Fig 2.
Fermentation kinetics and cell numbers in Chardonnay and Cabernet Sauvignon. Closed symbols, sugar consumption; open symbols, cell number.
Table 4.
Chemical composition of wines obtained by fermentation of Chardonnaya
| Component | AWRI1631 | AWRI2531 | AWRI2532 | AWRI2531 BDH1 | AWRI2532 BDH1 |
|---|---|---|---|---|---|
| Residual sugar (g/liter) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Malic acid (g/liter) | 4.8 ± 0.4 | 3.3 ± 0.1 | 4.2 ± 0.1 | 3.4 ± 0.2 | 4.1 ± 0.1 |
| Succinic acid (g/liter) | 3.2 ± 0.0 | 2.7 ± 0.0 | 2.6 ± 0.0 | 2.9 ± 0.1 | 3.2 ± 0.1 |
| Glycerol (g/liter) | 8.7 ± 0.1 | 31.1 ± 0.5 | 43.4 ± 0.1 | 30.3 ± 0.2 | 34.4 ± 0.4 |
| Acetic acid (g/liter) | 0.2 ± 0.0 | 0.2 ± 0.0 | 0.5 ± 0.0 | 0.3 ± 0.0 | 0.3 ± 0.0 |
| Ethanol (g/liter) | 124.3 ± 0.8 | 105.7 ± 0.7 | 96.9 ± 0.2 | 107.0 ± 1.1 | 105.6 ± 0.6 |
| Acetaldehyde (mg/liter) | 32 ± 1.8 | 290 ± 20 | 272 ± 22 | 175 ± 5 | 143 ± 29 |
| Acetoin (mg/liter) | 25 ± 8 | 478 ± 7 | 939 ± 94 | 105 ± 25 | 100 ± 26 |
| 2,3-Butanediol (mg/liter) | 14 ± 1.3 | 91 ± 21 | 81 ± 20 | 191 ± 23 | 148 ± 24 |
| Glycerol yield (g/g consumed sugar) | 0.03 ± 0.0 | 0.11 ± 0.0 | 0.13 ± 0.0 | 0.11 ± 0.0 | 0.10 ± 0.0 |
| Ethanol yield (g/g consumed sugar) | 0.48 ± 0.0 | 0.44 ± 0.0 | 0.42 ± 0.0 | 0.45 ± 0.0 | 0.46 ± 0.0 |
Values are averages and standard deviations from three replicates.
Table 5.
Chemical composition of wines obtained by fermentation of Cabernet Sauvignon
| Component | AWRI1631 | AWRI2531 | AWRI2532 |
|---|---|---|---|
| Residual sugar (g/liter) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Malic acid (g/liter) | 3.9 ± 0.2 | 6.9 ± 0.4 | 7.9 ± 0.0 |
| Succinic acid (g/liter) | 7.7 ± 0.4 | 7.9 ± 0.1 | 7.0 ± 0.2 |
| Glycerol (g/liter) | 9.0 ± 0.1 | 33.3 ± 0.6 | 42.2 ± 0.4 |
| Acetic acid (g/liter) | 0.1 ± 0.0 | 0.2 ± 0.0 | 0.5 ± 0.0 |
| Ethanol (g/liter) | 122.3 ± 0.6 | 103.7 ± 0.3 | 93.8 ± 0.6 |
| Acetaldehyde (mg/liter) | 29 ± 1.6 | 150 ± 16 | 227 ± 21 |
| Acetoin (mg/liter) | 33 ± 1 | 2495 ± 83 | 5923 ± 99 |
| 2,3-Butanediol (mg/liter) | 9.4 ± 0.6 | 226 ± 36 | 212 ± 42 |
| Glycerol yield (g/g sugar) | 0.04 ± 0.0 | 0.15 ± 0.0 | 0.19 ± 0.0 |
| Ethanol yield (g/g sugar) | 0.55 ± 0.0 | 0.48 ± 0.0 | 0.44 ± 0.0 |
Values are averages and standard deviations from three replicates.
