Abstract
Among the adaptive responses of bacteria to rapid changes in environmental conditions, those of the cell envelope are known to be the most crucial. Therefore, several mechanisms with which bacteria change their cell surface and membranes in the presence of different environmental stresses have been elucidated. Among these mechanisms, the release of outer membrane vesicles (MV) in Gram-negative bacteria has attracted particular research interest because of its involvement in pathogenic processes, such as that of Pseudomonas aeruginosa biofilm formation in cystic fibrosis lungs. In this study, we investigated the role of MV formation as an adaptive response of Pseudomonas putida DOT-T1E to several environmental stress factors and correlated it to the formation of biofilms. In the presence of toxic concentrations of long-chain alcohols, under osmotic stress caused by NaCl, in the presence of EDTA, and after heat shock, cells of this strain released MV within 10 min in the presence of a stressor. The MV formed showed similar size and charge properties, as well as comparable compositions of proteins and fatty acids. MV release caused a significant increase in cell surface hydrophobicity, and an enhanced tendency to form biofilms was demonstrated in this study. Therefore, the release of MV as a stress response could be put in a physiological context.
INTRODUCTION
The adaptation of bacteria to a rapid change of environmental conditions is a basic survival strategy. The bacterial membrane especially, as a complex interface with the environment, is very sensitive to stress. Therefore, several mechanisms have evolved to stabilize membrane viscosity, e.g., changes in the membrane fatty acid content (14, 17, 19). Due to the fact that Gram-negative bacteria have an outer and an inner membrane with different compositions and important biological functions, both membranes have to remain functional even under changing conditions. As of now, mechanisms affecting the inner membrane have been well studied (34, 35, 42), but the outer membrane and cell surface have received little consideration (5, 11, 31).
The best-investigated mechanism of Gram-negative bacteria, which is related to a change of the outer membrane, is the formation of outer membrane vesicles (MV) (24). MV were often associated with the human pathogen Pseudomonas aeruginosa that releases virulence factors with MV to attack the human lung epithelium (20). Since its discovery, this mechanism has been found in many Gram-negative bacteria, and it has become clear that the process of MV formation is highly conserved.
MV are connected to interspecies communication (30), the delivery of proteins, toxins, and DNA (30). Furthermore, the formation of toluene-containing MV was discussed as an adaptation mechanism to transport toxic compounds away from the cells (22). In addition, an important function of MV release is their involvement in biofilm formation (3). Bacteria growing in biofilms are known to have considerable advantages over the planktonic lifestyle. Furthermore, bacteria living in biofilms or microcolonies are significantly more tolerant of antibiotics, biocides, and other forms of environmental stress (3, 5, 15). The close cell-cell contact also facilitates horizontal gene transfer and sharing of metabolic by-products within the biofilm community (3, 6, 25).
Although tremendous work has been carried out to elucidate the cellular mechanisms leading to the release of MV, there is still not much known about the regulation of this process. Previous research on P. aeruginosa has shown that the conversion of 2-heptyl-4-quinolone (HHQ) into 2-heptyl-3-hydroxy-4-quinolone, the so-called Pseudomonas quinoline signal (PQS), by the monooxygenase PqsH increases the release of MV (7, 37). However, neither the complete physiological role of MV nor the regulation of their formation has yet been described.
In this study, we investigated the role and physiological function of MV formation in P. putida in the presence of rapidly changing environmental stress conditions. The released MV were analyzed with regard to size, surface charge, protein content, and fatty acid composition. In addition, we demonstrated that cells which released MV become more hydrophobic, thus enhancing their ability to form biofilms as a protective mechanism against stress.
MATERIALS AND METHODS
Chemicals and culture conditions.
All chemicals were reagent grade and obtained from commercial sources. P. putida DOT-T1E (CECT 5312) (33) was cultivated in a mineral medium as described by Hartmans et al. (12), with 4 g/liter disodium succinate as both carbon and energy source. Cells were grown to exponential growth phase in 50-ml shake cultures at 30°C in a horizontally shaking water bath at 180 rpm. Then, either the stressor was added (1-octanol, 1.25 mM; NaCl, 2 M; or EDTA, 10 mM final concentration) or the temperature was increased. After 1 h of incubation, the cells were harvested by centrifugation for 10 min at 6,000 × g. The supernatant was used to isolate membrane vesicles as described below. The pelleted cells were washed and finally resuspended in 10 mM KNO3 solution for further analysis.
Isolation of MV.
Isolation of MV was done as previously described (21). The supernatant was filtered through 0.45-μm-pore-size membranes (Sartorius AG, Goettingen, Germany). Subsequently, MV were harvested by centrifugation at 100,000 × g for 3 h at 4°C (L-90K ultracentrifuge; Beckman Coulter). Pelleted vesicles were resuspended in either 50 mM HEPES buffer or 10 mM KNO3 for Zetasizer analysis.
Cryo-TEM.
For cryo-transmission electron microscopy (cryo-TEM), 1 droplet of the isolated MV was applied to a copper grid covered by holey poly-l-lysine-coated carbon film (Quantifoil R3.5/1; Micro Tools GmbH, Jena, Germany), and excess liquid was blotted automatically for 1 s between two strips of filter paper. Subsequently, the samples were rapidly plunged into liquid ethane (cooled to −180°C) in a cryobox (Carl Zeiss NTS GmbH, Oberkochen, Germany). Excess ethane was removed with a piece of filter paper. The samples were transferred with a cryo-transfer unit (Gatan 626-DH) into the precooled cryo-electron microscope (Philips CM 120) operated at 120 kV and viewed under low-dose conditions. The images were recorded with a 1-k charge-coupled-device (CCD) camera (FastScan F114; TVIPS, Gauting, Germany).
