Abstract
Bin toxin from Bacillus sphaericus acts on Culex quinquefasciatus larvae by binding to Cqm1 midgut-bound receptors, and disruption of the cqm1 gene is the major cause of resistance. The goal of this work was to screen for a laboratory-selected resistance cqm1REC allele in field populations in the city of Recife, Brazil, and to describe other resistance-associated polymorphisms in the cqm1 gene. The cqm1REC allele was detected in the four nontreated populations surveyed at frequencies from 0.001 to 0.017, and sequence analysis from these samples revealed a novel resistant allele (cqm1REC-D16) displaying a 16-nucletotide (nt) deletion which is distinct from the 19-nt deletion associated with cqm1REC. Yet a third resistant allele (cqm1REC-D25), displaying a 25-nt deletion, was identified in samples from a treated area exposed to B. sphaericus. A comparison of the three deletion events revealed that all are located within the same 208-nt region amplified during the screening procedure. They also introduce equivalent frameshifts in the sequence and generate the same premature stop codon, leading to putative transcripts encoding truncated proteins which are unable to locate to the midgut epithelium. The populations analyzed in this study contained a variety of alleles with mutations disrupting the function of the corresponding Bin toxin receptor. Their locations reveal a hot spot that can be exploited to assess the resistance risk through DNA screening.
INTRODUCTION
The utilization of biolarvicides based on Bacillus sphaericus requires monitoring strategies which can predict or prevent potential resistance selection among exposed mosquito populations. The binary (Bin) crystal toxin, which is the major active insecticidal factor found in commercial B. sphaericus strains, acts on mosquito larvae after ingestion, processing and binding to specific receptors located on the midgut epithelium (5, 24). Bin toxin displays high activity against larvae of the Culex pipiens complex and B. sphaericus has an excellent persistence under field conditions, which make this an effective biolarvicide for controlling these species in urban areas (17). However, the mode of action of Bin toxin relies entirely on its binding to a single class of midgut receptors which are glycosylphosphatidylinositol (GPI)-anchored α-glucosidases named Cpm1 and Cqm1 for Culex pipiens and Culex quinquefasciatus, respectively (7, 29, 30). Failure of toxins to bind to their midgut receptors has been described, in a wide range of target insects, as the primary resistance mechanism to insecticidal proteins from entomopathogenic bacteria (11, 16, 25). In the case of B. sphaericus, this is a critical aspect since resistance cases have also been reported after laboratory selection or field exposure (2, 25, 27, 36, 38, 44).
Investigation of the B. sphaericus resistance mechanisms has confirmed the essential role for the binding of Bin toxin to its receptors, since mutations within the cpm1/cqm1 genes, which are recessively inherited, are the major causes leading to the absence of functional receptors in the midgut and consequent high resistance levels (36). Resistance cases unrelated to receptor binding failure were reported; however, the mechanisms involved were not elucidated to date (25). On the other hand, molecular characterization of resistance linked to cpm1/cqm1 genes, performed in two laboratory-selected colonies and one field population, revealed four distinct alleles associated with this phenotype: cpm1GEO and cqm1REC, laboratory-selected alleles in California (GEO colony) and in Recife, Brazil (CqRL1/2362 colony), respectively, and cpm1BP and cpm1BP-del, both characterized in a field population from the south of France (6, 8, 29). Each allele displays distinct resistance-associated mutations which result in potential transcripts for truncated soluble proteins lacking the GPI anchor (cpm1GEO, cqm1REC, cpm1BP) or for truncated GPI-anchored proteins which are still unable to bind Bin toxin due to the loss of 66 amino acids (cpm1BP-del). The CqRL1/2362 colony, derived from eggs collected in the Recife Metropolitan Area (RMA; Brazil) and laboratory selected, displays high levels of resistance (resistance ratio [RR], >100,000) due to the failure of Bin toxin binding to microvillus receptors, and larvae from this colony were found to be homozygous for the cqm1REC allele (26, 27). Characterization of this allele showed a 19-nucleotide (nt) deletion which changes the frame of the protein coding sequence and originates a premature stop codon. The resulting predicted protein coded by this allele lacks part of its C-terminal end, including the GPI-anchor site. As a consequence, in individuals found to be homozygous for the cqm1REC allele, no functional polypeptide is present as a midgut membrane-bound receptor, a condition essential for Bin toxin binding (29). The characterization of this mutation allowed the development of a PCR amplification assay, which upon screening for cqm1REC in larva samples from RMA field populations, showed the presence of the associated deletion in a frequency on the order of 10−3 and 10−2 in nontreated and treated areas, respectively (4).
Utilization of B. sphaericus biolarvicides in RMA has been an important tool for controlling C. quinquefasciatus, which is the exclusive vector of the nematode Wuchereria bancrofti in Brazil (32). In view of the strategic role of this biolarvicide in RMA and the advantage provided by the knowledge available on the molecular basis of resistance, the goal of this work was to provide data on the frequency of the cqm1REC allele through DNA screening, as well as to describe novel polymorphisms of the cqm1 gene which can disrupt the expression of the Cqm1 protein as a target site for Bin toxin. The search for new resistance-mediating events is a key step for the development and continuous improvement of molecular methods for resistance monitoring, since most alleles identified are recessively inherited and cannot be directly tracked through bioassays.