Production of malic and succinic acids was grape juice dependent. In Chardonnay, AWRI2531 produced less malic and succinic acids than the parental strain, while in Cabernet Sauvignon, the concentration of these acids was higher than for the parent. AWRI2531 produced 3.6-fold more glycerol in both wines. This increase in glycerol production generated 15% less ethanol in both wines. Acetic acid production was similar to that of the parent in Chardonnay, while in Cabernet Sauvignon the concentration was slightly higher than for AWRI1631. In Chardonnay, acetaldehyde, acetoin, and 2,3-butanediol concentrations increased 9-fold, 19-fold, and 6.5-fold, respectively, whereas in Cabernet Sauvignon, these concentrations were increased 5.2-fold, 75.6-fold, and 22.5-fold, respectively.
AWRI2532 produced less malic and succinic acids than AWRI1631 in Chardonnay. In Cabernet Sauvignon, however, malic acid production was higher than for the parental strain, while the concentration of succinic acid was less than that produced by AWRI1631. AWRI2532 produced, on average, 4.9-fold more glycerol than the parent in both Chardonnay and Cabernet Sauvignon. Consequently, ethanol production by this strain was 22% less in both wines. Acetic acid production was higher in both Chardonnay and Cabernet Sauvignon but still lower than levels considered detrimental to wine quality (55). Acetaldehyde, acetoin, and 2,3-butanediol production in Chardonnay increased 8.5-fold, 37.5-fold, and 5.8-fold, respectively, whereas in Cabernet Sauvignon these concentrations increased 7.8-fold, 179-fold, and 22.6-fold, respectively.
As high concentrations of acetoin negatively impact the organoleptic properties of the resulting wine (16), AWRI2531 and AWRI2532 were genetically modified to convert acetoin into the sensorially neutral compound 2,3-butanediol. Expression of BDH1, which encodes 2,3-butanediol dehydrogenase, was therefore increased in both strains. Compared to AWRI2531, AWRI2531 BDH1 showed a significant decrease in both acetaldehyde and acetoin concentrations, with an accompanying increase in the concentration of 2,3-butanediol; levels of all other metabolites remained similar (Table 4). In contrast, while AWRI2532 BDH1 also showed significantly lower acetaldehyde and acetoin concentrations combined with a higher 2,3-butanediol concentration than AWRI2532, AWRI2532 BDH1 also produced considerably less glycerol and more ethanol than its parent.
DISCUSSION
There is growing interest in alcoholic beverage industries to reduce the level of alcohol in beers and wines. There are many strategies available to achieve this end, but all have serious limitations (49). It is widely believed that microbiological approaches have the capacity to deliver the best outcome. This paper describes a survey of a large number of target S. cerevisiae genes that have been manipulated to test their potential for the development of a low-ethanol-wine yeast strain.
Genes involved in glucose repression, glycolysis, the pentose phosphate pathway, and the TCA cycle and a heterologous (A. niger) gene involved in glucose oxidation were separately engineered in a wine yeast with the aim of reducing ethanol production. Specifically, genes were manipulated to divert carbon away from ethanol production to other endpoints.
One of the obvious strategies to lower ethanol production is to decrease the activity of the main enzyme responsible for ethanol formation. Deletion of ADH1, which encodes alcohol dehydrogenase, has been shown to decrease ethanol production in laboratory yeast strains (9, 12, 13). In the present work, however, deletion of ADH1 did not affect ethanol formation, suggesting that other alcohol dehydrogenase isozymes compensate for loss of this enzyme in AWRI1631 wine yeast background under the conditions used in this study. Indeed, Adh3p has been shown to mimic the function of Adh1p in a genetic background lacking ADH1, ADH2, ADH4, and ADH5 (12). However, deletion of both ADH1 and ADH3, together and individually, had no effect on ethanol production. This might indicate that one or more of the other Adh isozymes compensates for loss of ADH1 and ADH3 in the genetic background of the yeast strain used for this work. ADH2, which encodes an alcohol dehydrogenase that metabolizes ethanol (12), had no impact on ethanol yield when overexpressed.