Characterization of the isolated MV.
The electrophoretic mobility (μ) and the average size of MV suspensions in 10 mM KNO3 at pH 6.2 were determined by dynamic light scattering using a Zetasizer (Zetasizer Nano ZS; Malvern Instruments Ltd., Malvern, Worcestershire, United Kingdom) at 150 mV. The zeta potential (ζ), as an indirect measure of surface charge of the MV, was approximated from the electrophoretic mobility according to the method of Helmholtz and Smoluchowski (18).
To characterize the fatty acid profile, extraction and methylation of the fatty acids was done as described before (16). The gas chromatography was executed under the following conditions using a gas chromatograph (6890N; Agilent Technologies, Böblingen, Germany): the GC CP-Sil 88 column (Agilent Technologies) was held at 40°C for 2 min, heated at 8°C per min up to 220°C, and held at that temperature for 5 min. The carrier gas used was He at a flow rate of 2 ml min−1. Fatty acid methyl esters were identified by comparing their retention times to standards.
In order to discover the proteins contained in the isolated MV, equal amounts of supernatant were analyzed by SDS-PAGE. After Coomassie brilliant blue staining, MV protein bands were excised and inserted into microcentrifuge tubes. Then, the bands were destained and digested proteolytically overnight at 37°C using trypsin. The identification of the purified peptides was carried out as reported by Bastida et al. (1) using a UPLC-linear trap quadrupole (LTQ) Orbitrap tandem mass spectrometer (MS-MS), MASCOT (Matrix Science) as the search engine, and bacterial sequences in the NCBI nr (nonredundant) peptide sequence database as the criteria for taxonomy.
Characterization of bacterial cell surface properties.
The physicochemical cell surface properties of bacteria were investigated using standard methods as described by others (46). The water contact angle was used to determine the cell surface hydrophobicity, and the zeta potential to characterize the surface charge of MV.
Biofilm formation assay.
The quantification of biofilm formation was performed according to O'Toole and Kolter (32). Samples of 1 ml of the stressed cells were adjusted to an optical density at 560 nm (OD560) of 0.05 and incubated on polystyrene microtiter plates (Primaria multiwell; VWR International GmbH, Darmstadt, Germany) at 28°C for 18 h. Then, 250 μl of a 0.1% solution of crystal violet (CV) was added and the plates were incubated for additional 30 min at room temperature. Subsequently, the supernatant was removed and each well was washed three times with distilled water. After the plates were dried, 1 ml of ethanol was added twice to remove the bound CV and the absorption was determined at 590 nm.
RESULTS
Characterization of the released MV.
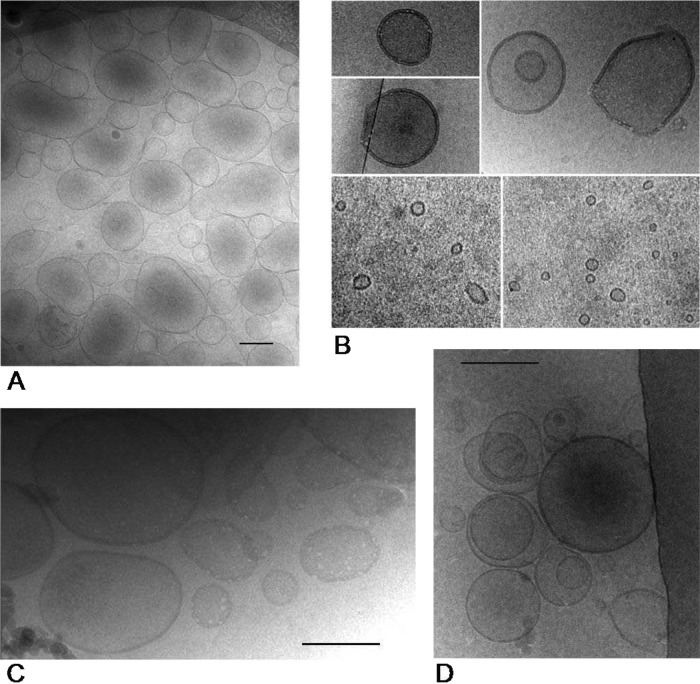
In order to investigate the influence of different stressors on the formation of MV, cells of P. putida were pretreated with 1-octanol, heat shock, NaCl, or EDTA. Afterwards, the vesicles formed were isolated and analyzed. By cryo-TEM and dynamic light scattering, we could show that different conditions resulted in different MV structures. In contrast to that, no or very few vesicles were detected in the supernatant of nonstressed cells. Figure 1 shows cryo-TEM images of MV isolated from the supernatant. While the vesicles isolated after treatment with 1-octanol or heat shock were similar in size, vesicles isolated after incubation with sodium chloride or EDTA appeared to be smaller (Fig. 1). Due to the limitations of cryo-TEM preparation and to get a partially oriented separation of the MV sizes, the MV were analyzed by dynamic light scattering using a Zetasizer in order to obtain the charge and a better size distribution. Regarding the size, one peak in the range of 110 nm up to 200 nm was observed for all conditions. In addition, after the treatment with EDTA and sodium chloride, another peak appeared in the range of 20 nm to 50 nm. In the case of the zeta potential, sodium chloride-induced MV had a considerably higher value than MV exposed to the other conditions (Table 1). Thus, most probably, salt ions were attached to the surface of the vesicles, causing this effect.
Fig 1.
