Abstract
Chloramphenicol and florfenicol are broad-spectrum antibiotics. Although the bacterial resistance mechanisms to these antibiotics have been well documented, hydrolysis of these antibiotics has not been reported in detail. This study reports the hydrolysis of these two antibiotics by a specific hydrolase that is encoded by a gene identified from a soil metagenome. Hydrolysis of chloramphenicol has been recognized in cell extracts of Escherichia coli expressing a chloramphenicol acetate esterase gene, estDL136. A hydrolysate of chloramphenicol was identified as p-nitrophenylserinol by liquid chromatography-mass spectroscopy and proton nuclear magnetic resonance spectroscopy. The hydrolysis of these antibiotics suggested a promiscuous amidase activity of EstDL136. When estDL136 was expressed in E. coli, EstDL136 conferred resistance to both chloramphenicol and florfenicol on E. coli, due to their inactivation. In addition, E. coli carrying estDL136 deactivated florfenicol faster than it deactivated chloramphenicol, suggesting that EstDL136 hydrolyzes florfenicol more efficiently than it hydrolyzes chloramphenicol. The nucleotide sequences flanking estDL136 encode proteins such as amidohydrolase, dehydrogenase/reductase, major facilitator transporter, esterase, and oxidase. The most closely related genes are found in the bacterial family Sphingomonadaceae, which contains many bioremediation-related strains. Whether the gene cluster with estDL136 in E. coli is involved in further chloramphenicol degradation was not clear in this study. While acetyltransferases for chloramphenicol resistance and drug exporters for chloramphenicol or florfenicol resistance are often detected in numerous microbes, this is the first report of enzymatic hydrolysis of florfenicol resulting in inactivation of the antibiotic.
INTRODUCTION
Resistance to natural antibiotics or their synthetic derivatives has been observed frequently from various sources, including antibiotic producers or antibiotic nonproducers (15). Resistance to antibiotics is a growing public concern owing to the emergence of multiple-antibiotic resistance in human and animal pathogens. However, the emergence and the genetic determinants of antibiotic resistance mechanisms are not fully understood and there may be several modes of resistance to different antibiotics. Because only a minor fraction of bacteria (less than 0.3%) in soils are culturable (1), the currently known antibiotic resistance mechanisms may be biased, as studies have been conducted in cultured microorganisms only (14).
Chloramphenicol (Cm), a representative amphenicol antibiotic, was considered to be a promising broad-spectrum antibiotic effective in both human and veterinary medicine. However, since 1994, its application in foods and animals has been prohibited by the European Union, because Cm residues in carcasses of food animals could potentially cause adverse side effects to meat consumers (25). Cm residues in meat can cause irreversible, fatal aplastic anemia in humans, and the condition is not dependent upon Cm concentrations. Therefore, Cm application for food animals has also been banned in many other countries, including the United States, Canada, Australia, Japan, China, and Korea. In contrast, florfenicol (Ff), a synthetic Cm derivative with a fluoro substitution at C3 and the replacement of a nitro group (-NO2) by a sulfomethyl group (-SO2CH3), is approved for the treatment of bacterial respiratory infections in cattle and pigs, because it does not cause any adverse side effects (in particular, aplastic anemia in animals) (29, 31). Both Cm and Ff bind to the bacterial ribosome and inhibit translation by interacting with the peptidyl transferase centers of ribosomes (25).
Resistance to Cm has been detected in a large number of microbes and is due to a variety of mechanisms. Most commonly, Cm acetyltransferase (CAT) inactivates Cm by specifically acetylating the hydroxyl group at C3 (27). Phosphotransferase also inactivates Cm by O-phosphorylation in Streptomyces venezuelae, the Cm-producing actinomycete (18). Otherwise, many drug exporters mediate Cm resistance in a bacterial system by specifically or nonspecifically driving out the antibiotic (2, 22, 25). Of note, the known CAT is unable to inactivate Ff, due to the specific fluoro substitution at C3 (7, 31). Only a small number of the Cm resistance genes identified to date mediate cross-resistance to Ff; these mainly encode drug efflux pumps. They include cmlA from Escherichia coli (33), floR from Salmonella enterica (5), fexA from Staphylococcus lentus (11), and pexA from uncultured bacteria (14). In particular, cfr from Staphylococcus sciuri encodes a Cm-Ff resistance function that is currently unidentified and differs from known acetyltransferase or efflux proteins (26).
In a previous study, we analyzed a soil metagenome and identified two different genes encoding bacterial esterases of the hormone-sensitive lipase family that were capable of reactivating Cm by deacetylation of Cm acetates (32). By characterizing these two Cm acetate esterases (CAE), we found that EstDL26 displayed regioselectivity by hydrolyzing 3-acetyl Cm in preference to 1-acetyl Cm. In contrast, EstDL136 revealed a remarkable preference toward fatty acid acetate and it deacetylated all Cm acetates (32). The involvement of two bacterial esterases in Cm acetate hydrolysis leads us to predict that the metagenome clone carrying a gene for CAE activity may also encode enzymes for Cm metabolism. The screening of functional enzymes for Cm metabolism revealed a subclone of pDL136 carrying estDL136 (32), pUDL136B, which encoded Cm hydrolysis. In this study, we discovered that the process of Cm inactivation by EstDL136 also hydrolyzes Ff, which confers a cross-resistance to Cm and Ff when estDL136 is expressed heterologously in E. coli. While some genes encoding drug exporters and cfr are known to be associated with Ff resistance, the hydrolytic inactivation of Ff described in this study has not been reported previously.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and DNA manipulation.