Several studies have shown that deletion or overexpression of specific genes involved in the TCA cycle affect not only organic acid production but also ethanol yields (1, 37, 51). Deletion of KGD1, KGD2, or FUM1 decreased production of ethanol in a lab strain (51), whereas ACO1 deletion resulted in a modest decrease in ethanol formation in a sake yeast (1).
In the work described here, manipulation of most of the genes involved in the oxidative or reductive branches of the TCA cycle affected the formation of organic acids but did not impact ethanol production. At least in some cases, this differs from what has been previously reported, perhaps reflecting differences in genetic backgrounds and/or different environmental conditions. Only increased expression of MDH2 and FRD1, involved in the reductive branch, led to a decrease in ethanol concentration, but this was rather modest. Increased expression of the cytosolic malate dehydrogenase (MDH2) can increase malate, fumarate, and citrate production (38), but the authors of that study noted that the activity of pyruvate carboxylase (encoded by PYC1 and PYC2) appeared to be rate limiting for malate synthesis. Therefore, increasing expression of more than one of the genes involved in the reductive branch of the TCA cycle simultaneously might divert more carbon away from ethanol production.
Another strategy with the potential to decrease ethanol production is to lift glucose repression from genes encoding enzymes involved in respiration and therefore burn carbon that would otherwise feed into ethanol production. Glucose repression involves a great number of genes and several signal transduction pathways (28, 47). Nevertheless, deletions of HXK2, GRR1, and MIG1 have each been shown to decrease ethanol production by redirecting carbon to biomass formation in continuous cultures (44). In the work described here, the only genetic modifications targeting glucose repression found to decrease ethanol formation were the deletions of HXK2 and MIG1. Regardless, the minor effect of these gene deletions on ethanol production, even in the presence of oxygen, suggests that in the AWRI1631 genetic background, other pathways related to glucose repression prevent respiration during wine fermentation.
The pentose phosphate pathway represents another potential sink for carbon. However, Cadiere et al. (5) found that yeast strains exhibiting higher carbon fluxes through this pathway showed no effect on ethanol production. Mutants lacking PGI1 and therefore channeling carbon through the pentose phosphate pathway were not able to sustain growth even when overexpressing ZWF1, indicating that another reaction or factor limits flux through this pathway (24). Therefore, it is perhaps not surprising that increased expression of ZWF1 and GND1 showed unchanged ethanol production in this study.
Expression in S. cerevisiae of the A. niger glucose oxidase gene has been previously shown to decrease ethanol production by diverting sugar metabolism toward gluconic acid formation (33). In the present work, this approach only showed a modest decrease in ethanol formation; additionally, hydrogen peroxide, a product of the glucose oxidase reaction, was most likely responsible for darkening the medium color. Therefore, to avoid perturbations to wine color when expressing glucose oxidase, other strategies, such as coexpressing a catalase, should be explored.
Glycerol proved to be the best carbon sink in the current study. All modifications intended to increase the formation of glycerol (TPI1 deletion, FPS1 modification, and GPD1 overexpression) made a substantial impact on ethanol production.
Although yeast strains lacking TPI1 have been shown to grow in rich medium (7), neither AWRI1631 ΔTPI1 nor AWRI1631 GPD1 ΔTPI1 was able to ferment high concentrations of sugar. Interestingly, however, these strains showed the greatest yields of glycerol on glucose relative to all other mutants tested, which is consistent with previous reports (8). In order to decrease Tpi1p activity without affecting fermentation performance, we explored two strategies, decreasing expression of TPI1 and lowering Tpi1p activity. Deleting REB1-, RAP1-, and GCR1-binding sites in the promoter region of TPI1 has been shown to decrease TPI1 expression in yeast (50). However, deletion of RAP1- and GCR1-binding sites had no impact on glycerol or ethanol concentrations. Replacement of glutamic acid-165 with aspartic acid in the active site of the chicken Tpi1p has been shown to considerably decrease the activity of the enzyme (54). Yeast Tpi1p also contains a glutamic acid in position 165; when this residue was replaced with aspartic acid, we observed an increase in glycerol formation and a decrease in ethanol production but no effect on fermentation performance.