Representative cryo-transmission electron microscopy images of MV formed by P. putida DOT-T1E after incubation with 1-octanol (A), NaCl (B), heat shock (C), and EDTA (D). Bars, 200 nm.
Table 1.
Zeta potentials and sizes of MV induced by different stressesa
| Stressor | Zeta potential (mV) | Size (nm) of peak: |
|
|---|---|---|---|
| 1 | 2 | ||
| 1-Octanol | −31.3 ± 3.7 | 172.2 ± 17.1 | |
| Heat shock | −27.1 ± 6.4 | 154.6 ± 5.8 | |
| NaCl | −13.4 ± 3.6 | 199.1 ± 45.4 | 45.7 ± 10.7 |
| EDTA | −28.3 ± 9 | 116 ± 13.3 | 27.3 ± 11.9 |
Values are means standard ± deviations.
Due to the obvious differences of the released MV, their fatty acid and protein compositions were analyzed. The results of the fatty acid methyl ester (FAME) analysis showed an approximately 5-fold enrichment of stearic acid (18:0) compared to the amount in the cells (Table 2). This enrichment of stearic acid was previously reported for outer MV (44) and is herewith confirmed. All in all, the degree of saturation of the MV was also increased and was even higher for sodium chloride- and EDTA-induced MV. The lesser degree of saturation for MV released after 1-octanol treatment or heat shock can be partially explained by cell lysis releasing membrane lipids into the medium.
Table 2.
Fatty acid composition of MV and stressed cells
| Fatty acid | % FAME present after indicated treatment |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| MV |
Cells |
||||||||
| 1-Octanol | 55°C | NaCl | EDTA | Control | 1-Octanol | 55°C | NaCl | EDTA | |
| 16:0 | 35.8 | 36.8 | 39.7 | 40.1 | 31.4 | 35.2 | 38.1 | 32.2 | 29.4 |
| 16:1 Δ9trans | 4.8 | 0.3 | 0.0 | 0.0 | 1.2 | 8.4 | 2.3 | 5.6 | 0.0 |
| 16:1 Δ9cis | 12.5 | 17.4 | 10.7 | 14.3 | 20.5 | 13.4 | 18.6 | 17.1 | 20.9 |
| 18:0 | 9.1 | 7.0 | 16.7 | 11.4 | 1.1 | 2.0 | 2.1 | 1.4 | 1.2 |
| 18:1 Δ11trans | 2.5 | 0.1 | 0.0 | 0.0 | 0.2 | 3.8 | 0.3 | 2.9 | 0.0 |
| 18:1 Δ11cis | 32.2 | 38.4 | 32.9 | 34.2 | 45.6 | 36.7 | 39.0 | 40.8 | 47.8 |
| DoSa | 0.8 | 0.8 | 1.3 | 1.1 | 0.6 | 0.6 | 0.7 | 0.5 | 0.4 |
DoS, degree of saturation.
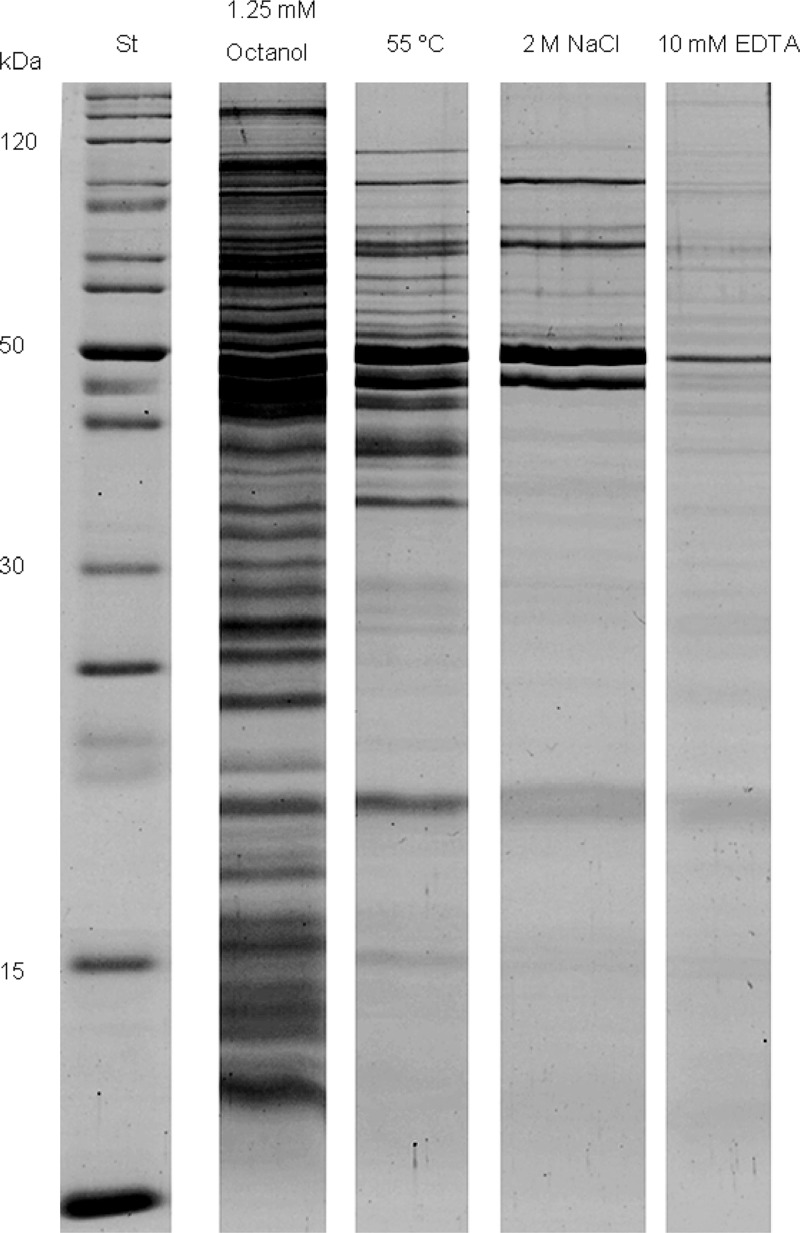
MV were also analyzed for their protein content. SDS-PAGE (Fig. 2) showed the protein profiles of the vesicles induced under different conditions. One consideration was that treatment with 1-octanol causes enhanced membrane permeability, which could lead to an easier release of cytoplasmic proteins. Due to the high sensitivity of mass spectrometry, the resulting data had to be analyzed very carefully. Therefore, only proteins appearing under more than one condition are listed in Table 3. It is noticeable that mostly outer membrane proteins were detected after every treatment.