E. coli DH5α, EPI100, and BL21 strains were routinely cultured at 37°C either on Luria-Bertani (LB) agar or in LB broth supplemented with the appropriate antibiotics. The antibiotic concentrations used for E. coli strains were 25 μg/ml for Cm and 100 μg/ml for ampicillin. General DNA manipulation was performed as described previously (24). A recombinant plasmid, pUDL136B, carried estDL136 and a putative gene with unknown function (32). Deletion of the putative gene (348 bp at 3′ terminus) from pUDL136B was performed through digestion with NcoI, and the resulting fragment was then self-ligated to generate pUEst136, carrying estDL136 only.
TLC.
E. coli DH5α carrying either pUC119 (negative control) or pUEst136 was cultured at 37°C in LB broth supplemented with 100 μg/ml ampicillin. After 1 day of incubation, E. coli strains were harvested by centrifugation and the cell pellets were lysed using a sonic dismembrator (model 500; Fisher Scientific, Waltham, MA). After centrifugation, Cm (80 μg/ml) was added to the cell extracts, which were then stored at 30°C for a period of 2 h. To terminate the reactions, the solution was extracted with an equal volume of butanol and then concentrated to dryness and resuspended with butanol, and the residues were used for thin-layer chromatography (TLC) analysis using silica gel 60 F254 (Merck, Darmstadt, Germany) with methanol/chloroform (1:3).
Chemical identification.
EstDL136 was overexpressed and purified as a His-tagged fusion protein, as described previously (32). Large-scale Cm hydrolysate production was performed by incubating Cm (50 mg) with EstDL136 (3 mg) at 37°C in 20 mM Tris-Cl buffer suspension (pH 8.0). After 24 h of incubation, the hydrolysis reaction was terminated by extracting the solution with equal volumes of ethyl acetate and butanol. The resultant extracts were concentrated, and the Cm hydrolysate was isolated by preparative TLC using hexane/ethyl acetate in the ratio of 1:1. The hydrolysate was then analyzed using reverse-phase high-performance liquid chromatography (HPLC) and mass spectroscopy (MS). Chromatography separation was performed using a Symmetry C18 column (150 mm by 4.6 mm; Waters) with a 10-to-30% linear gradient of methanol containing 0.1% trifluoroacetic acid for 10 min. Mass spectra were performed in the positive-ion mode with electrospray ionization (ESI) by scanning to m/z 1,000 s−1. A nitrogen atmospheric pressure ionization-electrospray vaporizer was used for this experiment, and the drying gas temperature was 350°C. The flow rate of the drying gas was 4 liter/min, the fragmentor voltage was 80 V, and the corona current intensity was 4 μA. ESI-MS results were recorded on a single-quadrupole mass spectrometer equipped with an ESI (MSD1100, Hewlett-Packard Co., CA). Proton nuclear magnetic resonance (1H-NMR) spectra were recorded on a Bruker AMX-500 (500 MHz) spectrometer (Bruker Analytische Messtechnik GmbH, Rheinstetten, Germany) using deuterated methanol (CD3OD) as a solvent and tetramethylsilane for an internal standard.
Detection of Cm and Ff hydrolysis.
The catalytic efficiency of purified EstDL136 on Cm and Ff was investigated by incubating the antibiotics (100 μg) with 30 μg of the enzyme under the conditions described above. Aliquots were collected at regular intervals between 1 and 5 h and subsequently extracted twice with an equal volume of butanol. The two extracts were combined and concentrated to dryness. The residues were dissolved in methanol and then used for HPLC analysis to examine the antibiotics and their hydrolysates. HPLC operation was performed using the Agilent 1100 series HPLC system (Agilent Technologies, Santa Clara, CA). A C18 column (SB C18, 3.5 μm, 150 by 4.6 mm; Zorbax) with a 0.9-ml/min flow rate was used for Cm derivatives, with a 10-to-30% linear gradient of methanol for the 0- to 10-min time period, increasing to a 30-to-60% gradient for the 10- to 18-min time period. Detection was achieved at 270 nm using a photodiode array (PDA) detector (Agilent Technologies). For the detection of Ff, a C18 column (C18, 5 μm, 250 by 4.6 mm; Phenomenex) with a 1-ml/min flow rate was used, with a 35-to-40% linear gradient of methanol performed for the first 6 min and a 40% linear gradient of methanol for a further 9 min. Ff and its hydrolysate were detected at 224 nm using a PDA detector.
Cm and Ff resistance test.
To determine the amphenicol antibiotic resistance of E. coli expressing estDL136 (the Cm hydrolase gene), E. coli DH5α carrying pUEst136 was cultured at 37°C in 100 ml LB broth supplemented with different concentrations of Cm or Ff (0 to 128 μg/ml). A negative-control experiment was performed using E. coli DH5α carrying pUC119 exposed to 100 μg/ml of ampicillin and 16 μg/ml of Cm or Ff; a second control using the pUEst136 strain exposed to 100 μg/ml of ampicillin only was also conducted using the above-mentioned method. Bacterial growth was monitored continuously by measuring optical density at 600 nm with a spectrophotometer (Beckman Coulter, Brea, CA).
Nucleotide sequence analysis.
In pDL136, a DNA fragment of 14.6 kb containing estDL136 was obtained by using the primer-walking strategy to analyze the DNA sequences of the genes flanking estDL136. DNA sequencing and primer synthesis were performed commercially at the DNA sequencing facility of GenoTech Corp. (Daejeon, Republic of Korea). DNA sequences were analyzed with the Basic Local Alignment Search Tool (BLAST) program provided by the National Center for Biotechnology Information (NCBI). Determination of the open reading frames (ORFs) was performed by using the ORF Finder software at the NCBI (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The start codon was determined by the presence of conserved Shine-Dalgarno sequences located 4 to 10 bp ahead of an ATG codon for the individual ORF. Putative homologues of each ORF were identified by using BLAST. The G+C content of each assembled nucleotide sequence was calculated by using the Seqool program (http://www.biossc.de/seqool/index.html).