MATERIALS AND METHODS
Mosquito colonies.
Three colonies were used in this study: CqSF, a Culex quinquefasciatus colony susceptible to Bacillus sphaericus that was established from egg rafts collected in the Recife Metropolitan Area (RMA) in Brazil; CqRL1/2362, a C. quinquefasciatus colony derived from CqSF and laboratory selected with B. sphaericus strain 2362 that displays a high level of resistance (>100,000-fold) to this entomopathogen (27); and RecLab, an Aedes aegypti colony established from egg samples also collected in RMA (21). Colonies have been maintained in the insectarium of the Centro de Pesquisas Aggeu Magalhães (CPqAM-FIOCRUZ), under controlled conditions, for at least 8 years. Larvae were reared in dechlorinated tap water and fed with cat biscuits. Adults were fed on 10% sucrose solution, and females were also fed with chicken blood. Insects were maintained at 26 ± 1°C, 70% humidity, and a photoperiod of 12 h of light and 12 h of darkness.
Culex quinquefasciatus population.
Four nontreated populations and one treated population from RMA were investigated. Nontreated areas were Ipojuca (IPO) and Jaboatão (JAB), located 60 and 20 km from Recife, respectively, and Roda de Fogo (ROD) and Azeitona (AZE), districts within the city. Samples from IPO and JAB consisted of egg rafts, collected in 2010, using around 30 oviposition traps (3) randomly placed in households within each area. Samples from ROD and AZE, consisting of larva batches collected directly from breeding sites in these areas in 1999, were stored at −70°C. The treated area of Água Fria (AGU) has been exposed to B. sphaericus since 2003 (32), and egg rafts were collected in 2010 according to the methodology described above. Eggs were used to establish subcolonies maintained under laboratory conditions, and larvae from F1 or F2 progenies were stored at −70°C until use.
Bioassays.
Multiple concentration bioassays were performed to establish the lethal concentrations of the B. sphaericus 2362 lyophilized powder SPH-88 (Institut Pasteur), after 48 h, for 50% (LC50) and 90% (LC90) of exposed larvae according to the standard procedure (35). Lethal concentrations were determined through probit analysis using the software SPSS 10.0 for Windows. Diagnostic dose bioassays were performed to discriminate susceptible and highly resistant individuals based on the exposure of individual fourth-instar larva samples to a high concentration of B. sphaericus 2362 for 48 h (1). Briefly, larvae were treated with 125 mg/liter of the biomass sample (sample no. 0448/09; CPqAM-FIOCRUZ) stored at −20°C, at a concentration more than 1,000-fold higher than the LC90 to the CqSF-susceptible colony, in a final volume of 2 ml of distilled water in 24-well plates. This high diagnostic dose was chosen based on the fact that all cqm1 resistance alleles already characterized confer total refractoriness to Bin toxin (25, 27, 38, 44).
AS-PCR.
For DNA isolation, individual fourth-instar larva samples were homogenized in DNAzol (Invitrogen), as recommended by the manufacturer, followed by precipitation with ethanol and DNA recovering in Tris-EDTA buffer. Allele-specific PCR (AS-PCR) was performed using specific primers, described in Chalegre et al. (4), and reactions were carried out for 35 cycles with an annealing temperature of 60°C using a Biometra thermocycler. Amplification products were separated by electrophoresis on 2.5% agarose gels, and each assay included no-DNA samples and A. aegypti DNA samples as negative controls. All AS-PCR products potentially amplified from the cqm1REC allele (fragments of less than 208 bp were expected due to the 19-nt deletion) and some fragments amplified from cqm1 (standard fragments of 208 bp) were subjected to automatic sequencing in an ABI Prism 3100 genetic analyzer (Applied Biosytems) to confirm their identity.
Cloning and sequencing of cqm1 alleles.
Genomic DNA from fourth-instar larvae was extracted as described, and PCRs were carried out with primers flanking the full-length coding sequence of cqm1 (see Table S1 in the supplemental material) using Platinum Taq high-fidelity DNA polymerase (Invitrogen). To obtain the partial sequence of the cqm1REC-D25 allele described in this study, a second set of primers were also used (see Table S1). PCR products were purified with the GFX DNA and gel band purification (GE Healthcare) kits. They were then ligated into the vector pGEM-T Easy (Promega) and subsequently transformed into the One Shot TOP10 chemically competent Escherichia coli cells (Invitrogen). Twelve clones from each sample were subjected to minipreparations in Luria-Bertani medium supplemented with ampicillin (100 μg/ml) and further purified with the QIAprep spin miniprep kit (Qiagen). After purification, the DNA samples were quantified and submitted for automatic sequencing. Alignment and assembly of the resulting nucleotide and amino acid sequences were performed with the DNASTAR software package, and manual refinement was done when needed.