Fig 2.
Comparative overview of SDS-PAGE results of the isolated MV. Lane 1, standard; lane 2, 1-octanol-induced MV; lane 3, heat shock-induced MV; lane 4, NaCl-induced MV; lane 5, EDTA-induced MV.
Table 3.
Proteins identified in MV released from P. putida DOT-T1E after different treatments
| Protein (source of homologous protein) | NCBI sequence no. | Molecular mass (kDa) | Probability scores after indicated treatmenta |
|---|---|---|---|
| TonB-dependent receptor (Pseudomonas stutzeri A1501) | gi148548751 | 88.8 | NaCl (206.3), 1-oct (55.7), HS (106.6), EDTA (183.1) |
| Ferripyoverdine receptor (Pseudomonas aeruginosa ) | gi170282622 | 85.1 | NaCl (243), 1-oct (179.8), HS (107.8), EDTA (72.6) |
| Elongation factor G (Pseudomonas mendocina ymp) | gi146308926 | 78.8 | NaCl (89.4), 1-oct (632.8), HS (391.1) |
| Outer membrane protein Opr86 (Pseudomonas aeruginosa PAb1) | gi296387845 | 72.9 | NaCl (158.9), 1-oct (91), HS (67.6), EDTA (134) |
| Putative lipoprotein (Pseudomonas fluorescens Pf-5) | gi152987388 | 69.6 | NaCl (60.9), 1-oct (66.3), HS (74.7), EDTA (111.5) |
| Chain A, quinoprotein ethanol dehydrogenase (Pseudomonas aeruginosa ) | gi10120672 | 64,0 | NaCl (152.8), 1-oct (768.1), HS (461.4), EDTA (375.2) |
| CopA family copper resistance protein (Pseudomonas aeruginosa 152504) | gi313107602 | 61.8 | HS (62.3), EDTA (214.3) |
| Chaperonin GroEL (Pseudomonas mendocina NK-01) | gi152986611 | 57.0 | NaCl (165.9), 1-oct (1168), HS (127.9), EDTA (265.4) |
| OprD family porin (Pseudomonas entomophila L48) | gi104781455 | 49.4 | NaCl (109.8), 1-oct (136.3), HS (131.3), EDTA (99.7) |
| Putative outer membrane protein precursor (Pseudomonas aeruginosa UCBPP-PA14) | gi116049240 | 45.5 | NaCl (136.7), HS (77.4), EDTA (97.7) |
| OprE3 (Pseudomonas aeruginosa ) | gi107102299 | 44.8 | NaCl (171.3), 1-oct (72.7), EDTA (135.5) |
| Elongation factor Tu (Pseudomonas putida KT2440) | gi26987181 | 43.5 | 1-oct (239.1), HS (161.4), EDTA (136.7) |
| Bmp family protein (Pseudomonas syringae pv. actinidiae strain M302091) | gi32455881 | 38.7 | HS (103.6), EDTA (79.3) |
| Extracellular solute-binding protein (Pseudomonas mendocina ymp) | gi146308575 | 36.7 | NaCl (141.6), EDTA (136.7) |
| OprF (Pseudomonas sp. MFY72) | gi37704628 | 34.5 | NaCl (113.1), 1-oct (122.7), HS (161.3), EDTA (133.5) |
| Periplasmic binding protein, putative (Pseudomonas putida KT2440) | gi26990659 | 33.2 | 1-oct (94), EDTA (75.6) |
| Small protease (Pseudomonas aeruginosa ) | gi54072636 | 20.8 | HS (105.9), EDTA (95) |
| Outer membrane protein OmpH (Pseudomonas fluorescens WH6) | gi152985326 | 19.1 | HS (91.6), EDTA (107.4) |
| Peptidoglycan-associated lipoprotein (Pseudomonas fluorescens Pf-5) | gi70732082 | 17.7 | NaCl (168.5), 1-oct (117.1), HS (123.8), EDTA (150.6) |
| Lipopolysaccharide transport periplasmic protein LptA (Pseudomonas fulva 12-X) | gi107099955 | 17.0 | NaCl (59.7), 1-oct (62.2), HS (149.3) |
| Riboflavin synthase subunit beta (Pseudomonas mendocina ymp) | gi146308868 | 16.5 | NaCl (76.5), 1-oct (84.2), HS (89.6), EDTA (79.2) |
| OprL (Pseudomonas fluorescens) | gi259090537 | 16,5 | NaCl (78.7), 1-oct (131.7) |
| 50S ribosomal protein L22 (Pseudomonas aeruginosa PAO1) | gi15599454 | 11.9 | NaCl (93.7), 1-oct (205), HS (121), EDTA (240.1) |
NaCl, 2 M; HS, heat shock (55°C); 1-oct, 1.25 mM 1-octanol; EDTA, 10 mM.