Nucleotide sequence accession number.
The nucleotide sequence of pDL136 has been deposited in GenBank with the accession number JN242251.
RESULTS
Chloramphenicol is hydrolyzed by EstDL136.
Investigation of Cm metabolism by TLC using cell extracts of E. coli carrying subclones of pDL26 and pDL136 identified a pDL136-derived plasmid, pUDL136B, carrying a gene for the metabolism of Cm (data not shown). Repetitive TLC analyses using pUEst136 led to the discovery that EstDL136 hydrolyzed Cm (Fig. 1). TLC analyses of the reaction mixture revealed that Cm (Rf = 0.87) was completely hydrolyzed in cell extracts of E. coli carrying pUEst136, while Cm retained its original structure in the negative-control pUC119 extract. Meanwhile, a new compound (Rf = 0.23) was concurrently produced during Cm hydrolysis in the pUEst136 extract (Fig. 1, lane 4), suggesting the enzymatic hydrolysis of Cm by EstDL136.
Fig 1.
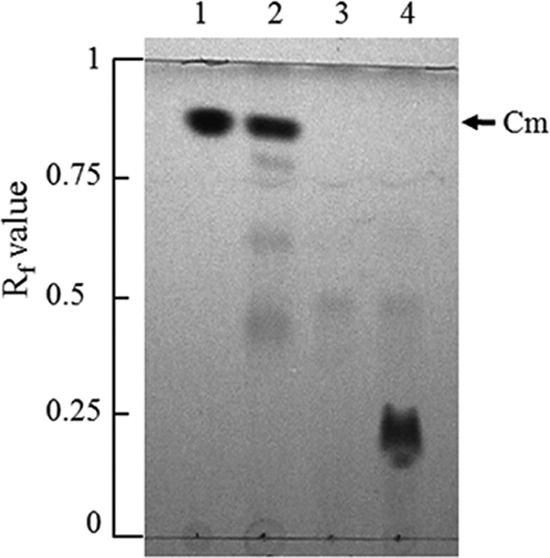
Degradation of chloramphenicol (Cm) by the cell extract of Escherichia coli expressing estDL136. Lane 1, Cm standard; lane 2, Cm (80 μg/ml) incubated with the cell extract of E. coli DH5α carrying pUC119; lanes 3 and 4, crude extracts of E. coli DH5α carrying pUEst136 incubated without and with Cm, respectively. For Cm incubated with cell lysate, reactions were terminated by using the extraction method with butanol. Butanol extracts were concentrated and used for thin-layer chromatography analysis with a developing solvent of methanol/chloroform (1:3). Rf value represents retardation factor, which is the fraction of an analyte in the mobile phase in chromatography.
Identification of the Cm hydrolysate.
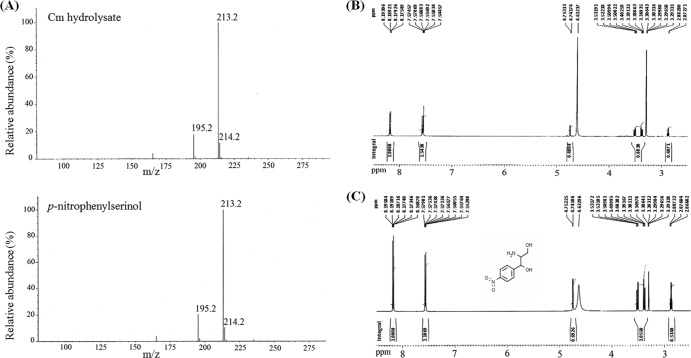
Large volumes of Cm (50 mg) were incubated with EstDL136, and the Cm hydrolysate (a newly produced compound detected on TLC; Rf = 0.23) was detected mainly in the butanol extract of the reaction mixture, wherein 14.2 mg of the hydrolysate was isolated further by preparative TLC. The Cm hydrolysate isolated was subjected to liquid chromatography (LC)-MS analysis, and the MS of the Cm hydrolysate was apparently identical to that of p-nitrophenylserinol (Sigma-Aldrich, St. Louis, MO), which was at m/z 213.2 [M+H]+; its dehydrated product was at m/z 195.2 [M+H-H2O]+ (Fig. 2A). In addition, 1H-NMR analysis demonstrated that the chemical shift of the Cm hydrolysate and p-nitrophenylserinol peaks were identical (Fig. 2B and C). These results with LC-MS and NMR strongly suggested that the structures of Cm hydrolysate and p-nitrophenylserinol are identical; therefore, we concluded that the hydrolysate of Cm is p-nitrophenylserinol, which is generated by hydrolyzing the amide linkage of Cm.
Fig 2.
Identification of the chloramphenicol (Cm) hydrolysate. (A) Comparative analysis of the liquid chromatography-mass spectroscopy profile of purified Cm hydrolysate and p-nitrophenylserinol standard. (B and C) Proton nuclear magnetic resonance (1H-NMR) analysis of the Cm hydrolysate (B) and p-nitrophenylserinol standard (C).
In vitro detection of Cm and Ff hydrolysis.
