Abstract
The distribution of viruses inhabiting the coral mucus remains undetermined, as there is no suitable standardized procedure for their separation from this organic matrix, principally owing to its viscosity and autofluorescence. Seven protocols were tested, and the most efficient separations were obtained from a chemical treatment requiring potassium citrate.
TEXT
Viruses have been probably the least studied among the biological entities that live in the mucus and form the coral holobiont (dinoflagellates, fungi, bacteria, archaea, cyanobacteria, etc.) (1, 4, 10, 11), until now. Speculation on their presence in coral lacks a scientific basis, and only a few scattered metagenomic studies have provided any new information (6, 12). It is now known that, although viruses can infect any of the microorganisms in the coral holobiont, they are mostly comprised of bacteriophages (6). Moreover, there is no quantitative estimate of their density (per unit of mass of mucus or per unit area) in the literature, and their prevalence in the surface mucus layer is based only on qualitative observations (2, 9). A standard reliable method is thus required for estimating their abundance levels. In this study, seven previously described protocols were tested, and their suitabilities for counting viruses reliably from the coral mucus were compared.
The mucus was collected from six scleractinian species in the Cap d'Agde aquarium (France)—Fungia sp., Turbinaria sp., Alveopora sp., Favia sp., Lobophyllia sp., and Platygyra sp.—and the soft coral Heteroxenia sp. Briefly, corals were taken out of the water and exposed to air for 1 to 3 min (7). This stress caused the mucus to be secreted, forming long gel-like threads dripping from the coral surface that were directly collected in cryotubes, fixed with formaldehyde (final concentration of 3%), and stored at −80°C until staining. Mucus samples were processed in triplicates as follows.
Chemical methods. (i) Potassium citrate method.
For the potassium citrate method (14), a total of 100 μl of fixed mucus was eluted into 900 μl of 0.02-μm-pore-size-filtered, pH 7 solution of 1% citrate potassium with 10 g potassium citrate, 1.44 g Na2HPO4 · 7H2O, and 0.24 g KH2PO4 per liter. All tubes were then vortexed at a moderate speed before particles were stained and enumerated (see below).
(ii) Methanol method.
For the methanol method (5), a total 160 μl of fixed mucus was incubated with a solution of 0.02-μm-pore-size-filtered methanol (20% final concentration) at 35°C for 15 min and sonicated for 30 s (amplitude of 20).
(iii) Sodium pyrophosphate method.
For the sodium pyrophosphate method (1a), a total of 100 μl of fixed mucus was inoculated with tetrasodium pyrophosphate (10 mM, final concentration), vortexed for 1 min, and incubated at ambient temperature for 15 min. The sample was then diluted (1:3) in Milli-Q filtered water and sonicated for 30 s.
(iv) Polysorbate 80 method.
For the polysorbate 80 method (15), a total of 100 μl of fixed mucus was diluted in 900 μl of 0.02-μm-pore-size-filtered Milli-Q water and incubated with 20% Tween 80 (polysorbate 80) at 35°C for 15 min and sonicated for 30 s.
Mechanical method.
The mechanical method was a homogenization method (6) based on a 1:10 dilution of the mucus after fixing (100 μl) in 0.02-μm-pore-size-filtered Milli-Q water, followed by 1 min of power homogenization (Polytron; PT 10 35 GT) at 5,000 rpm. A combination of this protocol and that based on the potassium citrate protocol was tested by diluting 100 μl of the mucus in 900 μl of 1% potassium citrate solution prior to homogenization.
Enzymatic method.
The trypsin method (3) involved 100 μl of fixed mucus eluted into a solution of EDTA (10 mM, final concentration), vortexed for 1 min, and sonicated for 3 min, followed by 1% trypsin incubation for 30 min at 37°C.
Staining procedure.
Viral abundances were determined in each mucus sample (treated and untreated) by using the standard technique with nucleic acid dye SYBR Gold and epifluorescence microscopy (8).
Statistical analyses.
The treatment effectiveness (TE) was calculated based on the difference in viral abundances between the treated (T) and untreated control (C) mucus samples: TE (%) = [(T − C)/C] × 100. The significance of treatments and coral species on viral abundance was tested using multifactorial analysis of variance (ANOVA) and Tukey-Kramer mean post hoc comparison tests (alpha level = 0.05).
The reliability of each protocol was simply inferred from the levels of abundance detected for viruses, which are the net result of parameters such as brightness, contrast, background fluorescence, presence of remaining aggregates, staining quality, even distribution of particles on membrane, etc. Thus, the protocol resulting in the highest values was considered the most effective for separation and enumeration of viruses.