RESULTS
Available data on the frequency of resistance alleles in field populations without a history of previous spraying remain scarce, and the lack of baseline information is one of the most limiting factors to evaluate resistance selection and to introduce management strategies. For this reason, an AS-PCR assay for the detection of cqm1REC, designed and evaluated previously (4), was applied to identify genotypes of C. quinquefasciatus larvae from nontreated populations of RMA. First, B. sphaericus susceptibility was investigated in field populations from IPO and JAB using in vivo bioassays. Multiple dose-response assays showed that larvae from both samples were susceptible since only a discrete increase in the LC50, with a resistance ratio (RR) around 3- and 4-fold for IPO and JAB, respectively, was observed and the LC90 showed similarities with the reference colony (Table 1). Diagnostic dose bioassays were also performed in an attempt to identify larva survivors from a high concentration of B. sphaericus that could potentially be homozygous-allele-resistant individuals. Samples of 1,680 and 720 larvae for IPO and JAB, respectively, were individually exposed, and there was no detection of survivors from these bioassays. Full mortality was already achieved after 24 h of B. sphaericus exposure in the treated groups, while the nontreated larvae from the control groups showed 2.9 and 1.7% mortality for IPO and JAB, respectively, after the standard period of 48 h of exposure. ROD and AZE larva susceptibilities were not analyzed since these samples were collected in 1999 and stored at −70°C without further evaluation.
Table 1.
Toxicity of B. sphaericus strain 2362 against larvaea
| Sample | No. of larvae | LC50 |
LC90 |
||
|---|---|---|---|---|---|
| Mean (95% fiducial limits) | RR | Mean (95% fiducial limits) | RR | ||
| CqSF | 360 | 0.004 (0.003–0.005) | 1.0 | 0.029 (0.019–0.049) | 1.0 |
| IPO | 1,480 | 0.013 (0.010–0.017) | 3.3 | 0.029 (0.022–0.044) | 1.0 |
| JAB | 1,480 | 0.017 (0.013–0.020) | 4.3 | 0.026 (0.023–0.049) | 0.9 |
| AGU | 1,140 | 0.024 (0.021–0.028) | 6.0 | 0.050 (0.042–0.059) | 1.7 |
Larvae were fourth-instar Culex quinquefasciatus from a susceptible laboratory colony (CqSF), two nontreated populations (IPO, JAB), and one treated population exposed to B. sphaericus (AGU). Shown are lethal concentrations (mg/liter) for 50% (LC50) or 90% (LC90) of larvae after 48 h. RR, resistance ratio; LC for the population tested/LC for the CqSF reference colony.
The AS-PCR performed in this study is based on the fact that according to the size of the DNA fragment amplified, using two primers flanking the 19-nt deletion which characterizes the cqm1REC allele, it is possible to identify fragments derived from either cqm1 or the cqm1REC resistant allele, corresponding to 208 or 189 bp, respectively (4). Here we define cqm1 as all alleles other than cqm1REC, taking into account that it is not possible to exclude the existence of unknown resistance mutations which are located outside the region under evaluation or do not alter the size of the amplified fragment. All populations analyzed here, which had no history of B. sphaericus spraying, nonetheless showed the presence of the cqm1REC allele. Frequencies of 0.003, 0.001, and 0.002 were detected in IPO, JAB, and AZE, respectively, whereas ROD showed a higher frequency of 0.017 (Table 2). In these populations, the cqm1REC allele was always found in heterozygous individuals, whereas most individuals were homozygous for the cqm1 allele (Fig. 1; Table 2).
Table 2.
Frequency of cqm1 alleles determined by PCRa
| Sample | Yr | Total no. of larvae | No. of larvae with genotype and frequency of each allele |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
cqm1 |
cqm1REC |
cqm1REC-D16 |
cqm1REC-D25 |
|||||||||
| SS | SR | RR | F | SR | RR | F | SR | RR | F | |||
| IPO | 2010 | 501 | 498 | 3 | 0 | 0.003 | 0 | 0 | 0 | 0 | 0 | 0 |
| JAB | 2010 | 510 | 507 | 1 | 0 | 0.001 | 1 | 1 | 0.003 | 0 | 0 | 0 |
| ROD | 1999 | 230 | 222 | 8 | 0 | 0.017 | 0 | 0 | 0 | 0 | 0 | 0 |
| AZE | 1999 | 240 | 236 | 1 | 0 | 0.002 | 3 | 0 | 0.006 | 0 | 0 | 0 |
| AGU | 2010 | 269 | 252 | 14 | 2 | 0.033 | 0 | 0 | 0 | 1 | 0 | 0.002 |
Frequency of cqm1 alleles in Culex quinquefasciatus larvae from four nontreated populations (IPO, JAB, ROD, AZE), as well as one treated population exposed to B. sphaericus (AGU). SS, homozygous for cqm1; SR and RR, heterozygous or homozygous for one of the resistance (r) alleles, respectively; F, allele frequency.
Fig 1.