The hydrolysis of Cm and Ff by the EstDL136 enzyme was investigated over time using HPLC, showing that EstDL136 gradually hydrolyzed Cm over time. The Cm peak with a retention time of 15.3 min decreased and had nearly disappeared in the 5-h reaction sample (Fig. 3A). In parallel with the disappearance of Cm, the presence of the hydrolysate with a retention time of 4.1 min, which is the same as that of p-nitrophenylserinol, gradually increased (Fig. 3A). Because Ff structurally and functionally resembles Cm, though it is resistant to CAT due to the fluoro substitution at C3 (27), we have examined whether EstDL136 is capable of hydrolyzing Ff. The hydrolysis of Ff by EstDL136 over time was apparent through HPLC analysis (Fig. 3B). Ff, with a retention time of 6.9 min, was completely hydrolyzed by EstDL136 in the 5-h sample compared with the control sample, and a new compound with a retention time of 2.7 min was produced simultaneously (Fig. 3B). We surmise that the newly produced compound is Ff hydrolysate, although it has not been structurally identified.
Fig 3.
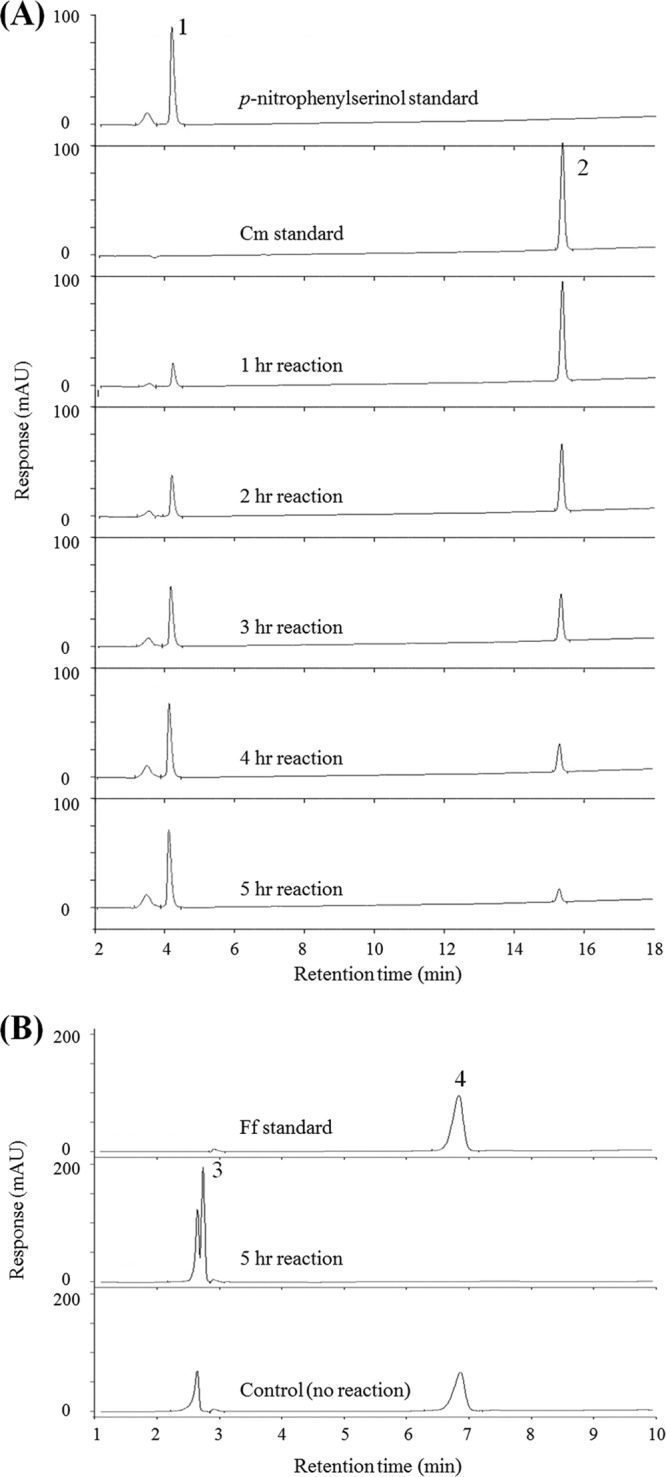
High-performance liquid chromatography (HPLC) analysis of the hydrolysates of chloramphenicol (Cm) and florfenicol (Ff) by purified EstDL136. (A) Detection of Cm hydrolysis. Peaks 1 and 2 indicate p-nitrophenylserinol and Cm, respectively. (B) Hydrolysis of Ff was achieved following incubation with EstDL136. Peaks 3 and 4 represent the Ff hydrolysate and Ff standard, respectively. mAU, milli-absorbance unit.
Heterologous expression of estDL136 conferred Cm and Ff resistance on E. coli.