Although FPS1 deletion has been shown to decrease glycerol yield and increase ethanol formation (60), increased expression of FPS1 failed to deliver the opposite phenotype (9). Indeed, glycerol transport, and hence glycerol production, is controlled by a short regulatory domain in the N-terminal extension of Fps1p (56, 57). When this domain is deleted, the channel is hyperactive, remaining open for glycerol efflux and resulting in continuous glycerol leakage from the cell, which is compensated for by increased glycerol production (45, 56). We observed that this increased glycerol production, as a consequence of deleting the regulatory domain of Fps1p lowered ethanol formation considerably.
Increased expression of GPD1 delivered the greatest impact on ethanol production. Moreover, increasing the number of copies of highly expressed GPD1 further decreased ethanol yield. However, increased glycerol production leads to increased acetic acid formation (10, 46), which is detrimental to wine quality. Two strategies were tried in the current work to decrease acetic acid production, ALD6 deletion and increased expression of ACS1. Only the deletion of ALD6 was effective at decreasing acetic acid concentration; this approach was shown previously to lower acetic acid production when glycerol 3-phosphate dehydrogenase is overexpressed (6, 15).
After all modified strains had been evaluated in rich medium, it was crucial to assess the most promising modified yeasts in synthetic must and then in grape juice. To the best of our knowledge, this is the first time low-alcohol-producing GM strains have been used to ferment must from different grape varieties. AWRI 2531 and AWRI 2532 showed similar metabolic profiles in Chardonnay and Cabernet Sauvignon musts. However, AWRI 2532 was slower to complete fermentation in both grape juices than AWRI 2531. In addition, AWRI 2532 was unable to complete fermentation in chemically defined juice, suggesting that this strain might struggle in highly clarified musts. Compared to the parental strain, AWRI 2531 generated wines with an ethanol concentration that was 2.4% (vol/vol) lower on average (from 15.6% [vol/vol] to 13.2% [vol/vol]), while AWRI 2532 produced wines that were 3.6% [vol/vol] lower on average (from 15.6% [vol/vol] to 12% [vol/vol]). Acetic acid concentrations in wines fermented with both strains were within the range considered acceptable for wine quality (55). However, consistent with previous reports (6, 15), other metabolites were increased in wines produced with these modified strains. Acetaldehyde concentration was over its sensory threshold described in wine (100 mg/liter) (52), eliciting a “bruised-apple” smell in these wines, which is detrimental to their sensory properties. Levels of acetoin, because of its low sensory threshold, are also likely to affect negatively the organoleptic properties of wine; nevertheless, acetoin can be converted into the sensorially neutral compound 2,3-butanediol by increasing expression of BDH1, which encodes 2,3-butanediol dehydrogenase (16). In the current work, increasing BDH1 expression not only enabled the conversion of acetoin to 2,3-butanediol but also resulted in lower acetaldehyde concentration. Unexpectedly, increasing BDH1 expression altered the production of glycerol and ethanol in AWRI2532, most likely driven by changes in redox balance. Although acetaldehyde and acetoin production was significantly decreased in wines fermented with strains AWRI2531 BDH1 and AWRI2532 BDH1, acetaldehyde concentrations still exceeded its sensory threshold, while acetoin concentrations were below the sensory threshold described in the literature (150 mg/liter) (17) and hence, at least in the case of acetaldehyde, may still affect negatively the sensory properties of the resulting wine.