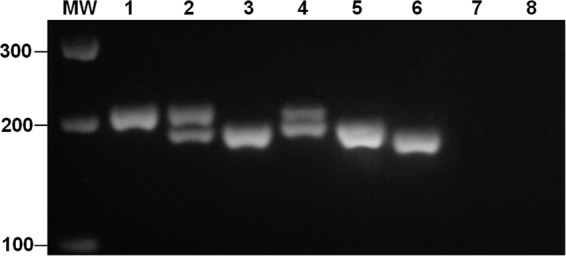
Fragments amplified from alleles of the Culex quinquefasciatus cqm1 gene. PCR produces profiles of homozygous for cqm1 (lane 1), heterozygous for cqm1REC (lane 2), homozygous for cqm1REC (lane 3), heterozygous for cqm1REC-D16 (lane 4), homozygous for cqm1REC-D16 (lane 5), and homozygous for cqm1REC-D25 (lane 6). No fragments were amplified from samples with Aedes aegypti DNA (lane 7) or without DNA (lane 8). Molecular size markers (molecular weight [MW]) in base pairs are shown on the left.
Screening for the cqm1REC allele involved sequencing of all amplified diagnostic fragments (<208 bp) potentially corresponding to this allele in order to confirm their identity. The analysis of the resulting sequences revealed not only the targeted cqm1REC 19-nt deletion but also a second deletion located in the same region encompassed by the amplified fragment. The new polymorphism found in these nontreated populations consists of a 16-nt deletion (nt 1306 to 1321), located 12 nucleotides downstream of the cqm1REC deletion (nt 1276 to 1294), and this allele was denominated cqm1REC-D16 (Fig. 2). Visual inspection of diagnostic fragments provided by AS-PCR does not allow a reliable distinction between products amplified from cqm1REC or cqm1REC-D16 alleles (Fig. 1). The cqm1REC-D16 allele was detected in both JAB and AZE samples. In JAB, its frequency of 0.003 was based on the finding of one heterozygote and one homozygote larva sample for this allele detected among 510 individuals analyzed, whereas in AZE, its frequency was 0.006, carried by only one heterozygous larva sample found among 240 larva samples (Table 2). The frequency of each cqm1REC or cqm1REC-D16 allele in 1,481 larva samples from all nontreated populations was 0.004 or 0.002, respectively, while the frequency of both was 0.006.
Fig 2.
Representation of the cqm1 gene encoding the Cqm1 receptor in Culex quinquefasciatus larvae (GenBank accession number DQ333335). (A) Full-length sequence of 1,848 nucleotides (nt) containing two introns of 50 nt (nt 1169 to 1218) and 55 nt (nt 1655 to 1709). (B) Nucleotide sequence from the region (nt 1221 to 1490) encompassing the polymorphisms found in alleles from Recife populations are indicated as follows: deletion of 19 nt corresponding to the cqm1REC allele is underlined; the six extra bases corresponding to the cqm1REC-D25 are boxed; and the 16-nt deletion from the cqm1REC-D16 allele, located 11 bases beyond the 19-nt deletion, is in italics and shaded in gray. Deletions start with a CGA trinucleotide motif in bold. The premature translation stop codon originating from the deletions described here is boxed in bold. For the allele-specific PCR, primers 1 and 2 were used for the 5′ and 3′ ends, respectively. These deletions were identified based on the sequencing of one strand of multiple DNA clones, which yielded identical results.
The DNA segment where the two deletions described above were found may be a hot spot for these kinds of mutations since a third deletion was detected in individuals from AGU, a treated area exposed to B. sphaericus. Resistance ratios for AGU larvae were 6- and 1.7-fold at LC50 and LC90, respectively. The AS-PCR screening based on a sample of 269 larvae showed a frequency of 0.033 for cqm1REC, and this allele was carried by heterozygous and homozygous larvae. The sequence analysis of the set of smaller-sized diagnostic fragments amplified from this AGU sample showed, besides the fragments containing the 19-nt deletion, one heterozygous individual for an allele containing a 25-nt deletion encompassing the 19-nt deletion for the cqm1REC allele, plus the six subsequent bases (Fig. 2). The allele presenting this new deletion, comprising nucleotides 1276 to 1300, was denominated cqm1REC-D25, and visual inspection of the corresponding fragment generated by AS-PCR also does not allow a reliable distinction compared to alleles containing the cqm1REC or cqm1REC-D16 deletion (Fig. 1).
The DNA extracted from larvae carrying the cqm1REC-D16 allele was used for cloning and sequencing of the entire cqm1 coding sequence in order to confirm its identity and analyze the whole sequence. For this purpose, 45 clones from 5 larva samples were analyzed, and among them, 21 were positive for the cqm1REC-D16 allele. The final sequence from the individuals carrying such a copy contained the two known introns of 50 and 55 bp, with an open reading frame of 1,727 bp in length. A total of 43 other single nucleotide differences were found throughout the sequence. Among them, 7 led to amino acid substitutions in the deduced protein (see Table S2 in the supplemental material); however, none of these is known to be associated with the capacity of Cqm1 to bind Bin toxin. The full-length sequence of the cqm1REC-D25 allele could not be amplified from the same larva genomic DNA which originally generated the PCR fragment of 183 nt containing the 25-nt deletion. Despite extensive trials using the primers available for amplifying the full-length sequence, and even additional primers designed for this purpose (see Table S1 in the supplemental material), only the wild-type cqm1 sequences without any deletions were found after analyzing the sequences of over 100 clones. Nevertheless, PCR assays using yet another set of primers (see Table S1, primers F4 and R4) resulted in the amplification of an 889-nt fragment whose sequence contained the 25-nt deletion. This sequence corresponded to about 50% of the full-length gene (positions 506 to 1419) and included the first intron; however, contrary to the sequence established for the cqm1REC-D16 allele, single nucleotide polymorphisms compared to the previously described cqm1 sequence (GenBank accession number DQ333335) were not found. Despite the high conservation of this 889-bp fragment, the failure to amplify a full-length copy of the cqm1REC-D25 allele suggests the existence of polymorphisms in other regions of its sequence which would prevent annealing of the available primers.