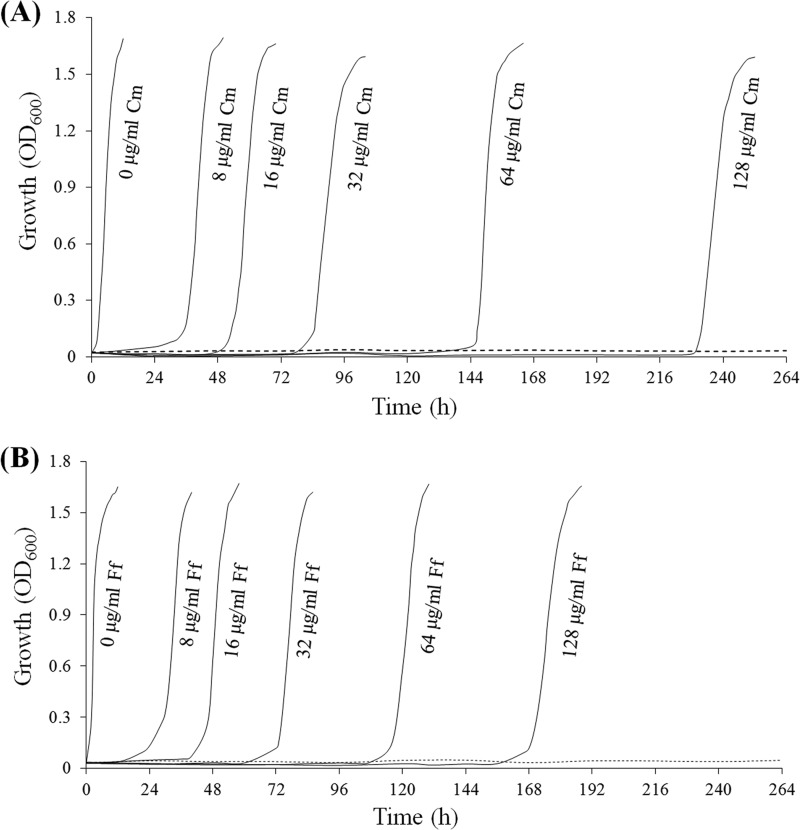
Both in vivo and in vitro, EstDL136 hydrolyzed Cm and Ff and generated functionally inactive derivatives (Fig. 1 and 3), suggesting that EstDL136 can cause amphenicol antibiotics to be ineffective. To investigate this possibility, a susceptibility test was performed with E. coli DH5α carrying pUEst136, which was known to be tolerant of the normally effective concentration (15 μg/ml) of Cm and Ff. When a susceptibility assay was performed with different concentrations of the antibiotics, E. coli displayed an extended lag phase to survive the antibiotic challenge (Fig. 4). Although E. coli DH5α carrying pUC119 was susceptible to Cm (16 μg/ml) and Ff (16 μg/ml) for the longest incubation period in this study (11 days), E. coli carrying pUEst136 recovered following antibiotic exposure (16 to 128 μg/ml) of various durations (Fig. 4). Based on this observation, we can conclude that a high concentration of an antibiotic will initially inhibit bacterial growth. Nevertheless, EstDL136 is apparently produced and hydrolyzes the antibiotic until the concentration is reduced to below the MIC, at which point the bacterial population can continue to grow normally. In addition, the faster recovery of the bacteria from Ff inhibition indicates the higher catalytic efficiency of EstDL136 toward Ff than Cm (Fig. 4).
Fig 4.
Growth of Escherichia coli carrying pUEst136 with different concentrations of chloramphenicol (Cm) or florfenicol (Ff). (A) Bacterial growth during culture with Cm (0 to 128 μg/ml). (B) Bacterial growth during culture with Ff (0 to 128 μg/ml). The dashed line indicates the negative control of E. coli carrying pUC119 cultured with 16 μg/ml of Cm or Ff. OD600, optical density at 600 nm.
Organization of the gene cluster flanking estDL136.
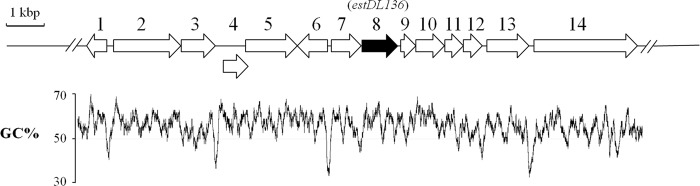
DNA sequence analysis of the 14.6 kb of pDL136 revealed the presence of 14 different putative ORFs, including estDL136 (orf8), based on their DNA sequences and deduced amino acid sequence similarity to known genes in GenBank (Table 1). The G+C content of the DNA sequence of 14,618 bp was 53.4%, although several AT-rich regions (30 to 40% G+C) were apparent in the intergenic area, which might be the promoter regions (Fig. 5). All 14 ORFs showed the highest similarity to corresponding similar proteins from the bacterial phylum Proteobacteria; in particular, the most closely related proteins for 11 of the 14 ORFs were from the bacterial family Sphingomonadaceae (Table 1). Most ORFs located near estDL136 encoded proteins that were similar to a variety of functional enzymes, including amidohydrolase, dehydrogenase/reductase, major facilitator transporter, esterase, and oxygenase. In addition, genes encoding the transcriptional regulator and membrane proteins were also identified (Table 1 and Fig. 5). Of the 14 ORFs, several (orf2, -3, -5, -8, -11, -12, and -14) were identified as having the highest similarity to genes from the genome of Novosphingobium aromaticivorans DSM 12444, which is known to degrade a wide range of xenobiotic and aromatic hydrocarbons (4). Although the homologues were not organized in the same cluster in N. aromaticivorans DSM 12444, they were located in close proximity to each other. Because EstDL136 inactivated Cm and Ff by enzymatic hydrolysis and enabled the cross-resistance of E. coli, we investigated whether the major facilitator transporter homologue (ORF5), which expels drugs from bacteria, may be involved in Cm resistance. Therefore, a subclone carrying a single orf5 gene was constructed and then tested for Cm resistance. However, E. coli carrying the subclone was susceptible to Cm and Ff challenge (data not shown). Using simple methods, we also tested whether E. coli carrying pDL136 can completely metabolize Cm or Ff; however, so far, the results regarding this mechanism are inconclusive (data not shown).
Table 1.