In summary, of all strategies aimed at decreasing ethanol production evaluated in this study, those intended to increase glycerol formation were the most effective. The efficiencies of several strategies that have been shown to alter ethanol formation, including diverting sugar to lactate production (11), manipulating hexose transporter genes (23), and expressing a H2O-forming NADH oxidase (25), as well as novel strategies, such as diverting carbon to the formation of reserve carbohydrates, remain to be tested.
ACKNOWLEDGMENTS
The AWRI, a member of the Wine Innovation Cluster in Adelaide, is supported by Australia's grape growers and winemakers through their investment body the Grape and Wine Research Development Corporation with matching funds from the Australian Government.
We acknowledge Charlie Boone from University of Toronto for kindly providing PCR deletion cassettes.
Footnotes
Published ahead of print 22 June 2012
REFERENCES
- 1. Arikawa Y, et al. 1999. Effect of gene disruptions of the TCA cycle on production of succinic acid in Saccharomyces cerevisiae. J. Biosci. Bioeng. 87: 28–36 [DOI] [PubMed] [Google Scholar]
- 2. Ausubel FM, et al. 2007. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, NY [Google Scholar]
- 3. Biyela BNE, du Toit WJ, Divol B, Malherbe DF, van Rensburg P. 2009. The production of reduced-alcohol wines using Gluzyme Mono (R) 10.000 BG-treated grape juice. S. Afr. J. Enol. Viticult. 30: 124–132 [Google Scholar]
- 4. Borneman AR, Forgan AH, Chambers PJ, Pretorius IS. 2008. Comparative genome analysis of a Saccharomyces cerevisiae wine strain. FEMS Yeast Res. 8: 1185–1195 [DOI] [PubMed] [Google Scholar]
- 5. Cadiere A, Ortiz-Julien A, Camarasa C, Dequin S. 2011. Evolutionary engineered Saccharomyces cerevisiae wine yeast strains with increased in vivo flux through the pentose phosphate pathway. Metab. Eng. 13: 263–271 [DOI] [PubMed] [Google Scholar]
- 6. Cambon B, Monteil V, Remize F, Camarasa C, Dequin S. 2006. Effects of GPD1 overexpression in Saccharomyces cerevisiae commercial wine yeast strains lacking ALD6 genes. Appl. Environ. Microbiol. 72: 4688–4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Campagno C, et al. 2001. Alterations of the glucose metabolism in a triose phosphate isomerase-negative Saccharomyces cerevisiae mutant. Yeast 18: 663–670 [DOI] [PubMed] [Google Scholar]
- 8. Compagno C, Boschi F, Ranzi BM. 1996. Glycerol production in a triose phosphate isomerase deficient mutant of Saccharomyces cerevisiae. Biotechnol. Progr. 12: 591–595 [DOI] [PubMed] [Google Scholar]
- 9. Cordier H, Mendes F, Vasconcelos I, Francois JM. 2007. A metabolic and genomic study of engineered Saccharomyces cerevisiae strains for high glycerol production. Metab. Eng. 9: 364–378 [DOI] [PubMed] [Google Scholar]
- 10. de Barros Lopes M, et al. 2000. Fermentation properties of a wine yeast over-expressing the Saccharomyces cerevisiae glycerol 3-phosphate dehydrogenase gene (GPD2). Aust. J. Grape Wine Res. 6: 208–215 [Google Scholar]
- 11. Dequin S, Barre P. 1994. Mixed lactic acid alcoholic fermentation by Saccharomyces cerevisiae expressing the Lactobacillus casei L(+)-LDH. Biotechnology 12: 173–177 [DOI] [PubMed] [Google Scholar]
- 12. de Smidt O, du Preez JC, Albertyn J. 2011. Molecular and physiological aspects of alcohol dehydrogenases in the ethanol metabolism of Saccharomyces cerevisiae. FEMS Yeast Res. 12: 33–47 [DOI] [PubMed] [Google Scholar]