The two deletion events found in the cqm1REC-D16 and cqm1REC-D25 alleles change the reading frame of the succeeding amino acids, and both originate a premature stop codon at position 1362, which is also the same stop codon created by the 19-nt deletion from the cqm1REC allele (Fig. 2). The resulting sequence from cqm1REC-D16 and cqm1REC-D25 alleles potentially encodes a truncated (437 amino acids long) protein. Both the 16- and 25-nt deletions can confer resistance, in homozygous individuals for any of these alleles, since they will not code for full GPI-anchored Cqm1 proteins, available in the midgut epithelium for the Bin toxin to bind. Comparative analysis of the three deletion events affecting the cqm1 gene highlighted the fact that while the 19- and 25-nt deleted segments share the same initial insertion point in the sequence (at nucleotide 1276), all three also start with a common CGA trinucleotide motif.
DISCUSSION
In this study, a DNA screening was performed to detect the cqm1REC allele in populations of C. quinquefasciatus from the Recife Metropolitan Area (RMA). Susceptibility to B. sphaericus in two nontreated populations of IPO and JAB showed that RR values at the LC90 were similar to that of the reference colony, and the slight variations in RRs found at the LC50 are comparable to previous RRs reported for other nontreated mosquito populations, which have demonstrated the existence of natural variations in their B. sphaericus susceptibilities (4, 20, 34, 36, 37). The status of two other nontreated populations of ROD and AZE could not be analyzed; nevertheless, they were expected to be susceptible since B. sphaericus had not been used in these areas and they were also geographically isolated from the only two exposed areas in RMA at the time the samples were collected (28, 31). The cqm1REC allele was found in all nontreated populations analyzed, despite being originally identified in a laboratory-selected colony (4, 29), highlighting the strategic importance of monitoring for this allele in RMA. This result contrasts with those observed for laboratory-selected cadherin alleles associated with Cry1Ac resistance and whose screening in field populations has not led to a positive detection (13, 33). The frequency of the cqm1REC allele found among the populations analyzed, which was on the order of 10−3 (0.001 to 0.003), is consistent with the previous screening of two nontreated RMA populations (4) and with studies on Bacillus thuringiensis (Bt) resistance genes in Lepidoptera that have estimated the initial frequency of such alleles in nonexposed populations as 0.0015 (15). However, the higher frequency observed for the ROD population indicates that variations in pretreatment frequency can occur and should be taken into account for evaluating the resistance risk prior to spraying. Recent surveys of Bt resistance genes in lepidopteran field populations from Bt cotton areas in China also showed a wide range of frequencies from 10−4 to 10−1 (14, 18, 19, 40, 45), with the latter being considered the first substantial increase in resistance gene frequency among the areas under study. The treated population of AGU evaluated here showed a higher cqm1REC frequency than those from the nontreated populations, which is consistent with B. sphaericus exposure in that area. However, this frequency has not increased compared to previous screenings performed in AGU (4, 32), suggesting that the selection pressure might be low. This could be related with the introduction of Bacillus thuringiensis serovar israelensis (B. thuringiensis subsp. israelensis) to replace B. sphaericus in certain stages of this control program (C. M. F. Oliveira, personal communication). Considering that B. thuringiensis subsp. israelensis does not display cross-resistance with B. sphaericus and it is able to eliminate resistant genotypes (27, 36, 43), this could be one reason for reducing the selection pressure in that area.
Molecular biology-based methods can be useful for monitoring early selection of resistance in field populations since known resistance alleles carried by heterozygous individuals can be directly identified. Screening performed in this study revealed two novel polymorphisms in the cqm1 gene, 16- and 25-nt deletions found at the same region which encompasses the 19-nt deletion originally found in cqm1REC. The finding of one homozygous larva sample for cqm1REC-D16 in the JAB population was not expected, considering its status of being a nontreated population, since data from a previous screening of B. sphaericus and Bt resistance alleles have shown such alleles only in heterozygous individuals under such conditions (4, 42, 45). The resistance phenotype conferred by cqm1REC-D16 and cqm1REC-D25 alleles could not be experimentally confirmed; nevertheless, the functional effect of these deletions on larva susceptibility is likely similar to that of cqm1REC, since they all provoke frameshifts and introduce the same premature stop codon in the sequence, which prevents the expression of full-length GPI-anchored proteins (6, 8, 29). Regardless of the fact that the Bin binding epitope on the Cqm1 protein is still unknown, the loss of the GPI anchor prevents its location on the midgut epithelium and, consequently, its function as the Bin toxin receptor.