List of open reading frames from a metagenomic clone, pDL136, and closely related proteins in GenBank
| ORF | No. of amino acids | Closely related protein (accession no., organism) | % Identity |
|---|---|---|---|
| 1 | 187 | Hypothetical protein PP1Y_Mpl2761 (YP_004538189, Novosphingobium sp. PP1Y) | 54 |
| 2 | 580 | Amidohydrolase 3 (YP_001166016, Novosphingobium aromaticivorans DSM 12444) | 77 |
| 3 | 294 | Hypothetical protein Saro_3630 (YP_001166015, Novosphingobium aromaticivorans DSM 12444) | 68 |
| 4 | 214 | Short-chain dehydrogenase/reductase (YP_001264122, Sphingomonas wittichii RW1) | 44 |
| 5 | 442 | Major facilitator transporter (YP_001166021, Novosphingobium aromaticivorans DSM 12444) | 51 |
| 6 | 260 | Short-chain dehydrogenase/reductase (ZP_02892805, Burkholderia ambifaria IOP40-10) | 55 |
| 7 | 255 | Short-chain dehydrogenase/reductase (YP_001263771, Sphingomonas wittichii RW1) | 60 |
| 8a | 310 | α/β-Hydrolase domain-containing protein (YP_001166038, Novosphingobium aromaticivorans DSM 12444) | 58 |
| 9 | 129 | Hypothetical protein H16_B0647 (YP_728809, Ralstonia eutropha H16) | 30 |
| 10 | 246 | Short-chain dehydrogenase/reductase (YP_003277770, Comamonas testosteroni CNB-2) | 54 |
| 11 | 159 | AsnC family transcriptional regulator (YP_001166091, Novosphingobium aromaticivorans DSM 12444) | 68 |
| 12 | 163 | AsnC family transcriptional regulator (YP_001165999, Novosphingobium aromaticivorans DSM 12444) | 63 |
| 13 | 359 | Rieske (2Fe-2S) domain-containing protein (YP_001263791, Sphingomonas wittichii RW1) | 58 |
| 14 | 887 | TonB-dependent receptor (YP_495816, Novosphingobium aromaticivorans DSM 12444) | 53 |
orf8 encodes EstDL136, and the DNA sequence determined in pDL136 was partial.
Fig 5.
Genetic organization of ORFs located close to estDL136, and G+C content analysis. Putative ORFs are listed from 1 to 14, and orf8 is estDL136. The G+C content (percentage) of the DNA sequence was calculated using the Seqool program with a 100-bp window; the average G+C content value is estimated to be 53.4%. A DNA scale is also shown (top left).
DISCUSSION
The inactivation of Cm by numerous bacteria has been detected repeatedly (25); however, it is not clear whether the Cm resistance detected in various environments is a direct consequence of the misuse or abuse of this broad-spectrum antibiotic. Although its use in human and veterinary medicine has been reduced substantially, dissemination of the resistance genes has been detected (3, 17), and some of these mediate cross-resistance to the synthetic, fluoro-substituted derivate, Ff (5, 8, 14). In comparison to Cm inactivation, only a few Ff resistance mechanisms have been reported to date. In this study, a new mechanism of Cm and Ff inactivation was detected from a novel esterase, EstDL136, which was encoded by a gene identified from a soil metagenome. EstDL136 has promiscuous amidase activity, hydrolytically inactivating Cm and Ff and resulting in the cross-resistance of E. coli to these antibiotics.
The hydrolytic inactivation of antibiotics has been demonstrated for many antimicrobial agents, among which the β-lactam resistance mediated by β-lactamase is the most well known (6, 21). In addition, the macrolide esterase and a fosfomycin-specific epoxide hydrolase were also shown to mediate the related drug resistance (10, 12, 23). The Cm hydrolase activity has been reported in the Cm-producing actinomycetes; however, its exact mechanism remains unclear (16, 19). This is in contrast to CAT and drug efflux pumps, which are widespread in numerous bacteria and are well characterized (25). EstDL136 catalyzed Cm hydrolysis and generated p-nitrophenylserinol when it was produced in E. coli or was coincubated with Cm in vitro (Fig. 1 and 3). This result supports our prediction that EstDL136 is capable of hydrolyzing the amide linkage in Cm to inactivate its function as an antibiotic, as p-nitrophenylserinol has no antimicrobial activity (9). Regarding Ff hydrolysis, although we did not identify the Ff hydrolysate, HPLC revealed that it is hydrolyzed by EstDL136 and a new compound produced simultaneously (Fig. 3), suggesting that the sulfomethyl and fluoro substitution of Ff do not interfere with its hydrolytic inactivation by EstDL136.
E. coli expressing estDL136 displayed an extended lag phase in bacterial population growth which was directly correlated with the antibiotic concentration (Cm and Ff), and eventually, all cultures grew to similar optical densities after Cm and Ff were gradually inactivated by EstDL136 (Fig. 4), suggesting the cross-resistance of E. coli to these antibiotics. Similar observations have been seen in Flavobacterium sp. strain CB6, which slowly adsorbed and degraded Cm, although the mechanism remains unknown (30). In the latter case, N-dichloroacetyl-p-nitrophenylserine rather than p-nitrophenylserinol was identified as a product of the degradation, indicating that different mechanisms are involved in hydrolysis by Flavobacterium sp. strain CB6. Another example is Streptomyces venezuelae, which was sensitive to exogenous Cm in a Cm-nonproducing medium. However, the production of Cm hydrolase inactivated Cm and resulted in growth lag phases of various durations (16), which is highly similar to our results obtained from E. coli expressing estDL136. Nevertheless, it is unknown whether the S. venezuelae Cm hydrolase is able to hydrolyze other amphenicol antibiotics, such as Ff.