- 13. Drewke C, Thielen J, Ciriacy M. 1990. Ethanol formation in Adho mutants reveals the existence of a novel acetaldehyde reducing activity in Saccharomyces cerevisiae. J. Bacteriol. 172: 3909–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dukes BC, Butzke CE. 1998. Rapid determination of primary amino acids in grape juice using an o-phthaldialdehyde/N-acetyl-L-cysteine spectrophotometric assay. Am. J. Enol. Viticult. 49: 125–134 [Google Scholar]
- 15. Eglinton JM, et al. 2002. Decreasing acetic acid accumulation by a glycerol overproducing strain of Saccharomyces cerevisiae by deleting the ALD6 aldehyde dehydrogenase gene. Yeast 19: 295–301 [DOI] [PubMed] [Google Scholar]
- 16. Ehsani M, Fernandez MR, Biosca JA, Julien A, Dequin S. 2009. Engineering of 2,3-butanediol dehydrogenase to reduce acetoin formation by glycerol-overproducing, low-alcohol Saccharomyces cerevisiae. Appl. Environ. Microbiol. 75: 3196–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ferreira V, Lopez R, Cacho JF. 2000. Quantitative determination of the odorants of young red wines from different grape varieties. J. Sci. Food Agric. 80: 1659–1667 [Google Scholar]
- 18. Flikweert M, Deswaaf M, van Dijken J, Pronk J. 1999. Growth requirements of pyruvate-decarboxylase-negative Saccharomyces cerevisiae. FEMS Microbiol. Lett. 174: 73–79 [DOI] [PubMed] [Google Scholar]
- 19. Flikweert M, et al. 1996. Pyruvate decarboxylase: an indispensable enzyme for growth of Saccharomyces cerevisiae on glucose. Yeast 12: 247–257 [DOI] [PubMed] [Google Scholar]
- 20. Gawel R, Francis L, Waters E. 2007. Statistical correlations between the in-mouth textural characteristics and the chemical composition of Shiraz wines. J. Agric. Food Chem. 55: 2683–2687 [DOI] [PubMed] [Google Scholar]
- 21. Gawel R, van Sluyter S, Waters E. 2007. The effects of ethanol and glycerol on the body and other sensory characteristics of Riesling wines. Aust. J. Grape Wine Res. 13: 38–45 [Google Scholar]
- 22. Guth H, Sies A. 2001 Flavour of wines: towards and understanding by reconstitution experiments and an analysis of ethanol's effect on odour activity of key compounds, p 128–139 In Blair RJ, Williams PJ, Høj PB. (ed), Proceedings of Eleventh Australian Wine Industry Technical Conference. Australian Wine Industry Technical Conference Inc. [Google Scholar]
- 23. Henricsson C, et al. 2005. Engineering of a novel Saccharomyces cerevisiae wine strain with a respiratory phenotype at high external glucose concentrations. Appl. Environ. Microbiol. 71: 6185–6192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heux S, Cadiere A, Dequin S. 2008. Glucose utilization of strains lacking PGI1 and expressing a transhydrogenase suggests differences in the pentose phosphate capacity among Saccharomyces cerevisiae strains. FEMS Yeast Res. 8: 217–224 [DOI] [PubMed] [Google Scholar]
- 25. Heux S, Sablayrolles JM, Cachon R, Dequin S. 2006. Engineering a Saccharomyces cerevisiae wine yeast that exhibits reduced ethanol production during fermentation under controlled microoxygenation conditions. Appl. Environ. Microbiol. 72: 5822–5828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hohmann S. 1991. Characterization of PDC6, a third structural gene for pyruvate decarboxylase in Saccharomyces cerevisiae. J. Bacteriol. 173: 7963–7969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Johansson M, Sjostrom JE. 1984. Enhanced production of glycerol in an alcohol dehydrogenase (Adh1) deficient mutant of Saccharomyces cerevisiae. Biotechnol. Lett. 6: 49–54 [Google Scholar]