A comparable situation, involving a wide range of polymorphisms, has been investigated in Lepidoptera cotton pests which display multiple cadherin alleles associated with Cry1Ac toxin resistance (12, 22, 45). To date, 12 cadherin alleles were found to be genetically linked to Cry1Ac toxin resistance, one from a laboratory-selected strain of Heliothis virescens, three from Pectinophora gossypiella, and eight alleles detected in Helicoverpa armigera (the last two species are from field populations) (12, 22, 23, 39, 42, 45). From the functional point of view, many of them are considered null alleles since they are disrupted by events which result in the generation of premature stop codons or aberrant splicing events in their sequences, expected to encode truncated proteins lacking toxin binding sites or the transmembrane domain (39, 42, 45). In these cases, the final result is an inability of the toxin to bind to its target tissue in a fashion similar to that observed for the cqm1 resistance alleles described here (10, 45). In terms of monitoring tools, DNA screening of cadherin alleles has been considered a complex task due to the diversity and multiple locations of events found over genes that can be as large as 16 kb, as was seen for H. armigera (41). In contrast, the cqm1 gene is around 1.8 kb (29), a size which facilitates its amplification and sequencing and allows DNA screening of coexisting, resistance-linked alleles in field populations.
The finding of novel events in the cqm1 gene associated to B. sphaericus resistance and mapped to the same region where other resistance mutations have been identified suggests the existence of a hot spot for such events. Aside from the polymorphisms recorded in RMA in Brazil, the same gene region is also the target of the mutations which characterize the cpm1BP and cpm1BP-del resistance alleles, which were found to coexist in a C. pipiens population from France (6). Overall, five of six resistance alleles characterized in cpm1/cqm1 genes have mutations located within this region, and only a single nonsense mutation (T1706A), from the cpm1GEO allele (California), is mapped outside (8). From the evolutionary point of view, further studies are needed in order to clarify the mechanisms responsible for the rise of such events in this specific region of the cqm1 gene, as well as the impact of these alleles on the biological performance of the targeted insects. Although resistance in individuals from the CqRL1/2362 colony was related to a discrete reduction of some biological parameters (9), this colony has been maintained in the laboratory for more than 10 years and recent data have shown that the cqm1REC allele is able to compete with cqm1, at least under laboratory conditions (1). Similarly, cadherin genes, in view of null alleles found associated with Cry1Ac resistance, do not seem to be essential for the survival of H. virescens, P. gossypiella, and H. armigera (12, 22, 45). In conclusion, the findings from this work indicate a diversity of polymorphisms for the cqm1 gene which can lead to a loss of function as the receptor for the B. sphaericus Bin toxin. The events behind these polymorphisms, detected in individuals from field populations of RMA, are nevertheless located in a specific region of the cqm1 gene, which allows for easy screening of the multiple events and is useful for assessing the resistance risk.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by “Programa Estratégico de Apoio à Pesquisa em Saúde (PAPES)” from Fundação Oswaldo Cruz-FIOCRUZ, Conselho Nacional de Pesquisa (CNPq Brazil, grant 403488/2008-7), Fundação de Amparo à Ciência e Tecnologia do Estado de Pernambuco (FACEPE Brazil, grant APQ 0427-2.13/08).
We thank Andréa Neves Guedes de Souza and Liliane Barbosa Amorim (CPqAM-FIOCRUZ) for the insectarium support and the Program for Technological Development in Tools for Health PDTIS-FIOCRUZ for the use of its facilities.
Footnotes
Published ahead of print 6 July 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Amorim LB, et al. 2010. Stability of Culex quinquefasciatus resistance to Bacillus sphaericus evaluated by molecular tools. Insect Biochem. Mol. Biol. 40: 311–316 [DOI] [PubMed] [Google Scholar]
- 2. Amorim LB, Oliveira CMF, Rios EM, Regis L, Silva-Filha MHNL. 2007. Development of Culex quinquefasciatus resistance to Bacillus sphaericus strain IAB59 needs long term selection pressure. Biol. Control 42: 155–160 [Google Scholar]
- 3. Barbosa RM, Souto A, Eiras AE, Regis L. 2007. Laboratory and field evaluation of an oviposition trap for Culex quinquefasciatus (Diptera: Culicidae). Mem. Inst. Oswaldo Cruz 102: 523–529 [DOI] [PubMed] [Google Scholar]