We do not know the ultimate source of estDL136 (32). However, the Ff resistance of E. coli expressing estDL136 may suggest that estDL136 could be important in aquaculture or livestock farming for pigs and cattle, because the broad-spectrum antibiotic Ff is now frequently used to treat bacterial respiratory tract infections in cattle and pigs or in salmon farming (25). So far, we do not have any clear idea whether EstDL136 will be functional in the hydrolysis of Ff in the milieu of pulmonary tissues. Genuine hydrolysis of Ff by the bacterial pathogens expressing estDL136 should be further proven in the milieu of pulmonary tissues. It is not clear whether genes similar to estDL136 will be present in the animal or fish pathogenic bacteria, because the ultimate source of estDL136 is unknown. Although we do not know whether estDL136 homologues are widespread in nonselective environments, the discovery of estDL136, encoding a specific hydrolase for Cm and Ff, could still be a potential threat to livestock farming and aquaculture.
We investigated the nucleotide sequence of pDL136 and found that the genes neighboring estDL136 encoded proteins similar to amidohydrolase, dehydrogenase/reductase, major facilitator transporter, esterase, transcriptional regulator, oxygenase, and membrane proteins (Table 1 and Fig. 5). Of importance, 11 of the 14 ORFs were most closely related to genes of the bacterial family Sphingomonadaceae, which are well known for their biodegrading and biosynthetic capabilities (Table 1). The G+C content of the DNA analyzed was 53.4%, indicating that it probably did not originate from the bacterial phylum Actinomycetes, in which Cm hydrolase activity has been reported, because actinomycetes frequently have a high G+C content of more than 70%. The possibility that pDL136 originates from Cm-nonproducing bacteria may reveal that Cm hydrolase has another function. Considering that EstDL136 hydrolyzed Cm and Ff and that many studies in the literature report hydrolases to be involved in toxin detoxification (13, 20, 34), it is plausible to hypothesize that the genes linked to estDL136 in pDL136 may be involved in the complete mineralization of amphenicol antibiotics. Although our investigation of pDL136 showed no such indication, it must be investigated further, because the CAT that is encoded on fosmid partially protects Cm by acetylation and interferes with the further degradation of Cm by the gene cluster in pDL136. Although we have observed Cm susceptibility of E. coli with orf5 encoding the major facilitator transporter homologue, it is still unclear whether the major facilitator transporter is involved in Cm resistance along with EstDL136. This is because we have not really examined whether E. coli carrying orf5 expressed a functional protein. In addition, Cm has been reported to be a substrate functioning in the nitroreductase pathway in Haemophilus influenzae (28); however, the metabolite of Cm in this pathway differed from the one presented in our study.
In conclusion, we identified a bacterial esterase responsible for the hydrolytic inactivation of Cm and Ff. This is the first report to detail the novel inactivation mechanism of Ff by enzymatic hydrolysis, independent of other drug efflux pumps. Because Ff is not a natural antibiotic but is chemically modified from Cm, the hydrolytic inactivation of Ff by soil bacteria is rather interesting. Our results demonstrate the great potential of soil bacteria to remediate synthetic antibiotics. We speculate that these antibiotic resistance genes will become widespread in nature owing to horizontal gene transfer, which will result in the acceleration of bacterial resistance to amphenicol antibiotics. This is a current concern because Ff is frequently used to treat animal diseases (29). The extended lag phase of E. coli expressing estDL136 during growth with high concentrations of Cm and Ff demonstrates that EstDL136 continuously hydrolyzes the antibiotics to defend the host. This feature provides insight into the bacterial mechanisms that may be used to survive challenge from lethal toxins released by microbial producers or human activities within a natural environment, such as soil.
ACKNOWLEDGMENTS
This research was supported by a grant from the Next-Generation BioGreen 21 Program (grant no. PJ008201), Rural Development Administration, and by a grant (project no. 60900251) from the Screening Center for Disease Resistant Vegetable Crops of TDPAF, funded by MIFAFF, Republic of Korea.
Footnotes
Published ahead of print 29 June 2012
REFERENCES
- 1. Amann RI, Ludwig W, Schleifer KH. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59: 143–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arcangioli MA, Leroy-Seètrin S, Martel JL, Chaslus-Dancla E. 1999. A new chloramphenicol and forfenicol resistance gene flanked by two integron structures in Salmonella typhimurium DT104. FEMS Microbiol. Lett. 174: 327–332 [DOI] [PubMed] [Google Scholar]
- 3. Beaber JW, Hochhut B, Waldor MK. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 184: 4259–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bell SG, Wong LL. 2007. P450 enzymes from the bacterium Novosphingobium aromaticivorans. Biochem. Biophys. Res. Commun. 360: 666–672 [DOI] [PubMed] [Google Scholar]
- 5. Braibant M, Chevalier J, Chaslus-Dancla E, Pages JM, Cloeckaert A. 2005. Structural and functional study of the phenicol-specific efflux pump FloR belonging to the major facilitator superfamily. Antimicrob. Agents Chemother. 49: 2965–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bush K. 2002. The impact of β-lactamases on the development of novel antimicrobial agents. Curr. Opin. Investig. Drugs 3: 1284–1290 [PubMed] [Google Scholar]