- 28. Klein CJL, Rasmussen JJ, Ronnow B, Olsson L, Nielsen J. 1999. Investigation of the impact of MIG1 and MIG2 on the physiology of Saccharomyces cerevisiae. J. Biotechnol. 68: 197–212 [DOI] [PubMed] [Google Scholar]
- 29. Kutyna DR, Varela C, Henschke PA, Chambers PJ, Stanley GA. 2010. Microbiological approaches to lowering ethanol concentration in wine. Trends Food Sci. Technol. 21: 293–302 [Google Scholar]
- 30. Kutyna DR, et al. 2012. Adaptive evolution of Saccharomyces cerevisiae to generate strains with enhanced glycerol production. Appl. Microbiol. Biotechnol. 93: 1175–1184 [DOI] [PubMed] [Google Scholar]
- 31. Luyten K, et al. 1995. Fps1, a yeast member of the MIP family of channel proteins, is a facilitator for glycerol uptake and efflux and is inactive under osmotic stress. EMBO J. 14: 1360–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maier G, Roth C, Schmitt RK. 1985. Diastereoselektivität bei der Hydridreduktion acyclischer Diketone (1,2-, 1,3-, 1,4- und 1,5-Induktion). Chem. Ber. 118: 704–721 [Google Scholar]
- 33. Malherbe DF, du Toit M, Otero RRC, van Rensburg P, Pretorius IS. 2003. Expression of the Aspergillus niger glucose oxidase gene in Saccharomyces cerevisiae and its potential applications in wine production. Appl. Microbiol. Biotechnol. 61: 502–511 [DOI] [PubMed] [Google Scholar]
- 34. Michnick S, Roustan JL, Remize F, Barre P, Dequin S. 1997. Modulation of glycerol and ethanol yields during alcoholic fermentation in Saccharomyces cerevisiae strains overexpressed or disrupted for GPD1 encoding glycerol 3-phosphate dehydrogenase. Yeast 13: 783–793 [DOI] [PubMed] [Google Scholar]
- 35. Nevoigt E, Stahl U. 1996. Reduced pyruvate decarboxylase and increased glycerol-3-phosphate dehydrogenase [NAD+] levels enhance glycerol production in Saccharomyces cerevisiae. Yeast 12: 1331–1337 [DOI] [PubMed] [Google Scholar]
- 36. Overkamp K, et al. 2002. Metabolic engineering of glycerol production in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 68: 2814–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peleg Y, Rokem JS, Goldberg I, Pines O. 1990. Inducible overexpression of the FUM1 gene in Saccharomyces cerevisiae: Localization of fumarase and efficient fumaric acid bioconversion to l-malic acid. Appl. Environ. Microbiol. 56: 2777–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pines O, Shemesh S, Battat E, Goldberg I. 1997. Overexpression of cytosolic malate dehydrogenase (MDH2) causes overproduction of specific organic acids in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 48: 248–255 [DOI] [PubMed] [Google Scholar]
- 39. Piskur J, Rozpedowska E, Polakova S, Merico A, Compagno C. 2006. How did Saccharomyces evolve to become a good brewer? Trends Genet. 22: 183–186 [DOI] [PubMed] [Google Scholar]
- 40. Pretorius IS. 2000. Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast 16: 675–729 [DOI] [PubMed] [Google Scholar]
- 41. Pretorius IS, Curtin C, Chambers PJ. 2012. The winemaker's bug: From ancient wisdom to opening new vistas with frontier yeast science. Bioeng. Bugs 3: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pretorius IS, Høj PB. 2005. Grape and wine biotechnology: challenges, opportunities and potential benefits. Aust. J. Grape Wine Res. 11: 83–108 [Google Scholar]
- 43. Pronk J. 1996. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12: 1607–1633 [DOI] [PubMed] [Google Scholar]