- 4. Chalegre KD, et al. 2009. Detection of an allele conferring resistance to Bacillus sphaericus binary toxin in Culex quinquefasciatus populations by molecular screening. Appl. Environ. Microbiol. 75: 1044–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Charles JF, Nielsen-LeRoux C, Delecluse A. 1996. Bacillus sphaericus toxins: molecular biology and mode of action. Annu. Rev. Entomol. 41: 451–472 [DOI] [PubMed] [Google Scholar]
- 6. Darboux I, Charles JF, Pauchet Y, Warot S, Pauron D. 2007. Transposon-mediated resistance to Bacillus sphaericus in a field-evolved population of Culex pipiens (Diptera: Culicidae). Cell. Microbiol. 9: 2022–2029 [DOI] [PubMed] [Google Scholar]
- 7. Darboux I, Nielsen-LeRoux C, Charles JF, Pauron D. 2001. The receptor of Bacillus sphaericus binary toxin in Culex pipiens (Diptera: Culicidae) midgut: molecular cloning and expression. Insect Biochem. Mol. Biol. 31: 981–990 [DOI] [PubMed] [Google Scholar]
- 8. Darboux I, et al. 2002. Loss of the membrane anchor of the target receptor is a mechanism of bioinsecticide resistance. Proc. Natl. Acad. Sci. U. S. A. 99: 5830–5835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Oliveira CM, Filho FC, Beltran JE, Silva-Filha MH, Regis L. 2003. Biological fitness of a Culex quinquefasciatus population and its resistance to Bacillus sphaericus. J. Am. Mosq. Control Assoc. 19: 125–129 [PubMed] [Google Scholar]
- 10. Fabrick JA, Mathew LG, Tabashnik BE, Li X. 2011. Insertion of an intact CR1 retrotransposon in a cadherin gene linked with Bt resistance in the pink bollworm, Pectinophora gossypiella. Insect Mol. Biol. 20: 651–665 [DOI] [PubMed] [Google Scholar]
- 11. Ferré J, Van Rie J. 2002. Biochemistry and genetics of insect resistance to Bacillus thuringiensis. Annu. Rev. Entomol. 47: 501–533 [DOI] [PubMed] [Google Scholar]
- 12. Gahan LJ, Gould F, Heckel DG. 2001. Identification of a gene associated with Bt resistance in Heliothis virescens. Science 293: 857–860 [DOI] [PubMed] [Google Scholar]
- 13. Gahan LJ, Gould F, Lopez JD, Jr, Micinski S, Heckel DG. 2007. A polymerase chain reaction screen of field populations of Heliothis virescens for a retrotransposon insertion conferring resistance to Bacillus thuringiensis toxin. J. Econ. Entomol. 100: 187–194 [DOI] [PubMed] [Google Scholar]
- 14. Gao Y, Wu K, Gould F. 2009. Frequency of Bt resistance alleles in H. armigera during 2006–2008 in Northern China. Environ. Entomol. 38: 1336–1342 [DOI] [PubMed] [Google Scholar]
- 15. Gould F, et al. 1997. Initial frequency of alleles for resistance to Bacillus thuringiensis toxins in field populations of Heliothis virescens. Proc. Natl. Acad. Sci. U. S. A. 94: 3519–3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heckel DG, et al. 2007. The diversity of Bt resistance genes in species of Lepidoptera. J. Invertebr Pathol. 95: 192–197 [DOI] [PubMed] [Google Scholar]
- 17. Lacey LA. 2007. Bacillus thuringiensis serovariety israelensis and Bacillus sphaericus for mosquito control. J. Am. Mosq. Control Assoc. 23: 133–163 [DOI] [PubMed] [Google Scholar]
- 18. Liu F, et al. 2008. Resistance allele frequency to Bt cotton in field populations of Helicoverpa armigera (Lepidoptera: Noctuidae) in China. J. Econ. Entomol. 101: 933–943 [DOI] [PubMed] [Google Scholar]
- 19. Liu F, et al. 2010. Evidence of field-evolved resistance to Cry1Ac-expressing Bt cotton in Helicoverpa armigera (Lepidoptera: Noctuidae) in northern China. Pest Manag. Sci. 66: 155–161 [DOI] [PubMed] [Google Scholar]
- 20. Liu H, Cupp EW, Guo A, Liu N. 2004. Insecticide resistance in Alabama and Florida mosquito strains of Aedes albopictus. J. Med. Entomol. 41: 946–952 [DOI] [PubMed] [Google Scholar]
- 21. Melo-Santos MA, Araujo AP, Rios EM, Regis L. 2009. Long lasting persistence of Bacillus thuringiensis serovar. israelensis larvicidal activity in Aedes aegypti (Diptera: Culicidae) breeding places is associated to bacteria recycling. Biol. Control 49: 186–191 [Google Scholar]
- 22. Morin S, et al. 2003. Three cadherin alleles associated with resistance to Bacillus thuringiensis in pink bollworm. Proc. Natl. Acad. Sci. U. S. A. 100: 5004–5009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morin S, et al. 2004. DNA-based detection of Bt resistance alleles in pink bollworm. Insect Biochem. Mol. Biol. 34: 1225–1233 [DOI] [PubMed] [Google Scholar]
- 24. Nielsen-Leroux C, Charles JF. 1992. Binding of Bacillus sphaericus binary toxin to a specific receptor on midgut brush-border membranes from mosquito larvae. Eur. J. Biochem. 210: 585–590 [DOI] [PubMed] [Google Scholar]
- 25. Nielsen-Leroux C, et al. 2002. High resistance to Bacillus sphaericus binary toxin in Culex pipiens (Diptera: Culicidae): the complex situation of west Mediterranean countries. J. Med. Entomol. 39: 729–735 [DOI] [PubMed] [Google Scholar]