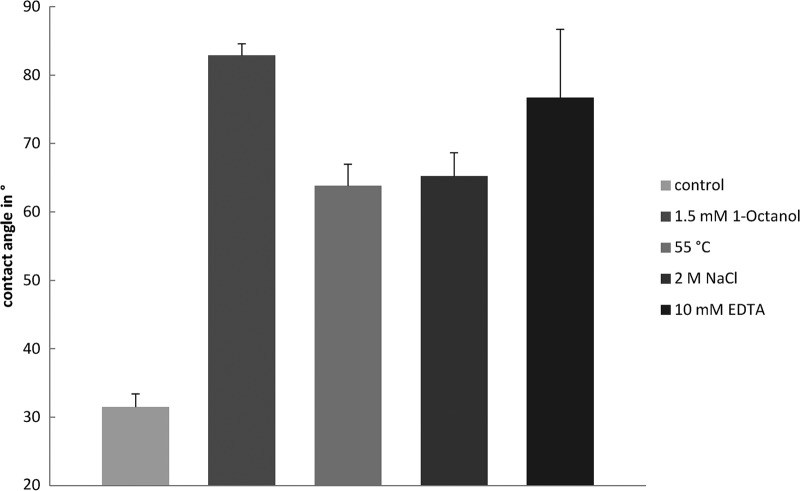
Rapid changes in cell surface properties in response to different stressors.
Since the release of MV is known to be related to a change in cell surface properties of P. putida (2), different conditions were tested for a similar response (Fig. 3). Interestingly, treatment of the cells with EDTA or high concentrations of sodium chloride caused results analogous to those induced by the addition of 1-octanol or heat shock (55°C). For all conditions, the kinetics showed the same progress, indicating that the process was almost finished after the cells had been incubated for 10 min in the presence of a stressor (see Fig. S1 in the supplemental material). The same reaction was observed when the cells were pretreated with 1 mM chloramphenicol, demonstrating that this response is independent of de novo protein biosynthesis. In contrast, neither 2,4-dichlorophenol nor 2,4-dinitrophenol caused any notable effect (data not shown).
Fig 3.
Effects of 1-octanol (A), heat shock (B), NaCl (C), and EDTA (D) on water contact angles of growing cells of P. putida DOT-T1E. Error bars show standard deviations.
Biofilm formation of stressed cells.
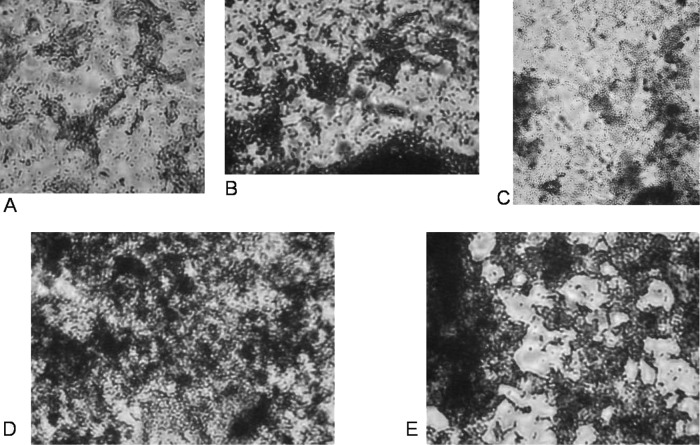
In order to investigate possible advantages of the release of MV, the ability of pretreated cells to form biofilms was investigated. It was observed that the growing cells did not cover the polystyrene surface homogenously but built more complex structures (Fig. 4). This is in agreement with previous studies (25).
Fig 4.
Microscopic images of biofilms formed by P. putida on polystyrene microtiter wells after incubation with HgCl2 (A), 1-octanol (B), heat shock (C), NaCl (D), and EDTA (E).
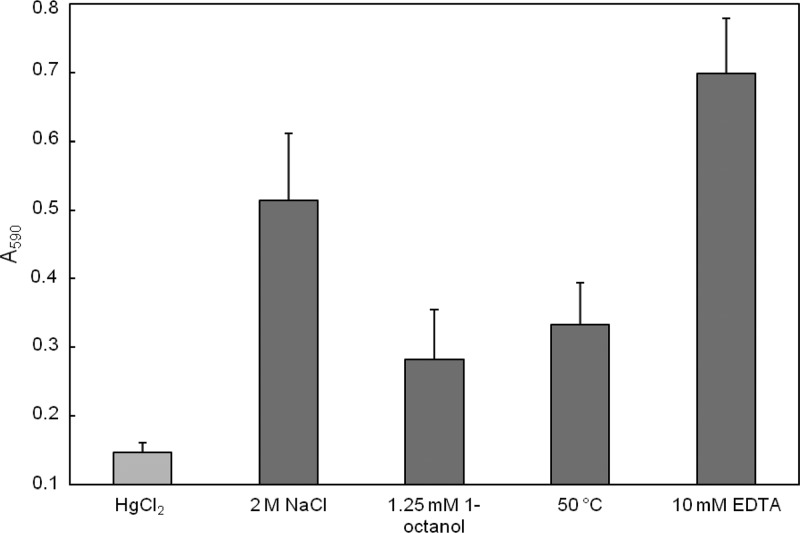
The biofilm formation was also quantified. Because every treatment caused the release of MV, as well as an inhibition of growth rates, HgCl2-treated cells were used as a control (2). The addition of 0.1 mM HgCl2 slightly reduced the growth rate without changing the outer membrane properties. In comparison to this control, all other samples showed an increased capability of forming biofilms (Fig. 5), showing that the release of MV as a stress response leads to an increased hydrophobicity and, in consequence, to enhanced biofilm formation.