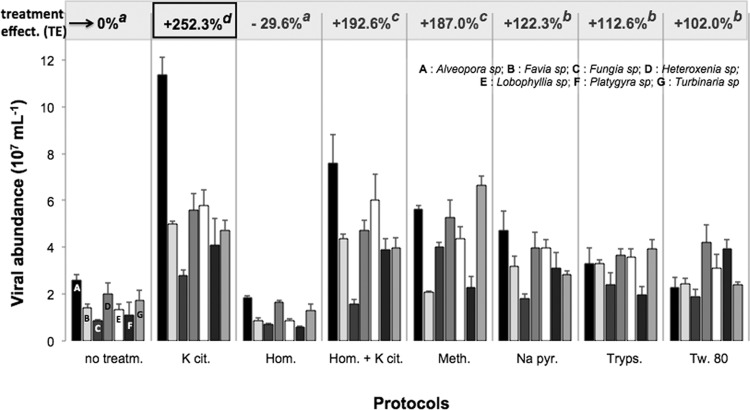
In this study, the 1% potassium citrate solution was the best overall eluent for the mucus samples, regardless of the coral species. Direct counts of viruses following their extraction with this compound ranged from 2.8 × 107 VLP ml−1 of mucus for Fungia sp. to 11.4 × 107 VLP ml−1 for Alveopora sp. (Fig. 1). The treatment effectiveness reached up to 252.3% and was significantly higher than that of the other chemical, mechanical, or enzymatic treatments (Tukey-Kramer, P < 0.05). Potassium citrate has been successfully used for more than 10 years to desorb viral particles from porous substrates, such as sediments and soils (13, 14). By raising the pH of a solution, it increases the electrostatic repulsion between the viruses and the substrate to which they are attached. Furthermore, unlike the other chemical eluents tested in this study, potassium citrate also greatly reduced the autofluorescence of the mucus, which made counting by epifluorescence much more reliable (Fig. 2). Last, the method is very fast, as the effect of the reagent is immediate and does not require any additional incubation. It is also inexpensive, as only a few milligrams are required of a stock at ca. 60 euros per 500 g of potassium citrate (VWR).
Fig 1.
Abundance of viruses as measured after their extraction from coral mucus using the various protocols, shown on the x axis. Error bars represent the standard deviations calculated with three values. The treatment effectiveness (TE), expressed as a percentage, represents the ratio between abundances measured for treated and untreated samples. An index (a, b, c, d) is assigned to each treatment protocol; two TEs with the same index are not significantly different (multifactorial analysis of variance, and Tukey-Kramer post hoc comparison tests, P < 0.05). Treatm., treatment; K cit., potassium citrate; Hom., homogenization; Meth., methanol; Na pyr., sodium pyrophosphate; Tryps., trypsin; Tw. 80, Tween 80.
Fig 2.
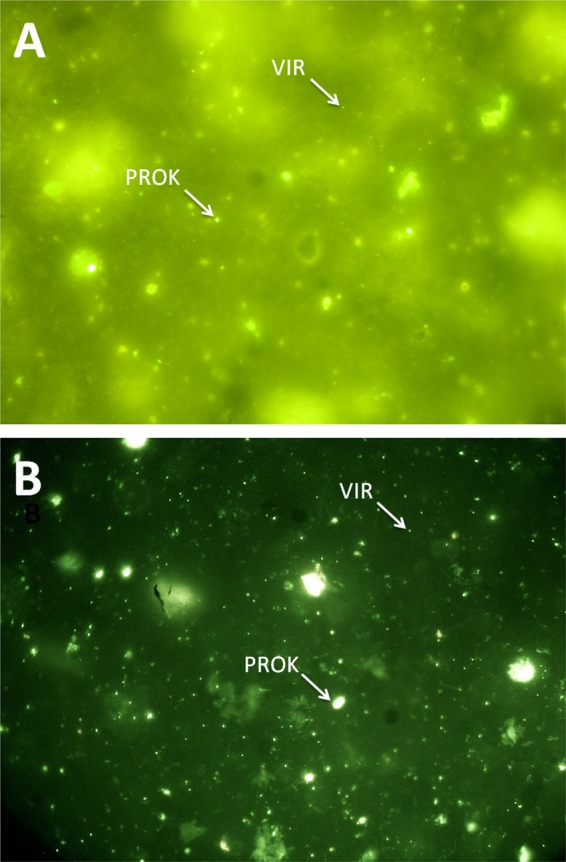
Epifluorescence microscopy image (×100 magnification) of mucus samples stained with SYBR Gold without any preliminary treatment (A) and after being treated with 1% potassium citrate (B). The small dots represent virus-like particles (VIR), the larger dots are prokaryotes (PROK).