- 26. Oliveira CM, et al. 2004. Inheritance and mechanism of resistance to Bacillus sphaericus in Culex quinquefasciatus (Diptera: Culicidae) from China and Brazil. J. Med. Entomol. 41: 58–64 [DOI] [PubMed] [Google Scholar]
- 27. Pei G, et al. 2002. A strain of Bacillus sphaericus causes slower development of resistance in Culex quinquefasciatus. Appl. Environ. Microbiol. 68: 3003–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Regis L, et al. 1995. Integrated control measures against Culex quinquefasciatus, the vector of filariasis in Recife. Mem. Inst. Oswaldo Cruz 90: 115–119 [DOI] [PubMed] [Google Scholar]
- 29. Romão TP, et al. 2006. A second independent resistance mechanism to Bacillus sphaericus binary toxin targets its alpha-glucosidase receptor in Culex quinquefasciatus. FEBS J. 273: 1556–1568 [DOI] [PubMed] [Google Scholar]
- 30. Silva-Filha MH, Nielsen-LeRoux C, Charles JF. 1999. Identification of the receptor for Bacillus sphaericus crystal toxin in the brush border membrane of the mosquito Culex pipiens (Diptera: Culicidae). Insect Biochem. Mol. Biol. 29: 711–721 [DOI] [PubMed] [Google Scholar]
- 31. Silva-Filha MH, Regis L, Oliveira CM, Furtado AE. 2001. Impact of a 26-month Bacillus sphaericus trial on the preimaginal density of Culex quinquefasciatus in an urban area of Recife, Brazil. J. Am. Mosq. Control Assoc. 17: 45–50 [PubMed] [Google Scholar]
- 32. Silva-Filha MHNL, et al. 2008. Culex quinquefasciatus field populations subjected to treatment with Bacillus sphaericus did not display high resistance levels. Biol. Control 44: 227–234 [Google Scholar]
- 33. Tabashnik BE, et al. 2006. DNA screening reveals pink bollworm resistance to Bt cotton remains rare after a decade of exposure. J. Econ Entomol. 99: 1525–1530 [DOI] [PubMed] [Google Scholar]
- 34. Vasquez MI, Violaris M, Hadjivassilis A, Wirth MC. 2009. Susceptibility of Culex pipiens (Diptera: Culicidae) field populations in Cyprus to conventional organic insecticides, Bacillus thuringiensis subsp. israelensis, and methoprene. J. Med. Entomol. 46: 881–887 [DOI] [PubMed] [Google Scholar]
- 35. WHO 1985. Informal consultation on the development of Bacillus sphaericus as a microbial larvicide. TDR/BCV/SPHAERICUS/85.3.1–24 WHO, Geneva, Switzerland [Google Scholar]
- 36. Wirth MC. 2010. Mosquito resistance to bacterial larvicidal toxins. Open Toxinology J. 3: 101–115 [Google Scholar]
- 37. Wirth MC, Ferrari JA, Georghiou GP. 2001. Baseline susceptibility to bacterial insecticides in populations of Culex pipiens complex (Diptera: Culicidae) from California and from the Mediterranean Island of Cyprus. J. Econ. Entomol. 94: 920–928 [DOI] [PubMed] [Google Scholar]
- 38. Wirth MC, Georghiou GP, Malik JI, Abro GH. 2000. Laboratory selection for resistance to Bacillus sphaericus in Culex quinquefasciatus (Diptera: Culicidae) from California, USA. J. Med. Entomol. 37: 534–540 [DOI] [PubMed] [Google Scholar]
- 39. Xu X, Yu L, Wu Y. 2005. Disruption of a cadherin gene associated with resistance to Cry1Ac {delta}-endotoxin of Bacillus thuringiensis in Helicoverpa armigera. Appl. Environ. Microbiol. 71: 948–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu Z, et al. 2009. Using an F(2) screen to monitor frequency of resistance alleles to Bt cotton in field populations of Helicoverpa armigera (Hubner) (Lepidoptera: Noctuidae). Pest Manag. Sci. 65: 391–397 [DOI] [PubMed] [Google Scholar]
- 41. Yang Y, Chen H, Wu S, Xu X, Wu Y. 2006. Identification and molecular detection of a deletion mutation responsible for a truncated cadherin of Helicoverpa armigera. Insect Biochem. Mol. Biol. 36: 735–740 [DOI] [PubMed] [Google Scholar]
- 42. Yang Y, Chen H, Wu Y, Wu S. 2007. Mutated cadherin alleles from a field population of Helicoverpa armigera confer resistance to Bacillus thuringiensis toxin Cry1Ac. App. Environ. Microbiol. 73: 6939–6944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yuan ZM, Pei GF, Regis L, Nielsen-Leroux C, Cai QX. 2003. Cross-resistance between strains of Bacillus sphaericus but not B. thuringiensis israelensis in colonies of the mosquito Culex quinquefasciatus. Med. Vet. Entomol. 17: 251–256 [DOI] [PubMed] [Google Scholar]
- 44. Yuan ZM, Zhang YM, Cai QX, Liu EY. 2000. High-level field resistance to Bacillus sphaericus C3-41 in Culex quinquefasciatus from Southern China. Biocontrol Sci. Technol. 10: 41–49 [Google Scholar]
- 45. Zhao J, Jin L, Yang Y, Wu Y. 2010. Diverse cadherin mutations conferring resistance to Bacillus thuringiensis toxin Cry1Ac in Helicoverpa armigera. Insect Biochem. Mol. Biol. 40: 113–118 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.