Fig 5.
Quantification of biofilm formation of P. putida after incubation with different stressors. Error bars show standard deviations.
DISCUSSION
The central finding of the present work is the release of MV as a reaction of the cells to several stress factors. This release of MV led to an increase in the cell surface hydrophobicity, which led to a strong tendency to form biofilms (3, 11, 25). This physiological sequence has never been described before.
In order to improve the knowledge about this stress response mechanism, the MV released were analyzed. Besides those of the usual size, with about a 150-nm radius, smaller vesicles occurred after treatment with osmotically effective concentrations of sodium chloride or EDTA. Both compounds probably disturb the interaction of calcium ions with lipopolysaccharide (LPS) by either complex formation (EDTA) or display of the calcium ions from the negative charged residues of the LPS. Consequently, this changed calcium-LPS composition could affect a potential mechanism involved in the formation of MV and leading to smaller vesicles. The zeta potential of the released MV correlates well with that of the cells (2). Because the released MV and the cell surface of nonstressed cells have similar charges, comparable surface compositions can be assumed. It was also previously shown that the zeta potential of P. putida changes after organic solvent stress (31).
The analysis of the fatty acid components demonstrates that the MV contained mainly C16:0 and C18:1Δ11cis. However, a considerable enrichment of stearic acid (18:0) was found in the MV compared to the amount in the cell membranes. This is consistent with other studies (44) and indicates that the released vesicles are formed from the outer membrane of P. putida .
The proteomic data of the isolated MV likewise illustrate a composition similar to the outer membrane of P. putida . Generally, the different outer membrane proteins identified were consistent with the results of other studies (4), e.g., OprD and OprF. Several unusual proteins were identified in vesicles after every treatment. Chain A of the quinoprotein ethanol dehydrogenase is known as a redox active enzyme using a special cofactor, pyrroloquinoline quinine (PQQ). It is supposed that all enzymes belonging to this class are located in the periplasm (9). In addition, other periplasmatic proteins were abundant in the MV. Recently, a sorting mechanism to pack proteins into MV was proposed (13). Interestingly, ribosomal proteins and elongation factors occur in the proteome of MV. Either these proteins are released by the cells after the exposure to the stressor due to an enhanced permeability of the membrane, including partial cell lysis, or they are specifically included in MV. The latter hypothesis is supported by the fact that P. aeruginosa -derived vesicles contain nucleic acids (23, 40).
The kinetics of MV release observed in our study indicates that it takes around 10 min until this process is completed. Therefore, transcriptional induction can be excluded. Instead, the system is most probably continuously expressed and can be allosterically regulated simply by activating an already existing cellular process. Such an assumption is in agreement with the results of other studies of both organisms, P. putida and P. aeruginosa (2, 26).
Since it was noticed that membrane vesicles play an important role in bacterial life, several mechanisms for their formation were proposed (29, 30). Considering our knowledge of the membrane as a mixture of so-called lipid rafts (10), the different types of the LPS layer might be organized in this way. Furthermore, it is assumed that lipid rafts fulfill an important function in signaling and vesicle formation in eukaryotic cells (41). It was shown that the PQS molecule interacts specifically with outer membrane lipids (28, 38). Thus, an enrichment of PQS or another signaling molecule in the outer membrane could potentially rearrange the lipid rafts, leading to a local accumulation of long-chain LPS (28, 36). This fractional enrichment could cause the formation of blebs on the outer site of the outer membrane and can be proposed as the initial step of the formation of MV (30). So far, the precursor HHQ but not PQS has been detected in P. putida (7, 8). Thus, although the enzyme responsible for PQS formation from HHQ in P. aeruginosa is not yet described in P. putida , it seems reasonable that PQS or a very similar quorum-sensing molecule is involved in the release of MV in P. putida (7, 8). This hypothesis is supported by the fact that exogenous PQS enhances MV production in P. putida (43).
The biological importance of the formation of MV is well described (39, 45). Very recently, Manning and Kuehn (27) demonstrated that E. coli releases MV as a fast response to envelope stress, leading to increased survival of the bacteria. Furthermore, the connection to biofilm formation was assumed before (3) and appears to be confirmed by the results of this study.
In conclusion, this study indicates that cells of P. putida sensing stress release MV, which leads directly to a highly hydrophobic cell surface. Consequently, they attach easily to each other, as well as to surfaces. This mechanism is probably conserved in most Gram-negative bacteria and involved in biofilm formation as an important mechanism to survive environmental stress conditions (Fig. 6).
Fig 6.