- 7. Cannon M, Hartford S, Davies J. 1990. A comparative study on the inhibitory actions of chloramphenicol, thiamphenicol, and some fluorinated derivates. J. Antimicrob. Chemother. 26: 307–317 [DOI] [PubMed] [Google Scholar]
- 8. Cloeckaert A, Baucheron S, Chaslus-Dancla E. 2001. Nonenzymatic chloramphenicol resistance mediated by IncC plasmid R55 is encoded by a floR gene variant. Antimicrob. Agents Chemother. 45: 2381–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cundliffe E, Demain AL. 2010. Avoidance of suicide in antibiotic-producing microbes. J. Ind. Microbiol. Biotechnol. 37: 643–672 [DOI] [PubMed] [Google Scholar]
- 10. Fillgrove KL, Pakhomova S, Newcomer ME, Armstrong RN. 2003. Mechanistic diversity of fosfomycin resistance in pathogenic microorganisms. J. Am. Chem. Soc. 125: 15730–15731 [DOI] [PubMed] [Google Scholar]
- 11. Kehrenberg C, Schwarz S. 2004. fexA, a novel Staphylococcus lentus gene encoding resistance to florfenicol and chloramphenicol. Antimicrob. Agents Chemother. 48: 615–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim YH, Cha CJ, Cerniglia CE. 2002. Purification and characterization of an erythromycin esterase from an erythromycin resistant Pseudomonas sp. FEMS Microbiol. Lett. 210: 239–244 [DOI] [PubMed] [Google Scholar]
- 13. Kneusel RE, Schiltz E, Matern U. 1994. Molecular characterization and cloning of an esterase which inactivates the macrolide toxin brefeldin A. J. Biol. Chem. 269: 3449–3456 [PubMed] [Google Scholar]
- 14. Lang KS, et al. 2010. Novel florfenicol and chloramphenicol resistance gene discovered in Alaskan soil using functional metagenomics. Appl. Environ. Microbiol. 76: 5321–5326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Levin BR, et al. 1998. Resistance to antimicrobial chemotherapy: a prescription for research and action. Am. J. Med. Sci. 315: 87–94 [DOI] [PubMed] [Google Scholar]
- 16. Malik VS, Vining LC. 1971. Metabolism of chloramphenicol by the producing organism. Some properties of chloramphenicol hydrolase. Can. J. Microbiol. 17: 1287–1290 [DOI] [PubMed] [Google Scholar]
- 17. Meunier D, et al. 2002. Salmonella enterica serotype Typhimurium DT104 antibiotic resistance genomic island I in serotype Paratyphi B. Emerg. Infect. Dis. 8: 430–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mosher RH, et al. 1995. Inactivation of chloramphenicol by O-phosphorylation. A novel resistance mechanism in Streptomyces venezuelae ISP5230, a chloramphenicol producer. J. Biol. Chem. 270: 27000–27006 [DOI] [PubMed] [Google Scholar]
- 19. Mosher RH, Ranade NP, Schrempf H, Vining LC. 1990. Chloramphenicol resistance in Streptomyces: cloning and characterization of a chloramphenicol hydrolase gene from Streptomyces venezuelae. J. Gen. Microbiol. 136: 293–301 [DOI] [PubMed] [Google Scholar]
- 20. Nishimura M, Ikeda K, Sugiyama M. 2006. Molecular cloning and characterization of gene encoding novel puromycin-inactivating enzyme from blasticidin S-producing Streptomyces morookaensis. J. Biosci. Bioeng. 101: 63–69 [DOI] [PubMed] [Google Scholar]
- 21. Poole K. 2004. Resistance to β-lactam antibiotics. Cell. Mol. Life Sci. 61: 2200–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rajamohan G, Srinivasan VB, Gebreyes WA. 2010. Molecular and functional characterization of a novel efflux pump, AmvA, mediating antimicrobial and disinfectant resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 65: 1919–1925 [DOI] [PubMed] [Google Scholar]
- 23. Rigsby RE, Fillgrove KL, Beihoffer LA, Armstrong RN. 2005. Fosfomycin resistance proteins: a nexus of glutathione transferases and epoxide hydrolases in a metalloenzyme superfamily. Methods Enzymol. 401: 367–379 [DOI] [PubMed] [Google Scholar]
- 24. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 25. Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A. 2004. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 28: 519–542 [DOI] [PubMed] [Google Scholar]
- 26. Schwarz S, Werckenthin C, Kehrenberg C. 2000. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemother. 44: 2530–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shaw WV. 1983. Chloramphenicol acetyltransferase, enzymology and molecular biology. Crit. Rev. Biochem. 14: 1–46 [DOI] [PubMed] [Google Scholar]
- 28. Smith AL, et al. 2007. Chloramphenicol is a substrate for a novel nitroreductase pathway in Haemophilus influenzae. Antimicrob. Agents Chemother. 51: 2820–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Soback S, Paape MJ, Filep R, Varma KJ. 1995. Florfenicol pharmacokinetics in lactating cows after intravenous, intramuscular and intramammary administration. J. Vet. Pharmacol. Ther. 18: 413–417 [DOI] [PubMed] [Google Scholar]
- 30. Sussmuth R, Haag R, Lingens F. 1979. Chloramphenicol resistance of three different Flavobacteria. J. Antibiot. (Tokyo) 32: 1293–1302 [DOI] [PubMed] [Google Scholar]
- 31. Syriopoulou VP, Harding AL, Goldmann DA, Smith AL. 1981. In vitro antibacterial activity of fluorinated analogs of chloramphenicol and thiamphenicol. Antimicrob. Agents Chemother. 19: 294–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tao W, et al. 2011. Characterization of two metagenome-derived esterases that reactivate chloramphenicol by counteracting chloramphenicol acetyltransferase. J. Microbiol. Biotechnol. 21: 1203–1210 [DOI] [PubMed] [Google Scholar]
- 33. White DG, et al. 2000. Characterization of chloramphenicol and florfenicol resistance in Escherichia coli associated with bovine diarrhea. J. Clin. Microbiol. 38: 4593–4598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang LH, Birch RG. 1997. The gene for albicidin detoxification from Pantoea dispersa encodes an esterase and attenuates pathogenicity of Xanthomonas albilineans to sugarcane. Proc. Natl. Acad. Sci. U. S. A. 94: 9984–9989 [DOI] [PMC free article] [PubMed] [Google Scholar]