Unlike its remarkable efficiency for extracting prokaryotes from mucus (3), trypsin did not give the highest viral counts, possibly because this enzyme might also cause substantial damage to the viral capsid proteins. The homogenization method, previously used for DNA extraction in coral mucus, also resulted in very poor quality observations. This mechanical treatment is a powerful mucus liquefier, but the resulting spreading over the entire membrane surface has seemingly increased the background fluorescence of mucus, thus reducing the contrast and brightness of the stained viruses.
Our results were obtained from cultured corals rather than from in situ scleractinian colonies and may not be applicable to natural habitats. Nonetheless, this study shows that the citrate potassium method represents an interesting tool to investigate the dynamic processes of colonization and the spatial and temporal variability of the viral community abundances within the coral holobiont. It also might help to cope with several methodological barriers that prevent single-cell studies of these biological entities, including flow cytometry and electron microscopy.
ACKNOWLEDGMENTS
The research reported here was financially supported by the French EC2CO project CORINE and the Total Foundation.
Footnotes
Published ahead of print on 22 June 2012.
REFERENCES
- 1. Bettarel Y, Thuy NT, Huy TQ, Hoang PK, Bouvier T. 2012. Observation of virus-like particles in thin sections of the bleaching scleractinian coral Acropora cytherea. J. Mar. Biol. Assoc. UK [Epub ahead of print.] doi:10.1017/S0025315411002062 [Google Scholar]
- 1a. Danovaro R, Middelboe M. 2010. Separation of free-virus particles from sediments in aquatic systems, p 74–81 in Wilhelm SW, Weinbauer MG, Suttle CA. (ed), Manual of aquatic viral ecology (MAVE). The American Society of Limnololgy and Oceanography, Waco, TX [Google Scholar]
- 2. Davy JE, Patten NL. 2007. Morphological diversity of virus-like particles within the surface microlayer of scleractinian corals. Aquat. Microb. Ecol. 47: 37–44 [Google Scholar]
- 3. Garren M, Azam F. 2010. New method for counting bacteria associated with coral mucus. Appl. Environ. Microbiol. 76: 6128–6133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garren M, Azam F. 2012. New directions in coral reef microbial ecology. Environ. Microbiol. 14: 833–844 [DOI] [PubMed] [Google Scholar]
- 5. Lunau M, Lemke A, Martens-Habbena WW, Simon M. 2005. An improved method for counting bacteria from sediments and turbid environments by epifluorescence microscopy. Environ. Microbiol. 7: 961–968 [DOI] [PubMed] [Google Scholar]
- 6. Marhaver KL, Edwards RA, Rohwer F. 2008. Viral communities associated with healthy and bleaching corals. Environ. Microbiol. 10: 2277–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Naumann MS, Richter C, el-Zibdah M, Wild C. 2009. Coral mucus as en efficient trap for picoplanktonic cyanobacteria: implication for pelagic-benthic coupling in the reef ecosystem. Mar. Eco. Prog. Ser. 385: 65–76 [Google Scholar]
- 8. Patel A, et al. 2007. Virus and prokaryote enumeration from planktonic aquatic environments by epifluorescence microscopy with SYBR Green I. Nat. Protoc. 2: 269–276 [DOI] [PubMed] [Google Scholar]
- 9. Patten NL, Harrison ML, Mitchell JG. 2008. Prevalence of virus-like particles within a staghorn scleractinian coral (Acropora muricata) from the Great Barrier Reef. Coral Reefs 27: 569–580 [Google Scholar]
- 10. Rosenberg E, Kushmaro A, Kramarsky-Winter E, Banin E, Loya Y. 2009. The role of microorganisms in coral bleaching. ISME J. 3: 139–146 [DOI] [PubMed] [Google Scholar]
- 11. Van Oppen MJH, Leong JO, Gates DR. 2009. Coral-virus interactions: a double-edged sword? Symbiosis 47: 1–8 [Google Scholar]
- 12. Vega Thurber RL, et al. 2008. Metagenomic analysis indicates that stressors induce production of herpes-like viruses in the coral Porites compressa. Proc. Natl. Acad. Sci. U. S. A. 105: 18413–18418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Williamson KE, Wommack KE, Radosevich M. 2003. Sampling natural viral communities from soil for culture-independent analyses. Appl. Environ. Microbiol. 69: 6628–6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Williamson KE, Wommack KE, Radosevich M. 2005. Abundance and diversity of viruses in six Delaware soils. Appl. Environ. Microbiol. 71: 3119–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yoon WB, Rosson RA. 1990. Improved method of enumeration of attached bacteria for study of fluctuation in the abundance of attached and free-living bacteria in response to diel variation in seawater turbidity. Appl. Environ. Microbiol. 56: 595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]