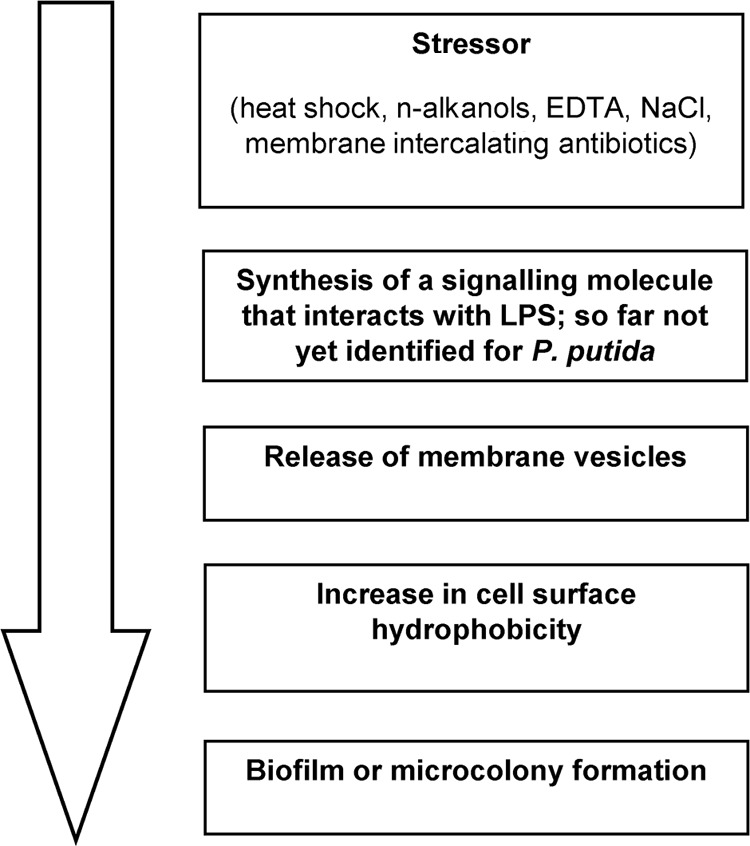
Possible sequence of biofilm formation in P. putida after exposure to a stressor.
Supplementary Material
ACKNOWLEDGMENTS
This work was partially supported by the European Commission within its Seventh Framework Program project BACSIN (contract no. 211684).
We thank Jana Reichenbach, Birgit Würz, Christine Schumann, and Kathleen Eismann (all UFZ) for their help with the experiments.
Footnotes
Published ahead of print 29 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Bastida F, et al. 2010. Elucidating MTBE degradation in a mixed consortium using a multidisciplinary approach. FEMS Microbiol. Ecol. 73: 370–384 [DOI] [PubMed] [Google Scholar]
- 2. Baumgarten T, et al. 2012. Alkanols and chlorophenols cause different physiological adaptive responses on the level of cell surface properties and membrane vesicle formation in Pseudomonas putida DOT-T1E. Appl. Microbiol. Biotechnol. 93: 837–845 [DOI] [PubMed] [Google Scholar]
- 3. Beveridge TJ, Makin SA, Kadurugamuwa JL, Li ZS. 1997. Interactions between biofilms and the environment. FEMS Microbiol. Rev. 20: 291–303 [DOI] [PubMed] [Google Scholar]
- 4. Choi DS, et al. 2011. Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa . Proteomics 11: 3424–3429 [DOI] [PubMed] [Google Scholar]
- 5. de Carvalho C, Wick LY, Heipieper HJ. 2009. Cell wall adaptations of planktonic and biofilm Rhodococcus erythropolis cells to growth on C5 to C16 n-alkane hydrocarbons. Appl. Microbiol. Biotechnol. 82: 311–320 [DOI] [PubMed] [Google Scholar]
- 6. de Kievit TR. 2009. Quorum sensing in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 11: 279–288 [DOI] [PubMed] [Google Scholar]
- 7. Diggle SP, et al. 2006. Functional genetic analysis reveals a 2-alkyl-4-quinolone signaling system in the human pathogen Burkholderia pseudomallei and related bacteria. Chem. Biol. 13: 701–710 [DOI] [PubMed] [Google Scholar]
- 8. Dos Santos VAPM, Heim S, Moore ERB, Stratz M, Timmis KN. 2004. Insights into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environ. Microbiol. 6: 1264–1286 [DOI] [PubMed] [Google Scholar]
- 9. Duine JA, Jongejan JA. 1989. Quinoproteins, enzymes with pyrrolo-quinoline quinone as cofactor. Annu. Rev. Biochem. 58: 403–426 [DOI] [PubMed] [Google Scholar]
- 10. Engelman DM. 2005. Membranes are more mosaic than fluid. Nature 438: 578–580 [DOI] [PubMed] [Google Scholar]
- 11. Flemming CA, Palmer RJ, Arrage AA, Van der Mei HC, White DC. 1999. Cell surface physico chemistry alters biofilm development of Pseudomonas aeruginosa lipopolysaccharide mutants. Biofouling 13: 213–231 [Google Scholar]
- 12. Hartmans S, Smits JP, van der Werf MJ, Volkering F, de Bont JAM. 1989. Metabolism of styrene oxide and 2-phenylethanol in the styrene-degrading Xanthobacter strain 124X. Appl. Environ. Microbiol. 55: 2850–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haurat MF, et al. 2011. Selective sorting of cargo proteins into bacterial membrane vesicles. J. Biol. Chem. 286: 1269–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Heipieper HJ, Diefenbach R, Keweloh H. 1992. Conversion of cis-unsaturated fatty acids to trans, a possible mechanism for the protection of phenol-degrading Pseudomonas putida P8 from substrate toxicity. Appl. Environ. Microbiol. 58: 1847–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heipieper HJ, Keweloh H, Rehm HJ. 1991. Influence of phenols on growth and membrane permeability of free and immobilized Escherichia coli. Appl. Environ. Microbiol. 57: 1213–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heipieper HJ, Loffeld B, Keweloh H, de Bont JAM. 1995. The cis/trans isomerization of unsaturated fatty acids in Pseudomonas putida S12: an indicator for environmental stress due to organic compounds. Chemosphere 30: 1041–1051 [Google Scholar]
- 17. Heipieper HJ, Neumann G, Cornelissen S, Meinhardt F. 2007. Solvent-tolerant bacteria for biotransformations in two-phase fermentation systems. Appl. Microbiol. Biotechnol. 74: 961–973 [DOI] [PubMed] [Google Scholar]
- 18. Hiementz PC. 1986. Principles of colloid and surface chemistry. Marcel Dekker, Inc., New York, NY [Google Scholar]
- 19. Isken S, de Bont JAM. 1998. Bacteria tolerant to organic solvents. Extremophiles 2: 229–238 [DOI] [PubMed] [Google Scholar]