- 44. Raghevendran V, Gombert AK, Christensen B, Kotter P, Nielsen J. 2004. Phenotypic characterization of glucose repression mutants of Saccharomyces cerevisiae using experiments with 13C-labelled glucose. Yeast 21: 769–779 [DOI] [PubMed] [Google Scholar]
- 45. Remize F, Barnavon L, Dequin S. 2001. Glycerol export and glycerol-3-phosphate dehydrogenase, but not glycerol phosphatase, are rate limiting for glycerol production in Saccharomyces cerevisiae. Metab. Eng. 3: 301–312 [DOI] [PubMed] [Google Scholar]
- 46. Remize F, Roustan J, Sablayrolles J, Barre P, Dequin S. 1999. Glycerol overproduction by engineered Saccharomyces cerevisiae wine yeast strains leads to substantial changes in by-product formation and to a stimulation of fermentation rate in stationary phase. Appl. Environ. Microbiol. 65: 143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Santangelo GM. 2006. Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70: 253–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schmidt SA, Dillon S, Kolouchova R, Henschke PA, Chambers PJ. 2011. Impacts of variations in elemental nutrient concentration of Chardonnay musts on Saccharomyces cerevisiae fermentation kinetics and wine composition. Appl. Microbiol. Biotechnol. 91: 365–375 [DOI] [PubMed] [Google Scholar]
- 49. Schmidtke LM, Blackman JW, Agboola SO. 2012. Production technologies for reduced alcoholic wines. J. Food Sci. 77: R25–R41 [DOI] [PubMed] [Google Scholar]
- 50. Scott EW, Baker HV. 1993. Concerted action of the transcriptional activators REB1, RAP1, and GCR1 in the high-level expression of the glycolytic gene TPI. Mol. Cell. Biol. 13: 543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Selecky R, Smogrovicova D, Sulo P. 2008. Beer with reduced ethanol content produced using Saccharomyces cerevisiae yeast deficient in various tricarboxylic acid cycle enzymes. J. Inst. Brewing 114: 97–101 [Google Scholar]
- 52. Shinohara H. 1984. L'importance des substances volatiles du vin. Formation et effets sur la qualité. Bull. OIV 641/642: 606–618 [Google Scholar]
- 53. Storici F, Resnick MA. 2006. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol. 409: 329–345 [DOI] [PubMed] [Google Scholar]
- 54. Straus D, Raines R, Kawashima E, Knowles JR, Gilbert W. 1985. Active site of triosephosphate isomerase: In vitro mutagenesis and characterization of an altered enzyme. P. Natl. Acad. Sci. U. S. A. 82: 2272–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Swiegers JH, Bartowsky EJ, Henschke PA, Pretorius IS. 2005. Yeast and bacterial modulation of wine aroma and flavour. Aust. J. Grape Wine Res. 11: 139–173 [Google Scholar]
- 56. Tamas MJ, et al. 2003. A short regulatory domain restricts glycerol transport through yeast Fps1p. Yeast 20: S253–S253 [DOI] [PubMed] [Google Scholar]
- 57. Tamas MJ, et al. 1999. Fps1p controls the accumulation and release of the compatible solute glycerol in yeast osmoregulation. Mol. Microbiol. 31: 1087–1104 [DOI] [PubMed] [Google Scholar]
- 58. Varela C, Pizarro F, Agosin E. 2004. Biomass content governs fermentation rate in nitrogen-deficient wine musts. Appl. Environ. Microbiol. 70: 3392–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wudl F, Aharon-Shalom E, Bertz SH. 1981. Perdeuteriotetramethyltetraselenafulvalene. J. Org. Chem. 46: 4612–4614 [Google Scholar]
- 60. Zhang A, Kong Q, Cao L, Chen X. 2007. Effect of FPS1 deletion on the fermentation properties of Saccharomyces cerevisiae. Lett. Appl. Microbiol. 44: 212–217 [DOI] [PubMed] [Google Scholar]