- 20. Kadurugamuwa JL, Beveridge TJ. 1995. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J. Bacteriol. 177: 3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kadurugamuwa JL, Lam JS, Beveridge TJ. 1993. Interaction of gentamicin with the A band and B band lipopolysaccharides of Pseudomonas aeruginosa and its possible lethal effect. Antimicrob. Agents Chemother. 37: 715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kobayashi H, Uematsu K, Hirayama H, Horikoshi K. 2000. Novel toluene elimination system in a toluene-tolerant microorganism. J. Bacteriol. 182: 6451–6455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kolling GL, Matthews KR. 1999. Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl. Environ. Microbiol. 65: 1843–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kulp A, Kuehn MJ. 2010. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 64: 163–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lopez D, Vlamakis H, Kolter R. 2010. Biofilms. Cold Spring Harbor Perspect. Biol. 2: a000398 doi:10.1101/cshperspect.a000398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Makin SA, Beveridge TJ. 1996. Pseudomonas aeruginosa PAO1 ceases to express serotype-specific lipopolysaccharide at 45°C. J. Bacteriol. 178: 3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Manning AJ, Kuehn MJ. 2011. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol. 11: 258 doi:10.1186/1471-2180-11-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mashburn-Warren L, et al. 2008. Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Mol. Microbiol. 69: 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mashburn-Warren L, McLean RJC, Whiteley M. 2008. Gram-negative outer membrane vesicles: beyond the cell surface. Geobiology 6: 214–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mashburn-Warren LM, Whiteley M. 2006. Special delivery: vesicle trafficking in prokaryotes. Mol. Microbiol. 61: 839–846 [DOI] [PubMed] [Google Scholar]
- 31. Neumann G, et al. 2006. Energetics and surface properties of Pseudomonas putida DOT-T1E in a two-phase fermentation system with 1-decanol as second phase. Appl. Environ. Microbiol. 72: 4232–4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28: 449–461 [DOI] [PubMed] [Google Scholar]
- 33. Ramos J, Duque E, Huertas M, Haidour A. 1995. Isolation and expansion of the catabolic potential of a Pseudomonas putida strain able to grow in the presence of high concentrations of aromatic hydrocarbons. J. Bacteriol. 177: 3911–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ramos JL, et al. 2002. Mechanisms of solvent tolerance in gram-negative bacteria. Annu. Rev. Microbiol. 56: 743–768 [DOI] [PubMed] [Google Scholar]
- 35. Ramos JL, et al. 2001. Responses of Gram-negative bacteria to certain environmental stressors. Curr. Opin. Microbiol. 4: 166–171 [DOI] [PubMed] [Google Scholar]
- 36. Schertzer JW, Boulette ML, Whiteley M. 2009. More than a signal: non-signaling properties of quorum sensing molecules. Trends Microbiol. 17: 189–195 [DOI] [PubMed] [Google Scholar]
- 37. Schertzer JW, Brown SA, Whiteley M. 2010. Oxygen levels rapidly modulate Pseudomonas aeruginosa social behaviours via substrate limitation of PqsH. Mol. Microbiol. 77: 1527–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schertzer JW, Whiteley M. 2012. A bilayer-couple model of bacterial outer membrane vesicle biogenesis. mBio 3: e00297–11 doi:10.1128/mBio.00297-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schooling SR, Beveridge TJ. 2006. Membrane vesicles: an overlooked component of the matrices of biofilms. J. Bacteriol. 188: 5945–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schooling SR, Hubley A, Beveridge TJ. 2009. Interactions of DNA with biofilm-derived membrane vesicles. J. Bacteriol. 191: 4097–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Simons K, Ikonen E. 1997. Functional rafts in cell membranes. Nature 387: 569–572 [DOI] [PubMed] [Google Scholar]
- 42. Sleator RD, Hill C. 2002. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 26: 49–71 [DOI] [PubMed] [Google Scholar]
- 43. Tashiro Y, Ichikawa S, Nakajima-Kambe T, Uchiyama H, Nomura N. 2010. Pseudomonas quinolone signal affects membrane vesicle production in not only Gram-negative but also Gram-positive bacteria. Microbes Environ. 25: 120–125 [DOI] [PubMed] [Google Scholar]
- 44. Tashiro Y, et al. 2011. Characterization of phospholipids in membrane vesicles derived from Pseudomonas aeruginosa . Biosci. Biotechnol. Biochem. 75: 605–607 [DOI] [PubMed] [Google Scholar]
- 45. Tashiro Y, Uchiyama H, Nomura N. 2012. Multifunctional membrane vesicles in Pseudomonas aeruginosa . Environ. Microbiol. 14: 1349–1362 [DOI] [PubMed] [Google Scholar]
- 46. van Loosdrecht MCM, Lyklema J, Norde W, Schraa G, Zehnder AJB. 1987. The role of bacterial cell wall hydrophobicity in adhesion. Appl. Environ. Microbiol. 53: 1893–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.